Abstract
We describe the synthesis of a novel type of mRNA template and its use in the preparation of mRNA–protein fusions. A light-induced psoralen crosslinking reaction was used to attach a puromycin-containing oligonucleotide to the 3′-end of an mRNA template. The photo-crosslinked template was found to undergo efficient mRNA–protein fusion formation in rabbit reticulocyte lysate. Fusion formation was subsequently tested with templates carrying puromycin linkers of different length and chemical composition. Short linkers with multiple triethyleneglycol phosphate building blocks allowed the most efficient fusion formation under a wide range of salt conditions. The present method simplifies the preparation of mRNA–protein fusions and thus significantly accelerates the in vitro protein evolution procedure which involves repetitive cycles of fusion production and selection.
INTRODUCTION
A recently described method for in vitro selection of peptides and proteins from large libraries of diverse sequences (1–3) relies on RNA–protein fusions, which consist of peptide or protein sequences covalently linked via their C-termini to the 3′-end of their own mRNA (PROfusion™). Libraries of RNA–protein fusion molecules can be subjected to repetitive rounds of in vitro selection to identify peptides and proteins with desired properties. The maximum library size that can be generated is limited only by the size and efficiency of the translation reaction and by the efficiency of fusion formation on the ribosome. At the present time, libraries containing up to 1014 different sequences have been generated and have been successfully used for the isolation of specific binders to a variety of different protein targets (for a review of recent applications see 4).
Until recently, mRNA templates for the preparation of mRNA–protein fusions were prepared by enzymatic ligation of a 3′-puromycin-terminated oligonucleotide to the 3′-end of the message. This enzymatic ligation requires a cumbersome gel purification of the ligated mRNA template, thus slowing down template synthesis. We have designed a novel type of mRNA template which can be readily prepared from mRNA and from a puromycin linker in a light-induced crosslinking reaction (Fig. 1). Fusion formation was tested with photo-crosslinked templates that encoded a short peptide (FLAG epitope) or a single chain antibody. Fusion yields were found to depend on the length and on the composition of the puromycin linker. The best fusion yields were found for short and flexible linkers and were roughly independent of the salt concentration in the buffer.
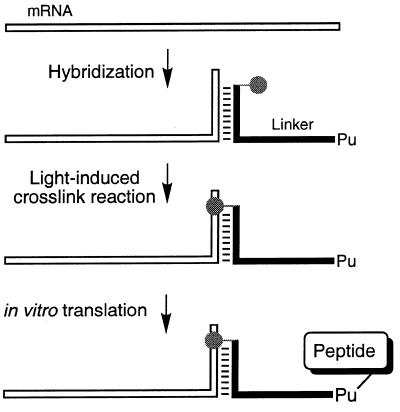
Figure 1.
Schematic outline of mRNA–peptide fusion synthesis. The mRNA was hybridized to a DNA linker carrying psoralen (gray circle) at the 5′-terminus and puromycin (Pu) at its 3′-end. Irradiation with UV light led to covalent crosslink formation between the linker and mRNA 3′-end. In vitro translation of the photo-crosslinked mRNA–puromycin template produced the mRNA–peptide fusion product.
MATERIALS AND METHODS
mRNA synthesis and purification
mRNAs (see Fig. 2a) were prepared by T7 RNA polymerase run-off transcription (Megashortscript transcription kit; Ambion, TX) of PCR DNA templates (5). After transcription, all RNAs were purified by electrophoresis on 6% TBE–urea polyacrylamide gels (Novex, CA). The product bands were visualized by UV shadowing, excised, crushed and soaked overnight in 0.3 M NaOAc. Following EtOH precipitation, the RNAs were resuspended and stored in H2O. Radiolabeled RNA was synthesized according to the same procedure by including [α-32P]UTP (Amersham, IL) in the transcription buffer.
