Abstract
GET Recombination, a simple inducible homologous recombination system for Escherichia coli, was used to target insertion of an EGFP cassette between the start and termination codons of the β-globin gene in a 200 kb BAC clone. The high degree of homology between the promoter regions of the β- and δ-globin genes also allowed the simultaneous generation of a δ-globin reporter construct with the deletion of 8.8 kb of intervening sequences. Both constructs expressed EGFP after transient transfection of MEL cells. Similarly, targeting of the EGFP cassette between the promoter regions of the γ-globin genes and the termination codon of the β-globin gene enabled the generation of reporter constructs for both Aγ- and Gγ-globin genes, involving specific deletions of 24 and 29 kb of genomic sequence, respectively. Finally the EGFP cassette was also inserted between the ɛ- and β-globin genes, with the simultaneous deletion of 44 kb of intervening sequence. The modified constructs were generated at high efficiency, illustrating the usefulness of GET Recombination to generate large deletions of specific sequences in BACs for functional studies. The establishment of stable erythropoietic cell lines with these globin constructs will facilitate the search for therapeutic agents that modify the expression of the individual globin genes in a physiologically relevant manner.
INTRODUCTION
To date, most functional studies are carried out by sub-cloning small fragments of genomic DNA into various reporter vectors. This approach has proved useful for the identification of promoters and other regulatory elements. However, many key regulatory sequences may escape detection in such assays, while it is difficult to extrapolate from such assays to the regulation of intact genomic loci in vivo. In an effort to overcome these limitations, a number of groups have utilised YACs for functional studies (1). The high efficiency of homologous recombination in yeast has facilitated the introduction of targeted modifications in YACs (2). However, the difficulty of isolation of YAC DNA, together with the inherent problems of instability and chimerism of YACs, have limited their use in functional studies.
With the rapid progress in the sequencing of human and other eukaryotic genomes and the increasing availability of fully sequenced clones containing intact functional loci from the various high quality PAC/BAC genomic DNA libraries (3,4), it is becoming more convenient to study the expression and regulation of genes in their native genomic environment.
A major factor favouring the use of small expression cassettes over PACs/BACs is the comparative ease of conventional DNA manipulation techniques for the generation of such cassettes. However, with the recent advances in the use of homologous recombination technology in Escherichia coli, it has now become possible to introduce site-specific modifications into genomic fragments in PAC/BAC clones (5–9), as well as on the E.coli chromosome (7,10,11).
We have found that the ET cloning system (7) could not be used to modify BACs in the E.coli DH10B host cell strain. We have modified the ET cloning system by the inclusion of the bacteriophage λ gam gene in a polycistronic operon placed under the same L-arabinose inducible promoter as the recE and recT genes (5). This minimised degradation of incoming double-stranded linear DNA fragments by transiently inhibiting the recBCD nuclease activity of E.coli DH10B without compromising its viability. Using this system, we were able to demonstrate the targeted insertion of a kanamycin gene into the backbone of pEBAC 148β, a second-generation BAC clone carrying the β-globin locus on a 185 kb genomic insert. Similarly, we were able to direct the in-frame fusion of a luciferase/kanamycin cassette into exon II of the β-globin gene to study the aberrant splicing exhibited by the common IVS I-110 G→A splicing mutation (5). We refer to this modification of ET cloning as the GET Recombination system. We have recently used the GET Recombination system in combination with a tetracycline counterselection cassette to direct the insertion of the IVS I-110 G→A splicing mutation in intron I of the β-globin gene, thus further extending the usefulness of the system for the functional analysis of BACs and the creation of accurate transgenic animal models (6).
We describe here further use of GET Recombination to engineer precisely EGFP reporter constructs into the β-, δ-, Aγ-, Gγ- and ɛ-globin genes within the β-globin locus. In addition, we have used the system to engineer specific deletions from 1.4 to 44 kb in length. Previously this type of modification could only be achieved by flanking the region to be deleted with loxP sites and utilising Cre recombinase to facilitate the deletion. Although this latter approach is widely used and has been utilised for numerous applications including study of LCR function in β-globin regulation (12), one must be mindful of the fact that the retention of foreign sequence artefacts may have untoward effects on the overall regulation of the system. This becomes evident when considering that even single base changes can often lead to severe disruption in gene regulation and function. We have also used GET Recombination to simultaneously generate recombinants at a number of homologous target sites in a single experiment, an approach which should greatly facilitate the functional mapping of key regulatory elements in large genomic fragments.
MATERIALS AND METHODS
Media
Escherichia coli strains were routinely grown in LB liquid culture, or on LB agar plates (15 g/l) containing the following antibiotics as indicated in the text: 100 µg/ml ampicillin (Amp); 12.5 µg/ml chloramphenicol (Cm); or 25 µg/ml kanamycin (Km).
Plasmids
pEBAC 148β, a BAC clone containing the entire β-globin locus located on a 185 kb genomic insert in a second generation BAC/PAC cloning vector, has been previously described (5).
Plasmid pEGFP-N22 used for the amplification of the EGFP-Neo/Kan cassette was derived from pEGFP-N2 (Clontech Laboratories, Palo Alto, CA) by deletion of the multicloning site and removal of the NotI site located downstream of the EGFP gene.
Plasmid pGETrec, a 6578 bp plasmid containing the E.coli recE and recT genes and the bacteriophage λ gam genes in a polycistronic operon, has been previously described (5).
