Abstract
Innate immunity functions as a rapid defense against broad classes of pathogenic agents. While the mechanisms of innate immunity in response to antigen exposure are well-studied, how pathogen exposure activates the innate immune responses and the role of genetic variation in immune activity is currently being investigated. Previously, we showed significant survival differences between the N2 and the CB4856 Caenorhabditis elegans isolates in response to Staphylococcus epidermidis infection. One of those differences was expression of the mab-5 Hox family transcription factor, which was induced in N2, but not CB4856, after infection. In this study, we use survival assays and RNA-sequencing to better understand the role of mab-5 in response to S. epidermidis. We found that mab-5 loss-of-function (LOF) mutants were more susceptible to S. epidermidis infection than N2 or mab-5 gain-of-function (GOF) mutants, but not as susceptible as CB4856 animals. We then conducted transcriptome analysis of infected worms and found considerable differences in gene expression profiles when comparing animals with mab-5 LOF to either N2 or mab-5 GOF. N2 and mab-5 GOF animals showed a significant enrichment in expression of immune genes and C-type lectins, whereas mab-5 LOF mutants did not. Overall, gene expression profiling in mab-5 mutants provided insight into MAB-5 regulation of the transcriptomic response of C. elegans to pathogenic bacteria and helps us to understand mechanisms of innate immune activation and the role that transcriptional regulation plays in organismal health.
Keywords: mab-5, immunity, gene expression, Staphylococcus epidermidis
Introduction
The innate immune system functions as an organism's first line of defense against pathogenic agents that overcome physical barriers of the host (e.g. cuticle/epidermis; Akira et al. 2006; Kumar et al. 2011). To protect against infections, an organism must not only recognize the invading microbe as foreign/pathogenic but also activate an appropriate systemic response to combat the pathogen. The “recognition-response” paradigm of the innate immune system is highly conserved across animal phyla (Ausubel 2005). In general, conserved microbial structures, termed pathogen-associated molecular patterns, are recognized by membrane-bound pathogen recognition receptors encoded by the host's genome (Kumar et al. 2011). Highly conserved components of signal transduction pathways [e.g. Toll-like receptors and nuclear factor kappa-ligh-chain-enhancer of activated B cells (NF-κB) transcription factors] translate the pathogen recognition signal into a cellular response via the activation of downstream effector molecules (Kopp and Ghosh 1995; Anderson 2000; Kawasaki and Kawai 2014). The specific effector molecules can vary between organisms but generally converge upon a set of evolutionarily conserved responses (e.g. reactive oxygen species generation, use of antimicrobial and antifungal peptides, and expression of bacterial-binding C-type lectins; McGreal et al. 2004; Muller et al. 2008; Kohchi et al. 2009; Koenderman et al. 2014).
The soil-dwelling nematode Caenorhabditis elegans is a tractable model system to examine host–pathogen interactions and investigate innate immune response to bacterial pathogens (Marsh and May 2012; Balla and Troemel 2013). C. elegans possess a robust immune response with evolutionarily conserved mechanisms for the detection and elimination of pathogens (Chavez et al. 2007; Schulenburg et al. 2008; Shivers et al. 2010; Marsh and May 2012; van der Hoeven et al. 2012; Dierking et al. 2016). However, core components of the innate immune response found in other organisms are either not involved in the innate immune response (Toll-like receptor) or are absent from the C. elegans genome entirely (NF-κB; Pujol et al. 2001; Couillault et al. 2004). The presence of a robust immune response suggests that C. elegans must utilize alternative pathways for immune activation, thus making nematodes an ideal model to study bacterial pathogenesis and identify additional genes that function in innate immunity.
Previous work has leveraged the genetic diversity of C. elegans wild isolates to investigate how genomic variation underlies pathogen avoidance and susceptibility (Schulenburg and Muller 2004; Reddy et al. 2009; Chang et al. 2011; Balla et al. 2015; Martin et al. 2017; Sterken et al. 2021). One such study found the Hawaiian wild isolate, CB4856, more susceptible to infection by the Gram-positive bacterium Staphylococcus epidermidis when compared with the lab-conditioned strain, N2 (var. Bristol; Lansdon et al. 2022). Additionally, gene expression analysis of CB4856 and N2 animals infected with S. epidermidis revealed that mab-5 expression was significantly increased in N2, but not CB4856 animals.
The mab-5 gene encodes for a Hox family transcription factor that is required for proper migration of Q neuroblasts (Chalfie et al. 1983; Kenyon 1986; Salser and Kenyon 1992; Maloof et al. 1999; Herman 2001; Chapman et al. 2008; Tamayo et al. 2013). In addition to their role in patterning the body plan, Hox genes are an important part of the C. elegans innate immune response. Notably, egl-5 function is important for the nematode response to the Gram-positive bacterium Staphylococcus aureus and Microbacterium nematophilum (Gravato-Nobre et al. 2005; Irazoqui et al. 2008; Nicholas and Hodgkin 2009). Evidence of mab-5 upregulation in N2 animals infected with S. epidermidis raised the possibility that MAB-5 may be involved in activation of the innate immune response.
In this study, we show that the mab-5 loss-of-function (LOF) mutations isolated in the N2 background are significantly more susceptible to S. epidermidis infection than wild-type animals or animals with a mab-5 gain-of-function (GOF) mutation, e1751. We used high-throughput RNA-sequencing (RNA-seq) to identify changes in gene expression in N2 and mab-5 GOF and mab-5 LOF animals exposed to S. epidermidis. The differentially expressed genes (DEGs) identified provide insight into MAB-5 regulation of the transcriptomic response of C. elegans to pathogenic bacteria. We believe that the results of this project will provide a better understanding of the mechanisms of innate immune activation and the role that transcriptional regulation plays in organismal health.
Methods
C. elegans and bacterial strains
C. elegans and bacterial strains used in this study are listed in Supplementary Table 1. All strains were maintained at 20°C on nematode growth medium (NGM) plates seeded with Escherichia coli OP50 or the EVL2000 isolate of S. epidermidis (Brenner 1974; Do et al. 2022). All bacteria strains were grown at 37°C with shaking in low-salt Luria Bertani medium (10 g Bacto-tryptone, 5 g yeast extract, and 5 g NaCl).
Survival assays
NGM plates were seeded with 20 µL of bacteria (E. coli or S. epidermidis EVL 2000) and incubated overnight at 37°C. Plates were acclimated to room temperature, and 30 late-stage L4 worms were transferred to the plate for each trial. Survival assays were performed at 20°C while transferring living worms to fresh plates and recording the number of dead worms daily. For each survival assay, a minimum of 3 replicates were collected for each worm strain and each pathogen.
Exposure of nematodes to pathogens
NGM plates were seeded with 200 µL of bacteria. Approximately 1,000–2,000 L4 stage worms reared on E. coli OP50 were transferred to an NGM plate seeded with bacteria and incubated at 20°C for 24 h. After 24 h, worms were washed with M9 buffer 3 times to remove excess bacteria. Worms were suspended in 100 µL of M9 buffer and mechanically disrupted in liquid nitrogen using a ceramic mortar and pestle. Frozen tissue was transferred to a 1.5-mL microcentrifuge tube containing 1 mL of TRIZOL and flash frozen in liquid nitrogen and stored at −80°C until RNA isolation. For each worm and bacterial strain, 3 biological replicates were collected.
RNA isolation and RNA-seq
Worm tissue was thawed on ice, vortexed for 5 s, and incubated at room temperature for 5 min. To each sample, 470 µL of chloroform was added, mixed by inversion, and phase separated for 2 min at room temperature. After centrifugation at 15,000 rpm at 4°C, the aqueous layer containing RNA (∼550 µL) was transferred to an RNase-free Eppendorf tube. Total RNA extraction was performed using the Monarch RNA Cleanup Kit (New England Biolabs) according to the manufacturer's instructions. Total RNA was quantified using the Qubit (ThermoFisher Scientific), and RNA quality was assessed using the TapeStation 4150 (Agilent Technologies). Sequence libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs) and sequenced using paired-end 2 × 100-bp sequencing on the Illumina NextSeq 2000.
RNA-Seq analysis and quantification of DEG
The quality of the 100-bp paired-end reads generated using the Illumina NextSeq 2000 was assessed using FastQC (Andrews 2010; v0.11.9). Illumina sequencing adapters and low-quality bases (Phred score <30) were trimmed from reads using Fastp (Chen et al. 2018; v0.20.0) with the “–correction” flag enabled. Reads were aligned to the C. elegans genome assembly (WormBase WS273 release) using HISAT2 (Kim et al. 2019; v2.1.0) with default settings. Aligned reads were mapped to the C. elegans annotated genome (WormBase WS273 release), and transcript abundances were quantified using featureCounts (Liao et al. 2014; v.2.0.0). Data normalization and DEG analysis were performed with DESeq2 (Love et al. 2014; v1.34.0) in R (v4.1.0) using a false discovery rate (FDR)-adjusted P-value < 0.05 and fold change ≥2 as cutoffs. Geneset enrichment analysis was performed with WormCat (Holdorf et al. 2020) using an FDR-adjusted P-value < 0.05 as the significance cutoff.
Identification of MAB-5–binding sites near DEGs
Genomic coordinates of DEGs were obtained through WormBase (Howe et al. 2016). The coordinates were extended 1-kb upstream and downstream of the gene start and end sites to encompass potential MAB-5 binding sites outside of the coding region. Genomic coordinates of MAB-5 binding sites from mixed-stage (embryonic) and L2 animals were obtained from the ModERN database (Kudron et al. 2018). Analysis of genomic coordinates to detect overlap between DEGs and MAB-5 binding sites was performed with Microsoft Excel.
Statistical analysis
All statistical analysis was carried out using SPSS Statistics 28.0.0.0 (IBM). The generalized Wilcoxon method was used for pairwise comparisons of survival curves and to calculate median survival (LT50). Chi-square test was used for assessing the statistical significance of the overlap in gene expression. Statistically significant differences are defined in the figure legends and noted in the figures with asterisks.
Results
mab-5 LOF mutants exhibit increased susceptibility to S. epidermidis infection
To explore the role of mab-5 in C. elegans innate immunity, we conducted survival analyses with the Gram-positive pathogen S. epidermidis using 2 mab-5 LOF alleles, e1239 and gk670, and the mab-5 GOF allele, e1751 (Kenyon 1986; Salser and Kenyon 1992; C. elegans Deletion Mutant Consortium 2012). Infection with the EVL2000 S. epidermidis strain found N2 and mab-5 GOF animals to be similarly susceptible to infection with a median survival (LT50) of 15.3 ± 1.4 and 15 ± 0.6 days, respectively (Fig. 1a, Supplementary Table 2). In contrast, mab-5 LOF animals were significantly shorter-lived after S. epidermidis infection relative to N2. mab-5(e1239) animals had a median lifespan of 10.5 ± 1.9 days (P < 0.001), whereas 50% of mab-5(gk670) animals were killed after an average of 11.5 ± 1.6 days (P < 0.001; Fig. 1a, Supplementary Table 2).