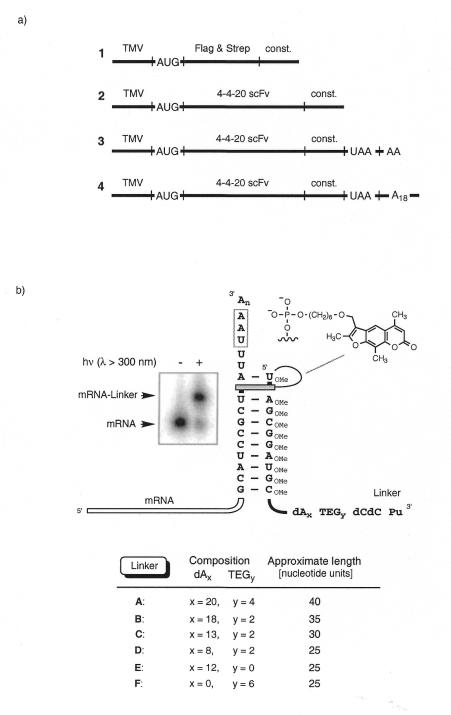
Figure 2.
(a) Design of the mRNAs used for photo-crosslink formation. TMV, a portion of the tobacco mosaic virus 5′-UTR (19) with a good initiation codon context (20); AUG, translation initiation codon; Flag & Strep, Flag epitope DYKDDDDK (11) followed by a Strep-Tag II sequence WSHPQFEK (12); 4-4-20 scFv, anti-fluorescein single chain antibody template (21,22); const, the linker hybridization sequence 5′-GCAUCCGCUAUU including the photo-crosslink site 5′-UA; UAA, termination codon; dA18, tag sequence. (b) Duplex formed between the constant 3′-sequence of the mRNA (const.) and the photolinker showing the putative psoralen intercalation site. Also shown is the structure of psoralen, which was attached through a C6 alkyl chain to the 5′-phosphate of the linker. The variables x and y determine the number of dA nucleotides and TEG units in the linker, respectively. The autoradiograph on the left depicts gel electrophoretic analysis of the photo-crosslinking reaction between mRNA 1 and linker B.
Puromycin linker synthesis and purification
Syntheses of the linkers (see Fig. 2b) were performed on an Expedite Synthesizer Model 8909 (PerSeptive Biosystems, MA). Puromycin–CPG, DNA phosphoramidites, 2′-OMe-RNA phosphoramidites, psoralen C6 phosphoramidite and triethyleneglycol (TEG) phosphoramidites (spacer 9) were used according to the recommended protocols (Glen Research, VA). Following deprotection in concentrated ammonium hydroxide for 8 h at 55°C, the linkers were purified by reverse phase HPLC on a C18 Spheri-5 column (Perkin-Elmer, CA) with 50 mM triethylammonium acetate in 5% (v/v) acetonitrile as buffer A and 50 mM triethylammonium acetate in 70% (v/v) acetonitrile as buffer B at a flow rate of 1.5 ml/min. A linear gradient of 15–60% buffer B over 45 min was used for elution. After drying, the linkers were resuspended and stored in H2O.
Photo-crosslinking reactions
The linkers (5 µM) were annealed to the target mRNAs (2.5 µM) in 25 mM Tris–HCl buffer, pH 7, 100 mM NaCl by heating to 85°C for 30 s followed by cooling to 4°C over 5 min. The reaction mixture was irradiated for 15 min at room temperature in borosilicate glass vials (Kimble/Kontes, NJ), using a hand held multi-wavelength UV lamp model UVGL-25 (UVP, CA) set to long wavelength (λ > 300 nm). The product mixture of the photo-crosslinking reaction between radiolabeled mRNA 1 and linker B was analyzed on a denaturing 6% TBE–urea gel (Novex) and visualized on a phosporimaging system (Molecular Dynamics, CA) (Fig. 2b). The photo-crosslink product mixtures generally contained <20% unreacted and >80% photo-crosslinked mRNA and were used directly for in vitro translation and subsequent mRNA–protein fusion formation.
