Abstract
Fungi biosynthesize diverse secondary metabolites, small organic bioactive molecules with key roles in fungal ecology. Fungal secondary metabolites are often encoded by physically clustered genes known as biosynthetic gene clusters (BGCs). Fungi in the genus Penicillium produce a cadre of secondary metabolites, some of which are useful (e.g. the antibiotic penicillin and the cholesterol-lowering drug mevastatin) and others harmful (e.g. the mycotoxin patulin and the immunosuppressant gliotoxin) to human affairs. Fungal genomes often also encode resistance genes that confer protection against toxic secondary metabolites. Some Penicillium species, such as Penicillium decumbens, are known to produce gliotoxin, a secondary metabolite with known immunosuppressant activity. To investigate the evolutionary conservation of homologs of the gliotoxin BGC and of genes involved in gliotoxin resistance in Penicillium, we analyzed 35 Penicillium genomes from 23 species. Homologous, lesser fragmented gliotoxin BGCs were found in 12 genomes, mostly fragmented remnants of the gliotoxin BGC were found in 21 genomes, whereas the remaining 2 Penicillium genomes lacked the gliotoxin BGC altogether. In contrast, broad conservation of homologs of resistance genes that reside outside the BGC across Penicillium genomes was observed. Evolutionary rate analysis revealed that BGCs with higher numbers of genes evolve slower than BGCs with few genes, suggestive of constraint and potential functional significance or more recent decay. Gene tree–species tree reconciliation analyses suggested that the history of homologs in the gliotoxin BGC across the genus Penicillium likely involved multiple duplications, losses, and horizontal gene transfers. Our analyses suggest that genes encoded in BGCs can have complex evolutionary histories and be retained in genomes long after the loss of secondary metabolite biosynthesis.
Keywords: comparative genomics, evolutionary biology, secondary metabolic gene clusters, duplication and loss, plant pathogen, secondary metabolism, specialized metabolism
Introduction
Gliotoxin is a secondary metabolite produced by certain fungi, including the major opportunistic human pathogen Aspergillus fumigatus (Gardiner and Howlett 2005; Raffa and Keller 2019). Secondary metabolites are bioactive molecules of low molecular weight that are not required for organismal growth but aid survival in harsh environments (Raffa and Keller 2019). Genes that participate in the biosynthesis of secondary metabolites, including gliotoxin, typically reside next to each other in fungal genomes and form biosynthetic gene clusters (BGCs) (Brown et al. 1996; Rokas et al. 2020).
Gliotoxin is a epidithiodioxopiperazine (ETP)-type fungal secondary metabolite that, in A. fumigatus, is biosynthesized by a 13-gene BGC (Scharf et al. 2012). Production of gliotoxin is implicated in A. fumigatus pathogenicity because gliotoxin suppresses the immune response of the mammalian host through diverse mechanisms, including by inhibiting protein complexes necessary for the generation of antimicrobial reactive oxygen species, decreasing cytotoxic activities of T lymphocytes, and preventing integrin activation (Yamada et al. 2000; Dolan et al. 2015; Schlam et al. 2016; Raffa and Keller 2019). The role of gliotoxin in modulating host biology suggests that it is a virulence factor and, potentially, 1 major component, or “card”, of virulence in a larger “hand” that fungi possess (Casadevall 2007; Raffa and Keller 2019). For example, virulence is attenuated in certain animal models of disease when gliP, the nonribosomal peptide synthetase gene involved in gliotoxin biosynthesis, is deleted (Cramer et al. 2006; Kupfahl et al. 2006; Sugui et al. 2007).
Fungi that produce gliotoxin require resistance to the toxin. Genes contributing to resistance include gliT, which encodes a thioredoxin reductase located within the gliotoxin BGC (Schrettl et al. 2010). Deletion of gliT in A. fumigatus results in hypersensitivity and lower resistance to the toxin (Owens et al. 2015). Other resistance genes include transcription factors, transporters, and oxidoreductases, all of which reside outside the BGC and—like gliT—are found in both gliotoxin-producing and nonproducing species (Castro et al. 2022). For example, the transcription factor RglT, which is the primary regulator of gliT (Ries et al. 2020), occurs both in Aspergillus species known to biosynthesize gliotoxin, such as A. fumigatus, as well as in species that do not biosynthesize gliotoxin, such as Aspergillus nidulans (Castro et al. 2022). Seven other genes are known to be regulated by rglT and contribute to gliotoxin resistance: gtmA (encodes a bis-thiomethyltransferase, AFUA_2G11120), kojR (transcription factor, AFUA_5G06800), abcC1 (ABC-transporter, AN7879/AFUA_1G10390), mtrA (methyltransferase, AN3717/AFUA_6G12780), AN9051 (oxidoreductase, AFUA_7G00700), AN1472 (MFS transporter, AFUA_8G04630), and AN9531 (NmrA-like family transcription factor, AFUA_7G06920) (Castro et al. 2022).
Previous research on Aspergillus showed that gliotoxin resistance genes are more widely conserved than gliotoxin BGC genes; for example, non-gliotoxin-producing species typically lack the entire BGC but harbor gliotoxin resistance genes (Ries et al. 2020; Steenwyk et al. 2020b; Castro et al. 2022). Some Penicillium species, such as P. decumbens, can produce gliotoxin (Feng et al. 2018), but the distribution and evolution of gliotoxin BGCs in other filamentous fungal lineages, such as Penicillium, remains unknown. To address this question, this study examined the evolutionary conservation of homologs of the gliotoxin BGC and resistance genes in Penicillium, a genus of fungi that is closely related to Aspergillus.