Fig. 1.
LOF mab-5 mutants exhibit greater susceptibility to S. epidermidis exposure relative to N2. Survival of wild-type (N2) mab-5 GOF mutant [mab-5(e1751)] and mab-5 LOF mutants [mab-5(e1239); mab-5(gk670)] on NGM with a lawn of S. epidermidis a) or E. coli b). In each experiment, 30 worms were placed on the bacterial lawn and transferred daily while scored for survival. Curves represent a minimum of 3 independent experiments for each genotype. For each bacteria, statistical analysis was carried out by generalized Wilcoxon (mutant vs N2). ***P < 0.001.
We also surveyed lifespan on nonpathogenic E. coli OP50, the standardized food for laboratory-reared C. elegans, to assess any general lifespan differences in mab-5 mutants (Brenner 1974). On E. coli, the median lifespan of N2 animals was 10 ± 1.0 days. Perturbation of mab-5 expression did not significantly affect lifespan on E. coli OP50, with 50% of the animals dying at 8.7 ± 0.7, 8.7 ± 1.5, and 11.0 ± 1.0 days for mab-5(e1751), mab-5(gk670), and mab-5(e1239), respectively (P > 0.05 vs N2; Fig. 1b, Supplementary Table 2).
We have previously reported findings whereby maintenance on the EVL2000 strain of S. epidermidis increases lifespan in N2 animals compared with those reared on E. coli OP50 (Lansdon et al. 2022), Not only did we corroborate this finding (N2 LT50: OP50 = 10 ± 1.0 days, EVL2000 = 15.3 ± 1.4; P < 0.001; Supplementary Table 2), but we observed mab-5 GOF mutants to have a similar increase in lifespan on S. epidermidis compared with E. coli. mab-5(e1751) animals fed S. epidermidis had a median survival time of 15 ± 0.6 days, whereas 50% of animals reared on OP50 died after 8.7 ± 0.7 days (P < 0.001; Supplementary Table 2). Conversely, we found no statistically significant difference in mab-5 LOF animals when comparing lifespan on E. coli with S. epidermidis. mab-5(e1239) animals had median lifespans of 11 ± 1.0 and 10.5 ± 1.9 days when reared on E. coli and S. epidermidis, respectively (P = 0.153), whereas the LT50 for mab-5(gk670) animals was 8.7 ± 1.5 and 11.5 ± 1.6 days when exposed to either E. coli or S. epidermidis, respectively (P = 0.103; Supplementary Table 2). Together, our results indicate that loss of mab-5 function is detrimental to C. elegans survival after S. epidermidis exposure and that this decrease in survival is not due to a general decrease in lifespan. Since mab-5(e1239) exhibited the greatest difference in survival between N2 and mab-5 LOF mutants, follow-up experiments used this allele.
DEG among N2, mab-5 LOF, and mab-5 GOF exposed to S. epidermidis
Lifespan assays suggested that mab-5 is important for C. elegans survival after S. epidermidis infection. To identify gene expression changes, N2 animals and mab-5 mutants were exposed to either S. epidermidis or E. coli OP50 for 24 h followed by RNA-seq.
We first identified gene expression changes in response to S. epidermidis across strains (for all analyses, DEGs were defined as those with an FDR-adjusted P-value ≤ 0.05 and fold change ≥2, infected vs control). In total, we identified 401, 100, and 2778 DEGs by treatment (S. epidermidis vs E. coli) for N2, mab-5(e1751), and mab-5(e1239), respectively. In each genotype, we found that most genes were upregulated following S. epidermidis exposure compared with OP50 [N2, 370/401 = 92.3%; mab-5(e1751), 85/100 = 85%; mab-5(e1239), 2408/2778 = 86.7%; Tables 1–3, Supplementary Tables 3–5].
Table 1.
DEGs in N2 animals following 24-h exposure to S. epidermidis.
| Condition | Total DEGsa | Total upregulated | Total downregulated | Total genes expressedb |
|---|---|---|---|---|
| S. epidermidis | 401 | 370 | 31 | 15,980 |
| E. coli OP50 | — | — | — | 15,394 |
a Genes with a FDR-adjusted P-value ≤0.05 and log2 fold change ≥1 relative to E. coli OP50.
b Total number of genes with >0 counts in each replicate.
Table 3.
DEGs in mab-5(e1239) LOF animals following 24-h exposure to S. epidermidis.
| Condition | Total DEGsa | Total upregulated | Total downregulated | Total genes expressedb |
|---|---|---|---|---|
| S. epidermidis | 2,778 | 2,408 | 370 | 17,144 |
| E. coli OP50 | — | — | — | 16,797 |
a Genes with a FDR-adjusted P-value ≤0.05 and log2 fold change ≥1 relative to E. coli OP50.
b Total number of genes with >0 counts in each replicate.
Table 2.
DEGs in mab-5(e1751) GOF animals following 24-h exposure to S. epidermidis.
| Condition | Total DEGsa | Total upregulated | Total downregulated | Total genes expressedb |
|---|---|---|---|---|
| S. epidermidis | 100 | 85 | 15 | 16,950 |
| E. coli OP50 | — | — | — | 16,539 |
a Genes with a FDR-adjusted P-value ≤0.05 and log2 fold change ≥1 relative to E. coli OP50.
b Total number of genes with >0 counts in each replicate.
Next, we compared the lists of DEGs between N2 and mab-5 GOF animals exposed to S. epidermidis to assess the degree of overlap in DEG between genotypes that had similar survival times following infection. Comparing upregulated genes between N2 and mab-5 GOF animals fed S. epidermidis, a minority, 31 in total, overlapped between the 2 genotypes (N2-only, 339/370 = 92%; mab-5 GOF-only 54/85 = 64%). Furthermore, the lack of overlap in differential expression was statistically significant (Χ2 = 46.35; P < 0.0001). In comparing downregulated genes between N2 and mab-5 GOF animals fed S. epidermidis, again a minority, 7 in total, overlapped between the 2 genotypes (N2-only, 24/31 = 77%; mab-5 GOF-only 8/15 = 53%). However, the lack of overlap in differential expression was not statistically significant (Χ = 2.77; P = 0.096; Fig. 2a, Supplementary Table 6). Notably, all DEGs shared between the 2 genotypes were expressed in the same direction. We also examined the correlation of gene expression using the log2 fold changes after infection. Although a minority of DEGs overlapped between N2 and mab-5 GOF animals, expression levels were positively correlated between the 2 genotypes (ρ = 0.491; P < 0.001; Fig. 2b).
Fig. 2.
Expression differences in N2- and mab-5(e1751)-infected animals. a) Intersection of upregulated (top) and downregulated (bottom) genes in N2 and mab-5(e1751) animals infected with S. epidermidis EVL2000. b) Correlation between log2 fold changes in N2- and mab-5(e1751)-infected animals. Spearman's rho value and P-value are presented in the upper left corner. Gray (Non-DE) and black dots (DE Genes) represent expressed gene and DEG, respectively.
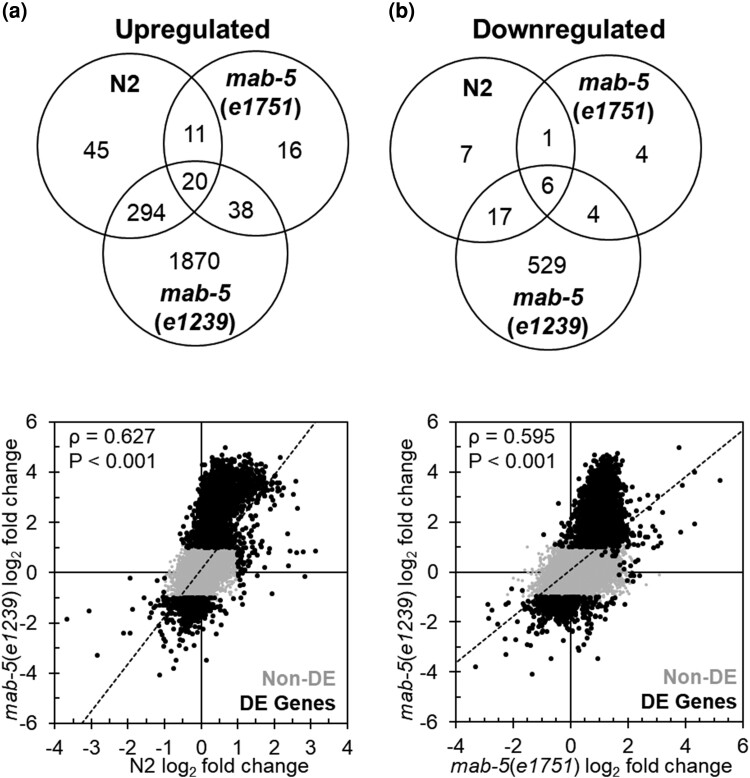
We performed tripartite Venn analysis on N2, mab-5 GOF, and mab-5 LOF animals after S. epidermidis exposure, again separating DEGs into upregulated and downregulated. In comparing upregulated genes, a majority of genes for both N2 and mab-5 GOF animals overlapped with mab-5 LOF (N2, 314/370 = 85%; mab-5 GOF, 58/85 = 68%; N2 overlap, Χ = 847.78, P < 0.001; mab-5 GOF overlap, Χ = 637.82, P < 0.001). In comparing downregulated genes among the 3 genotypes, we also observed a majority of genes shared between either N2 or the mab-5 GOF mutants with the mab-5 LOF mutant (N2, 23/31 = 74%; mab-5 GOF, 10/15 = 67%; N2 overlap, Χ = 198.05, P < 0.001; mab-5 GOF overlap, Χ = 176.17, P < 0.001; Fig. 3a, Supplementary Table 7). Within any overlap among genotypes, DEGs were expressed in the same direction, with the exception of one gene, ech-9, which was expressed in opposing directions (Supplementary Table 7). More specifically, ech-9 was upregulated in N2, downregulated in mab-5 LOF, and not differentially expressed in mab-5 GOF animals. We also examined the correlation of gene expression for both N2 and mab-5 GOF animals compared with mab-5 LOF animals using the log2 fold changes after infection. In both instances, expression levels were strongly correlated between the 2 genotypes (N2 and mab-5 LOF: ρ = 0.627, P < 0.001; mab-5 GOF and mab-5 LOF: ρ = 0.595, P < 0.001; Fig. 3b). Together, these results highlight the dramatic differences in gene expression between N2, mab-5 GOF, and mab-5 LOF animals exposed to S. epidermidis.
