Abstract
Whitebark pine (WBP, Pinus albicaulis) is a white pine of subalpine regions in the Western contiguous United States and Canada. WBP has become critically threatened throughout a significant part of its natural range due to mortality from the introduced fungal pathogen white pine blister rust (WPBR, Cronartium ribicola) and additional threats from mountain pine beetle (Dendroctonus ponderosae), wildfire, and maladaptation due to changing climate. Vast acreages of WBP have suffered nearly complete mortality. Genomic technologies can contribute to a faster, more cost-effective approach to the traditional practices of identifying disease-resistant, climate-adapted seed sources for restoration. With deep-coverage Illumina short reads of haploid megagametophyte tissue and Oxford Nanopore long reads of diploid needle tissue, followed by a hybrid, multistep assembly approach, we produced a final assembly containing 27.6 Gb of sequence in 92,740 contigs (N50 537,007 bp) and 34,716 scaffolds (N50 2.0 Gb). Approximately 87.2% (24.0 Gb) of total sequence was placed on the 12 WBP chromosomes. Annotation yielded 25,362 protein-coding genes, and over 77% of the genome was characterized as repeats. WBP has demonstrated the greatest variation in resistance to WPBR among the North American white pines. Candidate genes for quantitative resistance include disease resistance genes known as nucleotide-binding leucine-rich repeat receptors (NLRs). A combination of protein domain alignments and direct genome scanning was employed to fully describe the 3 subclasses of NLRs. Our high-quality reference sequence and annotation provide a marked improvement in NLR identification compared to previous assessments that leveraged de novo-assembled transcriptomes.
Keywords: genome assembly, whitebark pine, Pinus albicaulis, annotation, conifer, gymnosperm
Whitebark pine (Pinus albicaulis), a white pine of western North American subalpine regions, has become critically threatened throughout its range by white pine blister rust fungus (WPBR), mountain pine beetle, wildfire, and maladaptation from changing climate—vast acreages have suffered nearly complete mortality from WPBR. As genomic data can contribute to faster, cost-effective approaches for identifying disease-resistant, climate-adapted seed sources for restoration, Neale et al. present a high-quality reference sequence and annotation—a marked improvement in candidate WPBR-disease-resistance gene identification compared to previous assessments.
Introduction
Whitebark pine (Pinus albicaulis) is a 5-needle pine of subgenus Strobus, section Quinquefoliae, subsection Strobus. Sugar pine (Pinus lambertiana) is a closely related member of the same subsection whose genome was previously sequenced (Stevens et al. 2016). Whitebark pine is found in subalpine regions in the Western contiguous United States and Canada and is most often the tree-line tree species where it occurs. Whitebark pine is of significant and somewhat unique ecological importance. Its wingless seeds are harvested, dispersed, and cached by the Clark's nutcracker (Nucifraga columbiana). Thus, there is a mutualism between the tree and the bird to the extent they have coevolved (Tomback et al. 2001). In areas of joint whitebark pine, red squirrel (Tamiasciurus hudsonicus), and grizzly bear (Ursus arctos horribilis) habitat, whitebark pine seeds from cones cached in squirrel middens are an important food source for the bears (Mattson and Reinhart 1997). In addition, whitebark pine trees provide shade to the winter snowpack that helps extend the length of the annual snowmelt.
Unfortunately, for all its ecological importance to the subalpine environment, whitebark pine has become critically threatened throughout a significant part of its natural range (Tomback et al. 2001). The primary threat is mortality due to the introduced fungal pathogen white pine blister rust (WPBR) (Cronartium ribicola). Additional threats include mountain pine beetle (Dendroctonus ponderosae), wildfire, and maladaptation due to changing climate (Tomback and Achuff 2010). At some locations in the Northern Rockies and Canada, vast acreages of whitebark pine have suffered nearly complete mortality. In December 2022, after years of conservation efforts by the Whitebark Pine Ecosystem Foundation (whitebarkfound.org) and American Forests (americanforests.org), the United States Fish and Wildlife Service listed whitebark pine as a threatened species (US FWS 2022).
There is now an urgent need to conserve and restore whitebark pine throughout its natural range. This can be effectively accomplished if a very large number of WPBR-resistant and climate-adapted seed sources can be identified and if planting stock can be produced from those sources. Forest resource managers have for many years been developing such resources using phenotypically based approaches. Identifying WPBR-resistant sources involves finding putatively resistant trees in natural stands, collecting seeds from those trees, producing seedlings, and artificially inoculating seedlings with blister rust (Sniezko et al. 2008). This approach has been effective in several white pine species, notably sugar pine and western white pine (Pinus monticola); however, the discovery process is lengthy and expensive. Likewise, identifying climate-adapted sources employs long-term genetic testing in common gardens that can take decades to complete (Bower and Aitken 2008). Thus, any new technology that could speed up and reduce the cost of identifying seed sources for restoration would be highly desired. Genomic technologies offer one such solution. Just as has been done for human disease screening and for agronomically important traits in domestic crops and livestock, the specific genes underlying these traits must first be discovered. This is the long-term goal of our research. However, this discovery is profoundly enhanced by having a well-assembled and annotated reference genome sequence. To that end, in this paper, we report on the first reference genome sequence for whitebark pine.
Materials and methods
Reference tree
An approximately 150-year-old tree was selected from the Deschutes National Forest near Bend, Oregon by a United States Department of Agriculture (USDA) Forest Service geneticist. The exact identification number and location of the tree are held in confidence to maintain its security. Scion from the tree was collected and grafted to rootstock; clones are maintained at the USDA Forest Service Dorena Genetic Resource Center in Cottage Grove, Oregon. Tissue from these clones can be obtained upon request. Cones and needle tissue were collected from the reference tree in 2006 and 2021, respectively.
DNA isolation
The protocol used to isolate the haploid megagametophyte tissue from a single fertilized whitebark pine seed was similar to previous conifer genome sequencing projects (Neale et al. 2014; Zimin et al. 2014). Haploid genomic DNA was extracted from a single megagametophyte with the Omega Biotek E.Z.N.A. SP Plant DNA Kit. The extraction followed the manufacturer's protocol with the following modifications: polyvinylpyrrolidone (0.01 g) was added to the tissue prior to lysis, and the lysis time was extended to 1.5 h. The extracted DNA was quantified on a Qubit 2.0 (42.2 ng/μL), a Nanodrop ND-1000 (A260/280: 1.83; A260/230: 2.11), and quality was evaluated on an electrophoresis gel (fragment sizes > 20,000 bp).
DNA sequencing
Illumina short read
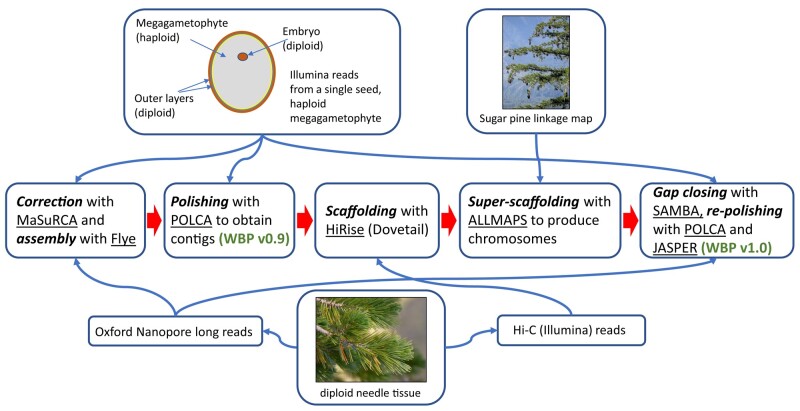
DNA was sequenced at the DNA Technologies and Expression Analysis Core at the UC Davis Genome Center. First, DNA libraries were prepared for whole-genome shotgun sequencing with no unique molecular identifiers using 400-ng DNA and the QIAseq FX DNA Library Kit from Qiagen. Then, sequencing was conducted on 3.5 lanes of a NovaSeqS4 with Illumina 150-bp paired-ends sequencing with an approximate insertion size of 400 bp, nonoverlapping ends, and 75× coverage. See Fig. 1 for the flow chart of the sequencing and assembly steps.
Fig. 1.
Flow chart for sequencing and assembly steps for the whitebark pine genome. The center row presents the sequence of activities (in boldface italic type) and software tools (underlined). The top and bottom rows describe the starting tissues, sequencing platforms, and sequence read and linkage map inputs and the thin arrows indicate where in the assembly process these inputs entered. The intermediate whitebark pine assembly (v0.9) emerges at the second step in the middle row, while the final assembly (v1.0) emerges at the end step of the middle row. WBP, whitebark pine. Photo credits: Sugar pine inset photograph by Mitch Barre via Wikimedia under Creative Commons Atribution-Share Alike 2.0 Generic license; Whitebark pine needles inset photograph by co-author Patrick McGuire.