Translation in vitro and mRNA–protein fusion formation
Translation reactions were performed in rabbit reticulocyte lysate (Ambion) for 30 min at 30°C. Reactions contained 100 pmol photo-crosslinked mRNA (see above), 10 mM creatine phosphate, 150 mM KOAc, 0.5 mM MgCl2, 0.1 mM each amino acid except methionine, 150 µCi [35S]methionine (>1000 Ci/mmol; Amersham) and 67% (v/v) lysate in a total volume of 300 µl. The total concentration of methionine in the lysate was 3.8 µM (including endogenous methionine). mRNA–protein fusion formation was promoted by the addition of KCl and MgCl2 to final concentrations of 590 and 50 mM, respectively, in a 500 µl volume. Incubation was continued for another 60 min at 20°C. Differing concentrations of KCl and MgCl2 were tested to explore salt dependence of fusion formation (see Fig. 4).
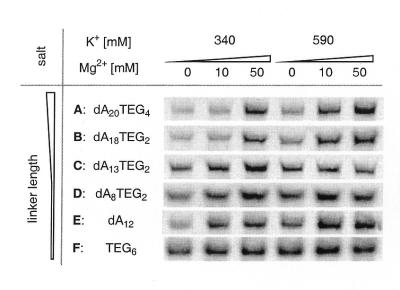
Figure 4.
Dependence of fusion yield on linker composition and on salt concentration. mRNA 3 was photo-crosslinked to linkers A–F and subjected to in vitro translation and to fusion formation under different salt concentrations. Gel electrophoresis was used to analyze the amount of mRNA–protein fusion product produced.
The products were isolated by diluting the lysate into 10 ml of binding buffer (100 mM Tris–HCl, pH 8.0, 10 mM EDTA, 1 M NaCl, 0.25% v/v Triton X-100) and by adding to the mixture 10 mg oligo(dT)–cellulose type 7 (Pharmacia, NJ). Samples were rotated for 60 min at 4°C. The solid support was then washed with 5 ml of ice-cold binding buffer, followed by elution with 100 µl aliquots of deionized H2O. The amount of mRNA–protein fusion isolated was determined by scintillation counting of the incorporated [35S]methionine. The product was analyzed by electrophoresis on 4–12% NuPage gels using MES running buffer (Novex). Gels were dried after extensive washing to remove excess [35S]methionine and bands were visualized on a phosphorimager system (Molecular Dynamics).
Flag immunoprecipitation and StrepTactin binding
A solution of 10 µl of 35S-labeled mRNA–peptide fusion (prepared from mRNA 1 with linker B) was added to 20 µl of Anti-Flag M2 affinity gel (Sigma, MO) in 300 µl of buffer containing 50 mM Tris–HCl, pH 7.4, 1% NP-40, 150 mM NaCl, 1 mM EDTA, 1 mM Na3VO4 and 1 mM NaF. A second precipitation experiment was carried out by adding the same fusion product to 20 µl StrepTactin Sepharose (Genosys, TX) in 300 µl of buffer containing 100 mM Tris–HCl, pH 7.1, 1 mM EDTA and 0.5 mg/ml yeast tRNA. Both precipitation mixtures were processed in parallel under identical conditions. Mixtures were rotated for 1 h at 4°C and then transferred to an Ultrafree-MC filter unit (0.45 µm; Millipore, MA). The buffer was removed by centrifugation and the residue washed with 5 × 300 µl ice-cold buffer. The residues were analyzed by scintillation counting and fusion binding was determined to be 57 and 62% for the Anti-Flag M2 matrix and the StrepTactin matrix, respectively. A control reaction with protein A–agarose (Sigma) showed no detectable binding to the matrix.