Fig. 3.
Expression differences in N2-, mab-5(e1751)-, and mab-5(e1239)-infected animals. a) Intersection of upregulated (left) and downregulated (right) genes in N2, mab-5(e1751), and mab-5(e1239) animals infected with S. epidermidis EVL2000. b) Correlation between log2 fold changes in N2- and mab-5(e1239)-infected animals (left) and mab-5(e1751)- and mab-5(e1239)-infected animals (right). Spearman's rho value and P-value are presented in the upper left corner. Gray (Non-DE) and black dots (DE Genes) represent expressed gene and DEG, respectively.
Although N2 and mab-5 GOF worms were equivalently susceptible to S. epidermidis, their respective transcriptomic responses were relatively divergent, with a minority of genes in common. In contrast, S. epidermidis infection resulted in DEG profiles with considerable overlap between mab-5 LOF animals and N2 and mab-5 GOF animals, even though mab-5 LOF animals were significantly more susceptible to S. epidermidis than animals with intact mab-5 function.
Geneset enrichment analysis among N2, mab-5 GOF, and mab-5 LOF exposed to S. epidermidis
Since mab-5 LOF mutants exhibited survival deficiencies when compared with N2 animals or mab-5 GOF mutants, we were interested in identifying biological processes that were enriched after S. epidermidis exposure. We first performed geneset enrichment analysis on DEGs that were exclusive to N2 (N2-only) and mab-5(e1751) (mab-5 GOF-only) and genes that were differentially expressed in both N2 and mab-5 GOF animals (N2 and mab-5 GOF), separating genes based on whether they were upregulated or downregulated. Using upregulated genes, we found an enrichment in biological functions pertaining to the organismal stress response and immunity in both N2 and mab-5 GOF animals (Table 4, Supplementary Table 8). The Gene Ontology (GO) terms “Major sperm protein,” “Extracellular material: collagen,” “Signaling: phosphatase,” “Cytoskeleton: microtubule,” and “Transmembrane protein” were enriched in N2-exlcusive DEGs, whereas genes corresponding to the GO term “Metabolism: lipid” were overrepresented in DEGs exclusive to mab-5 GOF animals. Using downregulated genes, we found an enrichment of genes involved in lipid metabolism exclusive to N2 animals and genes encoding detoxification enzymes enriched in both N2 and mab-5 GOF animals. No GO terms were enriched in the set downregulated genes exclusive to mab-5 GOF animals. Together, these results indicate a common enrichment of biological processes related to stress response and innate immunity in N2 and mab-5(e1751) GOF animals.
Table 4.
Enriched GO terms in N2 and mab-5(e1751) worms exposed to S. epidermidis.
| Upregulated genes | ||||
|---|---|---|---|---|
| GO term | Number of genes | percenta | FDR-adjusted P-valueb | |
| N2-only | Major sperm protein | 21 | 67.7 | 5.3 × 10−26 |
| Extracellular material: collagen | 25 | 13.6 | 1.3 × 10−16 | |
| Signaling: phosphatase | 17 | 8.7 | 1.7 × 10−8 | |
| Cytoskeleton: microtubule | 14 | 10.9 | 3.6 × 10−8 | |
| Unassigned | 132 | 2.1 | 5.7 × 10−8 | |
| Transmembrane protein: unassigned | 37 | 2.2 | 7.3 × 10−3 | |
| mab-5(e1751)-only | Metabolism: lipid | 7 | 2.7 | 2.1 × 10−3 |
| N2 and mab-5(e1751) | Stress response: pathogen | 4 | 2.1 | 1.0 × 10−3 |
| Downregulated genes | ||||
| N2-only | Metabolism: lipid | 7 | 2.7 | 6.4 × 10−6 |
| mab-5(e1751)-only | No enrichment of GO terms | |||
| N2 and mab-5(e1751) | Stress response: detoxification | 2 | 1.0 | 7.5 × 10−3 |
a Percentage of genes annotated with the indicated GO term, which are differentially expressed.
b FDR-adjusted P-value is a measure of enrichment (Fisher exact test; Bonferroni correction) of the GO term among DEGs.
We also performed geneset enrichment analyses on DEGs exclusive to N2, mab-5(e1751), or mab-5(e1239) animals, genes that were differentially expressed in all 3 genotypes, or DEGs identified in a combination of genotypes (N2 and mab-5 GOF, N2 and mab-5 LOF, and mab-5 GOF and mab-5 LOF) after S. epidermidis exposure, again separating genes based on upregulation or downregulation relative to an uninfected control (E. coli OP50). In upregulated genes exclusive to N2, the GO terms “Stress response: pathogen” and “Stress response: heavy metal” were enriched. In genes exclusive to mab-5 LOF animals, the GO terms “Signaling: phosphatase,” “Transmembrane Protein,” “Extracellular material: collagen,” Signaling: Tyrosine kinase,” “Proteolysis, metallopeptidase,” and “Major sperm protein” were enriched. No GO terms were enriched for DEGS exclusive to mab-5 GOF animals. The GO term “Stress response” was enriched in DEGs common to N2 and mab-5 GOF animals, whereas the GO terms “Major sperm protein,” “Extracellular material: collagen,” “Signaling: phosphatase,” and “Cytoskeleton: microtubule” were enriched in N2 and mab-5 LOF animals. A single GO term, “Metabolism: lipid,” was enriched in mab-5 GOF and mab-5 LOF animals, whereas there was no enrichment of GO terms in genes upregulated in all 3 genotypes (Table 5, Supplementary Table 9).
Table 5.
Enriched GO terms in upregulated N2, mab-5(e7151), and mab-5(e1239) worms exposed to S. epidermidis.
| GO term | Number of genes | percenta | FDR-adjusted P-valueb | |
|---|---|---|---|---|
| N2-only | Stress response: pathogen | 4 | 2.1 | 3.9 × 10−3 |
| Stress response: heavy metal | 2 | 12.5 | 5.3 × 10−3 | |
| mab-5(e1751)-only | No enrichment of GO terms | |||
| mab-5(e1239)-only | Unassigned | 747 | 11.8 | 8.3 × 10−45 |
| Signaling: phosphatase | 81 | 41.3 | 7.6 × 10−33 | |
| Transmembrane protein | 259 | 15.3 | 5.7 × 10−32 | |
| Extracellular material: collagen | 63 | 34.2 | 5.4 × 10−22 | |
| Cytoskeleton: microtubule | 53 | 41.4 | 1.7 × 10−21 | |
| Signaling: Y kinase | 29 | 39.7 | 3.1 × 10−11 | |
| Proteolysis: metallopeptidase | 28 | 22.8 | 3.2 × 10−6 | |
| Major sperm protein | 10 | 32.3 | 8.1 × 10−3 | |
| N2 and mab-5(e1751) | Stress response | 4 | 0.5 | 2.9 × 10−3 |
| N2 and mab-5(e1239) | Major sperm protein | 21 | 67.7 | 2.8 × 10−27 |
| Extracellular material: collagen | 25 | 13.6 | 4.8 × 10−18 | |
| Signaling: phosphatase | 17 | 8.7 | 1.9 × 10−9 | |
| Unassigned | 120 | 1.9 | 3.1 × 10−8 | |
| Cytoskeleton: microtubule | 13 | 10.2 | 5.6 × 10−8 | |
| mab-5(e1751) and mab-5(e1239) | Metabolism: lipid | 6 | 1.1 | 2.1 × 10−3 |
| N2 and mab-5(e1751) and mab-5(e1239) | No enrichment of GO terms | |||
a Percentage of genes annotated with the indicated GO term, which are differentially expressed.
b FDR-adjusted P-value is a measure of enrichment (Fisher exact test; Bonferroni correction) of the GO term among DEGs.
In downregulated genes exclusive to N2, the GO term “Lipid: metabolism” was enriched. In genes exclusive to the mab-5 LOF, the GO terms “Proteolysis proteasome: E3” and “Extracellular material: cuticlin” were enriched. There was no enrichment of genes exclusive to mab-5 GOF animals. We also identified enrichment of genes related to lipid metabolism in N2 and mab-5 LOF animals, whereas there was no enrichment of GO terms in genes shared by N2 and mab-5 GOF animals or genes shared by mab-5 GOF and mab-5 LOF animals. An enrichment of C-type lectins was observed in genes downregulated in all 3 genotypes, N2, mab-5(e1751), and mab-5(e1239; Table 6, Supplementary Table 9). Thus, when considering genes that are upregulated in either N2 animals or mab-5 GOF mutants but not mab-5 LOF mutants following S. epidermidis infection, our results indicate an enrichment of biological processes related to stress response and innate immunity. When considering genes that are downregulated, our results indicate an enrichment of C-type lectins in all 3 genotypes.
Table 6.
Enriched GO terms in downregulated genes identified in N2, mab-5(e1751), and mab-5(e1239) worms exposed to S. epidermidis.
| GO term | Number of genes | percenta | FDR-adjusted P-valueb | |
|---|---|---|---|---|
| N2-only | Metabolism: lipid | 3 | 0.6 | 2.0 × 10−3 |
| mab-5(e1751)-only | No enrichment of GO terms | |||
| mab-5(e1239)-only | Proteolysis proteasome: E3 | 81 | 13.7 | 2.6 × 10−39 |
| Extracellular material: cuticlin | 8 | 22.9 | 4.2 × 10−5 | |
| Unassigned | 167 | 2.6 | 8.5 × 10−5 | |
| N2 and mab-5(e1751) | No enrichment of GO terms | |||
| N2 and mab-5(e1239) | Metabolism: lipid | 4 | 0.8 | 2.9 × 10−3 |
| mab-5(e1751) and mab-5(e1239) | No enrichment of GO terms | |||
| N2 and mab-5(e1751) and mab-5(e1239) | Stress response: C-type lectin | 2 | 0.8 | 9.0 × 10−3 |
aPercentage of genes annotated with the indicated GO term, which are differentially expressed
b FDR-adjusted P-value is a measure of enrichment (Fisher exact test; Bonferroni correction) of the GO term among DEGs.
