Abstract
The repeated occurrence of genes in each other’s neighbourhood on genomes has been shown to indicate a functional association between the proteins they encode. Here we introduce STRING (search tool for recurring instances of neighbouring genes), a tool to retrieve and display the genes a query gene repeatedly occurs with in clusters on the genome. The tool performs iterative searches and visualises the results in their genomic context. By finding the genomically associated genes for a query, it delineates a set of potentially functionally associated genes. The usefulness of STRING is illustrated with an example that suggests a functional context for an RNA methylase with unknown specificity. STRING is available at http://www.bork.embl-heidelberg.de/STRING
INTRODUCTION
The availability of complete genome sequences has stimulated the development of new methods for protein function prediction (1–6). In contrast to classical, homology-based function assignment, these methods do not predict the function of proteins, but rather the functional association between proteins, based on the genomic association of their genes. One approach is based on the observation that genes that repeatedly occur in each other’s proximity on genomes (in potential operons) tend to encode functionally interacting proteins, e.g. the proteins are part of the same protein complex or metabolic pathway (1–3,7–9). Here we introduce a web-server that retrieves for a given query gene all the genes that repeatedly occur within potential operons. The server is named STRING (search tool for recurring instances of neighbouring genes). It also retrieves, by an iterative approach, the genes that are indirectly (via other genes) associated with the query gene. The web-interface (http://www.bork.embl-heidelberg.de/STRING ) visualises the results in their genomic context (Fig. 1).
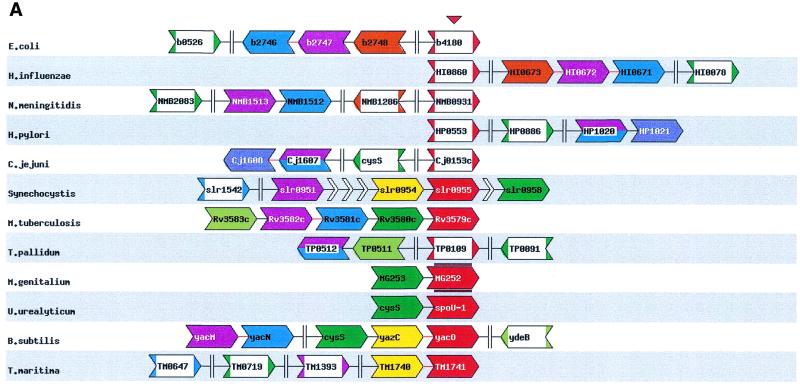
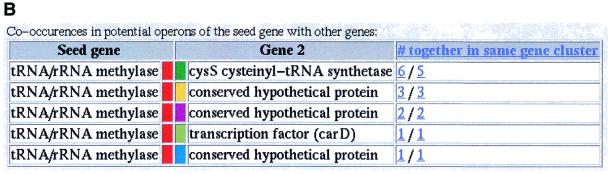
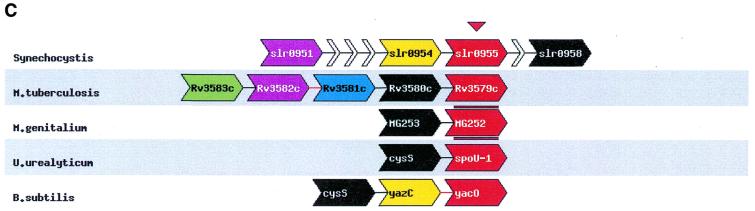
Figure 1.