Oxford Nanopore long read
For nanopore sequencing, a protocol similar to previous conifer genome sequencing projects was used (Scott et al. 2020; Neale et al. 2022). Because sequencing by the Oxford Nanopore Technologies (ONT) platform requires more DNA per run and cannot be amplified to maintain read length, needle tissue was used for DNA extraction and sample preparation. High molecular weight DNA was extracted following the protocol described in Workman et al. (2018). Briefly, tissue was ground in liquid nitrogen with a mortar and pestle for 20 min to properly disrupt tissue. This is followed by lysis in a nuclear isolation buffer (NIB) containing spermine, spermidine, triton, and β-mercaptoethanol in a 50-mL Falcon tube (Supplementary Table 1), with end-over-end rotation of the tube at 4°C for 15 min. The resulting lysed sample is filtered through a Steriflip and then centrifuged 1,900 × g for 20 min at 4°C. The supernatant was decanted, and the pellet was resuspended in 1 mL of NIB with a paintbrush. The resuspension was brought to a total volume of 15-mL NIB and centrifuged 1,900 × g for 10 min at 4°C. These steps were repeated (discard supernatant, resuspend pellet, and wash) until the supernatant was clear, usually 2–3 times. The final pellet was resuspended into 1-mL 1× HB buffer per gram of initial tissue. Nuclei can then be spun at 7,000 × g for 5 min, supernatant was removed, and pellets were snap frozen in liquid nitrogen and stored at −80°C for later DNA extraction.
Extracted nuclei were then lysed and gDNA precipitated using the Circulomics Nanobind Plant Nuclei Big DNA Kit, alpha version (EXT-PLH-001). DNA was sheared to 25 kb with the Megaruptor 2, and library preparation was performed according to the ligation sequencing kit (LSK109, ONT). Then, 1 μg of purified genomic DNA was input into the ligation sequencing kit (LSK108-LSK109, ONT). Samples were sequenced on R9.4 flowcells on either the minION or PromethION and then base-called using guppy 4.011-5.0.13 depending on the time of sequencing.
Assembly
The initial contig assembly utilized both ONT and Illumina data with a hybrid approach, where the ONT reads were first corrected using the Illumina reads, and then the corrected reads were assembled. Following the strategy used in our previous work assembling loblolly pine (Zimin et al. 2014) and other conifers, the whole-genome Illumina libraries were prepared from haploid megagametophyte tissue collected from a single seed (Fig. 1). This resulted in the reduction of the effective genome size, lowered the resource requirements on the hardware, and produces a more accurate assembly overall.
The contigs were assembled with MaSuRCA v4.0.6 (Zimin et al. 2017a). MaSuRCA used the “super-reads” technique to compress high-coverage Illumina reads into low (2× to 3×) coverage of much longer super-reads by first constructing a k-mer graph from k-mers (k = 99 here) found in the Illumina reads. The k-mers become nodes in a k-mer graph, and exact overlaps of k − 1 bases between k-mers are the edges. The super-reads technique used the graph to extend each Illumina read in 5′ and 3′ directions as far as possible, as long as the extension was unambiguous. The extended read is called a super-read. Many Illumina reads extend to the same super-read. The super-reads were then used to error-correct the ONT reads, essentially producing miniassemblies for each ONT read by using the ONT read as a template. This process yielded highly accurate “mega-reads,” with typically one or a few mega-reads covering each ONT read. The mega-reads were then assembled with a modified version of the Flye assembler (Kolmogorov et al. 2019).
Table 1 lists the data that were used for the initial contig assembly of the whitebark pine genome along with the sizes of intermediate super-reads and mega-reads. The Flye assembler has an internal limitation of a total input sequence of 549 Gb. To stay within this limit, a subset of the longest mega-reads was used as input to the Flye assembler. The Flye assembly process was also modified. The assembler was interrupted after the initial contig (called disjointig in the Flye paper terminology) building stage to skip the initial contig consensus. This was necessary because the Flye consensus algorithm would otherwise attempt to create a >50-Tb file of alignments of mega-reads to the contigs and eventually fail on data of this size. The consensus step was not needed because the mega-reads supplied to Flye were highly accurate. After skipping the consensus, the assembly continued with the repeat resolution and scaffolding steps. This process is automated in MaSuRCA (as of v4.0.7 and higher). The new versions automatically perform the necessary steps when the detected genome size is over 10 Gb. The statistics for this initial contig assembly (v0.1) are listed in Table 2. The key metric in Tables 1 and 2 is N50, a measure of contiguity of sequencing reads or assembled contigs. It is defined as the length of the shortest sequence for which longer and equal-length sequences contain at least half of the total sequence in the read data or assembly. For assemblies, N50 is a weighted average of the contig or scaffold lengths. For long-read technologies, which generate reads with widely varying lengths, N50 is a weighted average of the read lengths.
Table 1.
Quantitative statistics of the initial sequencing data and intermediate processed reads.
| Total sequence (bp) | Count | N50 size (bp) | |
|---|---|---|---|
| Original sequence data | |||
| Illumina reads | 2,511,282,622,124 | 18,120,442,068 | 151 |
| Nanopore reads | 571,078,527,938 | 53,691,131 | 19,989 |
| Nanopore ultralong reads | 322,256,363,438 | 15,170,331 | 42,785 |
| Derived data | |||
| Super-reads | 85,674,725,052 | 163,046,437 | 1,228 |
| Mega-reads (subset used for Flye assembly) | 548,999,989,868 | 23,818,550 | 23,140 |
Super-reads were produced from Illumina reads. Mega-reads were built from super-reads using ONT reads as templates. Each ONT read yielded 1 or several nonoverlapping mega-reads.
Table 2.
Quantitative statistics of the intermediate and final assembly steps.
| Assembly version | Total sequence (bp) | Number of contigs | N50 contig size (bp) | Number of scaffolds | N50 scaffold size (bp) | Consensus quality (%) |
|---|---|---|---|---|---|---|
| v0.1 | 26,961,471,748 | 194,849 | 389,205 | 194,178 | 397,606 | 99.97 |
| v0.9 | 27,687,627,594 | 101,182 | 727,847 | 100,511 | 735,520 | >99.999 |
| v1.0 | 27,605,955,854 | 92,740 | 537,007 | 34,176 | 2,005,774,401 | >99.999 |
The initial assembly (v0.1) was performed with the MaSuRCA assembler. That initial assembly was followed by scaffolding with SAMBA and polishing with POLCA to yield assembly v0.9. That assembly was filtered for redundancy, scaffolded by the HiRise scaffolder, and then super-scaffolded into chromosome-sized scaffolds with the ALLMAPS software, followed by SAMBA gap closing and polishing with JASPER to yield the final assembly (v1.0). N50 contig size decreased going from v0.9 assembly to v1.0 assembly because HiRise scaffolder breaks contigs that are inconsistent with the HiC data. Consensus quality was evaluated with POLCA software.
The initial contig assembly was followed by long-read contigging/scaffolding with SAMBA scaffolder (Zimin and Salzberg 2022). The original, uncorrected ONT reads that were 10 kb or longer were used for SAMBA scaffolding. Some of these reads may have been omitted in the contig assembly because of the input size limitation of the Flye assembler. The first iteration of SAMBA was very conservative, requiring ONT reads to match for a minimum of 9 kb to the ends of 2 contigs to join them. In the second iteration, that requirement was reduced to 4 kb. The scaffolder merges contigs and computes the consensus sequence filling the gap using the sequence of multiple ONT reads spanning the gap. Therefore, the “patches” that filled the gaps may have a higher error rate.
The final step of the contig assembly was polishing the assembly with Illumina data in 2 passes using POLCA (Zimin and Salzberg 2020). The initial quality of the contigs after the SAMBA scaffolding was estimated to be 99.988% or QV39. After 2 rounds of POLCA polishing, the consensus quality was 99.999% or QV50, corresponding to an estimated error rate of 1/100,000 bases. These steps resulted in assembly v0.9 (Fig. 1), with statistics shown in Table 2.
Next, the contigs were scaffolded with OmniC reads sequenced from the needle tissue (a variant of the HiC proximity ligation technique) with the HiRise scaffolder (Putnam et al. 2016) at Dovetail Genomics (now part of Cantata Bio). After the HiRise scaffolding, redundant duplicate contigs were identified. These exist because assemblers frequently leave extra copies of repeats or extra copies of alternative haplotype sequences already represented in the contigs as short contigs in the assembly. All 20,661 “short” contigs that were shorter than 10,000 bp were aligned to the rest of the assembly with nucmer aligner. These contigs contained 96,950,513 bp of sequence with N50 of 5,302 bp. Any contig that was shorter than 10,000 bp and that aligned to an interior of another contig with >95% identity over >95% of its length was removed from the set of short contigs. The remaining 1,371 short contigs containing 6,341,804 bp were added back to the assembly.