RESULTS
Photocrosslink formation between mRNA and puromycin linkers
Figure 2 illustrates the design of the mRNAs and puromycin linkers used in the crosslinking reactions. All mRNAs carried a 3′-terminal 10 nt hybridization site coding for the amino acid sequence ASA and ending with the dinucleotide sequence 5′-UA for psoralen photocrosslinking (6,7). In addition, mRNAs 3 and 4 contained a downstream stop codon (UAA) to induce mRNA release from the ribosome after translation of residual uncrosslinked template (vide infra). A poly(A) tail was attached to mRNA 4 for purification by oligo(dT). The puromycin linkers (A–F) were prepared on a DNA synthesizer using conventional solid supported phosphoramidite chemistry. A psoralen moiety was attached through a C6 alkyl chain to the 5′-phosphate of the linker (8). Flexible TEG phosphate spacers and polynucleotide sequences of various lengths were used to tether 5′-dCdC-puromycin to the 3′-end (see table in Fig. 2). The linker hybridization sequence was prepared from 2′-OMe-RNA phosphoramidites to enhance the pairing stability of the stem structure (9,10). Each linker was HPLC purified before use in the photo-crosslinking reaction.
For crosslink formation the linkers were hybridized to the target mRNA and irradiated with UV light of λ > 300 nm (8). The reaction between linker B and radiolabeled mRNA 1 was complete after 15 min, yielding >80% crosslinked product (Fig. 2b). The photo-crosslink product of mRNA 1 could be separated from free mRNA 1 and from excess linker using denaturing gel electrophoresis. For the longer mRNA substrates 2–4 (>800 nt) the relative size difference between mRNA and crosslinked mRNA became too small to be separated on a gel. In these cases the crude photo-crosslinking reaction mixtures were directly added to the lysate for fusion formation without any further purification.
In vitro translation and mRNA–protein fusion synthesis
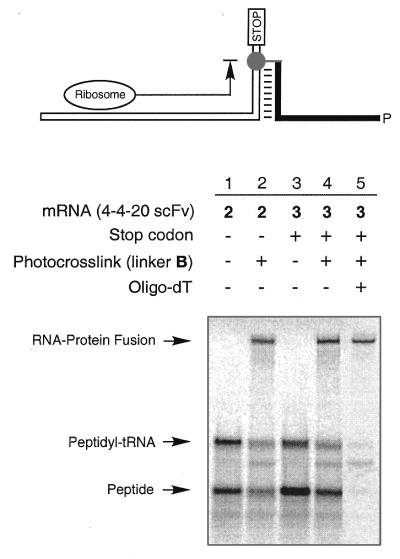
Translation and fusion formation were tested with mRNA 2 in the following experiments. First, scFv mRNA 2 was tested for in vitro translation in rabbit reticulocyte lysate. Reactions were carried out for 30 min at 30°C, followed by addition of KCl and MgCl2 to final concentrations of 590 and 50 mM, respectively (1,3). Gel analysis showed two bands that correspond to the peptidyl-tRNA and the free peptide (Fig. 3, lane 1). When translation was carried out with photo-crosslinked mRNA 2 a third and slower migrating band appeared, thus indicating successful mRNA–protein fusion formation (Fig. 3, lane 2). Increased yields of free protein and fusion (∼20% more) were obtained with mRNA 3, which was identical in coding sequence to mRNA 2 but carried a stop codon downstream of the photo-crosslink site (Fig. 3, lanes 3 and 4). Relative band intensities indicated that 30% of the total amount of synthesized protein was converted into mRNA–protein fusion (Fig. 3, lane 4). The mRNA–scFv fusion was purified by binding of the A18 linker regions to oligo(dT)–cellulose followed by washing with binding buffer (Fig. 3, lane 5) (1,3). The fusion product could be isolated at 1.3% yield based on the amount of photo-crosslinked input mRNA. Physical properties [gel mobility, binding to oligo(dT)–cellulose and selective peptide binding to affinity reagents] of the fusions prepared from photo-crosslinked mRNA were found to be identical to those of the fusion product obtained from enzymatically ligated mRNA templates (data not shown). In order to confirm the composition of the peptide portion of the fusion, fusions prepared from mRNA template 1 crosslinked to B, which encoded the Flag and Strep-Tag II epitopes, were tested for protein binding. Immunoprecipitation with Anti-Flag M2 antibody (11) on agarose and with StrepTactin Sepharose (12) showed 54 and 62% binding, respectively. Non-specific binding to protein A–agarose was <0.5%.