PCR reactions
PCR primers used for the amplification of the EGFP-Neo/Kan cassette from pEGFP-N22 were obtained from Sigma Genosys (Sydney, Australia) and were additionally subject to RPC cartridge purification. Targeting of the cassette to the β-globin gene was achieved using primers HbbEGFP (5′-ACATTTGCTTCTGACACAACTGTGTTCACTAGCAACCTCAAACAGACACCatggtgagcaagggcgaggagc-3′) and HbbNeo (5′-TTACCCTTCAAAGGAACCTTTAATAGAAATTGGACAGCAAGAAAGCGAGCccagagtcccgctcagaag-3′) (Fig. 1). Upper case characters (50 nt) relate to the homology targeting arms of the β-globin genomic sequences, and lower case characters to those used to prime amplification of the EGFP-Neo/Kan cassette. PCR reactions were performed in 25 µl reactions for 30 cycles (94°C, 30 s; 60°C, 30 s; 72°C, 2 min) with the Expand High Fidelity PCR System (Boehringer Mannheim, Mannheim, Germany) using the manufacturer’s specifications. A 2861 bp PCR product was obtained and gel purified to remove template plasmid before use for homologous recombination.
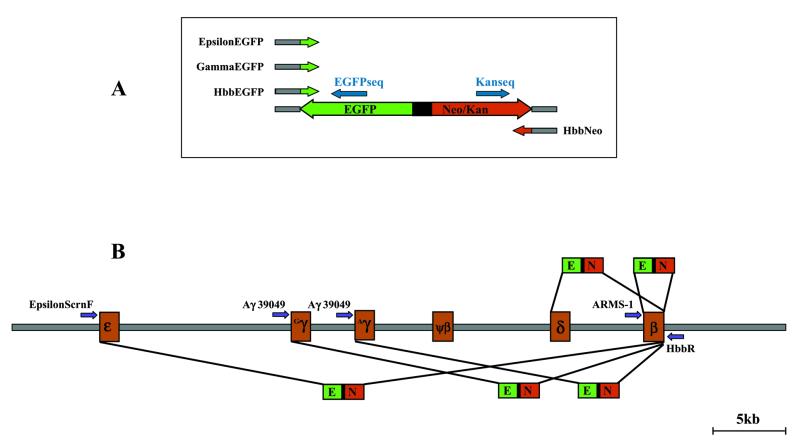
Figure 1.
(A) Diagrammatical representation of the EGFP-Neo/Kan cassette used in the homologous recombination experiments. The cassette was amplified from plasmid pEGFP-N22 with primers HbbNeo at the 3′-end and either HbbEGFP, GammaEGFP or EpsilonEGFP at the 5′-end. The gene-specific 50 nt homology arms at the 5′-ends of these primers are represented by grey boxes, while the 3′-ends of these primers are targeted to amplify the EGFP-Neo/Kan cassette. Primers EGFPseq and Kanseq (blue arrows) were used to sequence across the recombination junctions of the EGFP-Neo/Kan cassette and confirm the specificity of the recombinant clones. (B) Diagram of the β-globin locus drawn to scale, illustrating the targeting of the EGFP-Neo/Kan cassette to each one of the functional globin genes. The insertion of the EGFP-Neo/Kan cassette (2.7 kb) is accompanied by deletions ranging from 1.4 to 44 kb of the intervening sequences. Correct insertion of the EGFP-Neo/Kan cassette at each one of the genes was confirmed with primers HbbR at the 3′-end and either ARMS-1, Aγ39049 or EpsilonScrnF at the 5′-end (purple arrows). Primers LUG1 and LUG2 (not shown) amplify a 440 bp fragment from the β-globin gene while primer Aγ39512 (not shown) amplifies a 464 bp product from each of the γ-globin genes with primer Aγ39049.
PCR screening across the recombination junctions was performed with primers ARMS-1 (5′-TACGGCTGTCATCACTTAGACCTCACCCTG-3′) and HbbR (5′-GGCAGAATCCAGATGCTCAA-3′) (Fig. 1) in 25 µl reactions, using the same PCR conditions described above. These primers yield a PCR product of 1708 bp from the normal β-globin gene, while insertion of the EGFP-Neo/Kan cassette should yield a PCR product of 3044 bp.
Targeting of the EGFP cassette to the start codon of each of the γ-globin genes was achieved with primers GammaEGFP (5′-CTCGCTTCTGGAACGTCTGAGATTATCAATAAGCTCCTAGTCCAGACGCCatggtgagcaagggcgaggagc-3′) and HbbNeo, using the same cycling conditions (Fig. 1). Screening for γ-globin recombinants utilised primers Aγ39049 (5′-CCTCACTGGAGCTACAGACAAGA-3′), which anneals upstream of both γ-globin promoters, and HbbR to yield a PCR product of 3303 bp in recombinant clones and no PCR product in wild-type clones. To distinguish between recombination events occurring at either of the γ-globin genes, PCR reactions were performed using primers Aγ39049 and Aγ39512 (5′-CCTTGTCCTCCTCTGTGAAATGAC-3′), which amplify a 464 bp region across both γ-globin promoter regions. Recombinants where both γ-globin genes had been successfully deleted failed to yield a PCR product, whereas deletion of only the Aγ-gene gave the 464 bp product.