Following scaffolding with the OmniC data and redundancy filtering, 2 linkage maps (Weiss et al. 2020; De La Torre 2023) for the closely related sugar pine genome were utilized to super-scaffold the assembly to obtain chromosome-sized scaffolds. The 2 maps had a total of 7,767 markers (mostly short sequences). Of these markers, 2,959 mapped uniquely to the whitebark pine scaffolds. ALLMAPS software (Tang et al. 2015) was utilized to produce chromosome-sized scaffolds using the alignments of markers to the scaffolds and marker positions in the map. The 2 final steps following the scaffolding were additional gap closing with the SAMBA tool (Zimin and Salzberg 2022) using the ONT reads followed by polishing with the JASPER polisher (Guo et al. 2023) that used the Illumina data (Fig. 1). This additional polishing was needed because in the places where gaps in the scaffolds were filled, consensus computed only from the ONT reads that spanned these gaps would have resulted in low-quality sequence. The statistics of this final assembly (v1.0) are listed in Table 2.
Annotation and comparative genomics
Transcriptomic evidence
A combination of public RNA-seq (Illumina PE) data from mixed tissue types was employed for the first stage of annotation (PRJNA703422 and PRJNA352055). Illumina short reads were aligned to the v0.9 reference genome with HISAT2 v2.2.1, including the following flag to accommodate long introns --max-intronlen 25,00,000 (Kim et al. 2019). All libraries with mapping rates that exceeded 95% alignment and contained a minimum of 20 million reads were retained for evaluation (Supplementary Table 2). In addition, a set of recently de novo-assembled libraries (Illumina NovaSeq 150-bp PE), from needle tissue of 6 individuals from a single half-sib family from Shadow Lake 39, Mount Rainier National Park collected and flash frozen by the USDA Forest Service Dorena Genetic Resource Center (BioProject PRJNA933606) were used as further evidence (Supplementary Table 3). These were assembled with the Oyster River Protocol (ORP) workflow (v2.2.5; MacManes 2018), a combined pipeline that works with Trinity v2.9.1 (Haas et al. 2013), rnaSPAdes v3.13 (Bushmanova et al. 2019), and TransABySS v2.0.1 (Robertson et al. 2010) assemblers to generate a single reference assembly. This assembly was subsequently clustered at 90% with USearch (v9.0.2132; Edgar 2010), frame-selected with Transdecoder (v5.5.0; https://github.com/TransDecoder/TransDecoder), and filtered with eggNOG (v4.1; Huerta-Cepas et al. 2019). This transcriptome was further filtered for short fragments (<300 bp) with SeqKit (v2.2.0; Shen et al. 2016) and aligned to the v0.9 genome reference via Minimap2 ([-ax splice:hq -uf]; v2.24; Li 2018). Secondary alignments produced by Minimap2 were removed via SAMtools (v1.9; Danecek et al. 2021).
Structural annotation of the v0.9 genome
The v1.0 genome contains the same sequence as v0.9 with the only difference being that the contigs and scaffolds were rescaffolded into chromosome-sized scaffolds in v1.0, and a few scaffolds sequences were split. The v0.9 genome sequence was available earlier and the annotation was performed on that sequence given the scale of the assessment for the nucleotide-binding leucine-rich repeat receptor (NLR) classification. See Fig. 2 for the flow chart of the annotation steps. Initial assessment of the v0.9 reference genome was conducted with BUSCO v5.2.2 with the embryophyta database (odb10; Manni et al. 2021). Subsequently, repeat sequences were identified de novo with a combination of self-to-self comparisons and structural identification with RepeatModeler v2.01 (Flynn et al. 2020). The twice soft-masked genome was used as input to BRAKER v2.1.5 as well as the aligned RNA-Seq reads from NCBI (Brůna et al. 2021). The set of predicted proteins was filtered with eggNOG v5.0.2 and evaluated with QUAST v5.2.0 (Gurevich et al. 2013) and BUSCO (embryophyta). In parallel, StringTie2 v2.2.1 was run using different sets of transcriptomic input (Kovaka et al. 2019). The first gene space assembly utilized only the HISAT2 aligned short reads as input to StringTie2, while the second assembly was run in hybrid mode including both the HISAT2 alignments and full-length assembled transcripts assembly aligned to the genome with Minimap2 (Li 2018). Protein-coding sequences were generated from all StringTie2 runs with Gffread v0.12.1 (Pertea and Pertea 2020) and frame-selected and filtered with Transdecoder v5.5.0 and eggNOG v5.0.2. Supplemental Transdecoder scripts were used to obtain the coordinates of the frame-selected transcripts in the context of the genome. Transcripts that did not have a corresponding genome alignment after this filtering step were removed from the final coding sequence and protein sequence files. The final proteins were evaluated with BUSCO (embryophyta), EnTAP v0.10.8, and AGAT v1.0.0 (Hart et al. 2020; Dainat et al. 2022). EnTAP was run as a reciprocal BLAST search to estimate the alignment rate at 50/50 coverage between the query sequence and target databases (NCBI's RefSeq v208 and UniProt). AGAT was employed to provide basic filtering for structural anomalies and quantify statistics regarding structural aspects of the protein-coding regions (Dainat et al. 2022). After EnTAP annotation, transcripts without a similarity search or eggNOG match were scanned for protein domains using InterProScan, and those lacking any identifiable protein domains were removed.
Fig. 2.
Flow chart for annotation steps. Oval rectangles present the activities (in boldface italic type) and the software tools (underlined). Protein coding annotations v0.9 and v1.0 utilized the same input RNA libraries and alignments via HiSAT2. The first version of the annotation (v0.9) relied primarily on StringTie2 to resolve transcripts and incorporated additional models from high-quality NLRs curated from an independent BRAKER2 run. The second version of the annotation (v1.0) was conducted with EASEL that integrates direct evidence-based evaluations and high-quality ab initio predictions. Both annotations were functionally annotated with EnTAP and evaluated with benchmarks generated by BUSCO and AGAT.
Annotating the v1.0 genome
Initial assessment of the v1.0 genome was conducted with BUSCO v5.4.5 with the embryophyta database. Realignment of RNA evidence against the scaffolded reference (v1.0) was provided as input to the EASEL pipeline with a filtering threshold of 0.65 to generate a set of protein-coding gene predictions (Webster et al. 2023). The final proteins were evaluated with BUSCO (embryophyta), functionally annotated with EnTAP, and summarized with AGAT (Fig. 2).
NLR identification on the v0.9 genome
NLR proteins are a major family of plant disease resistance genes, which are categorized into the TNL, CNL, and RNL subfamilies based on their N-terminal domain. Three methods were utilized to generate a more complete representation of potential NLRs in whitebark pine: InterProScan, RGAugury, and NLR-Annotator. NLRs were identified from a de novo-assembled transcriptome, whole-genome scanning, and the genome annotation to provide comparison across the available genomic resources.
InterProScan v5.35-74.0 and RGAugury v1.0 identified NLRs from the protein sequences of the genome annotation through protein domain scanning (Li et al. 2016; Paysan-Lafosse et al. 2023). InterProScan was used to identify the NB-ARC, TIR, coiled-coil (CC), RPWB, and LRR domains using the Pfam, Gene3D, SUPERFAMILY, PRINTS, SMART, and CDD databases. The GFF3 file produced by InterProScan was filtered using a custom Python script to remove all entries without at least 1 NLR domain, to speed up the identification and classification steps downstream. Custom R scripts were employed to identify the NLRs and classify them into their subfamilies based on the N-terminal domain. Those with a TIR domain are TNLs, those with a CC domain are CNLs, and those with an RPW8 domain are RNLs. Subfamilies included both complete NLRs (containing N-terminal, NB-ARC, and LRR domains) and those missing just the LRR domain. Sequences without an N-terminal domain (NB-ARC only and NB-ARC-LRR) were considered unclassified. The RGAugury pipeline is quite similar, but it first implements a filtering step based on sequence similarity to the Resistance Gene Analog database before performing domain scanning with InterProScan. RGAugury was better able to identify CNL-type NLRs than InterProScan, which struggled to identify the CC N-terminal domain.
NLR-Annotator v2.0 was used to identify potential NLRs directly from the genome sequence using NLR-associated DNA motifs (Steuernagel et al. 2020). From the genome annotation, genes overlapping at least 80% of the predicted NLRs based on the NLR-Annotator boundaries were selected as potential NLRs with BEDTools v2.29 (Quinlan and Hall 2010) (Fig. 2). Custom R scripts were employed to combine NLR annotation results from the 3 methods and identify which annotations were unique to each method. To reintroduce gene models from the BRAKER annotation, gene predictions that overlapped at least 90% of the boundaries of a complete NLR (CNLs and TNLs) were retained and included in the primary genome annotation.
Results and discussion
Sequencing
Previously developed sequencing methods to analyze other conifers (Scott et al. 2020) were used to generate a combination of short-read (Illumina) and long-read (ONT) sequencing data in whitebark pine. This fusion of technologies brings together the advantages of both approaches: leveraging long nanopore reads to span repetitive sequences commonly found in conifers producing a highly contiguous genome assembly (Fig. 1). Although the error rate of nanopore sequencing is steadily improving, it still poses challenges for the final assembly. By integrating these long reads with highly accurate, albeit shorter, Illumina reads, a more precise assembly was produced while maintaining a high level of contiguity.