DEG in mab-5 mutants relative to wild-type N2 animals on E. coli OP50
It is possible that perturbations in MAB-5 function are inducing expression of host response genes in the absence of a pathogen and, as a result, would not be detected in our earlier RNA-seq analysis. Using wild-type N2 animals as a baseline control, we identified gene expression changes in mab-5 GOF and mab-5 LOF mutants reared on nonpathogenic E. coli OP50 (for all analyses, DEGs were defined as those with an FDR-adjusted P-value ≤ 0.05 and fold change ≥2, mab-5 mutant vs N2). In total, we identified 1472 and 167 DEGs for mab-5(e1751) and mab-5(e1239) mutants, respectively. In both GOF and LOF mutants, we found that most genes were upregulated compared with N2 [mab-5(e1751), 1240/1472 = 84%; mab-5(e1239), 144/167 = 86%; Table 7, Supplementary Tables 10 and 11].
Table 7.
DEGs in mab-5 mutants relative to wild-type N2 animals on E. coli OP50.
| Genotype | Total DEGsa | Total upregulated | Total downregulated | Total genes expressedb |
|---|---|---|---|---|
| mab-5(e1751) | 1,472 | 1,240 | 232 | 16,539 |
| mab-5(e1239) | 167 | 144 | 23 | 16,797 |
| N2 | — | — | — | 15,394 |
a Genes with a FDR-adjusted P-value ≤0.05 and log2 fold change ≥1 relative to E. coli OP50.
b Total number of genes with >0 counts in each replicate.
We then performed geneset enrichment analyses on DEGs in mab-5 GOF or mab-5 LOF mutants, separating genes by updownregulation or downregulation relative to N2. In mab-5(e1751) DEGs, we did not observe enrichment of GO terms related to the host stress response or immune defense. Upregulated genes were enriched for the GO terms “Extracellular material: collagen,” “Signaling: phosphatase,” “Cytoskeleton: microtubule,” “Major sperm protein,” “Transmembrane protein,” “Signaling: Tyrosine Kinase,” and “Proteolysis general: metallopeptidase,” whereas downregulated genes were enriched for “Proteolysis proteasome: E3” and “Transcription factor: Zinc-finger” (Table 8, Supplementary Table 12). In mab-5 LOF mutants, we found enrichment of the GO terms “Extracellular material: collagen” and “Stress response: pathogen” in upregulated genes, whereas there was no enrichment of GO terms in downregulated genes (Table 9, Supplementary Table 13). Together, our results suggest that loss of MAB-5 function activates a subset of host immune response genes in the absence of infection by S. epidermidis.
Table 8.
Enriched GO terms in uninfected mab-5(e1751) worms.
| GO term | Number of genes | percenta | FDR-adjusted P-valueb | |
|---|---|---|---|---|
| Upregulated genes | Extracellular material: collagen | 78 | 42.4 | 9.6 × 10−44 |
| Signaling: phosphatase | 72 | 36.7 | 4.1 × 10−37 | |
| Unassigned | 479 | 7.6 | 4.9 × 10−27 | |
| Cytoskeleton: microtubule | 49 | 38.3 | 5.8 × 10−26 | |
| Major sperm protein | 28 | 90.3 | 3.6 × 10−22 | |
| Transmembrane protein: unassigned | 163 | 9.6 | 8.8 × 10−19 | |
| Signaling: Y kinase | 24 | 32.9 | 2.1 × 10−11 | |
| Proteolysis general: metallopeptidase | 16 | 13.0 | 8.1 × 10−3 | |
| Downregulated genes | Proteolysis proteasome: E3 | 41 | 7.0 | 1.6 × 10−22 |
| Transcription factor: ZF | 6 | 11.3 | 2.5 × 10−4 | |
| Unassigned | 77 | 1.2 | 9.2 × 10−3 |
a Percentage of genes annotated with the indicated GO term, which are differentially expressed.
b FDR-adjusted P-value is a measure of enrichment (Fisher exact test; Bonferroni correction) of the GO term among DEGs.
Table 9.
Enriched GO terms in uninfected mab-5(e1239) worms.
| GO term | Number of genes | percenta | FDR-adjusted P-valueb | |
|---|---|---|---|---|
| Upregulated | Extracellular material: collagen | 12 | 6.5 | 1.2 × 10−8 |
| Stress response: pathogen | 9 | 4.7 | 2.6 × 10−5 | |
| Downregulated | No enrichment of GO terms | |||
a Percentage of genes annotated with the indicated GO term, which are differentially expressed.
b FDR-adjusted P-value is a measure of enrichment (Fisher exact test; Bonferroni correction) of the GO term among DEGs.
Expression differences and geneset enrichment analysis in N2-infected animals and uninfected mab-5(e1239) mutants
Because uninfected mab-5 LOF mutants showed an enrichment in host defense genes, we were interested in examining the overlap in transcriptional responses between N2 animals exposed to S. epidermidis and uninfected mab-5 LOF animals. Comparing upregulated genes between infected N2 and uninfected mab-5 LOF animals, a minority, 52 in total, overlapped between the 2 (N2-only, 318/370 = 86%; mab-5 GOF-only, 92/144 = 64%). Further the lack of overlap in gene expression was statistically significant (Χ = 31.25; P < 0.0001). In comparing downregulated genes, only 2 genes overlapped between the 2 genotypes (N2-only, 29/31 = 94%; mab-5 GOF-only, 21/23 = 91%). However, the lack of overlap in differential expression was not statistically significant (Χ = 0.10; P = 0.756; Fig. 4a, Supplementary Table 14). Notably, all DEGs shared between infected N2 animals and uninfected mab-5(e1239) mutants were expressed in the same direction. We also examined the correlation of gene expression using the log2 fold changes for each condition. Although a minority of DEGs overlapped between N2 and mab-5 LOF animals, expression levels were positively correlated between the 2 genotypes (ρ = 0.440; P < 0.001; Fig. 4b).
Fig. 4.
Expression differences in N2-infected animals compared with mab-5(e1239)-uninfected animals. a) Intersection of upregulated (top) and downregulated (bottom) genes in N2 animals infected with S. epidermidis and mab-5(e1239)-uninfected animals. b) Correlation between log2 fold changes in N2-infected and mab-5(e1239)-uninfected animals. Spearman's rho value and P-value are presented in the upper left corner. Gray (Non-DE) and black dots (DE Genes) represent expressed gene and DEG, respectively.
Separating genes based on upregulation or downregulation, we then performed geneset enrichment on DEGs exclusive to infected N2 animals (N2-only), exclusive to uninfected mab-5 LOF mutants [mab-5(e1239)-only] or differentially expressed in both conditions [N2 and mab-5(e1239)].
In upregulated genes exclusive to N2-infected animals, we observed an enrichment of genes corresponding to the GO terms “Major sperm protein,” “Signaling: phosphatase,” “Extracellular material: collagen,” and “Cytoskeleton: microtubule”. In genes exclusive to uninfected mab-5 LOF animals, we observed an enrichment in biological functions pertaining to the organismal stress response and immunity. In DEGs shared by infected N2 and uninfected mab-5 LOF animals, we saw an enrichment of collagens and C-type lectins. Using downregulated genes, we found an enrichment of genes involved in lipid metabolism exclusive to N2 animals and genes encoding acid phosphatase enzymes enriched in both infected N2 and uninfected mab-5 LOF animals. No GO terms were enriched in the set of downregulated genes exclusive to uninfected mab-5 LOF animals (Table 10, Supplementary Table 15). Together, these data indicate that there are a handful of host immune genes whose expression is induced by loss of mab-5 function in the absence of S. epidermidis infection, but the expression of these genes is not altered in wild-type N2 animals infected with S. epidermidis.
Table 10.
Enriched GO terms when comparing gene expression in uninfected mab-5(e1239) animals with N2 worms exposed to S. epidermidis.
| Upregulated genes | ||||
|---|---|---|---|---|
| GO term | Number of genes | percenta | FDR-adjusted P-valueb | |
| N2-only | Major sperm protein | 21 | 67.7 | 1.6 × 10−26 |
| Signaling: phosphatase | 17 | 8.7 | 7.3 × 10−9 | |
| Extracellular material: collagen | 16 | 8.7 | 2.4 × 10−8 | |
| Cytoskeleton: microtubule | 13 | 10.2 | 1.7 × 10−7 | |
| Unassigned | 120 | 1.9 | 1.3 × 10−6 | |
| mab-5(e1239)-only | Stress response: pathogen | 6 | 3.1 | 1.4 × 10−3 |
| N2 and mab-5(e1239) | Extracellular material: collagen | 9 | 4.9 | 1.8 × 10−9 |
| Stress response: C-type lectin | 5 | 2.0 | 1.5 × 10−3 | |
| Downregulated genes | ||||
| N2-only | Metabolism: lipid | 8 | 1.5 | 1.5 × 10−6 |
| mab-5(e1239)-only | No enrichment of GO terms | |||
| N2 and mab-5(e1239) | Lysosome: acid phosphatase | 1 | 3.2 | 6.1 × 10−3 |
a Percentage of genes annotated with the indicated GO term, which are differentially expressed.
b FDR-adjusted P-value is a measure of enrichment (Fisher exact test; Bonferroni correction) of the GO term among DEGs.
Overlap of known MAB-5–binding sites with pathogen-related DEGs
We next examined whether any of the DEGs corresponding to the enriched GO terms “Stress response: C-type lectin” or “Stress response: Pathogen” could be direct targets of MAB-5. In total, 23 genes with innate immune function were differentially expressed across our different gene expression analyses. For each gene, we obtained genomic coordinates comprising the coding region and 1-kb upstream and downstream of the start and end sites (Table 11). These coordinates were cross-referenced to known MAB-5–binding sites obtained from the ModERN database (Kudron et al. 2018). From this analysis, we found that none of the genomic coordinates of the 23 innate immune genes overlapped with MAB-5–binding sites cataloged in the ModERN database.
Table 11.