Targeting of the EGFP cassette to the start codon of the ɛ-globin gene was achieved with primers EpsilonEGFP (5′-TCTGCTTCCGACACAGCTGCAATCACTAGCAAGCTCTCAGGCCTGGCATCatggtgagcaagggcgaggagc-3′) and HbbNeo (Fig. 1). Recombinant clones yielded a PCR product of 2904 bp by screening with primers EpsilonScrnF (5′-CAGCTGCAATCACTAGCAAG-3′) and HbbR.
Primers LUG1 (5′-ACAAGACAGGTTTAAGGAGACCA-3′) and LUG2 (5′-GTCTGTTTCCCATTCTAAACTGTA-3′) amplify a 440 bp sequence within the β-globin gene, and were used either to show the presence or absence of β-globin sequence in unmodified or recombinant clones, respectively, or for preparation of the LUG probe for Southern blot hybridisations. PCR reactions using this primer pair were routinely performed in 25 µl reactions for 30 cycles (94°C, 30 s; 60°C, 30 s; 72°C, 30 s).
Purification of PCR products
The LUG PCR product used as a hybridisation probe, and the EGFP-Neo/Kan PCR product used either as a hybridisation probe or for homologous recombination, were extracted from a 1% agarose gel and purified using the Qiagen gel extraction kit (Qiagen, Hilden, Germany).
Sequencing reactions
Direct DNA sequencing of recombinant BAC clones was performed using the Big Dye Terminator kit (PE Applied Biosystems, Foster City, CA). Sequencing primers EGFPseq (5′-GATGAACTTCAGGGTCAG-3′) and Kanseq (5′-ATATTGCTGAAGAGCTTGG-3′) annealed at sites located ~150 bp internally from either end of the EGFP-Neo/Kan cassette, to enable sequencing outwards in both directions across the homology junctions (Fig. 1).
Homologous recombination
GET Recombination was routinely performed as previously described (5). Briefly, ~300–500 ng of purified PCR product was electroporated (Gene-Pulser II; Bio-Rad, Hercules, CA) into 40 µl of electrocompetent E.coli DH10B cells carrying both pEBAC 148β and pGETrec.
The cells were prepared for electroporation by growing 250 ml cultures, inoculated from an overnight culture at 30°C, in LB containing Cm and Amp until an OD600 of 0.4 was reached. Expression of the recE, recT and the gam genes was then induced by the addition of L-arabinose to a final concentration of 0.2% (w/v) for a further 40 min. The cells were then harvested and made electrocompetent (Gibco BRL, Gaithersburg, MD). Cells electroporated with PCR product were subsequently incubated in 1 ml of SOC medium for 1 h, spread onto LB–Agar plates containing Cm and Km, and colonies allowed to form over a 20 h period.
Single colonies were randomly picked, grown in 1 ml mini-cultures in LB containing both Cm and Km, and 1 µl aliquots of culture were used directly for PCR screening for recombinant clones.
Transient in vivo expression assays
Murine erythroleukaemia (MEL IV 3A4) cells were maintained in continuous culture in DMM medium supplemented with 10% FCS. Aliquots containing 2 × 106 cells were electroporated in 400 µl culture medium with 100 µg CsCl-purified DNA using Bio-Rad Gene-Pulser II under the following conditions: capacitance 1050 µF, voltage 260 V, pulse time 30 ms (13). Cells were split into two equal aliquots, plated into 6-well fibronectin (Boehringer Mannheim) coated tissue culture plates, and allowed to attach overnight at 37°C in 5% CO2. The cells were induced to differentiate 24 h following electroporation by supplementing the culture media with DMSO to a final concentration of 2% and hemin to a final concentration of 50 µM. Cells were examined for EGFP expression by fluorescence microscopy, 72 h following induction.
RESULTS
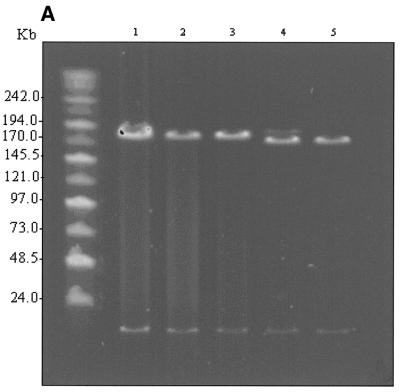
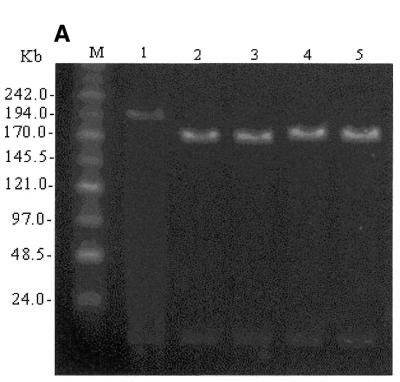
Amplification of the EGFP-Neo/Kan cassette with primers HbbEGFP/HbbNeo and electroporation of the purified PCR product into DH10B cells carrying both pEBAC 148β and the pGETrec plasmid under Cm and Km selection resulted in the isolation of a number of clones resistant to both antibiotics. Sixteen clones were screened by PCR with primers LUG1/LUG2 for the deletion of the β-globin gene. Three clones appeared to be non-recombinant, since the Km resistance appeared to arise from carry-over of the pEGFP-N22 template plasmid (data not shown). The remaining 13 clones failed to give a PCR product, indicating a deletion of the β-globin gene, while they were all positive for the presence of the EGFP gene using internal primers (data not shown). However, only five of these clones appeared to be positive for the insertion of the EGFP-Neo/Kan cassette into the β-globin gene using the external primers ARMS-1 and HbbR (data not shown). As the promoter regions of the β- and δ-globin genes are highly homologous with only four mismatches over the 50mer region used for targeting, we considered the possibility that some of the recombinant clones arose by targeting the promoter region of the δ-globin gene. Insertion of the EGFP-Neo/Kan cassette between the start codon of the δ-globin gene and the termination codon of the β-globin gene would delete a sequence of 8814 bp and replace it with 2761 bp, leading to a reduction in size of ~6 kb (Fig. 1). Two clones from each group were selected at random for more detailed mapping. Digestion of pEBAC 148β DNA with NotI releases the 185 kb genomic insert from the 17 kb vector backbone when resolved on a 1% PFGE gel (Fig. 2A, lane 1). Replacement of the β-globin gene with the EGFP-Neo/Kan cassette is expected to give rise to an increase in size of 1337 bp which is not resolved under these conditions in clones positive for this type of replacement (Fig. 2A, lanes 2 and 3). However, a clear reduction in size of the genomic insert by ~6 kb is obvious in the remaining two clones (Fig. 2A, lanes 4 and 5).