First, short-read Illumina sequencing data were generated from DNA of a megagametophyte. The haploid megagametophyte DNA precludes the typical difficulties associated with diploid DNA and natural genetic variation between alleles. From this DNA, ∼2.5 Tb of sequence was generated for an estimated ∼100× coverage (Table 1).
It has previously been found that short-read sequencing, especially in conifers, results in low contiguity as the highly repetitive areas typical to these genomes are impossible to assemble with short reads alone. Complicating this issue, existing long-read sequencing methods require relatively large amounts of DNA and achieving the long-read length precludes the use of PCR. As an alternative, high-molecular-weight genomic DNA from needle tissue was extracted from the same tree (Workman et al. 2018). Using a combination of cryogenic tissue grinding and nuclei extraction, high-quality DNA was obtained, which was then subjected to either long-read (N50 20 kb, 571 Gb, ∼23×) or ultralong-read (N50 42.8 kb, 322 Gb, 13×) nanopore sequencing.
Assembly
The MaSuRCA assembler transformed the Illumina reads into super-reads (see Methods and Fig. 1). Table 1 shows that the super-read transformation turned over 18 billion 151-bp Illumina reads into about 163 million super-reads. Half of the sequence in the super-reads was in sequences of 1,228 bp or longer. MaSuRCA then used super-reads to correct the ONT reads by building miniassemblies of overlapping super-reads for each ONT read. These miniassemblies are produced using the ONT reads as templates, and they are called mega-reads. Mega-reads are long and they have a very low error rate, less than 0.5%. The mega-reads algorithm resulted in producing about 24 million mega-reads with an N50 size of 23,140 bp. The MaSuRCA assembly (v0.1) (Table 2) was followed with scaffolding with SAMBA and polishing with POLCA, resulting in assembly v0.9 (Table 2). The v0.9 assembly was then scaffolded with HiRise with OmniC data and super-scaffolded with ALLMAPS using the alignments of markers to the scaffolds and marker positions from the sugar pine map. Figure 3 shows the alignment of the markers from the sugar pine maps to the whitebark pine super-scaffolds. Some discrepancies between the scaffolds and the map were observed in chromosomes 1, 2, 3, 5, 6, and 11. These discrepancies could be due to interchromosomal rearrangements between the sugar pine and whitebark pine genomes. However, they could also be due to misassemblies in the scaffolds of whitebark pine, which cannot be resolved with the currently available data. Scaffolding with ALLMAPS resulted in 24,069,114,767 bp of sequence anchored to the chromosomes of which 23,671,235,725 bp was also oriented. Additional gap closing was then applied to the scaffolds with the SAMBA tool that used original uncorrected ONT reads to fill gaps in the scaffolds. SAMBA closed 1,484 gaps in the assembly, adding 9,065,412 bp of sequence to the assembly. Finally, the JASPER tool was applied to polish the assembly with the Illumina reads. The final polished assembly (v1.0) (Table 2) has an error rate of less than 1 error in 100,000 bases, and it contains 27,605,955,854 bp of sequences in 34,176 scaffolds with N50 contig size of 537,007 bp. Approximately 87.2% (24,072,309,274 bp) of the total sequence was placed on the 12 chromosomes.
Fig. 3.
Alignment of the sugar pine linkage map markers to the whitebark pine super-scaffolds. The individual chromosome plots are produced by the ALLMAPS software. The vertical bars in the middle of each of the 12 panels represent the chromosomes. The individual scaffolds of a chromosome are indicated in white or gray shading within those vertical bars. The 2 linkage maps are shown alongside each chromosome representation with marker alignments indicated with fine lines from the central chromosome representation to the linkage maps.
Annotation
Identifying and masking repetitive regions
Prior to the alignment of the transcriptomic short reads, repeat identification with RepeatModeler generated a custom library of 2,576 unique repeat sequences, of which 558 could be classified. This repeat library was used with RepeatMasker to softmask 77.6% of the genome sequence (Fig. 2; Table 3 and Supplementary Table 4). The overall repetitive content was comparable to Pinus taeda at 74% and P. lambertiana at 79% (Stevens et al. 2016). The majority of the repetitive elements were LTRs, which comprised almost 42% of the genome, and roughly 32% of the genome was unclassified repetitive sequences (Supplementary Table 4). The high proportion of unclassified elements is likely due to RepeatModeler being unable to classify many of the repeats in the generated custom repeat library that was used to mask the genome, as LTRs comprised only 55% of the repeat content in whitebark pine where they usually contribute around 70% of the TE content in conifers (De La Torre et al. 2014; Stevens et al. 2016; Fujino et al. 2023).
Table 3.
Statistics on the structural annotation of the whitebark pine reference genome assembly.
| v0.9 assembly | v1.0 assembly | |
|---|---|---|
| Completeness (C = complete; S = single copy; D = duplicated; F = fragmented; M = missing) | ||
| Genome BUSCO v5 | C:55.3% (S: 45.5%; D: 9.8%), F: 24.3%, M: 20.4% | C: 65.5% (S: 57.1%; D: 8.4%), F: 19.1%, M: 15.4% |
| Annotation BUSCO v5 | C: 70.6% (S: 43.2%; D: 27.4%), F: 15.1%, M: 14.3% | C: 73.9% (S: 21.7%; D: 52.2%), F: 5.5%, M: 20.6% |
| Protein-coding genes | ||
| Total number of genes | 27,010 | 27,555 |
| Number of single-exon genes | 4,836 | 6,9789 |
| Number of multi-exon genes | 22,174 | 20,577 |
| Mono:multi ratio | 0.22 | 0.33 |
| Total number of transcripts | 47,911 | 58,831 |
| Transcript N50 | 1,578 bp | 1,590 bp |
| Longest intron | 1.39 Mb | 2.45 Mb |
| Average number of exons | 6 | 7.5 |
| Functional annotation | ||
| EnTAP annotation rate (gene family) | 92.8% | 99.10% |
| EnTAP annotation rate (sequence similarity search) | 86.5% | 71.45% |
| Repeat detection | ||
| Softmasked % (LTR %) | 77.6% (42%) | 77.4% (41.5%) |
RNA sequence data for annotation and transcriptome assembly
A total of 12 Illumina RNA-seq libraries (Supplementary Table 2) were mapped to the whitebark pine reference v0.9 genome following quality control. The final set of selected Illumina libraries ranged from 23.9 to 66.9 M reads and aligned well to the reference (94.8–96%). These alignments were used with BRAKER and StringTie2 to generate the draft genome assembly (Kovaka et al. 2019; Brůna et al. 2021). Two RNA-seq libraries (SRR13823648 and SRR13823649) generated from megagametophyte tissue were used for a de novo transcriptome assembly that was utilized for NLR annotation (Supplementary Table 2). This transcriptome assembly consisted of 37,586 transcripts and had a BUSCO (embryophyta) completeness of 92.6% (S: 88.8%; D: 3.8%).
Preliminary annotation of the v0.9 genome
The preliminary protein-coding predictions generated from BRAKER amounted to an overestimate with 636.6 K initial models (BUSCO: C: 45.0% [S: 35.6%; D: 9.4%]; Supplementary Table 5). Following basic gene family level filtering with eggNOG, a total of 219.5 K transcripts (BUSCO: C: 45.0% [S: 35.7%; D: 9.3%]; N50: 1,164 bp; longest intron: 140 kb) were retained. The short reads processed by StringTie2 produced a total of 63,123 transcripts (BUSCO: C: 70.9% [S: 43.3%; D: 27.6%]; N50 2,281 bp). These transcripts were filtered via Transdecoder/eggNOG, leaving a total of 48,567 transcripts (BUSCO: C: 70.5% [S: 43.2%; D: 27.3%]; N50 1,578 bp; longest intron: 1.39 Mb).
To improve upon challenges associated with short-read alignment against the complex and repetitive conifer genome, the de novo-assembled transcripts resulting from an independent transcriptomic sampling were aligned at 71% to the genome with Minimap2 (Supplementary Table 3). These transcripts were then used as long-read input for a hybrid long- and short-read transcriptome assembly using StringTie2. The hybrid run of StringTie2 generated a total of 62,936 transcripts (BUSCO: C: 71.4% [S: 37.4%; D: 34.0%]; N50 1,807 bp). These gene models were filtered via Transdecoder/eggNOG, resulting in a total of 45,380 transcripts (BUSCO: C: 70.4% [S: 45.7%; D: 24.7%]; N50 1,515 bp; longest intron: 1.02 Mb; Supplementary Table 5).