Figure 3.
In vitro translation and fusion formation. Protein synthesis from mRNA templates 2 and 3 is shown in lanes 1 and 3, respectively. Translation of mRNA templates photo-crosslinked to linker B produced mRNA–protein fusions (lanes 2 and 4). Template 3 carries a stop codon following the linker crosslink site. Lane 5 shows the fusion product after purification on oligo(dT)–cellulose. The products were analyzed by electrophoresis on a 4–12% NuPage gel using MES running buffer (Novex).
To test the effect of linker length and composition on fusion yield, mRNA 3 was photo-crosslinked to linkers A–F and each template was subsequently tested for fusion formation (Fig. 4). After incubation for 30 min at 30°C, the samples were divided into aliquots, to which varying amounts of KCl and MgCl2 were added. The samples were incubated for another 60 min at 20°C, then the fusion product was analyzed by gel electrophoresis. The best mRNA–protein fusion yields were obtained with the long linkers A and B (40 and 35 nt, respectively) under high salt conditions. A lower salt concentration resulted in a significant drop in fusion formation with linkers A and B. On the other hand, fusion yields for the shorter linkers C–F were less salt dependent. In general, the fusion yield increased with the number of flexible TEG spacers. Analysis of crude lysate revealed that up to 45% of the total protein was present as mRNA–protein fusions.
The fusion prepared from mRNA 3 with linker F cannot be purified on oligo(dT)–cellulose because linker F is missing the oligo(dA) tract. Therefore, mRNA 4 was prepared, which codes for the same protein (scFv) as mRNA 3 but carries an A18 stretch at its 3′-end. This 3′-terminal poly(dA) sequence allowed efficient purification of fusions with linker F on oligo(dT)–cellulose.
DISCUSSION
The current model for mRNA–protein fusion formation (1,3) involves pseudo-intramolecular (cis) amide bond formation between the mRNA template and the encoded protein on the ribosome. In this model, protein elongation proceeds to the end of the open reading frame (ORF), where the ribosome stalls. The absence of a stop codon at the mRNA 3′-end prevents the recruitment of release factors; consequently, the message and the nascent protein remain bound to the ribosome. Puromycin, attached through a poly(dA) linker to the ORF, accepts the nascent protein from the peptidyl-tRNA by ribosomal peptidyltransferase-catalyzed peptide bond formation to produce the mRNA–protein fusion molecule. Previously, the mRNA–puromycin template has been prepared by enzymatic ligation of a d(A27CC)-puromycin linker to the 3′-end of the mRNA using T4 DNA ligase (1,3). The problem with this enzymatic step is the need for a splint, which is difficult to design because run-off transcription with T7 polymerase can produce heterogeneous 3′-ends (5). Also, complete removal of the splint after ligation is important because residual splint can inhibit fusion formation (unpublished results). To circumvent these problems, we have developed a novel type of mRNA–puromycin template for fusion preparation. The template was synthesized in an efficient, light-induced interstrand crosslinking reaction (8). Our decision to use photochemistry was based in part on the finding that 2′-O-alkyl antisense oligonucleotides that have been psoralen crosslinked to mRNA lead to target-specific arrest of translation (13). We reasoned that a 3′-terminal interstrand crosslink on the RNA would lead to ribosome stalling, thus fulfilling the same role as the DNA linker in the enzymatically ligated mRNA–puromycin template.