Examination of the evolutionary conservation of homologs of the gliotoxin BGC and gliotoxin resistance genes among 35 strains of 23 Penicillium species revealed that most Penicillium genomes encode fragmented gliotoxin BGCs; 2 lacked gliotoxin BGCs altogether. In contrast, some Penicillium expansum strains encoded 2 homologous gliotoxin BGCs. Codon optimization analysis revealed that genes in Penicillium BGCs are lowly optimized, consistent with the observed BGC fragmentation. In contrast, gliotoxin resistance genes are codon-optimized, suggesting that Penicillium species encounter exogenous gliotoxin in their environments. Examination of evolutionary rates revealed that genes from highly fragmented gliotoxin BGCs evolved at significantly higher rates than genes from lesser fragmented BGCs, suggesting that more fragmented BGCs experienced relaxation of selective constraints for longer. Gene tree–species tree reconciliation analyses inferred that the evolution of homologs of genes in the gliotoxin BGC of the genus Penicillium was complex, involving gene duplications, losses, and horizontal transfers, even though the ability of these fungi for gliotoxin resistance presumably remained conserved.
Materials and methods
Data collection and quality assessment
The genomes and gene annotations of 35 Penicillium strains from 23 species as well as of 2 outgroups (A. fumigatus and Aspergillus fischeri) were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/) (Supplementary Table 1).
Genome assembly and annotation quality were examined to evaluate whether the dataset is sufficient for comparative genomics. The quality and characteristics of the genomes (N50, L50, assembly size, number of scaffolds, and gene count) were evaluated using BioKIT (v0.1.0) (Steenwyk et al. 2022) (Supplementary Fig. 1). The average N50 value was ∼1.85 Mb bases, where 46% of genomes consisted of N50 values greater than 1 Mb, and the lowest N50 value was 31,119 bases for P. expansum CMP 1. Gene annotation completeness was assessed using BUSCO (v5.0.0) (Waterhouse et al. 2018) (Supplementary Fig. 2). BUSCO uses a predetermined set of near-universally conserved single-copy genes (or BUSCO genes) to identify their presence in a query proteome (characterized as single-copy, duplicated, or fragmented) or absence. The 4,181 BUSCO genes from the Eurotiales OrthoDB dataset were used (Manni et al. 2021; Zdobnov et al. 2021). Nearly all the genomes have high BUSCO gene coverage (average: 95.9% ± 3.1%), with the lowest percentages being for Penicillium coprophilum (87.9%) and P. decumbens (85.3%).
Identification and characterization of gliotoxin BGC and resistance homologs
Identification of gliotoxin BGC and resistance homologs
The representative gliotoxin BGC (BGC0000361, download date: April 2022) from the Aspergillus fumigatus Af293 reference strain was downloaded from the Minimum Information about a Biosynthetic Gene Cluster (MiBIG) database (Kautsar et al. 2020). Command-line NCBI BLASTP (Camacho et al. 2009) searches for the Af293 gliotoxin BGC against the proteome of each species were executed. Highly similar sequences were identified using an expectation value threshold of 1e−4 and a query coverage of 50%. The resulting BLAST outputs were then cross-referenced with the NCBI feature table file, which contains genome location information for each gene, and parsed to identify clusters of homologs. Lesser fragmented BGCs are defined as having at least 7/13 genes from the query gliotoxin BGC present, including gliP, encoding the core nonribosomal peptide synthetase (Castro et al. 2022); more fragmented clusters are defined as having between 3 and 6/13 genes from the gliotoxin BGC without a requirement for this cluster to include gliP. When identifying BGCs, up to 4 genes between each pair of adjacent homologs were allowed using the A. fumigatus Af293 BGC from the MiBIG database (Kautsar et al. 2020) as reference (Castro et al. 2022).
To rule out gene annotation errors in cases where genes were inferred to be absent, command-line NCBI tBLASTn searches for the Af293 gliotoxin BGC against the genome sequences were conducted. Highly similar sequences were identified using an expectation value threshold of 1e−10. The resulting outputs were analyzed, and no new presence/absence information was found.
Sequence similarity searches were also conducted for homologs of 8 gliotoxin resistance genes (abcC1/AFUA_1G10390, mtrA/AFUA_6G12780, AFUA_7G00700, AFUA_8G04630, AFUA_7G06920, rglT/AFUA_1G09190, gtmA/AFU2G11120, kojR/AFUA_5G06800), 3 of which were transcription factors (AFUA_7G06920, rglT, kojR). The gene query sequences for all 8 of these gliotoxin resistance genes were retrieved from the A. fumigatus Af293 genome (Nierman et al. 2005). An expectation value threshold of 1e−3 and a query coverage threshold of 50% were used; a lower query coverage threshold of 40% was used for the 3 transcription factors.