As an additional metric for completeness, the de novo-assembled transcripts were aligned to the reference genome independently, resulting in a total of 66,233 unique alignments (BUSCO: C: 88.50% [S: 46.60%; D: 41.90%]; N50 2,217 bp; Supplementary Table 5). These alignments represent variation and gaps and do not directly translate to viable protein-coding models but can provide a benchmark for completeness.
Filtering the v0.9 genome annotation
The Transdecoder/eggNOG-filtered StringTie2 short-read predictions were selected as the best overall annotation. This annotation was further refined by removing transcripts without an EnTAP similarity search or eggNOG annotation that also lacked any protein domains identified using InterProScan, reducing the annotation by 683 genes. An additional 27 complete NLR genes identified from the genome using NLR-Annotator, and overlapping a gene model generated by BRAKER, were added to the annotation (Supplementary Table 6). This final set consisted of 27,010 genes and represented a total of 47,911 transcripts. The annotation had a BUSCO completeness of 70.6% (S: 43.2%; D: 27.4%) and an EnTAP similarity search annotation rate of 86.5%, and the longest intron recorded was 1.39 Mb in length (Table 3). The annotated gene space of whitebark pine is larger and more representative than those of P. lambertiana and P. taeda, which contained 13,936 and 9,024 high-confidence genes with BUSCO completeness of 53 and 30%, respectively (Stevens et al. 2016). The genome annotations of the spruce (Picea) species range from 35 to 49% completeness (Gagalova et al. 2022). More recent conifer genome assemblies report higher BUSCO completeness, such as Sequoia sempervirens at 65.5% completeness (Neale et al. 2022), Pinus tabuliformis at 84% (Niu et al. 2022), and Cryptomeria japonica at 91.4% (Fujino et al. 2023).
Compared to the 70.6% BUSCO completeness of the genome annotation, at the genome level, the whitebark pine genome accounted for only 55.3% BUSCO completeness using the embryophyta lineage, likely due to challenges associated with the predictions across long introns as well as the abundant pseudogenes and high repeat content (Table 3). This result is typical of conifer genomes and, despite this, the whitebark pine genome BUSCO completeness was slightly higher compared to that of several other recently assembled conifer genomes (sugar pine, spruce, coast redwood, and Chinese pine; Stevens et al. 2016; Gagalova et al. 2022; Neale et al. 2022; Niu et al. 2022, respectively).
Annotation of the v1.0 genome
A total of 27,555 genes and 58,831 transcripts were identified using the EASEL pipeline (Webster et al. 2023). The BUSCO completeness was slightly improved at 73.9% (S: 21.7%; D: 52.2%); the ratio of monoexonic to multiexonic genes was 0.33; and the longest intron was 2.45 Mb in length, nearly double the longest intron identified in the v0.9 annotation (Table 3).
NLR identification on the v0.9 genome
NLRs are a major class of disease resistance genes that recognize specific virulence factors. They have a characteristic domain structure with one of 3 canonical N-terminal domains, a nucleotide-binding domain, and a leucine-rich repeat domain. NLRs can be divided into subfamilies based on their N-terminal domain; TNLs contain a TIR domain, CNLs contain a CC domain, and RNLs contain an RPW8 domain (Van Ghelder et al. 2019). The combination of the 3 software methods used to identify NLRs from v0.9 of the genome, before scaffolding (InterProScan, RGAugury, and NLR-Annotator), was necessary to fully describe all 3 types of NLRs, as can be seen from the overlap between complete NLRs identified from the genome annotation by each method or lack thereof (Fig. 4a). The 3 methods were able to independently identify the majority of TNL-type NLRs. Thirty-four TNLs were identified by all 3 methods, 11 were identified by domain scanning methods only, 11 were unique to NLR-Annotator, and 1 was unique to InterProScan. Here, the domain scanning methods performed equally well, and the results of NLR-Annotator were a useful addition to the set of complete NLRs. For identifying RNLs, InterProScan was necessary as the other 2 programs cannot identify the RPW8 domain and would otherwise identify these as NLRs missing an N-terminal domain. For CNLs, RGAugury was necessary as NLR-Annotator and InterProScan were not as effective in identifying CNLs; 7 complete CNLs were identified using RGAugury only.
Fig. 4.
Results of NLR annotation methods. a) Within the genome annotation, complete NLRs identified by each method and annotations with support from multiple methods. In each cluster, the upper-left circle (yellow) represents NLRs identified only using InterProScan; the upper-right circle (coral/red) represents NLRs identified using only RGAugury; and the lower circle (pink) represents NLRs identified using only NLR-Annotator and supported by the genome annotation. b) NLRs identified by input type: a de novo-assembled transcriptome, the genome sequence, and the genome annotation. In each bar, the top rectangle represents the number of complete NLRs; the second-from-the-top rectangle represents the number of NLRs missing an LRR domain; the second-from-the-bottom rectangle represents the number of NLRs missing an N-terminal domain; and the bottom rectangle represents the number of NLRs identified only by the NB-ARC domain. c) Breakdown of total classified NLRs in the genome annotation with the addition of genes recovered from BRAKER and their contribution to the NLR classes. From left to right, the bars represent the TNL, CNL, and RNL classes of NLRs. For each class (bar), the top rectangle (gray) represents the number of complete NLRs; the next rectangle down (orange) represents the partial NLRs; and the bottom rectangle (blue, missing from the RNL bar) represents the NLRs recovered from BRAKER.
Integration of the gene models from BRAKER that overlapped with complete NLRs from genome scanning was the greatest contributor to the CNL subfamily, contributing 21 complete CNLs out of a total of 33. The ratio of TNLs, CNLs, and RNLs is as expected in conifers, with TNLs being the largest class followed by CNLs, and RNLs being the smallest class (Van Ghelder et al. 2019). RNLs are also more abundant in conifers and some members of the Rosaceae compared to most land plants, which typically have 10 or fewer RNLs (Van Ghelder et al. 2019). Without recovering complete CNL gene models from the ab initio BRAKER gene predictions of whitebark pine genes, there would have been double the number of complete RNLs identified compared to the number of CNLs. Based on NLR identifications in other conifers, the number of CNLs is generally 2 to 3 times as many as the number of RNLs (Van Ghelder et al. 2019). In the giant sequoia (Sequoiadendron giganteum) genome, there were 53 complete CNLs and 17 complete RNLs identified (Scott et al. 2020), following the expected ratio. In the whitebark pine genome, there were 33 complete CNLs to 19 complete RNLs identified. Since the gene models in the whitebark pine annotation were more dependent on RNA evidence, CNLs expressed at a lower level at the time of sampling could contribute to their reduced representation in the v0.9 genome annotation.
NLRs were also identified from a de novo transcriptome assembly as well as by directly scanning the v0.9 genome as a comparison to the v0.9 genome annotation. A total of 89 potential NLRs were identified from the de novo-assembled transcripts using InterProScan, RGAugury, and a modification of the motif-finding portion of NLR-Annotator (Fig. 4b; Supplementary Table 6). Of the 89 NLRs, 24 were considered complete, meaning they contained an N-terminal domain, an NB-ARC domain, and an LRR domain. From the genome annotation, 238 potential NLRs were identified using InterProScan, RGAugury, and the results of the NLR-Annotator scan on the whole v0.9 genome. Gene annotations with the gene model overlapping with at least 80% of the predicted NLR boundaries from the genome scan were identified as potential NLRs. These results were combined with those from the domain scanning methods, resulting in a total of 88 complete NLRs. About 3 times as many NLRs could be identified using the v0.9 genome annotation compared to the transcriptome with a noticeable increase in completeness (Fig. 4b; Supplementary Table 6). This likely results from a combination of challenges associated with de novo transcriptome assembly, such as fragmentation and fewer total number of transcripts (transcriptome: 37.5 K transcripts, N50: 744 bp; v0.9 genome annotation: 47.9 K transcripts, N50: 1,578 bp). The genome annotations reflect a combination of more transcriptomic input as well as transcripts assembled using the genome as guidance, which likely provides a more accurate representation of the gene space.
NLRs have been extensively studied in angiosperms, allowing for the creation of RefPlantNLR, which contains 481 NLRs with representatives from species across 31 genera, many of which have been experimentally validated (Kourelis et al. 2021). In comparison, NLRs have been cataloged in 8 conifer species across 6 genera using primarily transcriptomic resources (Van Ghelder et al. 2019; Scott et al. 2020; Bondar et al. 2022; Ence et al. 2022). A better understanding of NLRs in conifer species would help to explore the mechanisms of disease defense in conifers and provide candidates for disease resistance genes. The InterProScan method of NLR identification was adapted from a prior study that identified between 338 and 725 NLRs across 7 conifer species transcriptome assemblies, including 2 species of Pinus (Van Ghelder et al. 2019). At the lower end, this is 3 times the number of NLRs found in the whitebark pine transcriptome and more than the NLRs found in the genome annotation, indicating that the variation in the RNA sequences used as evidence, including variation in the total number of unique tissues and depth of sequencing, may have an impact on NLR identification. Utilizing a combination of NLR identification methods did improve the ability to identify NLRs compared to using InterProScan alone, and they were especially important for identifying the CNL class of NLRs.