Genomic coordinates of innate immune genes identified via geneset enrichment analysis.
| Gene ID | Gene name | Chromosome | Gene start (bp)a | Gene end (bp)a |
|---|---|---|---|---|
| WBGene00020326 | math-38 | II | 1576378 | 1580986 |
| WBGene00018274 | F41C3.8 | II | 4715046 | 4719536 |
| WBGene00011979 | T24B8.5 | II | 9081626 | 9084193 |
| WBGene00014046 | clec-60 | II | 10479043 | 10482874 |
| WBGene00008492 | F01D5.1 | II | 13995958 | 13998581 |
| WBGene00006627 | tsp-1 | III | 8237001 | 8240423 |
| WBGene00009526 | clec-169 | IV | 1260324 | 1265622 |
| WBGene00016669 | ilys-2 | IV | 2466678 | 2469879 |
| WBGene00018971 | clec-67 | IV | 3921165 | 3926039 |
| WBGene00021581 | clec-70 | IV | 3931708 | 3937307 |
| WBGene00021582 | clec-71 | IV | 3936458 | 3940979 |
| WBGene00021583 | clec-72 | IV | 3943816 | 3948362 |
| WBGene00008584 | irg-4 | IV | 12434833 | 12439236 |
| WBGene00014132 | ZK896.1 | IV | 12871776 | 12876360 |
| WBGene00010125 | dod-22 | IV | 12964374 | 12968392 |
| WBGene00010123 | F55G11.2 | IV | 12966445 | 12970289 |
| WBGene00010124 | F55G11.4 | IV | 12972624 | 12976603 |
| WBGene00010745 | dod-17 | IV | 12975303 | 12979343 |
| WBGene00022261 | clec-210 | V | 3117377 | 3121323 |
| WBGene00009432 | cld-9 | V | 13748181 | 13752891 |
| WBGene00011844 | T19C9.8 | V | 17238097 | 17241721 |
| WBGene00017892 | F28B4.3 | X | 3226501 | 3236408 |
| WBGene00017582 | F18G5.6 | X | 9261045 | 9265331 |
aGene start and end sites comprise the coding region and 1-kb upstream and downstream of the start and end sites to encompass potential noncoding regulatory elements.
The lack of binding sites suggests an indirect relationship between mab-5 function and expression of innate immune genes, perhaps through the differential expression of other transcription factors. We compared our DEG lists with a table of known C. elegans transcription factors (Supplementary File 1). In total, we identified 68 unique transcription factors that were differentially expressed, with 63 of 68 differentially expressed only in mab-5(e1239) LOF animals infected with S. epidermidis (Supplementary Table 16). Of the 68 transcription factors, 18 had binding site data in the ModERN database which we cross-referenced with the genomic coordinates of innate immune genes identified earlier (Table 11). Of 18, only one transcription factor, pqm-1, had any binding site overlap. Specifically, we observed pqm-1–binding sites overlapping the genomic coordinates of F41C3.8, clec-72, and T24B8.5. Together, these data suggest an indirect relationship between mab-5 function and the expression of these innate immune genes, rather than any of the immune genes serving as direct targets of MAB-5 or as direct targets of a transcription factor differentially expressed in mab-5 mutants.
Discussion
mab-5 is required for prolonging the lifespan following S. epidermidis exposure
In a previous study, we found that S. epidermidis was pathogenic to CB4856 animals and shortened lifespan, whereas the lifespan of N2 animals was prolonged (Lansdon et al. 2022). These results suggested that the genomic variation between these isolates resulted in vastly different outcomes when they encountered S. epidermidis. Comparing gene expression differences between N2 animals and the Hawaiian isolate, CB4856, we noted that mab-5 was upregulated in N2, but not CB4856. Upon reflection, mab-5 was a good candidate to potentially underlie the differences in survival between N2 and CB4856 as Hox genes are known to contribute to innate immunity in C. elegans (Gravato-Nobre et al. 2005; Irazoqui et al. 2008; Nicholas and Hodgkin 2009).
We hypothesized that if mab-5 was underlying the pathogenic outcomes observed in CB4586, then LOF mab-5 mutants would be as susceptible to infection as CB4856 wild-type animals. In contrast, we found a more nuanced outcome. Specifically, in our survival assays, we found that animals lacking mab-5 function had lost the lifespan extension effect of feeding S. epidermidis (compared with E. coli) observed in the wild-type N2 animals. In this context, it is important to note that all of the mab-5 mutations used here were isolated in an otherwise N2 genetic background.
First, we used the e1239 mutation, which was isolated in a forward genetic screen for male abnormal phenotypes. The e1239 mutation is a strong LOF that molecularly affects a splicing site, leading to decreased MAB-5 protein levels. We confirmed our results using the gk670 mutation, which was isolated in a different laboratory using a reverse genetic approach to create genetic deletion mutations in C. elegans genes. The gk670 allele removes a significant portion of the mab-5 coding region and introduces potential frameshifts that would prevent function of MAB-5 protein from being produced. The similarity in the susceptibility to S. epidermidis in the e1239 and gk670 animals indicates it is the LOF in mab-5 rather than potential background differences. Further, we found that a GOF mutation, e1751, which was also isolated in a screen for male abnormal phenotypes, exhibited the same lifespan extension effects after feeding on S. epidermidis as wild-type N2 animals.
Taken together, we confirmed that in the N2 genetic background, feeding on S. epidermidis extends lifespan compared with feeding on E. coli. We concluded that intact mab-5 function was required for the animals to derive the benefit of S. epidermidis. By itself, loss of mab-5 expression does not appear to be deleterious to these animals since lifespan on E. coli OP50 was not significantly different from N2 or mab-5 GOF animals. Finally, we cannot specifically conclude that a failure to upregulate mab-5 in the CB4856 background leads to the pathogenic outcome. Rather, there are likely to be additional genetic and/or genomic variation between these 2 isolates, which underlies the differences in survival.
S. epidermidis induces stress-like pathways in the N2 background
There has been limited investigation into the host–pathogen relationship between C. elegans and S. epidermidis. Some reports suggest S. epidermidis can be pathogenic, while others, including our work, suggest it can be either pathogenic or beneficial. On the pathogen side, it is suggested that S. epidermidis accumulates in the intestine and relies in part on biofilm formation to kill the host (Begun et al. 2007). On the host side, research has exclusively used wild-type animals, focusing either on the expression of select immune genes, namely atf-7 and mpk-1, or surveying for global transcriptional changes after S. epidermidis infection (JebaMercy et al. 2015; Lansdon et al. 2022).
We also used transcriptional analysis to better understand how mab-5 might contribute to the interactions we observed between S. epidermidis and C. elegans. We compared gene expression profiles within genotypes between treatments, e.g. N2 on E. coli vs S. epidermidis, etc. as well as between genotypes, e.g. N2 vs mab-5(e1239) on E. coli, etc. Subsequently, we grouped DEGs and performed geneset enrichment analysis on sets of genes that were either exclusive to N2, mab-5 GOF, or mab-5 LOF animals, genes that were differentially expressed in all 3 genotypes, or DEGs identified in a combination of genotypes.
When we examined the genes that were differentially expressed across genotypes, we observed enrichment in at least 3 categories of genes that provide evidence that C. elegans activate stress-response pathways when shifted from E. coli to S. epidermidis. First, in our upregulated geneset, we saw a significant enrichment of genes encoding major sperm proteins. Major sperm proteins are the primary cytoskeletal components that enable sperm motility and promote oocyte maturation prior to fertilization (Roberts and Stewart 1995; Kuwabara 2003). Accelerated yolk production and an increased fertility rate have been associated with pathogenic infection in C. elegans (DePina et al. 2011). Additionally, when animals are starved or exposed to pathogenic stressors, sperm production as well as the prevalence of males increases to promote genetic diversity (Morran et al. 2009; Sharika et al. 2018).
Our analysis also detected an enrichment of genes encoding extracellular matrix (ECM) proteins, specifically collagens, in upregulated DEGs shared by N2 and mab-5 LOF animals. Additionally, a separate set of collagen genes was enriched in upregulated DEGs exclusive to mab-5 LOF animals. Collagens comprise both the cuticle and basement membrane of the ECM and are an integral component of the C. elegans epidermal barrier (Kramer 1994). Further, collagens have demonstrated roles in the host stress response as changes in collagen composition and collagen gene expression have been reported in animals experiencing innate immune and osmotic stressors (Ermolaeva and Schumacher 2014; Taffoni and Pujol 2015; Dodd et al. 2018; Mesbahi et al. 2020; Sandhu et al. 2021; Chandler and Choe 2022; Lansdon et al. 2022). Our findings provide additional support for collagens being associated with or possibly important for the C. elegans innate immune response, though it is not clear whether they are responsible for any observed differences in survival.
Additionally, an enrichment of genes involved in lipid metabolism was also identified. We found sets of lipid metabolism genes enriched in DEGs exclusive to either N2 or mab-5 GOF animals. We also identified additional sets of lipid metabolism genes that were enriched in both the mab-5 GOF and LOF mutants as well as a geneset enriched in both N2 animals and the mab-5 LOF mutant. It is well-established that pathogenic infections can alter host metabolism and affect cellular pathways that control host nutritional status and lipid homeostasis (Anderson and Pukkila-Worley 2020). The enrichment of lipid metabolism in all 3 genotypes, albeit different sets of genes, suggests that the animals recognize S. epidermidis as a stressor and are modulating host metabolic processes in response. Looking specifically at N2 and mab-5 GOF animals, we observed altered expression of genes involved in the synthesis of polyunsaturated fatty acids (elo-6), lipid transport (lbp-8, vit-1, vit-3, vit-4), triglyceride mobilization (lipl-5), and fatty acid degradation (lipl-1, lipl-2, acdh-2, W03F9.4). We also observed differential expression of genes involved in sphingolipid metabolism, with an upregulation of genes encoding anabolic processes (gba-4) and a downregulation of genes encoding catabolic processes (asah-1). Lipid metabolism also has a demonstrated role in the induction of the innate immune response. Nematodes with LOF mutations in elongase (elo) and desaturase (fat) genes exhibit greater susceptibility to pathogenic microbes (Nandakumar and Tan 2008; Anderson et al. 2019). Lipase-like (lipl) genes are expressed in response to nutritional stress conditions, and in the case of lipl-5, expression is altered upon exposure to pathogenic bacteria (Mallo et al. 2002). Further, the suppression of sphingolipid catabolism and the synthesis of sphingolipid-derived signaling molecules have been shown to increase C. elegans immunity toward pathogenic microbes (Lee et al. 2020; Nasrallah et al. 2023).
Last, of the DEGs that overlapped in N2, mab-5 GOF, and mab-5 LOF animals, all but one was expressed in the same direction. The ech-9 gene encodes an enoyl-CoA hydratase that is essential for metabolizing fatty acids. It was found to be upregulated in N2 animals, downregulated in mab-5(e1239), and not differentially expressed in mab-5(e1751). Differential expression of ech-9 has been previously reported following exposure to the Gram-negative pathogen, Pseudomonas aeruginosa, as well as the Gram-positive pathogens, St. aureus and M. nematophilum (Irazoqui et al. 2010; Pellegrino et al. 2014). Our transcriptomic data indicate that expression of ech-9 may also impact lipid metabolism and the ability of C. elegans to survive following S. epidermidis infection.
Overall, the analysis of genes induced across animals fed S. epidermidis compared with E. coli suggests that the animals recognize S. epidermidis as a stressor and that the phenotypic outcomes are likely after the recognition event. Further, the differential expression of specific lipid metabolism genes exclusively in N2 and mab-5 GOF animals suggests a potential role for their involvement in mounting an immune response to S. epidermidis infection.