Estimating codon optimization
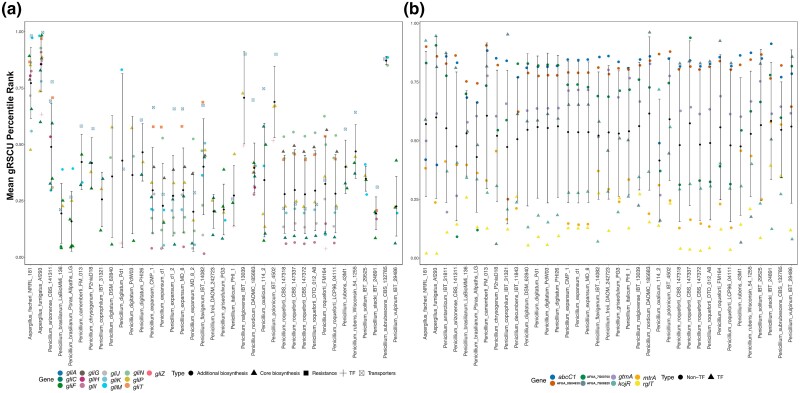
To estimate the potential functional significance of the partial gliotoxin BGCs present in Penicillium genomes, mean gene-wise relative synonymous codon usage (gRSCU) was determined for each clustered gli homolog across all proteomes using BioKIT (Steenwyk et al. 2022). This provides insight into how codon usage bias influences the expression level of a particular homolog. The percentile rankings of each of the present and clustered gli homologs were calculated using the R package dplyr (v1.0.9) (Wickham et al. 2022), and these values, for each species, were then plotted using the R package ggplot2 (Wickham 2016).
Synteny analysis
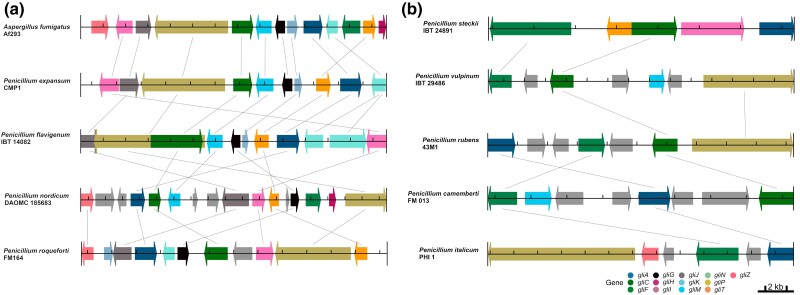
Alignments of representative Penicillium genomes with lesser and more fragmented gliotoxin BGCs were generated using a GenomeDiagram in Biopython (Cock et al. 2009). Five genomes (A. fumigatus Af293, Penicillium flavigenum IBT 14082, Penicillium roqueforti FM164, Penicillium nordicum DAOMC 185683, and P. expansum CMP1) with the largest number of different, homologous gli cluster genes above 7, and including gliP, were chosen to visualize the conservation of synteny of lesser fragmented gliotoxin BGCs across the phylogeny. Similarly, the 5 genomes (Penicillium steckii IBT 24891, Penicillium vulpinum IBT 29486, Penicillium rubens 43M1, Penicillium camemberti FM 013, and Penicillium italicum PHI 1) with the greatest number of different, homologous gli cluster genes above 3 and below 7, and not needing to include gliP, were chosen to visualize synteny of mostly fragmented BGCs across the phylogeny.
Phylogenetic analysis
Species tree inference
The evolutionary relationships of Penicillium species were obtained from a previous study (Steenwyk et al. 2019) using treehouse (Steenwyk and Rokas 2019). For 3 species with population-level data, within-species relationships were inferred using phylogenomics. To do so, protein sequences of BUSCO genes were first aligned using MAFFT (v7.490) with the –auto parameter (Katoh and Standley 2013). Codon-based alignments were generated by threading the corresponding DNA sequences onto the protein alignment with the thread_dna function in PhyKIT (v1.11.2) (Steenwyk et al. 2021). The resulting nucleotide alignments were trimmed using ClipKIT (v1.3.0) (Steenwyk et al. 2020a) with default parameters. The resulting aligned and trimmed sequences were concatenated into a supermatrix with 8,124,861 sites using the create_concat function in PhyKIT. The concatenated matrix was then analyzed with IQ-TREE 2 (v2.0.6), a software that implements a maximum likelihood framework for inferring phylogenies. All other evolutionary relationships between species were constrained following the relationships inferred in a previously published study (Steenwyk and Rokas 2019). The best-fitting substitution model (GTR + F + I + G4) was determined using ModelFinder (Kalyaanamoorthy et al. 2017).
Single-gene tree inference
To infer the evolutionary history of gli homologs, the translated amino acid sequences of individual gli genes (both clustered and unclustered) were compiled and aligned with MAFFT (v7.490) using the –auto parameter (Katoh and Standley 2013). The corresponding nucleotide sequences for each file were obtained from the CDS files for each species, using the faidx function of BioKIT (v0.1.0) (Steenwyk et al. 2022). These nucleotide sequences were then threaded onto the protein alignments using the thread_dna function of PhyKIT (Steenwyk et al. 2021), resulting in a codon-based alignment. All individual codon-based gene alignments were trimmed with ClipKIT (Steenwyk et al. 2020a) with default parameters. The trimmed alignments were used to construct a phylogeny using IQ-TREE 2 (Minh et al. 2020). The best-fitting substitution model was chosen for each gli gene using Bayesian information criteria (BIC) implemented in ModelFinder (Kalyaanamoorthy et al. 2017) from IQ-TREE 2. Branch support in each phylogenetic tree was assessed by 1,000 bootstraps using ultrafast bootstrapping approximation (Hoang et al. 2018). Tree visualization was carried out using the R packages ape (v5.6.2) (Paradis and Schliep 2019) and phytools (v1.0.3) (Revell 2012).
To characterize variation in the evolution of individual homologous genes of the gliotoxin BGC, the trimmed alignments and maximum-likelihood trees from IQ-TREE 2 were used as input into the evolutionary_rate, total_tree_length, and pairwise_identity functions of PhyKIT to estimate 2 tree-based measures of evolutionary rate and 1 sequence-based measure. Evolutionary rate is defined as the total tree length divided by the number of terminals (Telford et al. 2014; Steenwyk et al. 2021). The total tree length is the sum of all branches (Steenwyk et al. 2021).