Using NLR-Annotator to scan the v0.9 genome directly, 2,239 potential NLRs were identified, of which 595 were complete (Fig. 4b; Supplementary Table 6). However, only 151 of the 2,239 NLRs overlapped with a gene from the v0.9 genome annotation and 54 of them were considered complete. The partial NLRs identified through genome scanning without an overlapping genome annotation are most likely pseudogenes and nonfunctional NLRs. NLRs are under rapid evolution and often undergo tandem duplications and rearrangements or recombinations, and pseudogenes with significant deletions or missing domains can accumulate (Marone et al. 2013). Partial NLRs identified by NLR-Annotator that have support from the genome annotation could be from incomplete transcripts or truncated gene models, as the annotation only included gene models with RNA evidence before genes were recovered from BRAKER. Twenty-one complete CNLs and 6 complete TNLs identified by NLR-Annotator that were not in the genome annotation but were supported by gene models from BRAKER generated from the v0.9 genome were included in the overall v0.9 genome annotation, resulting in a total of 265 candidate NLRs, of which 116 were complete (Fig. 4c; Supplementary Table 6). This is far less than the 595 complete NLRs predicted using NLR-Annotator to scan the genome directly. Some “complete” NLRs may have only recently become nonfunctional and therefore less fragmented. Others may be real NLRs that were not expressed in any of the RNA-seq samples used for the annotation or predicted via ab initio methods. For comparison, 375 NLRs were identified in the giant sequoia reference genome examining the intersection between NLR-Annotator genome-scan predictions and the genome annotation. These gene models were primarily composed of BRAKER predictions that were supplemented with full-length transcript alignments. The pseudochromosomal assembly of giant sequoia also made it possible to identify the uneven distribution of the NLRs throughout the genome (Scott et al. 2020). With improved contiguity and completeness of both the genome and annotation, more NLRs are likely to be identified in whitebark pine.
This first in-depth classification of these elements in whitebark pine provides candidates for genes contributing to quantitative disease resistance against WPBR. NLRs have been identified as candidate genes for major disease resistance loci in P. lambertiana (Cr1) (Stevens et al. 2016), P. monticola (Cr2) (Liu et al. 2013), Pinus strobiformis (Cr3) (Liu, Schoettle, et al. 2022), and Pinus flexilis (Cr4) (Liu et al. 2021). Some NLRs have been identified as candidates for quantitative disease resistance in P. lambertiana (Weiss et al. 2020). A more recent study of the Cr1 locus developed the marker Cr1AM1 to identify SNPs in the region associated with Cr1 within the P. lambertiana genome (Wright et al. 2022). In v1.5 of the P. lambertiana genome, variants of this marker aligned with greatest identity to 2 locations within the 6.3-Mb fragscaff scaffold_6044. These alignments did not directly overlap with any annotated genes on the scaffold. Although the alignment suggests that Cr1 is an intergenic locus, it may be affecting the activation or expression of nearby genes resulting in the disease resistance response. There were 18 genes located on this scaffold and 7 were NLR genes, providing candidate genes for disease resistance to WPBR in P. lambertiana.
Among the North American white pines, P. albicaulis has demonstrated the greatest variation in resistance to WPBR across its extensive range. To date, patterns of major gene resistance have not been identified, suggesting a different mechanism from that of P. lambertiana, P. strobiformis, and P. flexilis (Liu, et al. 2022). Despite this, the putative Cr1AM1 marker sequence was aligned to the P. albicaulis v0.9 genome assembly. The best alignment, recorded at 93%, was to scaffold_64902 of length 327 kb with no annotated genes. Five genes were identified on scaffold_64902 from the BRAKER annotation, but none of these putative genes were homologous to or aligned near the genes annotated on scaffold_6044 in P. lambertiana. The scaffold identified in the P. albicaulis genome also exhibits little sequence similarity to the scaffold identified in P. lambertiana. Further studies are needed to provide a comprehensive representation of the NLR space in P. albicaulis and identify specific candidates for improved disease resistance.
Conclusion
This paper reports the first important step in developing genomic technologies that can be employed to more efficiently and rapidly identify genetic resources that can be used in the restoration of the threatened whitebark pine: a well-assembled and annotated reference genome sequence. The core research team of this project has previously generated reference genome sequences for 5 other conifer species (P. taeda, Neale et al. 2014; Zimin et al. 2014; Zimin et al. 2017b; P. lambertiana, Stevens et al. 2016; Crepeau et al. 2017; Pseudotsuga menziesii, Neale et al. 2017; S. giganteum, Scott et al. 2020; and S. sempervirens, Neale et al. 2022). In all these cases, DNA from a single tree was used to generate the reference genome sequence. Table 4 presents the quantitative statistics of these 5 conifer genomes along with those of 4 other recently published large conifer genomes. Comparison of genome size, gene number, and genome annotations among these genomes with that from whitebark pine reflects very strong similarity in gene and repetitive DNA content. However, at the phenotypic level, these conifers are quite different from each other in many ways (anatomy, morphology, life history, reproductive traits, adaptative traits, disease and insect susceptibility/resistance, etc.). These large phenotypic differences must be due in a large part to allelic variation among a common set of genes and the expression of these genes. Thus, research must now begin in whitebark pine to discover population-level allelic variation and variation in gene expression. As our team has done for our other conifer genome projects, we will now embark on genome-wide association studies and environmental association studies to discover natural variation and investigate its relationship with the vast amount of phenotypic and adaptive variation in populations of whitebark pine. Discovery from studies of this nature will lead to the development of applied genomic screening tools to be used in restoration programs.
Table 4.
Quantitative statistics of recently published large conifer genomes.
| Genome assembly (source)a | Total sequence (Gbp) | N50 contig size (bp) | N50 scaffold size (bp) | Number of contigs | Number of scaffolds | Sequencing strategy |
|---|---|---|---|---|---|---|
| Douglas-fir v1.0 (1) | 14.4 | 67,133 | 381,586 | 1,726,175 | 1,236,665 | Illumina |
| Sugar pine v1.5 (2) | 25.5 | 7,947 | 292,700 | 14,950,590 | 4,253,097 | Illumina + 10× |
| Loblolly pine v2.0 (3) | 20.5 | 28,106 | 107,038 | 2,724,159 | 1,760,464 | Illumina + PacBio RSII |
| Giant sequoia v2.0 (4) | 8.1 | 318,087 | 690,549,816 | 52,835 | 8,216 | Illumina + ONT + HiC |
| Coast redwood v1.0 (5) | 26.5 | 97,162 | 45,116,641 | 548,918 | 393,407 | Illumina + ONT + HiC |
| Siberian larch (6) | 7.9 | 1,064 | 6,479 | 12,480,170 | 11,197,034 | Illumina |
| White spruce (7) | 21.6 | 9,736 | 161,389 | 3,849,851 | 2,445,336 | Illumina + 10× |
| Japanese larch (8) | 13.0 | 447,849 | 447,849 | 65,219 | 65,219 | PacBio |
| Chinese pine (9) | 25.4 | 2,601,037 | 2,107,674,557 | 22,739 | 7,371 | Illumina + PacBio CLR + HiC |
| Engelmann spruce (10) | 20.7 | 17,914 | 403,791 | 2,394,260 | 946,236 | Illumina + ONT + 10× |
| Whitebark pine v1.0 (11) | 27.6 | 537,007 | 2,005,774,401 | 92,740 | 34,176 | Illumina + ONT + OmniC |
a Sources: 1 = Neale et al. (2017); 2 = Crepeau et al. (2017); 3 = Zimin et al. (2017b); 4 = Scott et al. (2020); 5 = Neale et al. (2022); 6 = Kuzmin et al. (2019); 7 = Gao et al. (2022); 8 = Sun et al. (2022); 9 = Niu et al. (2022); 10 = NCBI BioProject PRJNA504036; 11 = this paper.
Supplementary Material
Contributor Information
David B Neale, Department of Plant Sciences, University of California, Davis, CA 95616, USA; Whitebark Pine Ecosystem Foundation, Missoula, MT 59808, USA.
Aleksey V Zimin, Department of Biomedical Engineering and Center for Computational Biology, Johns Hopkins University, Baltimore, MD 21218, USA.
Amy Meltzer, Department of Biomedical Engineering and Center for Computational Biology, Johns Hopkins University, Baltimore, MD 21218, USA.
Akriti Bhattarai, Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, CT 06269, USA.
Maurice Amee, Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, CT 06269, USA.
Laura Figueroa Corona, School of Forestry, Northern Arizona University, Flagstaff, AZ 86011, USA.
Brian J Allen, Department of Plant Sciences, University of California, Davis, CA 95616, USA; University of California Cooperative Extension, Central Sierra, Jackson, CA 95642, USA.
Daniela Puiu, Department of Biomedical Engineering and Center for Computational Biology, Johns Hopkins University, Baltimore, MD 21218, USA.