Increased survival on S. epidermidis is associated with functional categories corresponding to pathogen detection and response
Our analysis identified an enrichment of genes encoding C-type lectins and proteins involved in pathogen response. C-type lectins are a superfamily of proteins that contribute to the recognition of pathogenic agents in both vertebrates and invertebrates with well over 200 family members in C. elegans (Zelensky and Gready 2005; Schulenburg et al. 2008). Lectins exhibit differential upregulation after pathogen exposure with only a few upregulated across multiple pathogen species, suggesting a response that is specific to the pathogen rather than the pathogen type (e.g. Gram-negative vs Gram-positive; Troemel et al. 2006; Alper et al. 2007; Wong et al. 2007; Schulenburg et al. 2008). We observed a downregulation of 2 lectins, clec-169 and clec-210. in all 3 genotypes after S. epidermidis exposure. The literature regarding clec-169 and clec-210 is limited but shows both lectins are induced in response to infection by the Gram-negative pathogen, Klebsiella pneumoniae (Yang et al. 2023). Their downregulation suggests a lack of involvement in the response to S. epidermidis exposure. Two additional lectins, clec-60 and clec-71, were upregulated exclusively in N2 and mab-5 GOF animals. Upregulation of clec-60 has been observed in response to infection with Gram-positive pathogens such as Enterococcus faecalis and St. aureus, whereas infection with Gram-negative pathogens such as P. aeruginosa leads to repression of clec-60 (O’Rourke et al. 2006; Wong et al. 2007; Irazoqui et al. 2010; Head and Aballay 2014). Changes in clec-71 expression have been observed following exposure to Cutibacterium acnes, a Gram-positive bacterium found on the surface of the skin and other epithelial linings (Huang et al. 2020; Mongaret et al. 2021).
The upregulation of clec-60 and clec-71 expression in both N2 and mab-5 GOF animals coupled with previous literature showing an induction of expression after infection with other Gram-positive pathogens raises the possibility that clec-60 and clec-71 are downstream targets of mab-5 and may enable C. elegans to recognize and respond to S. epidermidis.
Geneset enrichment analysis also identified 2 pathogen response genes, dod-22 and F55G11.2, that were upregulated in both N2 and mab-5 GOF animals but not differentially expressed in mab-5(e1239) mutants. Both dod-22 and F55G11.2 encode for proteins containing complement C1r/C1s, Uegf, Bmp1 (CUB)-like domains. CUB domain proteins possess a diverse array of functions, ranging from complement activation and the innate immune response to developmental patterning and cell signaling (Bork and Beckmann 1993; Perry et al. 2007; Bolz et al. 2010; Simonsen et al. 2012; Ermolaeva and Schumacher 2014; Fanelli et al. 2023). Additionally, dod-22 and F55G11.2 have previously been shown to be activated following exposure to Gram-negative pathogens, such as P. aeruginosa, Burkholderia pseudomallei, and Vibrio cholerae (Sahu et al. 2012; Lee et al. 2013; Liu et al. 2016). Our findings demonstrate that these immune genes are also upregulated in response to S. epidermidis, suggesting that they may contribute to defense against Gram-positive pathogens as well.
Loss of mab-5 induces expression of stress response genes in the absence of infection
We identified DEGs in both uninfected mab-5(e1239) and mab-5(e1751) animals on E. coli OP50 (vs N2 var. Bristol) and performed geneset enrichment analysis. In uninfected mab-5 GOF animals, we found enrichment of several categories identified in previous analyses, including enrichment of genes encoding collagens and major sperm proteins, but did not observe enrichment of GO categories corresponding to the organismal stress response. In contrast, uninfected mab-5 LOF animals exhibited an enrichment of genes, 15 in total, that are typically induced following exposure to pathogenic bacteria. In comparing expression of these 15 genes with DEGs from uninfected mab-5 GOF animals, we identified only 3 genes, clec-60, dod-17, and F55G11.4, that were also differentially expressed. Thus, in the absence of infection, loss of mab-5 function leads to upregulation of a subset of immune response genes. However, a similar change in expression of immune genes was not observed in mab-5 GOF animals, indicating that by itself, constitutive activation of mab-5 is not inducing a pathogen-related stress response.
By identifying DEGs using different baselines of gene expression (S. epidermidis vs E. coli OP50; mab-5 LOF vs N2, etc.), we identified several independent sets of immune genes enriched in different conditions, comprising a total of 23 genes. Of these genes, only 4, F18G5.6, F41C3.8, F01D5.1, and F28B4.3, were upregulated in either N2 or mab-5(e1751) animals infected with S. epidermidis but were not differentially expressed in mab-5(e1239) animals, regardless of infection status. These 4 genes remain functionally uncharacterized, and we were unable to find evidence in the literature of their involvement in the C. elegans immune response. Two genes, F01D5.1 and F18G5.6, encode short proteins (<150 amino acids), suggesting a potential role as antimicrobial peptides. To shed some light on their function, we used SignalP 6.0 to predict the presence of signal peptides as C. elegans antimicrobial peptides contain a signal peptide on the N-terminus of the protein (Dierking et al. 2016; Teufel et al. 2022). The protein sequence of F01D5.1 contained a cleavable signal peptide, whereas the sequence of F18G5.6 did not, suggesting that F01D5.1 may function as an antimicrobial peptide. The F28B4.3 gene encodes a much longer protein (>2000 amino acids) and is predicted to contain a C-type lectin domain and thus may be involved in pathogen detection. The F41C3.8 gene encodes 2 protein isoforms between 300 and 350 amino acids in length and is predicted to be membrane localized as evidenced by a transmembrane domain.
mab-5 may indirectly regulate genes downstream of S. epidermidis exposure
Using existing ChIP-Seq datasets from the ModERN database, we cross-referenced MAB-5–binding sites with genomic coordinates of the 23 DEGs corresponding to the enriched GO terms “Stress response: C-type lectins” or “Stress response: Pathogen.” However, we did not observe any overlap between those genomic coordinates and MAB-5–binding sites. One possibility is that mab-5 is indirectly regulating expression of these genes by working with other transcription factors to modulate the immune response following S. epidermidis exposure. To examine this possibility, we identified 68 transcription factors that were differentially expressed in animals infected with S. epidermidis, with 63 of those exclusively expressed in infected mab-5 LOF animals. Again using existing ChIP-Seq datasets, we found a single transcription factor, PQM-1, to have binding sites overlapping 3 genes, clec-72, T24B8.5, and F41C3.8. Both clec-72 and T24B8.5 are induced during infection with multiple pathogens, whereas differential expression of F41C3.8 after pathogen exposure has not been reported previously (Troemel et al. 2006; Bolz et al. 2010; Irazoqui et al. 2010; Campos et al. 2021). The pqm-1 gene was upregulated in both N2 and mab-5 GOF animals infected with S. epidermidis but was not differentially expressed in mab-5 LOF animals regardless of infection status. PQM-1 has previously been implicated in the C. elegans innate immune response, and its function is required for the transcription of genes encoding innate immune effectors and detoxification enzymes (Tepper et al. 2013; Dowen et al. 2016; Rajan et al. 2019; Borbolis et al. 2023). Thus, our analysis of transcription factor–binding sites suggests that mab-5 may be indirectly regulating expression of immune genes through intermediate transcription factors such as PQM-1. A limitation to our analysis is the availability of published ChIP-seq data and the experimental conditions used to obtain those data. Although we identified 68 differentially expressed transcription factors, only 18 had ChIP-Seq available for our analysis. It is entirely possible that some of the remaining 50 transcription factors could have binding sites overlapping innate immune genes similar to pqm-1. Additionally, the growth conditions used in the ChIP-Seq experiments could make the difference. The ChIP-Seq data used in this analysis were collected from N2 worms reared on a liquid culture diet of E. coli OP50 during either the embryonic or L2 larval stage of development (Niu et al. 2011). Our RNA-seq experiments used 1-day-old adults transferred from E. coli to S. epidermidis. It is possible that at earlier life stages and under nonpathogenic conditions, MAB-5, or any of the other transcription factors differentially expressed after S. epidermidis exposure, does not directly bind promoters of these immune genes but does so under different conditions.
Integration of metabolic and innate immune pathways during microbial infection
Our findings suggest that mab-5 is important for prolonging the lifespan of animals infected with S. epidermidis and may do so indirectly through intermediate transcription factors such as PQM-1. PQM-1 has prominent roles in the immune response and the metabolism of lipids in response to environmental stressors (Rajan et al. 2019; Heimbucher et al. 2020). Further, it is a key regulator of DAF-2–mediated longevity (Tepper et al. 2013). The involvement of PQM-1 raises the possibility that the lifespan extension following S. epidermidis exposure is due to a nutritional challenge rather than a robust immune response. This alternative hypothesis is supported by the differential expression of genes involved in lipid metabolism and the organismal stress response. However, it is difficult to say one way or another as microbial infections not only induce an innate immune response but also alter metabolism as the host seeks to maintain cellular homeostasis in response to the environmental stressor (Tepper et al. 2013). This cross-talk between innate immune and metabolic pathways is well documented in C. elegans, most notably in long-lived daf-2 mutants, which require the p38 mitogen-activated protein kinase PMK-1 pathway for pathogen resistance and lifespan extension (Troemel et al. 2006).
To summarize, we have shown that mab-5 is important for prolonging the lifespan of animals with an N2 genetic background following S. epidermidis exposure. Our transcriptomic analyses have identified genes previously implicated in immunity, such as ech-9, clec-60, clec-71, dod-22, and F55G11.2, as well as genes lacking functional annotation (F18G5.6, F41C3.8, F01D5.1, and F28B4.3) that are upregulated in animals expressing mab-5 but either are not differentially expressed or are downregulated in mab-5(e1239) LOF mutants. However, it is yet to be determined how these genes are contributing to the prolonged lifespan of mab-5–expressing animals infected with S. epidermidis. Examining S. epidermidis susceptibility in mutant strains that express mab-5 yet silence the expression of any of the aforementioned genes would help address this question. Further, determining the nutritional quality of S. epidermidis and assessing S. epidermidis susceptibility in insulin signaling mutants would help address whether mab-5 is functioning to enhance the innate immune response or is involved in the organismal response to dietary restriction. Additionally, performing ChIP-seq analysis on N2 animals and mab-5 mutants would serve to identify downstream targets of MAB-5 following S. epidermidis infection. By leveraging both of these approaches, we will be able to better understand the role of the MAB-5/Hox family transcription factor in the C. elegans innate immune response to microbial pathogens.