Tree topology testing
Diverse evolutionary scenarios can result in single genes that have evolutionary histories distinct from organismal history (Steenwyk et al. 2023). To shed light on the evolutionary history of the duplicated P. expansum BGCs, topology testing was conducted. Specifically, we tested if duplication occurred within the lineage of P. expansum or deeper in the tree before the diversification of P. expansum isolates. To do so, IQ-TREE 2 (Minh et al. 2020) was used to compute log-likelihoods of a constrained tree (monophyly of homologs) and the observed tree in which a polyphyly of homologs in both clusters is seen (inconsistent with the known species tree). For each comparison, 1,000 RELL replicates (Kishino et al. 1990) were performed. The AU test results (Shimodaira 2002) was used for comparison.
Gene tree–species tree reconciliation under gene duplication, transfer, and loss
Considering the observed variation in gli gene tree topologies relative to the species phylogeny (Steenwyk et al. 2019), we used GeneRax, a tool for species tree-aware maximum likelihood-based gene family tree inference under gene duplication, transfer and loss (Morel et al. 2020). Two models were employed to reconcile the gene family trees with the species tree; the Undated DTL model that accounts for duplications, losses, and transfers, and the UndatedDL model that accounts for duplications and losses only. Both were run using the SPR tree search algorithm. The main outputs from this analysis are the inferred gene trees reconciled with the species tree, in RecPhyloxml format, along with reconciliation likelihood scores (1 per model). We then visualized these trees via Thirdkind, a program that builds svg representations of gene trees that are reconciled with the species phylogeny through the inference of loss, duplication, and transfer events (Penel et al. 2022).
Results and discussion
The gliotoxin BGC is fragmented in Penicillium species
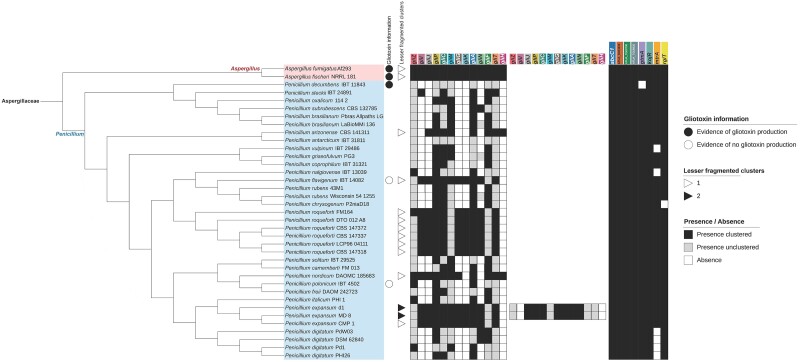
Presence/absence data of homologs of the 13 genes in the gliotoxin BGC among the 23 Penicillium species analyzed reveals that the cluster is largely fragmented in the genus Penicillium (Fig. 1). The genomes of 12 strains from 5 Penicillium species (Penicillium arizonense, P. flavigenum, P. roqueforti, P. nordicum, P. expansum), encoded lesser fragmented BGCs; the genomes of the other 23 strains spanning 18 Penicillium species encoded more fragmented BGCs (Supplementary Figs. 3–15). Two lesser fragmented BGCs, which contained 10/13 genes and 7/13 genes, were identified in P. expansum strains d1 and MD 8, respectively. Regardless of the number of lesser fragmented BGCs found, to our knowledge, none of the Penicillium species in question are known to produce gliotoxin, except P. decumbens (Feng et al. 2018), suggesting that the absence of clustering in this species may be due to strain heterogeneity and requires further exploration.
Fig. 1.
Phylogeny of Penicillium genomes. Different genera are depicted using different-colored boxes. The clades of the genera Aspergillus and Penicillium are indicated on the phylogeny. Shaded circles next to species/strain names indicate gliotoxin production information from the literature, or lack thereof (Fischer et al. 2000; Spikes et al. 2008; Knowles et al. 2020; Redrado et al. 2022). Shaded triangles in the second column depict number of clusters identified. Remaining color strips depict gene presence clustered (black), presence unclustered (gray), and absence (white) according to the requirements outlined in the Materials and methods section. Analyses of the evolutionary history of gli homologs using species tree-aware maximum likelihood-based gene family tree inference suggest that they have a complex evolutionary history characterized by gene duplications, gene losses, and horizontal transfers (Supplementary Figs. 24–36).
Gliotoxin resistance genes are broadly conserved
The presence/absence results of homologs of the 8 gliotoxin resistance genes showed that all species possessed abcC1, AFUA_8G04630, AFUA_7G00700, AFUA_7G06920, and kojR homologs (Fig. 1, Supplementary Figs. 16–23). In addition, only Penicillium species with mostly fragmented gliotoxin BGCs lacked at least 1 resistance gene, such as gtmA, mtrA, and rglT. Penicillium chrysogenum lacked both rglT and gliT, an observation consistent with the transcriptional dependency of gliT to rglT (Ries et al. 2020). These results raise the hypothesis that the genetic mechanisms likely involved in gliotoxin resistance have been largely maintained throughout Penicillium evolution, whereas BGCs are more readily lost.