Different parts of the result of an example search performed using STRING that detects a potential new functional interaction. The search was started with a gene from Mycoplasma genitalium that codes for a hypothetical tRNA/rRNA methylating enzyme (MG252) as query. The main graphic of the result (A) shows, after the iterations have converged, that the query (the red genes) occur repeatedly in the same gene cluster with cystenyl transfer RNA synthase (the green genes). The table of co-occurences in clusters (B) shows that this organisation is present in six species, twice in closely related species. It recently has been shown that, at least in some species, the cys-tRNA is modified (15,16). Based on the pattern of conserved gene clusters, we propose that MG252 plays a role in the reported modification of the cys-tRNA. The other retrieved genes co-occurring with each other and with our query gene are less frequently connected to MG252 and cys-tRNA synthase, and are absent from the Mycoplasmas. Although this pattern suggests a less intimate involvement with the proposed interaction, the molecular functions still support some sort of functional link: the gene family in light green is homologous to a ribonuclease, and the family in purple is homologous to sugar nucleotidyl transferase. In this example, the iterations provide us with insight into the conserved genomic organisation of the associated genes. The ribonuclease only repeatedly occurs with the query, while the sugar nucleotidyl transferase has itself a very tight association with an hypothetical protein (the blue genes). When one, in the table of co-occurences, clicks on the number of times our query and the cys-tRNA co-occur in the same cluster in distantly related species, the diagram that only displays these organisations is shown (C). The query gene family is in red, while the cys-tRNA is assigned black. These two colours are reserved to denote the genes that this diagram focuses on. Genes from the same orthologous family have the same colour. The red gene symbols aligned above and below MG252 are its orthologues in the other species. The truncated small white gene-like symbols are genes that are located between the genes retrieved via the conserved gene clusters, but that are themselves not conserved in that position. The gene symbols with two colours are assigned to different gene families because they are the result of fusions. An interruption symbol, such as between yacN and cysS, means that the two displayed stretches of the genome are not in the same gene cluster. The lines between the genes symbolise the stretches of DNA in between the genes, and are linked to the DNA sequence of that stretch, while the gene symbols are linked to their GenBank entries.
METHODOLOGY
The tool starts with a single seed gene. In the zero iteration it retrieves and displays the genes that repeatedly occur with this gene in clusters on the genome in multiple, phylogenetically distant species (for a definition see 10). We define gene clusters here as introduced by Overbeek et al. with the concept of a ‘run’, a stretch of genes on the same strand not interrupted by >300 bp (2). In addition we count two genes that are actually fused into one gene as being in the same run. In subsequent iterations the tool repeats this process using as seeds all the new genes retrieved in the previous iteration, thereby uncovering the set of genes that are indirectly linked to the seed gene. The iterations continue until the number of iterations set by the user is reached, or until no new genes are found (convergence). Normally the query gene is used as seed. If the query gene is not part of a conserved gene cluster itself, the tool uses orthologues of the query gene that are in conserved gene clusters as seed. When a protein sequence is submitted as query, the tool performs a blast search against the proteins from the published genomes (NCBI basic protein blast2.0 with a cut-off E-value of 10–5; 11). If a perfect match is found, that gene is used as seed. Otherwise the user can select a seed from the list of blast hits. With the results of the last iteration the tool also displays the genes that are not retrieved via conserved gene order but that are still present in the species of which other genes already have been retrieved. The presence or absence of these genes that are not in a conserved cluster complements the cluster information. The explicit focus on (iteratively) searching and displaying the integral conserved genomic organisation for a given gene is one of the defining features of this server, and set it apart from what is currently available at servers like KEGG (12). A conceptual similar approach is being developed independently at WIT (http://wit.integratedgenomics.com/IGwit/ ), which in principle allows one to obtain similar results. Apart from many small differences in the implementation and visualisation, the major difference seems to be that STRING is a specialised and dedicated server for this type of search.
Orthology is operationally defined as ‘bidirectional best, significant (E < 0.01), hit’, based on Smith–Waterman (13) comparisons of the complete genomes with one another, and including the possibility of gene fusion/fission (14). The iterative usage of these orthology relations can give rise to inconsistencies, due to unrecognised paralogy, unrecognised homology, and/or gene fusion. However, the quality of orthology prediction here is relatively high because of the additional requirement in STRING of conserved gene order (14).
DISPLAY
All the retrieved information is displayed in one graphic that features extra information about the genes and their context (Fig. 1A). The extra information includes additional non-conserved neighbouring genes, the gene order, the relative location of the gene clusters in the genome, and the relative direction of transcription of the genes. Also featured is a table that lists how often the seed gene occurs in the same run with each other gene, both in all genomes as well as only in phylogenetically distant genomes (Fig. 1B). This indicates the degree of genomic association between the two genes, and thereby the strength of functional association between their respective products. The number of co-occurrences of two genes in the same cluster is linked to a page that displays only the clusters in those species containing that specific organisation and highlighting the two specified genes (Fig. 1C). To assist in assessing the substructure of genomic associations between all the retrieved genes, the number of co-occurrences of genes in the same cluster for every pair of genes is shown in a separate matrix which can be accessed by clicking on its link.