Supplementary Material
Contributor Information
Christopher Kywe, Department of Molecular Biosciences, University of Kansas, Lawrence, KS 66045, USA.
Erik A Lundquist, Department of Molecular Biosciences, University of Kansas, Lawrence, KS 66045, USA.
Brian D Ackley, Department of Molecular Biosciences, University of Kansas, Lawrence, KS 66045, USA.
Patrick Lansdon, Department of Molecular Biosciences, University of Kansas, Lawrence, KS 66045, USA.
Data availability
The raw sequence files used for differential gene expression analysis are available in FASTQ format in the NCBI BioProject repository (BioProject accession number: PRJNA973087; https://www.ncbi.nlm.nih.gov/sra/PRJNA973087). Raw and processed counts of sequence reads, overlapping genes, and the test statistic and P-values for all genes tested can be found in a supplemental file on figshare: https://doi.org/10.25387/g3.25025117.
Supplemental material available at G3 online.
Funding
This study was funded by grants from the Kansas IDeA Network of Biomedical Research Excellence (P20GM103418) awarded to CK and PL, the National Institute of Neurological Disorders and Stroke (R01NS115467) awarded to EAL, and the National Institute of General Medical Sciences (P30GM145499) awarded to BDA.
Author contributions
CK, EAL, BDA, and PL acquired funding for the project; BDA and PL designed and conceived the project; CK, BDA, and PL acquired, analyzed, and interpreted the data. CK, EAL, BDA, and PL wrote the manuscript. All authors have approved the manuscript.
Literature cited
- Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell. 124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. 2007. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol. 27(15):5544–5553. doi: 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KV. 2000. Toll signaling pathways in the innate immune response. Curr Opin Immunol. 12(1):13–19. doi: 10.1016/s0952-7915(99)00045-x. [DOI] [PubMed] [Google Scholar]
- Anderson SM, Cheesman HK, Peterson ND, Salisbury JE, Soukas AA, Pukkila-Worley R. 2019. The fatty acid oleate is required for innate immune activation and pathogen defense in Caenorhabditis elegans. PLoS Pathog. 15(6):e1007893. doi: 10.1371/journal.ppat.1007893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SM, Pukkila-Worley R. 2020. Immunometabolism in Caenorhabditis elegans. PLoS Pathog. 16(10):e1008897. doi: 10.1371/journal.ppat.1008897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: A quality control tool for high throughput sequence data [Online]. Available from http://www.bioinformatics.babraham.ac.uk/projects/fastqc/2010
- Ausubel FM. 2005. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 6(10):973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- Balla KM, Andersen EC, Kruglyak L, Troemel ER. 2015. A wild C. elegans strain has enhanced epithelial immunity to a natural microsporidian parasite. PLoS Pathog. 11(2):e1004583. doi: 10.1371/journal.ppat.1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla KM, Troemel ER. 2013. Caenorhabditis elegans as a model for intracellular pathogen infection. Cell Microbiol. 15(8):1313–1322. doi: 10.1111/cmi.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun J, Gaiani JM, Rohde H, Mack D, Calderwood SB, Ausubel FM, Sifri CD. 2007. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 3(4):e57. doi: 10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolz DD, Tenor JL, Aballay A. 2010. A conserved PMK-1/p38 MAPK is required in Caenorhabditis elegans tissue-specific immune response to Yersinia pestis infection. J Biol Chem. 285(14):10832–10840. doi: 10.1074/jbc.M109.091629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbolis F, Ranti D, Papadopoulou MD, Dimopoulou S, Malatras A, Michalopoulos I, Syntichaki P. 2023. Selective destabilization of transcripts by mRNA decapping regulates oocyte maturation and innate immunity gene expression during ageing in C. elegans. Biology (Basel). 12(2):171. doi: 10.3390/biology12020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Beckmann G. 1993. The CUB domain. A widespread module in developmentally regulated proteins. J Mol Biol. 231(2):539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos JC, Wu Z, Rudich PD, Soo SK, Mistry M, Ferreira JC, Blackwell TK, Van Raamsdonk JM. 2021. Mild mitochondrial impairment enhances innate immunity and longevity through ATFS-1 and p38 signaling. EMBO Rep. 22(12):e52964. doi: 10.15252/embr.202152964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Deletion Mutant Consortium . 2012. Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda). 2(11):1415–1425. doi: 10.1534/g3.112.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Thomson JN, Sulston JE. 1983. Induction of neuronal branching in Caenorhabditis elegans. Science. 221(4605):61–63. doi: 10.1126/science.6857263. [DOI] [PubMed] [Google Scholar]
- Chandler LM, Choe KP. 2022. Extracellular matrix regulation of stress response genes during larval development in Caenorhabditis elegans. G3 (Bethesda). 12(11):jkac221. doi: 10.1093/g3journal/jkac221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Paek J, Kim DH. 2011. Natural polymorphisms in C. elegans HECW-1 E3 ligase affect pathogen avoidance behaviour. Nature. 480(7378):525–529. doi: 10.1038/nature10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JO, Li H, Lundquist EA. 2008. The MIG-15 NIK kinase acts cell-autonomously in neuroblast polarization and migration in C. elegans. Dev Biol. 324(2):245–257. doi: 10.1016/j.ydbio.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez V, Mohri-Shiomi A, Maadani A, Vega LA, Garsin DA. 2007. Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics. 176(3):1567–1577. doi: 10.1534/genetics.107.072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. 2004. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 5(5):488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- DePina AS, Iser WB, Park SS, Maudsley S, Wilson MA, Wolkow CA. 2011. Regulation of Caenorhabditis elegans vitellogenesis by DAF-2/IIS through separable transcriptional and posttranscriptional mechanisms. BMC Physiol. 11(1):11. doi: 10.1186/1472-6793-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierking K, Yang W, Schulenburg H. 2016. Antimicrobial effectors in the nematode Caenorhabditis elegans: an outgroup to the Arthropoda. Philos Trans R Soc Lond B Biol Sci. 371(1695):20150299. doi: 10.1098/rstb.2015.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do N, Ackley BD, Lansdon P. 2022. Draft genome sequence of novel Staphylococcus epidermidis strain EVL2000, exhibiting pathogenicity against Caenorhabditis elegans. Microbiol Resour Announc. 11(4):e0123921. doi: 10.1128/mra.01239-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd W, Tang L, Lone JC, Wimberly K, Wu CW, Consalvo C, Wright JE, Pujol N, Choe KP. 2018. A damage sensor associated with the cuticle coordinates three core environmental stress responses in Caenorhabditis elegans. Genetics. 208(4):1467–1482. doi: 10.1534/genetics.118.300827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen RH, Breen PC, Tullius T, Conery AL, Ruvkun G. 2016. A microRNA program in the C. elegans hypodermis couples to intestinal mTORC2/PQM-1 signaling to modulate fat transport. Genes Dev. 30(13):1515–1528. doi: 10.1101/gad.283895.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Schumacher B. 2014. Insights from the worm: the C. elegans model for innate immunity. Semin Immunol. 26(4):303–309. doi: 10.1016/j.smim.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanelli MJ, Welsh CM, Lui DS, Smulan LJ, Walker AK. 2023. Immunity-linked genes are stimulated by a membrane stress pathway linked to Golgi function and the ARF-1 GTPase. Sci Adv. 9(49):eadi5545. doi: 10.1126/sciadv.adi5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravato-Nobre MJ, Nicholas HR, Nijland R, O’Rourke D, Whittington DE, Yook KJ, Hodgkin J. 2005. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 171(3):1033–1045. doi: 10.1534/genetics.105.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head B, Aballay A. 2014. Recovery from an acute infection in C. elegans requires the GATA transcription factor ELT-2. PLoS Genet. 10(10):e1004609. doi: 10.1371/journal.pgen.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimbucher T, Hog J, Gupta P, Murphy CT. 2020. PQM-1 controls hypoxic survival via regulation of lipid metabolism. Nat Commun. 11(1):4627. doi: 10.1038/s41467-020-18369-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman M. 2001. C. elegans POP-1/TCF functions in a canonical Wnt pathway that controls cell migration and in a noncanonical Wnt pathway that controls cell polarity. Development. 128(4):581–590. doi: 10.1242/dev.128.4.581. [DOI] [PubMed] [Google Scholar]
- Holdorf AD, Higgins DP, Hart AC, Boag PR, Pazour GJ, Walhout AJM, Walker AK. 2020. WormCat: an online tool for annotation and visualization of Caenorhabditis elegans genome-scale data. Genetics. 214(2):279–294. doi: 10.1534/genetics.119.302919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KL, Bolt BJ, Cain S, Chan J, Chen WJ, Davis P, Done J, Down T, Gao S, Grove C, et al. 2016. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 44(D1):D774–D780. doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Pan W, Kim W, White A, Li S, Li H, Lee K, Fuchs BB, Zeng K, Mylonakis E. 2020. Caenorhabditis elegans mounts a p38 MAPK pathway-mediated defence to Cutibacterium acnes infection. Cell Microbiol. 22(10):e13234. doi: 10.1111/cmi.13234. [DOI] [PubMed] [Google Scholar]
- Irazoqui JE, Ng A, Xavier RJ, Ausubel FM. 2008. Role for beta-catenin and HOX transcription factors in Caenorhabditis elegans and mammalian host epithelial-pathogen interactions. Proc Natl Acad Sci U S A. 105(45):17469–17474. doi: 10.1073/pnas.0809527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui JE, Troemel ER, Feinbaum RL, Luhachack LG, Cezairliyan BO, Ausubel FM. 2010. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 6(7):e1000982. doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JebaMercy G, Prithika U, Lavanya N, Sekar C, Balamurugan K. 2015. Changes in Caenorhabditis elegans immunity and Staphylococcal virulence factors during their interactions. Gene. 558(1):159–172. doi: 10.1016/j.gene.2014.12.056. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Kawai T. 2014. Toll-like receptor signaling pathways. Front Immunol. 5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. 1986. A gene involved in the development of the posterior body region of C. elegans. Cell. 46(3):477–487. doi: 10.1016/0092-8674(86)90668-9. [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenderman L, Buurman W, Daha MR. 2014. The innate immune response. Immunol Lett. 162(2):95–102. doi: 10.1016/j.imlet.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Kohchi C, Inagawa H, Nishizawa T, Soma G. 2009. ROS and innate immunity. Anticancer Res. 29(3):817–821. [PubMed] [Google Scholar]
- Kopp EB, Ghosh S. 1995. NF-kappa B and rel proteins in innate immunity. Adv Immunol. 58:1–27. doi: 10.1016/s0065-2776(08)60618-5. [DOI] [PubMed] [Google Scholar]