Figure 2.
(A) NotI digestion of unmodified pEBAC 148β clone (lane 1), β-globin gene replaced with EGFP-Neo/Kan (two independent clones, lanes 2 and 3) and δβ-globin genes replaced with EGFP-Neo/Kan (two independent clones, lanes 4 and 5). The 185 kb genomic fragment was released by NotI digestion and separated from the vector backbone (17 kb) by PFGE. The expected 6 kb reduction in size of the genomic insert is clearly visible in the clones where the EGFP-Neo/Kan cassette replaced both the δ- and β-globin genes (lanes 4 and 5). (B) As in (A), except that the clones were digested with XhoI. The only detectable change in any of the fragments is the expected 6 kb reduction in the size of the 75 kb fragment in lanes 4 and 5, which contains the δ- and β-globin genes.
Digestion of pEBAC 148β using XhoI produces at least six fragments (Fig. 2B, lane 1). XhoI cuts only twice in the known sequence of the β-globin locus releasing a 4.9 kb fragment, but prediction of the sizes of the other fragments is not currently possible, since the sequence of the 185 kb fragment is not fully known. However, Southern blot analysis with a 32P-labelled LUG probe showed that the β- and δ-globin genes are located in the uppermost 75 kb fragment (data not shown). There is no obvious change in the size of this fragment when the EGFP-Neo/Kan cassette replaces the β-globin gene (Fig. 2B, lanes 2 and 3). However, there is a clear decrease in size by ~6 kb in lanes 4 and 5, consistent with simultaneous replacement of both the δ- and β-globin genes with the EGFP-Neo/Kan cassette. All other fragments released by XhoI digestion remain unchanged in size when compared to the unmodified pEBAC 148β clone, indicating the absence of any rearrangements outside the region which was targeted for homologous recombination.
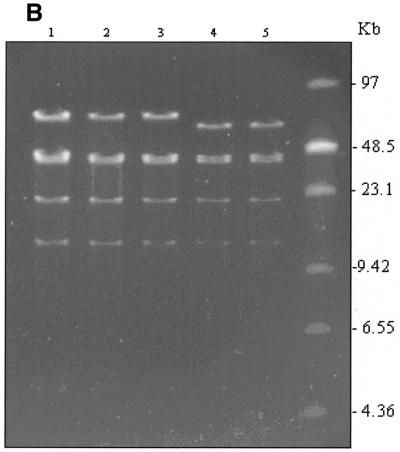
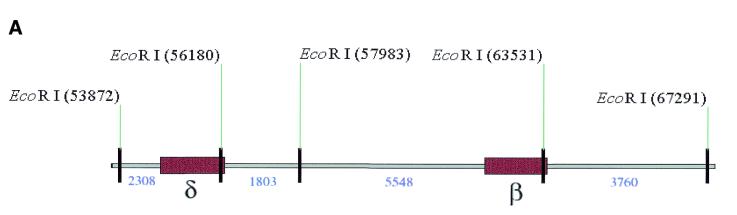
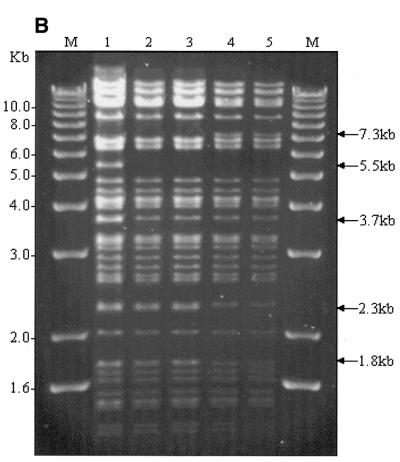
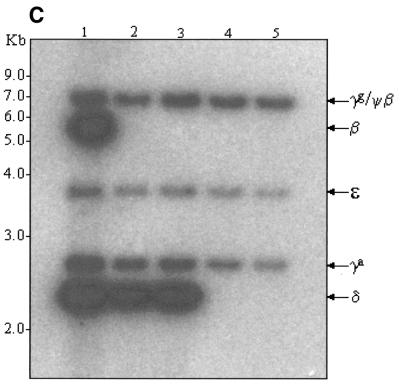
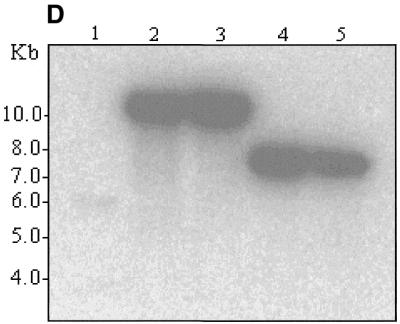
High-resolution mapping of the same five clones was carried out after EcoRI digestion (Fig. 3). Deletion of the β-globin gene and replacement with the EGFP-Neo/Kan cassette leads to loss of the EcoRI site within the β-globin gene (Fig. 3A) and disappearance of the 5.5 and 3.7 kb fragments (Fig. 3B, lanes 2 and 3). Since the EGFP-Neo/Kan cassette does not contain an EcoRI site, a new band should appear at ~10.5 kb. Although appearance of this new band on the ethidium bromide gel is masked by the presence of many other fragments in the same region, its presence was clearly demonstrated following Southern blot analysis and probing with a 32P-labelled EGFP-Neo/Kan PCR product, (Fig. 3D, lanes 2 and 3). Further evidence to support a β-globin gene deletion was obtained using Southern blot analysis and hybridisation with a 32P-labelled LUG probe under conditions of moderate stringency. The high degree of homology exhibited by all members of the β-globin-like genes with respect to the region amplified by the LUG primers allows simultaneous detection of all β-globin-like genes on a single Southern blot (Fig. 3C). Deletion of the 5.5 kb band corresponding to the β-globin gene can be clearly seen following recombination in these clones (Fig. 3C, lanes 2 and 3). In contrast to the clones in lanes 2 and 3, the clones in lanes 4 and 5 show not only absence of the 5.5 and 3.7 kb EcoRI fragments, but also deletion of the 2.3 and 1.8 kb fragments arising from the EcoRI site within the δ-globin gene (Fig. 