COVERAGE
STRING finds results for 24 768 out of the 59 416 genes in the presently included set of completely sequenced genomes. Although there is little operon structure in eukaryotes, to the extent that orthologues of their genes are present in prokaryotes, it is possible to predict functional associations for these genes. In this way we found results for 637 genes out of the 1681 yeast genes that have orthologues in the prokaryotes.
SELECTIVITY
We tested the probability that two genes repeatedly occur in one cluster by chance. In randomly shuffled genomes the probability that a given gene occurs with the same other gene in one cluster in two species is 0.02. For three species this probability is <0.002, and for four species or more it is <0.0005. The accuracy in terms of predicted functional relations is difficult to determine because of the broad definition of functional association, which includes a spectrum of possible protein relations ranging from direct ones such as physical interactions to more vague ones like the proteins being active in the same cellular process. Notice, however, that the functional link tends to be stronger when the conservation is stronger (6). Furthermore, the interpretation of the type of association is facilitated by what is known about the putative molecular functions of the proteins, that can be inferred from conventional homology (see the example of cys-tRNA in Fig. 1). In general, only the user can interpret the nature of the association by knowledge of the genes and organisms involved.
CONCLUDING REMARKS
STRING provides a platform for searching and interpreting conserved patterns in genome organisation with the aim of finding functional associations for a given gene. The iterations and visualisation of the thereby retrieved genes allow the analysis and delineation of the set of potential interaction partners.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank the members of the Bork group for helpful discussion and feedback. This work was supported by the DFG and the BMBF.
REFERENCES
- 1.Dandekar T., Snel,B., Huynen,M. and Bork,P. (1998) Trends Biochem. Sci., 23, 324–328. [DOI] [PubMed] [Google Scholar]
- 2.Overbeek R., Fonstein,M., D’Souza,M., Pusch,G.D. and Maltsev,N. (1998) In Silico Biol., 1, 0009. [PubMed] [Google Scholar]
- 3.Overbeek R., Fonstein,M., D’Souza,M., Pusch,G.D. and Maltsev,N. (1999) Proc. Natl Acad. Sci. USA, 96, 2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcotte E.M., Pellegrini,M., Ng,H.L., Rice,D.W., Yeates,T.O. and Eisenberg,D. (1999) Science, 285, 751–753. [DOI] [PubMed] [Google Scholar]
- 5.Enright A.J., Iliopoulos,I., Kyrpides,N.C. and Ouzounis,C.A. (1999) Nature, 402, 86–90. [DOI] [PubMed] [Google Scholar]
- 6.Pellegrini M., Marcotte,E.M., Thompson,M.J., Eisenberg,D. and Yeates,T.O. (1999) Proc. Natl Acad. Sci. USA, 96, 4285–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mushegian A.R. and Koonin,E.V. (1996) Trends Genet., 12, 289–290. [DOI] [PubMed] [Google Scholar]
- 8.Tamames J., Casari,G., Ouzounis,C. and Valencia,A. (1997) J. Mol. Evol., 44, 66–73. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe H., Mori,H., Itoh,T. and Gojobori,T. (1997) J. Mol. Evol., 44, S57–S64. [DOI] [PubMed] [Google Scholar]
- 10.Huynen M.A. and Snel,B. (2000) Adv. Protein Chem., 54, 345–379. [DOI] [PubMed] [Google Scholar]
- 11.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M. and Goto,S. (2000) Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith T.F. and Waterman,M.S. (1981) J. Mol. Biol., 25, 195–197. [Google Scholar]
- 14.Huynen M.A. and Bork,P. (1998) Proc. Natl Acad. Sci. USA, 95, 5849–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamann C.S., Sowers,K.R., Lipman,R.S. and Hou,Y.M. (1999) J. Bacteriol., 181, 5880–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipman S.A. and Hou,Y.M. (1998) Proc. Natl Acad. Sci. USA, 95, 13495–13500. [DOI] [PMC free article] [PubMed] [Google Scholar]