Jessica Wright, USDA Forest Service, Pacific Southwest Research Station, Davis, CA 95618, USA.
Amanda R De La Torre, School of Forestry, Northern Arizona University, Flagstaff, AZ 86011, USA.
Patrick E McGuire, Department of Plant Sciences, University of California, Davis, CA 95616, USA.
Winston Timp, Department of Biomedical Engineering and Center for Computational Biology, Johns Hopkins University, Baltimore, MD 21218, USA.
Steven L Salzberg, Department of Biomedical Engineering and Center for Computational Biology, Johns Hopkins University, Baltimore, MD 21218, USA; Departments of Computer Science and Biostatistics, Johns Hopkins University, Baltimore, MD 21218, USA.
Jill L Wegrzyn, Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, CT 06269, USA; Institute for Systems Genomics, University of Connecticut, Storrs, CT 06269, USA.
Data availability
The whitebark pine assemblies v0.9 and v1.0 and the annotation files for both assemblies are available at the TreeGenes repository (https://treegenesdb.org/FTP/temp/P_albicaulis/) (Wegrzyn et al. 2019). The raw reads are available at NCBI under BioProject PRJNA1003249 and the v0.9 assembly is also available under BioProject PRJNA1034085.
Supplemental material available at G3 online.
Funding
Funding for this study was provided by a grant to DBN at the University of California, Davis from the US Department of Agriculture (USDA) Forest Service Pacific Southwest Research Station grant 11-JV-11272135-005 and grants to DBN at the Whitebark Pine Ecosystem Foundation (WPEF) from the nonprofit conservation organization, American Forests and the family foundation, Krieger Charitable Trust. AVZ and SLS were supported in part by the US National Institutes of Health (NIH) Research Project grant R01-HG006677 and a US National Science Foundation (NSF) grant IOS-1744309. JLW acknowledges the Computational Biology Core within the Institute for Systems Genomics at the University of Connecticut’s High Performance Computing facility and the US NSF CAREER grant #1943371 for funding graduate students.
Literature cited
- Bondar EI, Feranchuk SI, Miroshnikova KA, Sharov VV, Kuzmin DA, Oreshkova NV, Krutovsky KV. 2022. Annotation of Siberian larch (Larix sibirica Ledeb.) nuclear genome—one of the most cold-resistant tree species in the only deciduous genus in Pinaceae. Plants (Basel). 11(15):2062. doi: 10.3390/plants11152062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower AD, Aitken SN. 2008. Ecological genetics and seed transfer guidelines for Pinus albicaulis (Pinaceae). Am J Bot. 95(1):66–76. doi: 10.3732/ajb.95.1.66. [DOI] [PubMed] [Google Scholar]
- Brůna T, Hoff KJ, Lomsadze A, Stanke M, Borodovsky M. 2021. Braker2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom Bioinform. 3(1):lqaa108. doi: 10.1093/nargab/lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushmanova E, Antipov D, Lapidus A, Prjibelski AD. 2019. rnaSPAdes: a de novo transcriptome assembler and its application to RNA-seq data. Gigascience. 8(9):giz100. doi: 10.1093/gigascience/giz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepeau MW, Langley CH, Stevens KA. 2017. From pine cones to read clouds: rescaffolding the megagenome of sugar pine (Pinus lambertiana). G3 (Bethesda). 7(5):1563–1568. doi: 10.1534/g3.117.040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dainat J, Hereñú D, Davis E, Crouch K, Sol L, Agostinho N. 2022. Another Gff analysis toolkit to handle annotations in any GTF/GFF format (Version v1.0). Zenodo. doi: 10.5281/zenodo.3552717. [DOI] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. GigaScience. 10(2):giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre AR. 2023. Updated Pinus lambertiana high-density linkage maps [Dataset]. Dryad. 10.5061/dryad.573n5tbdz [DOI]
- De La Torre AR, Birol I, Bousquet J, Ingvarsson PK, Jansson S, Jones SJM, Keeling CI, MacKay J, Nilsson O, Ritland K, et al. 2014. Insights into conifer giga-genomes. Plant Physiol. 166(4):1724–1732. doi: 10.1104/pp.114.248708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26(19):2460–2461. 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Ence D, Smith KE, Fan S, Gomide Neves L, Paul R, Wegrzyn J, Peter GF, Kirst M, Brawner J, Nelson CD, et al. 2022. NLR diversity and candidate fusiform rust resistance genes in loblolly pine. G3 (Bethesda). 12(2):jkab421. doi: 10.1093/g3journal/jkab421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 117(17):9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T, Katsushi Y, Yokoyama TT, Hamanaka T, Harazono Y, Kamada H, Kobayashi W, Ujino-Ihara T, Uchiyama K, Matsumoto A, et al. 2023. A chromosome-level genome assembly of a model conifer plant, the Japanese cedar, Cryptomeria japonica D. Don. bioRxiv 529822. 10.1101/2023.02.24.529822, preprint: not peer reviewed. [DOI]
- Gagalova KK, Warren RL, Coombe L, Wong J, Nip KM, Yuen MMS, Whitehill JGA, Celedon JM, Ritland C, Taylor GA, et al. 2022. Spruce giga-genomes: structurally similar yet distinctive with differentially expanding gene families and rapidly evolving genes. Plant J. 111(5):1469–1485. doi: 10.1111/tpj.15889. [DOI] [PubMed] [Google Scholar]
- Gao Y, Cui Y, Zhao R, Chen X, Zhang J, Zhao J, Kong L.. 2022. Cryo-treatment enhances the embryogenicity of mature somatic embryos via the lncRNA–miRNA–mRNA network in white spruce. Int J Molec Sci 23(3):1111. doi: 10.3390/ijms23031111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, Salzberg SL, Zimin AV.. 2023. Jasper: a fast genome polishing tool that improves accuracy of genome assemblies. PloS Comput Biol 19(3):e1011032. doi: 10.1371/journal.pcbi.1011032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AJ, Ginzburg S, Xu MS, Fisher CR, Rahmatpour N, Mitton JB, Paul R, Wegrzyn JL. 2020. EnTAP: bringing faster and smarter functional annotation to non-model eukaryotic transcriptomes. Mol Ecol Resour. 20:591–604. doi: 10.1111/1755-0998.13106. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, et al. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47(D1):D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37(5):540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- Kourelis J, Sakai T, Adachi H, Kamoun S. 2021. RefPlantNLR is a comprehensive collection of experimentally validated plant disease resistance proteins from the NLR family. PLoS Biol. 19(10):e3001124. doi: 10.1371/journal.pbio.3001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M. 2019. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20(1):278. doi: 10.1186/s13059-019-1910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin DA, Feranchuk SI, Sharov VV, Cybin AN, Makolov SV, Putintseva YA, Oreshkova NV, Krutovsky KV. 2019. Stepwise large genome assembly approach: a case of Siberian larch (Larix sibirica Ledeb). BMC Bioinformatics 20:37. 10.1186/s12859-018-2570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Quan X, Jia G, Xiao J, Cloutier S, You FM. 2016. RGAugury: a pipeline for genome-wide prediction of resistance gene analogs (RGAs) in plants. BMC Genomics. 17(1):852. doi: 10.1186/s12864-016-3197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-J, Johnson JS, Sniezko RA. 2022. Genomic advances in research on genetic resistance to white pine blister rust in North American white pines. In: De La Torre AR, editor. The Pine Genomes. Switzerland: Springer Nature. p. 163–191. doi: 10.1007/978-3-030-93390-6_8. [DOI] [Google Scholar]
- Liu J-J, Schoettle AW, Sniezko RA, Waring KM, Williams H, Zamany A, Johnson JS, Kegley A. 2022. Comparative association mapping reveals conservation of major gene resistance to white pine blister rust in southwestern white pine (Pinus strobiformis) and limber pine (P. flexilis). Phytopathology. 112(5):1093–1102. doi: 10.1094/PHYTO-09-21-0382-R. [DOI] [PubMed] [Google Scholar]
- Liu J-J, Schoettle AW, Sniezko RA, Williams H, Zamany A, Rancourt B. 2021. Fine dissection of limber pine resistance to Cronartium ribicola using targeted sequencing of the NLR family. BMC Genomics. 22(1):567. doi: 10.1186/s12864-021-07885-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-J, Sturrock RN, Benton R. 2013. Transcriptome analysis of Pinus monticola primary needles by RNA-seq provides novel insight into host resistance to Cronartium ribicola. BMC Genomics. 14:884. doi: 10.1186/1471-2164-14-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacManes MD. 2018. The Oyster River Protocol: a multi-assembler and kmer approach for de novo transcriptome assembly. PeerJ. 6:e5428. doi: 10.7717/peerj.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Molec Biol Evol. 38(10):4647–4654. doi: 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone D, Russo MA, Laidò G, De Leonardis AM, Mastrangelo AM. 2013. Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int J Mol Sci. 14(4):7302–7326. doi: 10.3390/ijms14047302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson DJ, Reinhart DP. 1997. Excavation of red squirrel middens by grizzly bears in the whitebark pine zone. J Appl Ecol. 24(4):926–940. doi: 10.2307/2405283. [DOI] [Google Scholar]