3A). Deletion of this site with the simultaneous insertion of the EGFP-Neo/Kan cassette generates a new 7.3 kb fragment. The appearance of this new fragment is clearly visible on the ethidium bromide stained gel (Fig. 3B, lanes 4 and 5) as well as following hybridisation of the Southern blot to the EGFP-Neo/Kan probe (Fig. 3D, lanes 4 and 5). Simultaneous deletion of both the β- and δ-globin genes in these two clones was also clearly demonstrated by Southern blot analysis with the 32P-labelled LUG probe (Fig. 3C, lanes 4 and 5). Thus all changes noted in the high-resolution EcoRI digest of the recombinant clones could be accounted for either by the replacement of the β-globin gene (Fig. 3, lanes 2 and 3), or by the replacement of both the β- and δ-globin genes (Fig. 3, lanes 4 and 5) with the EGFP-Neo/Kan cassette.
Figure 3.
(A) EcoRI restriction map in the region of the β- and δ-globin genes. (B) EcoRI digestion of unmodified and modified pEBAC 148β clones. Lane 1, unmodified pEBAC 148β; lanes 2 and 3, β globin gene replaced with the EGFP-Neo/Kan (two independent clones); lanes 4 and 5, δβ-globin genes replaced with the EGFP-Neo/Kan (two independent clones). (C) Southern blot analysis of the gel depicted in (B) and hybridised with a 32P-labelled LUG probe. (D) Southern blot analysis of the gel depicted in (B) and hybridised with a 32P-labelled EGFP-Neo/Kan probe.
Direct DNA sequencing on clones with both types of modifications was performed using the primers EGFPseq and Kanseq to provide sequence data across the homologous recombination junctions. Both primers yielded 350–400 bp of sequence outwards from the EGFP-Neo/Kan cassette in both directions and into the genomic sequence of each clone. Sequence data aligned with perfect homology to the regions both upstream and downstream of the β-globin gene in the clones carrying the β-globin replacement reporter construct. In a similar fashion, we demonstrated perfect alignment of the sequence data from the clones with the δβ-globin gene deletions to the regions located upstream of the start codon of the δ-globin gene and downstream of the termination codon of the β-globin gene (results not shown).
Using similar methodology, we were able to engineer targeted deletions of 24 and 29 kb of genomic sequence encompassing all sequences between and including the β-globin gene and either the Aγ- or Gγ-globin genes, respectively, with the concurrent insertion of the EGFP-Neo/Kan cassette. This was achieved using primer HbbNeo to target the cassette to the termination codon of the β-globin gene and primer GammaEGFP to target the cassette to the start codon of either of the γ-globin genes. Since the 50 nt γ-globin targeting sequence is identical in both γ-globin gene promoter regions except for a single base mismatch, it was envisaged that GET Recombination would lead to the insertion of the EGFP-Neo/Kan cassette at either the Aγ- or Gγ-globin genes. In a single electroporation experiment, 19 Km-resistant clones were obtained. PCR screening using primers Aγ39049F and HbbR yielded a 3303 bp product in 12 of the Km-resistant clones indicating that the required recombination had occurred at either of the two γ-globin genes. Further PCR analysis using primers Aγ39049F and Aγ39512R, which amplify a 464 bp region across both γ-globin gene promoters, indicated that six of the 12 positive clones contained the Gγ-gene, while the other six clones failed to yield a PCR product, indicating the deletion of both γ-globin genes (results not shown).