Gli homologs in Penicillium species had a complex history involving gene duplications, losses, and horizontal transfers
Duplication, loss, and horizontal gene transfer of fungal BGCs and their genes are well established (Wisecaver and Rokas 2015; Rokas et al. 2020). To examine whether gli homologs had more complex evolutionary histories involving duplication and horizontal transfer, we reconciled the history of each gene with the species history using 2 models in the GeneRax software: a model that reconciles gli gene histories to the species phylogeny by allowing duplications and losses (Undated DL model) and a model that allows duplications, transfers, and losses (Undated DTL model). The DTL model had a significantly better fit than the DL model (log likelihood score of −1837.01 vs −2437.53, respectively), suggesting that the history of gli homologs included gene duplications, losses, and horizontal gene transfers (Supplementary Figs. 24–36).
Penicillium species experienced changes in gliotoxin BGC synteny over time
Examination of the synteny of genes in the BGC showed that it is mostly conserved and similar to the arrangement of the A. fumigatus Af293 gliotoxin BGC across representative, lesser fragmented BGCs, such as P. flavigenum IBT 14082 and P. expansum CMP 1 (Fig. 2). In contrast, there is extensive divergence in synteny conservation among more fragmented BGCs (Fig. 2). More specifically, 12 out of 35 Penicillium species/strains were found to have a lesser fragmented, homologous BGC. Two strains of P. expansum (d1 and MD 8) were found to have 2 BGCs, indicating variation exists in gene presence/absence within the species. For example, P. roqueforti shows population variation in the presence of gliZ, a major transcriptional regulator of gliotoxin biosynthesis (Bok et al. 2006); 5 of the 6 strains of P. roqueforti examined lack gliZ. Thus, we hypothesize that the ancestor of P. roqueforti had a gliZ homolog, but the gene was lost over time in most of the strains, highlighting the importance of population-level sampling. Overall, it can be seen that the gliotoxin BGC has experienced relocations, duplications, and horizontal transfers of its genes, specifically in P. expansum strains d1 and MD 8, a finding consistent with observations from many other secondary metabolite-producing BGCs (Rokas et al. 2018).
Fig. 2.
Conservation of gliotoxin BGC synteny for representative Penicillium species. Synteny analysis of representative genomes with lesser fragmented a) and more fragmented b) gliotoxin BGCs. Each interval along the track represents 2 kb.
Contrasting codon optimization in gli genes vs gliotoxin resistance genes
Compared to the 2 outgroup Aspergillus species—A. fumigatus and A. fischeri—gliotoxin BGC genes found in Penicillium species have much lower gRSCU percentile values, a measure of codon optimization (Steenwyk et al. 2022) (Fig. 3). Specifically, the mean gRSCU percentile rank of gliotoxin BGC genes among the Aspergillus outgroups is 0.81, while that among the Penicillium species is 0.35; these scores suggest that gli genes from Aspergillus are more codon-optimized than gli genes from Penicillium. Regardless of mean gRSCU values, gliT and gliA homologs, when present, are ranked consistently with the top 3 to 4 clustered genes. However, when considering resistance genes, the spread and range of their gRSCU values are similar across all species. The mean gRSCU percentile rank of homologs of gliotoxin resistance genes among the Aspergillus outgroups is 0.58, while that among the Penicillium species is 0.53. The similar percentile ranking of transporter and resistance genes between Penicillium species and Aspergillus species known to be resistant to exogenous gliotoxin suggests that these Penicillium species may ecologically encounter exogenous gliotoxin. This finding further supports the hypothesis that gliotoxin resistance has been maintained throughout Penicillium evolution, whereas BGC production has not.
Fig. 3.
Gene-wise relative synonymous codon usage (gRSCU) for gliotoxin BGC and resistance genes. a) Percentile rankings of gene-wise relative synonymous codon usage (gRSCU) among gliotoxin BGC genes, in comparison to all other genes. Types/functionality of each gene of the gliotoxin BGC is depicted by shape in the categories of “Core biosynthesis”, “Additional biosynthesis”, “Resistance”, “Transcription Factor”, and “Transporter”. b) Percentile ranking of gene-wise relative synonymous codon usage (gRSCU) among gliotoxin resistance genes, in comparison to all other genes. Types/functionality of each resistance gene is depicted by shape in the categories of “Non-Transcription Factor and Transcription Factor”.
Homologs of gli genes in lesser fragmented clusters are evolving at a slower rate than more fragmented clusters
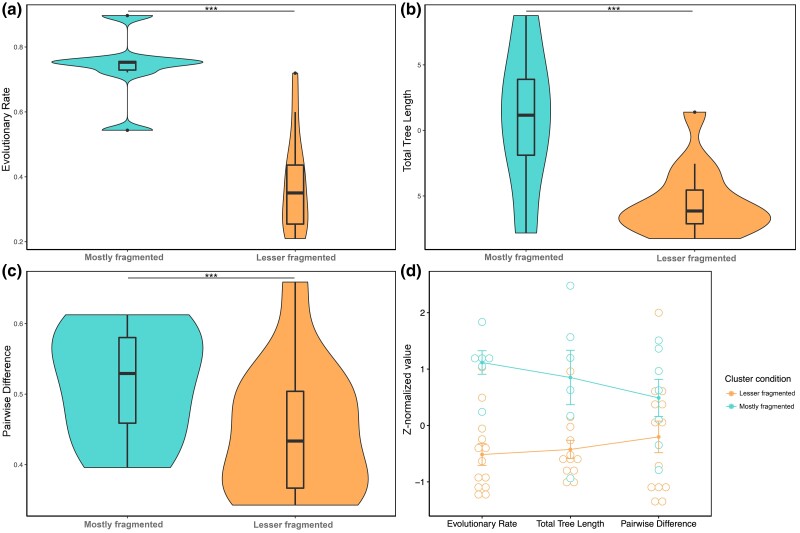
In the comparison of tree-based and sequence-based measures of evolutionary rate, homologs of gli genes from lesser fragmented clusters are evolving at a significantly slower pace (P < 0.0001; two-way ANOVA) than those from more fragmented clusters across all 3 metrics (Fig. 4, Supplementary Figs. 3–15).