The puromycin linkers were chemically prepared and carried a 5′-terminal psoralen moiety, followed by a hybridization region and by a flexible tether with puromycin at the 3′-end (Fig. 2). Hybridization of the puromycin linker to the 3′-end of the mRNA followed by light-induced crosslink formation produced the photo-crosslinked mRNA–puromycin template. We found that a 10 base complementary region between the mRNA 3′-end and the photolinker was sufficient to achieve efficient hybridization and photo-crosslink formation, with yields consistently exceeding 80%. The use of 2′-OMe-RNA in the linker hybridization sequence was intended to increase the stability of the 10 bp duplex (9,10); it also protected the mRNA from cleavage by any RNase H activity in the lysate (14). The hydroxymethyl trioxsalen moiety (psoralen) (8) was tethered to the 5′-phosphate through a hexamethylene spacer (psoralen C6). The use of a shorter spacer (psoralen C2) led to a significant drop in photo-crosslink efficiency (<40%; data not shown).
Reaction mixtures of photo-crosslinked mRNA template did not require further purification and could be used directly for efficient fusion formation in rabbit reticulocyte lysate (Fig. 3). The fusion yield for crosslinked mRNA template 3, which carries a stop codon after the crosslink site, was found to be ∼20% better than was the yield for crosslinked template 2, which does not carry a stop codon. This result not only suggested a translational arrest before the crosslink site but also demonstrated that a stop codon downstream of the crosslink site has a beneficial effect on translation yields of free and fused protein. The presence of ∼20% unmodified (not crosslinked) mRNA in the translation mixture might be responsible for this effect: mRNA 2 (without a stop codon) is not actively released after translation and can remain bound to the ribosome (15,16); in contrast, the stop codon in mRNA 3 will initiate the recruitment of release factors after translation of the ORF. Immediate release of the mRNA makes the ribosome available for another round of translation (17,18). An increased ratio of free peptide over peptidyl-tRNA further indicates multiple translation of mRNA template 3. Addition of an 8-fold excess of unmodified mRNA to photo-crosslinked mRNA led to increased production of free protein, but did not lower the fusion yield (data not shown). This finding supports the notion that the ribosome can undergo multiple rounds of translation before stalling on a photo-crosslinked template and thus producing the fusion molecule. The stop codon allows for efficient turnover of residual non-crosslinked mRNA and thus increases the chance of the ribosome binding and translating a crosslinked template to produce the mRNA–protein fusion. This mechanism does not apply to enzymatically ligated mRNA because a stop codon would initiate ribosome release from both unligated and ligated templates, thus abolishing fusion formation completely.
Previous investigations with enzymatically ligated mRNA templates suggested that fusion yield was highly dependent on linker length, linker composition and reaction conditions (3). We found an analogous behavior for photo-crosslinked mRNA–puromycin templates, which showed the best fusion yields for linkers A and B (35–40 nt units) in the presence of high salt concentrations. Shorter linkers and substitution of dA by TEG units resulted in reduced salt dependence. The best results were obtained with the TEG6 linker F, which resulted in good yields with less dependence on the concentrations of added K+ and Mg2+.
In summary, photo-crosslinked mRNA–puromycin conjugates are useful templates for efficient preparation of mRNA–protein fusions. They can be prepared easily by a light-induced crosslinking reaction from almost stoichiometric amounts of mRNA template and puromycin oligonucleotide. Unlike the enzymatic ligation protocol (3), the yield of crosslink formation is not hampered by heterogeneous mRNA 3′-ends produced by T7 run-off transcription (5). Also, no exogenous splint is required for efficient crosslink formation and the resulting mRNA–puromycin templates can be used for in vitro translation and fusion formation in rabbit reticulocyte lysate and possibly other in vitro translation systems without further purification. The use of a puromycin linker that contains multiple TEG units allows efficient fusion formation under a wide range of salt conditions. This flexibility allows the salt conditions to be adjusted to the buffer needs of the protein without compromising fusion efficiency. The present method simplifies and accelerates mRNA–protein fusion preparation compared to previously reported methods (1,3) and also makes fusion preparation more amenable to automation. We expect these improvements to be most useful in in vitro protein selection, which requires repetitive rounds of fusion library preparation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. Matteucci for helpful discussions and R. Kuimelis, D. Lipovsek, M. McPherson, R. Wagner and M. Wright for suggestions on the manuscript. The plasmids for 4-4-20 scFv were a gift from B. Seed.