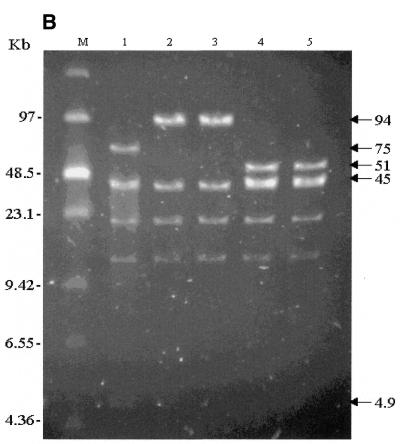
Restriction mapping of recombinant clones using NotI and XhoI digestions was carried out (Fig. 4). NotI digestion demonstrated a clear reduction in the size of the genomic inserts of the recombinant clones. A 26 kb reduction in size of the genomic insert is expected in the clones where the EGFP-Neo/Kan cassette replaces both γ-globin genes as well as the δ- and β-globin genes (Fig. 4A, lanes 2 and 3). Similarly a 21 kb reduction in size of the genomic insert is expected in the clones where the EGFP-Neo/Kan cassette replaces the Aγ-, δ- and β-globin genes (Fig. 4A, lanes 4 and 5).
Figure 4.
(A) NotI digestion of unmodified and modified pEBAC 148β clones. The genomic fragment was released by NotI digestion and separated from the vector backbone (17 kb) by PFGE. Lane 1, unmodified pEBAC 148β clone; lanes 2 and 3, GγAγδβ-globin genes replaced with EGFP-Neo/Kan (two clones); lanes 4 and 5, Aγδβ-globin genes replaced with EGFP-Neo/Kan (two clones). (B) As in (A), except that the clones were digested with XhoI. Changes in the restriction fragments in lanes 2 and 3 are consistent with insertion of the EGFP-Neo/Kan cassette between the start codon of the Gγ-globin gene and the termination codon of the β-globin gene, accompanied by a 29 kb deletion. Similarly, changes in restriction fragments in lanes 4 and 5 are consistent with a 24 kb deletion between the start codon of the Aγ-globin gene and the termination codon of the β-globin gene.
XhoI digestion of the same clones revealed loss of either the Aγ- or both γ-globin genes. All recombinant clones lack the 4.9 kb band that is released by XhoI digestion between the two γ-globin genes (Fig. 4B, lanes 2–5), as would be expected from deletion of either one or both γ-globin genes. Recombinant clones with an apparent deletion of both γ-globin genes (Fig. 4B, lanes 2 and 3) showed the disappearance of two additional bands (75 and 45 kb) and the appearance of a band at ~94 kb, as would be expected from a 29 kb AγGγδβ deletion with the simultaneous insertion of the 2.7 kb EGFP-Neo/Kan cassette. On the other hand, recombinant clones with an intact Gγ-globin gene (Fig. 4B, lanes 4 and 5) showed a reduction in size of the 75 kb fragment to ~51 kb, as would be expected from a 24 kb deletion between the start codon of the Aγ-globin gene and the termination codon of the β-globin gene, and the simultaneous insertion of the 2.7 kb EGFP-Neo/Kan cassette. High-resolution mapping using EcoRI digestion and hybridisation with the LUG probe confirmed these data and further demonstrated that no unwanted rearrangements had occurred in any of the recombinant clones. All changes in the restriction fragments due to the engineered deletions were in accordance with the generation of the desired modifications (results not shown).
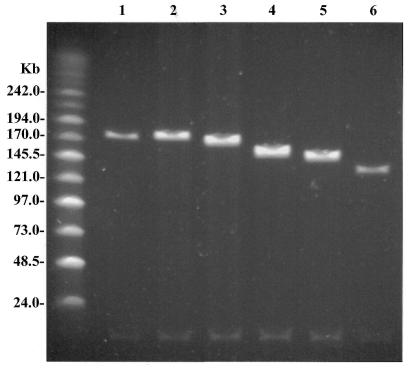
Given the good efficiency of obtaining recombinant clones with deletions up to 29 kb of genomic DNA, we were interested to explore further if there is an upper size limit to the extent of the deletions that can be engineered into BAC clones by GET Recombination. The EGFP-Neo/Kan cassette was therefore targeted between the start codon of the ɛ-globin gene and the termination codon of the β-globin gene using primers EpsilonEGFP and HbbNeo for the amplification of the cassette. This should result in the deletion of 44 kb of genomic sequence from the β-globin locus and its replacement by the 2.7 kb EGFP-Neo/Kan cassette (Fig. 1). Fifty-four clones from a single electroporation were analysed and all showed absence of the β-globin gene with primers LUG1/LUG2. Further analysis of six of these clones by EcoRI digestion showed that they were all missing the expected EcoRI fragments between the ɛ- and β-globin genes (data not shown). One of these clones was analysed by PFGE after NotI digestion, in parallel with modified clones representing each of the globin genes in the β-globin locus. A reduction in size of the genomic insert by ~40 kb is clearly evident in this clone (Fig. 5, lane 6). At the same time this gel clearly demonstrates the stepwise larger deletions that have been introduced into the β-globin locus as the target sites for GET Recombination have been extended from 1.4 to 44 kb apart.