Fig. 4.
Evolutionary rate comparison across gliotoxin BGCs. Multi-method comparison of evolutionary rates between lesser fragmented and mostly fragmented gliotoxin BGCs. Lesser fragmented clusters were required to contain a gliP homolog and at least 7 different genes of the cluster. Mostly fragmented clusters had no requirement to contain a gliP homolog and only needed to contain at least 3 different genes of the cluster. a) Comparison of evolutionary rates, as a function of total tree length divided by the number of taxa, between lesser fragmented and mostly fragmented gliotoxin BGCs. b) Comparison of total tree length between lesser fragmented and mostly fragmented gliotoxin BGCs. c) Comparison of pairwise identity between lesser fragmented and mostly fragmented gliotoxin BGCs. d) Comparison of the Z-normalized values of each metric between lesser fragmented and more fragmented gliotoxin BGC evolutionary rate values via a 3-way interaction plot.
A complex evolutionary history of the P. expansum gliotoxin BGCs
A tree topology test was conducted to illuminate the evolutionary origin of the multiple gliotoxin BGCs encoded in P. expansum (Supplementary Fig. 37). The maximum likelihood phylogeny suggests that a horizontal gene transfer or a combination of duplication and loss occurred between P. flavigenum and P. expansum. A combination of duplication and loss would be supported by monophyly of P. expansum gli gene homologs. After conducting tree topology tests comparing log likelihood values between the maximum likelihood phylogeny and an alternative tree wherein P. expansum homologs were constrained to be monophyletic, the likelihood scores of the unconstrained and constrained topologies were not significantly different, which suggests that the hypothesis that a combination of duplication and loss occurred within the P. expansum lineage cannot be rejected (P > 0.05, Approximately Unbiased test, Supplementary Table 2). In contrast, the GeneRax analyses (Supplementary Figs. 24–36) (Morel et al. 2020; Penel et al. 2022), support a scenario where genes present within the multiple gliotoxin BGCs in P. expansum were transferred largely between P. flavigenum and P. expansum d1, and between P. expansum d1 and P. expansum MD8 (e.g. Supplementary Fig. 31).
Conclusions
The evolution of gli homologs across the genus Penicillium likely involved multiple duplications, losses, and horizontal gene transfers. The presence/absence results of homologs of the 8 resistance genes suggest that their origins predate the Aspergillus and Penicillium genera suggesting that resistance has long been important among these species. The genes in Penicillium gliotoxin BGCs are less codon-optimized (gRSCU percentile rank mean: 0.35) compared to their Aspergillus counterparts (gRSCU percentile rank mean: 0.81) suggesting that gli genes may be more important for Aspergillus ecology. In contrast, resistance genes are similarly codon-optimized, suggesting that resistance is relevant to the lifestyle of both genera.
Although informative, this work only utilizes publicly available protein annotations of biotechnologically and medically relevant Penicillium fungi, making it important to expand upon the species/strains studied. Moreover, this same targeted gliotoxin analysis within a larger phylogeny of Aspergillus species, for which there is greater evidence of the production of this secondary metabolite, may be helpful. An analysis of gli homologs across the fungal kingdom, including careful delineation of gli orthologs and paralogs, would also provide us with more insight into the evolutionary mechanisms that gave rise to the gliotoxin BGC and to the diversity of gli homologs. In addition, expanding on the causes of conservation of lesser fragmented gliotoxin BGCs within a variety of Penicillium strains may be important, especially because evidence of production is lacking. However, it is important to note that, although there is no evidence of gliotoxin production for many of these species, this does not necessarily mean that they cannot produce the toxin (e.g. they may biosynthesize gliotoxin only under certain, yet to be identified, conditions).
Acknowledgments
We thank members of the Rokas Laboratory at Vanderbilt University for support and feedback on this work. We also thank the Vanderbilt Data Science Institute and the Beckman Scholars Program for their undergraduate enrichment opportunities. This work was performed in part using resources contained within the Advanced Computing Center for research and Education at Vanderbilt University in Nashville, TN.
Contributor Information
Charu Balamurugan, Department of Biological Sciences, Vanderbilt University, VU Station B #35-1634, Nashville, TN 37235, USA; Vanderbilt Evolutionary Studies Initiative, Vanderbilt University, Nashville, TN 37235, USA.
Jacob L Steenwyk, Department of Biological Sciences, Vanderbilt University, VU Station B #35-1634, Nashville, TN 37235, USA; Vanderbilt Evolutionary Studies Initiative, Vanderbilt University, Nashville, TN 37235, USA; Howards Hughes Medical Institute and the Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Gustavo H Goldman, Faculdade de Ciencias Farmacêuticas de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, São Paulo CEP 14040-903, Brazil.
Antonis Rokas, Department of Biological Sciences, Vanderbilt University, VU Station B #35-1634, Nashville, TN 37235, USA; Vanderbilt Evolutionary Studies Initiative, Vanderbilt University, Nashville, TN 37235, USA.
Data availability
All data necessary for confirming the article's conclusions are present within the article, figures, tables, and supplemental material (found at this Figshare repository: 10.6084/m9.figshare.23600772).
Funding
CB was supported as a Beckman Scholar by the Arnold and Mabel Beckman Foundation. JLS is a Howard Hughes Medical Institute Awardee of the Life Sciences Research Foundation. Research in AR's lab is supported by grants from the National Science Foundation (DEB-2110404), the National Institutes of Health/National Institute of Allergy and Infectious Diseases (R01 AI153356), and the Burroughs Wellcome Fund.