- Kramer JM. 1994. Structures and functions of collagens in Caenorhabditis elegans. FASEB J. 8(3):329–336. doi: 10.1096/fasebj.8.3.8143939. [DOI] [PubMed] [Google Scholar]
- Kudron MM, Victorsen A, Gevirtzman L, Hillier LW, Fisher WW, Vafeados D, Kirkey M, Hammonds AS, Gersch J, Ammouri H, et al. 2018. The ModERN resource: genome-wide binding profiles for hundreds of Drosophila and Caenorhabditis elegans transcription factors. Genetics. 208(3):937–949. doi: 10.1534/genetics.117.300657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. 2011. Pathogen recognition by the innate immune system. Int Rev Immunol. 30(1):16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- Kuwabara PE. 2003. The multifaceted C. elegans major sperm protein: an ephrin signaling antagonist in oocyte maturation. Genes Dev. 17(2):155–161. doi: 10.1101/gad.1061103. [DOI] [PubMed] [Google Scholar]
- Lansdon P, Carlson M, Ackley BD. 2022. Wild-type Caenorhabditis elegans isolates exhibit distinct gene expression profiles in response to microbial infection. BMC Genomics. 23(1):229. doi: 10.1186/s12864-022-08455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Escobar I, Jang Y, Kim W, Ausubel FM, Mylonakis E. 2020. In the model host Caenorhabditis elegans, sphingosine-1-phosphate-mediated signaling increases immunity toward human opportunistic bacteria. Int J Mol Sci. 21(21):7813. doi: 10.3390/ijms21217813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wong RR, Chin CY, Lim TY, Eng SA, Kong C, Ijap NA, Lau MS, Lim MP, Gan YH, et al. 2013. Burkholderia pseudomallei suppresses Caenorhabditis elegans immunity by specific degradation of a GATA transcription factor. Proc Natl Acad Sci U S A. 110(37):15067–15072. doi: 10.1073/pnas.1311725110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sellegounder D, Sun J. 2016. Neuronal GPCR OCTR-1 regulates innate immunity by controlling protein synthesis in Caenorhabditis elegans. Sci Rep. 6(1):36832. doi: 10.1038/srep36832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, Ewbank JJ. 2002. Inducible antibacterial defense system in C. elegans. Curr Biol. 12(14):1209–1214. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- Maloof JN, Whangbo J, Harris JM, Jongeward GD, Kenyon C. 1999. A Wnt signaling pathway controls Hox gene expression and neuroblast migration in C. elegans. Development. 126(1):37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]
- Marsh EK, May RC. 2012. Caenorhabditis elegans, a model organism for investigating immunity. Appl Environ Microbiol. 78(7):2075–2081. doi: 10.1128/AEM.07486-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N, Singh J, Aballay A. 2017. Natural genetic variation in the Caenorhabditis elegans response to Pseudomonas aeruginosa. G3 (Bethesda). 7(4):1137–1147. doi: 10.1534/g3.117.039057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGreal EP, Martinez-Pomares L, Gordon S. 2004. Divergent roles for C-type lectins expressed by cells of the innate immune system. Mol Immunol. 41(11):1109–1121. doi: 10.1016/j.molimm.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Mesbahi H, Pho KB, Tench AJ, Leon Guerrero VL, MacNeil LT. 2020. Cuticle collagen expression is regulated in response to environmental stimuli by the GATA transcription factor ELT-3 in Caenorhabditis elegans. Genetics. 215(2):483–495. doi: 10.1534/genetics.120.303125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongaret C, Velard F, Reffuveille F. 2021. Cutibacterium acnes: the urgent need to identify diagnosis markers. Infect Immun. 89(4):e00753-20. doi: 10.1128/IAI.00753-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran LT, Cappy BJ, Anderson JL, Phillips PC. 2009. Sexual partners for the stressed: facultative outcrossing in the self-fertilizing nematode Caenorhabditis elegans. Evolution. 63(6):1473–1482. doi: 10.1111/j.1558-5646.2009.00652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U, Vogel P, Alber G, Schaub GA. 2008. The innate immune system of mammals and insects. Contrib Microbiol. 15:21–44. doi: 10.1159/000135684. [DOI] [PubMed] [Google Scholar]
- Nandakumar M, Tan MW. 2008. Gamma-linolenic and stearidonic acids are required for basal immunity in Caenorhabditis elegans through their effects on p38 MAP kinase activity. PLoS Genet. 4(11):e1000273. doi: 10.1371/journal.pgen.1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah MA, Peterson ND, Szumel ES, Liu P, Page AL, Tse SY, Wani KA, Tocheny CE, Pukkila-Worley R. 2023. Transcriptional suppression of sphingolipid catabolism controls pathogen resistance in C. elegans. PLoS Pathog. 19(10):e1011730. doi: 10.1371/journal.ppat.1011730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas HR, Hodgkin J. 2009. The C. elegans Hox gene egl-5 is required for correct development of the hermaphrodite hindgut and for the response to rectal infection by Microbacterium nematophilum. Dev Biol. 329(1):16–24. doi: 10.1016/j.ydbio.2009.01.044. [DOI] [PubMed] [Google Scholar]
- Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, et al. 2011. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 21(2):245–254. doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. 2006. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 16(8):1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, Haynes CM. 2014. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. 516(7531):414–417. doi: 10.1038/nature13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SE, Robinson P, Melcher A, Quirke P, Buhring HJ, Cook GP, Blair GE. 2007. Expression of the CUB domain containing protein 1 (CDCP1) gene in colorectal tumour cells. FEBS Lett. 581(6):1137–1142. doi: 10.1016/j.febslet.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, Ewbank JJ. 2001. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 11(11):809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- Rajan M, Anderson CP, Rindler PM, Romney SJ, Ferreira Dos Santos MC, Gertz J, Leibold EA. 2019. NHR-14 loss of function couples intestinal iron uptake with innate immunity in C. elegans through PQM-1 signaling. Elife. 8:e44674 doi: 10.7554/eLife.44674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KC, Andersen EC, Kruglyak L, Kim DH. 2009. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 323(5912):382–384. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TM, Stewart M. 1995. Nematode sperm locomotion. Curr Opin Cell Biol. 7(1):13–17. doi: 10.1016/0955-0674(95)80039-5. [DOI] [PubMed] [Google Scholar]
- Sahu SN, Lewis J, Patel I, Bozdag S, Lee JH, LeClerc JE, Cinar HN. 2012. Genomic analysis of immune response against Vibrio cholerae hemolysin in Caenorhabditis elegans. PLoS One. 7(5):e38200. doi: 10.1371/journal.pone.0038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser SJ, Kenyon C. 1992. Activation of a C. elegans antennapedia homologue in migrating cells controls their direction of migration. Nature. 355(6357):255–258. doi: 10.1038/355255a0. [DOI] [PubMed] [Google Scholar]
- Sandhu A, Badal D, Sheokand R, Tyagi S, Singh V. 2021. Specific collagens maintain the cuticle permeability barrier in Caenorhabditis elegans. Genetics. 217(3):iyaa047. doi: 10.1093/genetics/iyaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H, Hoeppner MP, Weiner J III, Bornberg-Bauer E. 2008. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology. 213(3–4):237–250. doi: 10.1016/j.imbio.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Schulenburg H, Muller S. 2004. Natural variation in the response of Caenorhabditis elegans towards Bacillus thuringiensis. Parasitology. 128(4):433–443. doi: 10.1017/s003118200300461x. [DOI] [PubMed] [Google Scholar]
- Sharika R, Subbaiah P, Balamurugan K. 2018. Studies on reproductive stress caused by candidate Gram positive and Gram negative bacteria using model organism, Caenorhabditis elegans. Gene. 649:113–126. doi: 10.1016/j.gene.2018.01.088. [DOI] [PubMed] [Google Scholar]
- Shivers RP, Pagano DJ, Kooistra T, Richardson CE, Reddy KC, Whitney JK, Kamanzi O, Matsumoto K, Hisamoto N, Kim DH. 2010. Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet. 6(4):e1000892. doi: 10.1371/journal.pgen.1000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen KT, Gallego SF, Faergeman NJ, Kallipolitis BH. 2012. Strength in numbers: “Omics” studies of C. elegans innate immunity. Virulence. 3(6):477–484. doi: 10.4161/viru.21906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken MG, van Sluijs L, Wang YA, Ritmahan W, Gultom ML, Riksen JAG, Volkers RJM, Snoek LB, Pijlman GP, Kammenga JE. 2021. Punctuated loci on chromosome IV determine natural variation in orsay virus susceptibility of Caenorhabditis elegans strains Bristol N2 and Hawaiian CB4856. J Virol. 95(12):e02430-20. doi: 10.1128/JVI.02430-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taffoni C, Pujol N. 2015. Mechanisms of innate immunity in C. elegans epidermis. Tissue Barriers. 3(4):e1078432. doi: 10.1080/21688370.2015.1078432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo JV, Gujar M, Macdonald SJ, Lundquist EA. 2013. Functional transcriptomic analysis of the role of MAB-5/Hox in Q neuroblast migration in Caenorhabditis elegans. BMC Genomics. 14(1):304. doi: 10.1186/1471-2164-14-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ. 2013. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 154(3):676–690. doi: 10.1016/j.cell.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel F, Almagro Armenteros JJ, Johansen AR, Gislason MH, Pihl SI, Tsirigos KD, Winther O, Brunak S, von Heijne G, Nielsen H. 2022. Signalp 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol. 40(7):1023–1025. doi: 10.1038/s41587-021-01156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. 2006. P38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2(11):e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven R, McCallum KC, Garsin DA. 2012. Speculations on the activation of ROS generation in C. elegans innate immune signaling. Worm. 1(3):160–163. doi: 10.4161/worm.19767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. 2007. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 8(9):R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WH, Chen PH, Chang HH, Kwok HL, Stern A, Soo PC, Chen JH, Yang HC. 2023. Impaired immune response and barrier function in GSPD-1-deficient C. elegans infected with Klebsiella pneumoniae. Curr Res Microb Sci. 4:100181. doi: 10.1016/j.crmicr.2023.100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelensky AN, Gready JE. 2005. The C-type lectin-like domain superfamily. FEBS J. 272(24):6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence files used for differential gene expression analysis are available in FASTQ format in the NCBI BioProject repository (BioProject accession number: PRJNA973087; https://www.ncbi.nlm.nih.gov/sra/PRJNA973087). Raw and processed counts of sequence reads, overlapping genes, and the test statistic and P-values for all genes tested can be found in a supplemental file on figshare: https://doi.org/10.25387/g3.25025117.
Supplemental material available at G3 online.