REFERENCES
- 1.Roberts R.W. and Szostak,J.W. (1997) Proc. Natl Acad. Sci. USA, 94, 12297–12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nemoto N., Miyamoto-Sato,E., Husimi,Y. and Yanagawa,H. (1997) FEBS Lett., 414, 405–408. [DOI] [PubMed] [Google Scholar]
- 3.Liu R., Barrick,J.E., Szostak,J.W. and Roberts,R.W. (2000) Methods Enzymol., 318, 268–293. [DOI] [PubMed] [Google Scholar]
- 4.Roberts R.W. (1999) Curr. Opin. Chem. Biol., 3, 268–273. [DOI] [PubMed] [Google Scholar]
- 5.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinden R.R. and Hagerman,P.J. (1984) Biochemistry, 23, 6299–6303. [DOI] [PubMed] [Google Scholar]
- 7.Gamper H., Piette,J. and Hearst,J.E. (1984) Photochem. Photobiol., 40, 29–34. [DOI] [PubMed] [Google Scholar]
- 8.Pieles U. and Englisch,U. (1989) Nucleic Acids Res., 17, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue H., Hayase,Y., Imura,A., Iwai,S., Miura,K. and Ohtsuka,E. (1987) Nucleic Acids Res., 15, 6131–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majlessi M., Nelson,N.C. and Becker,M.M. (1998) Nucleic Acids Res., 26, 2224–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopp T.P., Prikett,K.S., Price,C., Libby,R.T., March,C.J., Cerretti,P., Urdal,D.L. and Conlon,P.J. (1988) Biotechnology, 6, 1205–1210. [Google Scholar]
- 12.Schmidt T.G., Koepke,J., Frank,R. and Skerra,A. (1996) J. Mol. Biol., 255, 753–766. [DOI] [PubMed] [Google Scholar]
- 13.Johansson H.E., Belsham,G.J., Sproat,B.S. and Hentze,M.W. (1994) Nucleic Acids Res., 22, 4591–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue H., Hayase,Y., Iwai,S. and Ohtsuka,E. (1987) FEBS Lett., 215, 327–330. [DOI] [PubMed] [Google Scholar]
- 15.Hanes J. and Pluckthun,A. (1997) Proc. Natl Acad. Sci. USA, 94, 4937–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He M. and Taussig,M.J. (1997) Nucleic Acids Res., 25, 5132–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tate W.P. and Brown,C.M. (1992) Biochemistry, 31, 2443–2450. [DOI] [PubMed] [Google Scholar]
- 18.Tuite M.F. and Stansfield,I. (1994) Mol. Biol. Rep., 19, 171–181. [DOI] [PubMed] [Google Scholar]
- 19.Gallie D.R., Sleat,D.E., Watts,J.W., Turner,P.C. and Wilson,T.M. (1988) Nucleic Acids Res., 16, 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak M. (1983) Microbiol. Rev., 47, 1–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bedzyk W.D., Weidner,K.M., Denzin,L.K., Johnson,L.S., Hardman,K.D., Pantoliano,M.W., Asel,E.D. and Voss,E.W.,Jr (1990) J. Biol. Chem., 265, 18615–18620. [PubMed] [Google Scholar]
- 22.Mallender W.D., Carrero,J. and Voss,E.J. (1996) J. Biol. Chem., 271, 5338–5346. [DOI] [PubMed] [Google Scholar]