Literature cited
- Bok JW, Chung D, Balajee SA, Marr KA, Andes D, Nielsen KF, Frisvad JC, Kirby KA, Keller NP. 2006. Gliz, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect Immun. 74(12):6761–6768. doi: 10.1128/IAI.00780-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, Adams TH, Leonard TJ. 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci U S A. 93(4):1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics. 10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A. 2007. Determinants of virulence in the pathogenic fungi. Fungal Biol Rev. 21(4):130–132. doi: 10.1016/j.fbr.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro PA, de Colabardini AC, Moraes M, Horta MAC, Knowles SL, Raja HA, Oberlies NH, Koyama Y, Ogawa M, Gomi K, et al. 2022. Regulation of gliotoxin biosynthesis and protection in Aspergillus species. PLoS Genet. 18(1):e1009965. doi: 10.1371/journal.pgen.1009965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 25(11):1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer RA, Gamcsik MP, Brooking RM, Najvar LK, Kirkpatrick WR, Patterson TF, Balibar CJ, Graybill JR, Perfect JR, Abraham SN, et al. 2006. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot Cell. 5(6):972–980. doi: 10.1128/EC.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan SK, O’Keeffe G, Jones GW, Doyle S. 2015. Resistance is not futile: gliotoxin biosynthesis, functionality and utility. Trends Microbiol. 23(7):419–428. doi: 10.1016/j.tim.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Feng H, Liu S, Su M, Kim EL, Hong J, Jung JH. 2018. Gliotoxin is antibacterial to drug-resistant piscine pathogens. Nat Prod Sci. 24(4):225–228. doi: 10.20307/nps.2018.24.4.225. [DOI] [Google Scholar]
- Fischer G, Müller T, Schwalbe R, Ostrowski R, Dott W. 2000. Species-specific profiles of mycotoxins produced in cultures and associated with conidia of airborne fungi derived from biowaste. Int J Hyg Environ Health. 203(2):105–116. doi: 10.1078/S1438-4639(04)70015-2. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Howlett BJ. 2005. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol Lett. 248(2):241–248. doi: 10.1016/j.femsle.2005.05.046. [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautsar SA, Blin K, Shaw S, Navarro-Muñoz JC, Terlouw BR, van der Hooft JJJ, van Santen JA, Tracanna V, Suarez Duran HG, Pascal Andreu V, et al. 2020. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 48(D1):gkz882. doi: 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino H, Miyata T, Hasegawa M. 1990. Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J Mol Evol. 31(2):151–160. doi: 10.1007/BF02109483. [DOI] [Google Scholar]
- Knowles SL, Mead ME, Silva LP, Raja HA, Steenwyk JL, Goldman GH, Oberlies NH, Rokas A. 2020. Gliotoxin, a known virulence factor in the major human pathogen Aspergillus fumigatus, is also biosynthesized by its nonpathogenic relative Aspergillus fischeri. mBio. 11(1):e03361-19. doi: 10.1128/mBio.03361-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfahl C, Heinekamp T, Geginat G, Ruppert T, Härtl A, Hof H, Brakhage AA. 2006. Deletion of the gliP gene of Aspergillus fumigatus results in loss of gliotoxin production but has no effect on virulence of the fungus in a low-dose mouse infection model. Mol Microbiol. 62(1):292–302. doi: 10.1111/j.1365-2958.2006.05373.x. [DOI] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Zdobnov EM. 2021. BUSCO: assessing genomic data quality and beyond. Current Protocols. 1(12):e323. doi: 10.1002/cpz1.323. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel B, Kozlov AM, Stamatakis A, Szöllősi GJ. 2020. GeneRax: a tool for species-tree-aware maximum likelihood-based gene family tree inference under gene duplication, transfer, and loss. Mol Biol Evol. 37(9):2763–2774. doi: 10.1093/molbev/msaa141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 438(7071):1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- Owens RA, O’Keeffe G, Smith EB, Dolan SK, Hammel S, Sheridan KJ, Fitzpatrick DA, Keane TM, Jones GW, Doyle S. 2015. Interplay between gliotoxin resistance, secretion, and the methyl/methionine cycle in Aspergillus fumigatus. Eukaryotic Cell. 14(9):941–957. doi: 10.1128/EC.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Schliep K. 2019. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 35(3):526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Penel S, Menet H, Tricou T, Daubin V, Tannier E. 2022. Thirdkind: displaying phylogenetic encounters beyond 2-level reconciliation. Bioinformatics. 38(8):2350–2352. doi: 10.1093/bioinformatics/btac062. [DOI] [PubMed] [Google Scholar]
- Raffa N, Keller NP. 2019. A call to arms: mustering secondary metabolites for success and survival of an opportunistic pathogen. PLoS Pathog. 15(4):e1007606. doi: 10.1371/journal.ppat.1007606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redrado S, Esteban P, Domingo MP, Lopez C, Rezusta A, Ramirez-Labrada A, Arias M, Pardo J, Galvez EM. 2022. Integration of in silico and in vitro analysis of gliotoxin production reveals a narrow range of producing fungal species. Journal of Fungi. 8(4):361. doi: 10.3390/jof8040361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evolution. 3(2):217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- Ries LNA, Pardeshi L, Dong Z, Tan K, Steenwyk JL, Colabardini AC, Ferreira Filho JA, de Castro PA, Silva LP, Preite NW, et al. 2020. The Aspergillus fumigatus transcription factor RglT is important for gliotoxin biosynthesis and self-protection, and virulence. PLoS Pathog. 16(7):e1008645. doi: 10.1371/journal.ppat.1008645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Mead ME, Steenwyk JL, Raja HA, Oberlies NH. 2020. Biosynthetic gene clusters and the evolution of fungal chemodiversity. Nat Prod Rep. 37(7):868–878. doi: 10.1039/C9NP00045C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Wisecaver JH, Lind AL. 2018. The birth, evolution and death of metabolic gene clusters in fungi. Nat Rev Microbiol. 16(12):731–744. doi: 10.1038/s41579-018-0075-3. [DOI] [PubMed] [Google Scholar]