Figure 5.
Comparison of all modified clones by PFGE. A single representative clone from each of the modified constructs was grown in overnight culture and mini prep DNA was used for NotI digestion to release the genomic insert. Digested DNA resolved by PFGE clearly shows the increasing reduction in size of the genomic insert of each of the modified clones whilst maintaining the same position of the backbone vector band on the gel. Lane 1, unmodified pEBAC 148β; lane 2, β-globin gene replaced with EGFP-Neo/Kan; lane 3, δβ-globin genes replaced with EGFP-Neo/Kan; lane 4, Aγδβ-globin genes replaced with EGFP-Neo/Kan; lane 5, GγAγδβ-globin genes replaced with EGFP-Neo/Kan; lane 6, ɛGγAγδβ-globin genes replaced with EGFP-Neo/Kan.
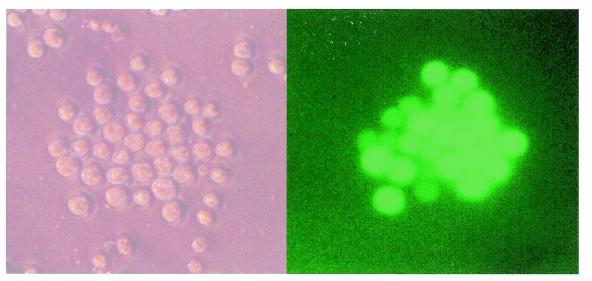
The insertion of the EGFP coding sequence in frame with the start codons of the β-, δ- Aγ-, Gγ- and ɛ-globin genes in pEBAC 148β should place EGFP expression under the normal regulatory elements of the β-globin locus for each one of these genes. In preliminary studies this was examined in a transient expression assay using MEL cells. Transfection with pEBAC 148βG, a version of pEBAC 148β carrying EGFP under the control of the CMV promoter on the backbone of the vector (unpublished data), was used as a control for transfection efficiency. Although transfection efficiency of MEL cells with the various pEBAC 148β constructs was low (data not shown), the Δβ-globin gene deletion/replacement construct showed significant expression of EGFP after induction of differentiation (Fig. 6). Further studies are in progress with the Δβ-, Δδβ-, ΔAγδβ-, ΔGγAγδβ- and ΔɛGγAγδβ-globin/EGFP fusion constructs to study the regulation and level of EGFP expression in this transient assay, as well as in stable cell lines.
Figure 6.
Transient expression from pEBAC 148β modified by fusion of an EGFP-Neo/Kan cassette to the β-globin start codon after electroporation into MEL cells and induction of differentiation with hemin/DMSO for 72 h. The image on the right shows a clone of EGFP expressing MEL cells following induction. The panel on the left shows a phase contrast image of the same field of view.
DISCUSSION
The β-globin locus has been extensively studied over the last 20 years, yet many aspects of its regulation are still poorly understood. Most studies to date involve sub-cloning of specific regions into small expression vectors. A number of studies with YACs carrying the intact β-globin locus have clearly demonstrated the benefits of performing functional studies on the intact β-globin locus under physiologically relevant conditions (1,2).
Our results clearly demonstrate the usefulness of GET Recombination to facilitate precise engineering of the β-globin locus in a 200 kb second-generation BAC clone in E.coli. We have precisely inserted a 2761 bp long EGFP-Neo/Kan cassette between the start codons of either the β-, δ-, Aγ-, Gγ- or ɛ-globin genes and the termination codon of the β-globin gene, in such a way that EGFP expression is directly under the control of the normal upstream regulatory elements of the corresponding globin genes. The use of these constructs in erythropoietic cell lines should greatly facilitate the identification of potential therapeutic agents that are capable of modulating the expression of each of the globin genes in a physiologically relevant manner.
Recombinant clones in this study were identified by the use of the Km-selectable marker. The need to include such a selectable marker with the reporter gene means, however, that the expression of the reporter gene is dissociated from the influence of any intragenic or 3′-end regulatory elements. This limitation may be overcome by insertion of the reporter sequence in-frame into an exon of the gene of interest, in such a way as not to leave behind any operational sequences. This could be achieved by two-stage GET Recombination (6): in the first stage a counter-selection cassette may be inserted into the target exon, while in the second stage the counter-selection cassette is replaced by the coding sequence of the reporter gene without its start and termination codons. Judicious selection of exposed sequences as targets for modification and the inclusion of linker sequences at both ends of the reporter gene may enable incorporation of the reporter gene in fusion proteins without disturbance of the overall protein structure or function.