- Neale DB, McGuire PE, Wheeler NC, Stevens KA, Crepeau MW, Cardeno C, Zimin AV, Puiu D, Pertea GM, Sezen UU, et al. 2017. The Douglas-fir genome sequence reveals specialization of the photosynthetic apparatus in Pinaceae. G3 (Bethesda). 7:3157–3167. doi: 10.1534/g3.117.300078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale DB, Wegrzyn JL, Stevens KA, Zimin AV, Puiu D, Crepeau MW, Cardeno C, Koriabine M, Holtz-Morris AE, Liechty JD, et al. 2014. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 15(3):R59. doi: 10.1186/gb-2014-15-3-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale DB, Zimin AV, Zaman S, Scott AD, Shrestha B, Workman RE, Puiu D, Allen BJ, Moore ZJ, Sekhwal MK, et al. 2022. Assembled and annotated 26.5 Gbp coast redwood genome: a resource for estimating evolutionary adaptive potential and investigating hexaploid origin. G3 (Bethesda). 12(1):jkab380. doi: 10.1093/g3journal/jkab380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu S, Li J, Bo W, Yang W, Zuccolo A, Giacomello S, Chen X, Han F, Yang J, Song Y, et al. 2022. The Chinese pine genome and methylome unveil key features of conifer evolution. Cell. 185(1):204–217.e14. doi: 10.1016/j.cell.2021.12.006. [DOI] [PubMed] [Google Scholar]
- Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Pinto BL, Salazar GA, Bileschi ML, Bork P, Bridge A, Colwell L, et al. 2023. InterPro in 2022. Nucleic Acids Res. 51(D1):D418–D427. doi: 10.1093/nar/gkac993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea G, Pertea M. 2020. Gff utilities: GffRead and GffCompare. F1000Res. 9:ISCB Comm J-304. doi: 10.12688/f1000research.23297.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, O’Connell BL, Stites JC, Rice BJ, Blanchette M, Calef R, Troll CJ, Fields A, Hartley PD, Sugnet CW, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26(3):342–350. doi: 10.1101/gr.193474.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, et al. 2010. De novo assembly and analysis of RNA-seq data. Nat Methods. 7(11):909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- Scott AD, Zimin AV, Puiu D, Workman R, Britton M, Zaman S, Caballero M, Read AC, Bogdanove AJ, Burns E, et al. 2020. A reference genome sequence for giant sequoia. G3 (Bethesda). 10(11):3907–3919. doi: 10.1534/g3.120.401612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Le S, Li Y, Hu F. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PloS One. 11(10):e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniezko RA, Kegley AJ, Danchok R. 2008. White pine blister rust resistance in North American, Asian and European species-results from artificial inoculartion trials in Oregon. Ann For Res. 51(1):53–66. doi: 10.15287/afr.2008.145. [DOI] [Google Scholar]
- Steuernagel B, Witek K, Krattinger SG, Ramirez-Gonzalez RH, Schoonbeek H-J, Yu G, Baggs E, Witek AI, Yadav I, Krasileva KV, et al. 2020. The NLR-annotator tool enables annotation of the intracellular immune receptor repertoire. Plant Physiol. 183(2):468–482. doi: 10.1104/pp.19.01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens KA, Wegrzyn JL, Zimin A, Puiu D, Crepeau M, Cardeno C, Paul R, Gonzalez-Ibeas D, Koriabine M, Holtz-Morris AE, et al. 2016. Sequence of the sugar pine megagenome. Genetics. 204(4):1613–1626. doi: 10.1534/genetics.116.193227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Xie YH, Li Z, Liu YJ, Sun XM, Li JJ, Quan WP, Zeng QY, Van de Peer Y, Zhang SG. 2022. The Larix kaempferi genome reveals new insights into wood properties. J Integr Plant Biol. 64(7):1364–1373. doi: 10.1111/jipb.13265. [DOI] [PubMed] [Google Scholar]
- Tang H, Zhang X, Miao C, Zhang J, Ming R, Schnable JC, Schnable PS, Lyons E, Lu J. 2015. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 16(1):3. doi: 10.1186/s13059-014-0573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomback DF, Achuff P. 2010. Blister rust and western forest biodiversity: ecology, values and outlook for white pines. For Pathol. 40(3-4):186–225. doi: 10.1111/j.1439-0329.2010.00655.x. [DOI] [Google Scholar]
- Tomback DF, Arno SF, Keane RE. 2001. Whitebark Pine Communities: Ecology and Restoration. Washington (DC): Island Press. [Google Scholar]
- US FWS . 2022. Endangered and threatened wildlife and plants; threatened species status with section 4(d) rule for whitebark pine (Pinus albicaulis). 87 FR 76882. https://www.govinfo.gov/content/pkg/FR-2022-12-15/pdf/2022-27087.pdf.
- Van Ghelder C, Parent GJ, Rigault P, Prunier J, Giguère I, Caron S, Stival Sena J, Deslauriers A, Bousquet J, Esmenjaud D, et al. 2019. The large repertoire of conifer NLR resistance genes includes drought responsive and highly diversified RNLs. Sci Rep. 9(1):11614. doi: 10.1038/s41598-019-47950-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C, Fetter K, Zaman S, Vuruputoor V, Bhattarai A, Chinta V, Wegrzyn J. 2023. EASEL. GitLab. https://gitlab.com/PlantGenomicsLab/easel. [accessed 2023 Nov 12]. [Google Scholar]
- Wegrzyn JL, Staton MA, Street NR, Main D, Grau E, Herndon N, Buehler S, Falk T, Zaman S, Ramnath R, et al. 2019. Cyberinfrastructure to improve forest health and productivity: The role of tree databases in connecting genomes, phenomes, and the environment. Front Plant Sci. 10:813. doi: 10.3389/fpls.2019.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M, Sniezko RA, Puiu D, Crepeau MW, Stevens K, Salzberg SL, Langley CH, Neale DB, De La Torre AR. 2020. Genomic basis of white pine blister rust quantitative disease resistance and its relationship with qualitative resistance. Plant J. 104(2):365–376. doi: 10.1111/tpj.14928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman R, Timp W, Fedak R, Kilburn D, Hao S, Liu K. 2018. High molecular weight DNA extraction from recalcitrant plant species for third generation sequencing. Protoc Exch. 2018:1–2. doi: 10.1038/protex.2018.059. [DOI] [Google Scholar]
- Wright JW, Stevens KA, Hodgskiss P, Langley CH. 2022. Snps in a large genomic scaffold are strongly associated with Cr1R, major gene for resistance to white pine blister rust in range-wide samples of sugar pine (Pinus lambertiana). Plant Dis. 106(6):1639–1644. doi: 10.1094/PDIS-08-21-1608-RE. [DOI] [PubMed] [Google Scholar]
- Zimin AV, Puiu D, Luo MC, Zhu T, Koren S, Marçais G, Yorke JA, Dvořák J, Salzberg SL. 2017a. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res. 27(5):787–792. http://www.genome.org/cgi/doi/10.1101/gr.213405.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Salzberg SL. 2020. The genome polishing tool POLCA makes fast and accurate corrections in genome assemblies. PLoS Comput Biol. 16(6):e1007981. doi: 10.1371/journal.pcbi.1007981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Salzberg SL. 2022. The SAMBA tool uses long reads to improve the contiguity of genome assemblies. PLoS Comput Biol. 18(2):e1009860. doi: 10.1371/journal.pcbi.1009860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A, Stevens KA, Crepeau MW, Holtz-Morris A, Koriabine M, Marçais G, Puiu D, Roberts M, Wegrzyn JL, de Jong PJ, et al. 2014. Sequencing and assembly of the 22-Gb loblolly pine genome. Genetics. 196(3):875–890. doi: 10.1534/genetics.113.159715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Stevens KA, Crepeau MW, Puiu D, Wegrzyn JL, Yorke JA, Langley CH, Neale DB, Salzberg SL. 2017b. An improved assembly of the loblolly pine mega-genome using long-read single-molecule sequencing. Gigascience. 6(1):1–4. doi: 10.1093/gigascience/giw016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- De La Torre AR. 2023. Updated Pinus lambertiana high-density linkage maps [Dataset]. Dryad. 10.5061/dryad.573n5tbdz [DOI]
Supplementary Materials
Data Availability Statement
The whitebark pine assemblies v0.9 and v1.0 and the annotation files for both assemblies are available at the TreeGenes repository (https://treegenesdb.org/FTP/temp/P_albicaulis/) (Wegrzyn et al. 2019). The raw reads are available at NCBI under BioProject PRJNA1003249 and the v0.9 assembly is also available under BioProject PRJNA1034085.
Supplemental material available at G3 online.