- Scharf DH, Heinekamp T, Remme N, Hortschansky P, Brakhage AA, Hertweck C. 2012. Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl Microbiol Biotechnol. 93(2):467–472. doi: 10.1007/s00253-011-3689-1. [DOI] [PubMed] [Google Scholar]
- Schlam D, Canton J, Carreño M, Kopinski H, Freeman SA, Grinstein S, Fairn GD. 2016. Gliotoxin suppresses macrophage immune function by subverting phosphatidylinositol 3,4,5-trisphosphate homeostasis. mBio. 7(2):e02242. doi: 10.1128/mBio.02242-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M, Carberry S, Kavanagh K, Haas H, Jones GW, O'Brien J, Nolan A, Stephens J, Fenelon O, Doyle S. 2010. Self-protection against gliotoxin—a component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog. 6(6):e1000952. doi: 10.1371/journal.ppat.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 51(3):492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Spikes S, Xu R, Nguyen CK, Chamilos G, Kontoyiannis DP, Jacobson RH, Ejzykowicz DE, Chiang LY, Filler SG, May GS. 2008. Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J Infect Dis. 197(3):479–486. doi: 10.1086/525044. [DOI] [PubMed] [Google Scholar]
- Steenwyk JL, Buida TJ III, Gonçalves C, Goltz DC, Morales G, Mead ME, LaBella AL, Chavez CM, Schmitz JE, Hadjifrangiskou M, et al. 2022. BioKIT: a versatile toolkit for processing and analyzing diverse types of sequence data. Genetics. 221(3):iyac079. doi: 10.1093/genetics/iyac079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Buida TJ III, Labella AL, Li Y, Shen X-X, Rokas A. 2021. PhyKIT: a broadly applicable UNIX shell toolkit for processing and analyzing phylogenomic data. Bioinformatics. 37(16):2325–2331. doi: 10.1093/bioinformatics/btab096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Buida TJ III, Li Y, Shen X-X, Rokas A. 2020a. ClipKIT: a multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biol. 18(12):e3001007. doi: 10.1371/journal.pbio.3001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Li Y, Zhou X, Shen X-X, Rokas A. 2023. Incongruence in the phylogenomics era. Nat Rev Genet. 24(12):834–850. doi: 10.1038/s41576-023-00620-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Mead ME, Knowles SL, Raja HA, Roberts CD, Bader O, Houbraken J, Goldman GH, Oberlies NH, Rokas A. 2020b. Variation among biosynthetic gene clusters, secondary metabolite profiles, and cards of virulence across Aspergillus species. Genetics. 216(2):481–497. doi: 10.1534/genetics.120.303549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Rokas A. 2019. Treehouse: a user-friendly application to obtain subtrees from large phylogenies. BMC Res Notes. 12(1):541. doi: 10.1186/s13104-019-4577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Shen X-X, Lind AL, Goldman GH, Rokas A. 2019. A robust phylogenomic time tree for biotechnologically and medically important fungi in the genera Aspergillus and Penicillium. mBio. 10(4):e00925-19. doi: 10.1128/mBio.00925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui JA, Pardo J, Chang YC, Zarember KA, Nardone G, Galvez EM, Müllbacher A, Gallin JI, Simon MM, Kwon-Chung KJ. 2007. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot Cell. 6(9):1562–1569. doi: 10.1128/EC.00141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford MJ, Lowe CJ, Cameron CB, Ortega-Martinez O, Aronowicz J, Oliveri P, Copley RR. 2014. Phylogenomic analysis of echinoderm class relationships supports Asterozoa. Proc R Soc Lond B Biol Sci. 281(1786):20140479. doi: 10.1098/rspb.2014.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: Elegant Graphics for Data Analysis. Switzerland: Springer. [Google Scholar]
- Wickham H, François R, Henry L, Müller K, Studio R. dplyr: A Grammar of Data Manipulation. R package version 1.0.9, https://github.com/tidyverse/dplyr, https://dplyr.tidyverse.org.2022.
- Wisecaver JH, Rokas A. 2015. Fungal metabolic gene clusters—caravans traveling across genomes and environments. Front Microbiol. 6:161. doi: 10.3389/fmicb.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A, Kataoka T, Nagai K. 2000. The fungal metabolite gliotoxin: immunosuppressive activity on CTL-mediated cytotoxicity. Immunol Lett. 71(1):27–32. doi: 10.1016/S0165-2478(99)00155-8. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Kuznetsov D, Tegenfeldt F, Manni M, Berkeley M, Kriventseva EV. 2021. OrthoDB in 2020: evolutionary and functional annotations of orthologs. Nucleic Acids Res. 49(D1):D389–D393. doi: 10.1093/nar/gkaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data necessary for confirming the article's conclusions are present within the article, figures, tables, and supplemental material (found at this Figshare repository: 10.6084/m9.figshare.23600772).