We have also demonstrated the potential of GET Recombination to generate deletions of various lengths in a single experiment by targeting homologous sequences that are repeated at various distances from a unique target site. Thus by targeting the insertion of the 5′-end of the EGFP-Neo/Kan cassette to the start codon of the β-globin gene, eight out of 13 recombinant clones we obtained had an insertion at the start codon of the δ-globin gene, with a deletion of 8.8 kb of intervening sequences. This was somewhat surprising, since there are four base pair mismatches between the 50mer targeting sequence in the HbbEGFP primer and the homologous sequence in the δ-globin gene. However, these mismatches are restricted in the first 20 bases from the 5′-end of the HbbEGFP primer, and thus may not interfere with homologous recombination over the rest of the targeting sequence. In contrast only five out of 13 clones showed recombination across the 1418 bp interval defined by the start and stop codons of the β-globin gene, indicating that GET Recombination across an 8.8 kb interval is at least as efficient as across a 1.4 kb interval. Similarly, targeting of the EGFP-Neo/Kan cassette to the start codon of the Aγ-globin gene with primer GammaEGFP which has only a single base mismatch from the homologous sequence in the Gγ-globin gene, resulted in an equal number of recombinant clones for each of the γ-globin genes. Since highly conserved repetitive sequences are found scattered throughout most BAC clones that have been sequenced, the targeting of such repetitive sequences during GET Recombination should generate clones with different length deletions in a single experiment. This should greatly facilitate the localisation and identification of distant tissue- and locus-specific regulatory elements.
Recombination of the EGFP-Neo/Kan cassette between the start codons of either the β-, δ-, Gγ-, Aγ- or ɛ-globin genes and the termination codon of the β-globin gene involves the deletion of 1.4–44 kb of intervening sequences. The creation of deletions as large as 44 kb during GET Recombination was at least as efficient as the creation of smaller deletions. Thus there is no clear upper limit to the extent of the deletions that may be induced by GET Recombination. This is in agreement with the recent findings of Hill et al. (14), who used a similar approach to trim the ends of overlapping BACs, with the precise deletion of overlaps as long as 70 kb.
In conclusion, these and earlier studies (5,6) demonstrate that GET Recombination allows the precise engineering of BAC clones in a number of useful ways. The manipulations vary from the insertion of single base disease causing mutations (6), to the insertion of deletions at least up to 44 kb long (this report) and the insertion of reporter genes and other selectable markers at various positions of interest. Such studies should greatly facilitate functional analysis of BAC clones and localise potentially important, and as yet unidentified, regulatory sequences in various genomic loci.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Mr Ricardo Villain for plasmid pEGFP-N22, and to Dr Kumaran Narayanan who carried out some of the preliminary work. M.O. would like to thank his wife Maria and daughter Emiliana for their support during the course of this work. This work was supported by a Centre Grant from the Thalassaemia International Federation and the Cyprus Thalassaemia Association, as well as by grants from the Brockhoff Foundation, the Ronald Geoffrey Arnott Foundation and the Thalassaemia Society of New South Wales.
REFERENCES
- 1.Peterson K.R. (1999) Methods Enzymol., 306, 186–203. [DOI] [PubMed] [Google Scholar]
- 2.Peterson K.R., Li,Q.L., Clegg,C.H., Furukawa,T., Navas,P.A., Norton,E.J., Kimbrough,T.G. and Stamatoyannopoulos,G. (1995) Proc. Natl Acad. Sci. USA, 92, 5655–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ioannou P.A., Amemiya,C.T., Garnes,J., Kroisel,P.M., Shizuya,H., Chen,C., Batzer,M.A. and de Jong,P.J. (1994) Nature Genet., 6, 84–89. [DOI] [PubMed] [Google Scholar]
- 4.Shizuya H., Birren,B., Kim,U.J., Mancino,V., Slepak,T., Tachiiri,Y. and Simon,M. (1992) Proc. Natl Acad. Sci. USA, 89, 8794–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narayanan K., Williamson,R., Zhang,Y., Stewart,A.F. and Ioannou,P.A. (1999) Gene Therapy, 6, 442–447. [DOI] [PubMed] [Google Scholar]
- 6.Nefedov M., Williamson,R. and Ioannou,P.A. (2000) Nucleic Acids Res., 28, e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Buchholz,F., Muyrers,J.P.P. and Stewart,A.F. (1998) Nature Genet., 20, 123–128. [DOI] [PubMed] [Google Scholar]
- 8.Angrad P.O., Daigle,N., van der Hoeven,F., Schöler,H.R. and Stewart,A.F. (1999) Nucleic Acids Res., 27, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muyrers J.J.P., Zhang,Y., Testa,G. and Stewart,A.F. (1999) Nucleic Acids Res., 27, 1555–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karoui M.E., Amundsen,S.K., Dabert,P. and Gruss,A. (1999) Nucleic Acids Res., 27, 1296–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Posfai G., Kolisnychenko,V., Bereczki,Z. and Blattner,F.R. (1999) Nucleic Acids Res., 27, 4409–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiering S., Epner,E., Kim,C.G., Telling,A., Robinson,K., Zhuang,Y. and Groudine,M. (1994) In Stamatoyannopoulos,G. (ed.), Molecular Biology of Hemoglobin Switching, Volume 1 of the Proceedings of the 9th Conference on Hemoglobin Switching. Intercept Ltd, UK, pp. 59–69.
- 13.Baum C., Forster,P., Hegewisch-Becker,S. and Harbers,K. (1994) Biotechniques, 17, 1058–1062. [PubMed] [Google Scholar]
- 14.Hill F., Benes,V., Thomasova,D., Stewart,A.F., Kafatos,F.C. and Ansorge,W. (2000) Genomics, 64, 111–113. [DOI] [PubMed] [Google Scholar]