Abstract
Data scarcity and data imbalance are two major challenges in training deep learning models on medical images, such as brain tumor MRI data. The recent advancements in generative artificial intelligence have opened new possibilities for synthetically generating MRI data, including brain tumor MRI scans. This approach can be a potential solution to mitigate the data scarcity problem and enhance training data availability. This work focused on adapting the 2D latent diffusion models to generate 3D multi-contrast brain tumor MRI data with a tumor mask as the condition. The framework comprises two components: a 3D autoencoder model for perceptual compression and a conditional 3D Diffusion Probabilistic Model (DPM) for generating high-quality and diverse multi-contrast brain tumor MRI samples, guided by a conditional tumor mask. Unlike existing works that focused on generating either 2D multi-contrast or 3D single-contrast MRI samples, our models generate multi-contrast 3D MRI samples. We also integrated a conditional module within the UNet backbone of the DPM to capture the semantic class-dependent data distribution driven by the provided tumor mask to generate MRI brain tumor samples based on a specific brain tumor mask. We trained our models using two brain tumor datasets: The Cancer Genome Atlas (TCGA) public dataset and an internal dataset from the University of Texas Southwestern Medical Center (UTSW). The models were able to generate high-quality 3D multi-contrast brain tumor MRI samples with the tumor location aligned by the input condition mask. The quality of the generated images was evaluated using the Fréchet Inception Distance (FID) score. This work has the potential to mitigate the scarcity of brain tumor data and improve the performance of deep learning models involving brain tumor MRI data.
Keywords: Latent diffusion model, Generative models, Brain tumor imaging, Synthetic data
1. INTRODUCTION
Recent advancements in image generative models1, 2 have led to significant improvements in the quality and realism of generated images. One of the most promising recent developments is the diffusion model,2–5 a type of generative model that can generate high-quality and diverse images. Unlike traditional approaches, the diffusion model learns the data distribution by iteratively adding noise to the original image and then gradually denoising it until the desired output is achieved. The recent strides in developing diffusion models mark a notable shift in the image generation domain and offer promising potential for various applications, especially in the domain of medical imaging. In medical imaging, the capacity to generate high-fidelity, diverse, and realistic images plays a crucial role in enhancing limited datasets, enabling more robust training of deep learning models for diverse tasks such as image segmentation, anomaly detection, and prediction. The integration of diffusion models into medical imaging also paves the way for improved diagnostics, patient care, and scientific understanding in the healthcare sector.
This study focuses on adapting 2D latent diffusion models (LDMs)2 to generate 3D multi-contrast brain tumor MRI data utilizing a tumor mask as a guidance factor. The framework consists of two main components: a 3D autoencoder model for perceptual compression and a conditional 3D Diffusion Probabilistic Model (DPM) for generating diverse and high-quality multi-contrast brain tumor MRI samples. This generation process is guided by a conditional tumor mask. Prior studies have focused on creating either 2D multi-contrast6 or 3D single-contrast MRI samples.7–9 Several challenges persist in developing conditional 3D LDMs for generating multi-contrast MRI data: (1) The complexity of multi-contrast 3D LDMs demands substantial computational resources and time for both training and inference due to the high dimensionality of MRI data; (2) Introducing a conditioning element into 3D LDMs increases model intricacy and extends the required training duration.
In this study, we developed conditional 3D LDMs to generate 3D multi-contrast brain tumor MRI data. Our models were trained using two brain tumor datasets: The Cancer Genome Atlas (TCGA) public dataset10 and an internal dataset from the University of Texas Southwestern Medical Center (UTSW). The resulting models were able to produce high-quality multi-contrast MRI samples of brain tumors with the tumor location aligned by the input condition mask. These generated samples offer augmented data for various tasks involving brain tumor data, such as brain tumor segmentation or molecular prediction.
2. METHODOLOGY
2.1. Datasets and Image Preprocessing
This study utilized retrospective brain tumor MRI data from two datasets: The Cancer Genome Atlas (TCGA)10 dataset downloaded from The Cancer Imaging Archive (TCIA)11 and the internal dataset collected from UT Southwestern Medical Center (UTSW). Data included in this study was required to have the preoperative MRI scans of all four sequences: T1-weighted (T1), post-contrast T1-weighted (T1C), T2-weighted (T2), and T2-weighted fluid-attenuated inversion recovery (FLAIR). The dataset utilized for training our LDMs comprised 583 glioma patients, segregated into 466 samples for training and 117 samples for validation.
The MRI scans from both datasets were preprocessed using the federated tumor segmentation (FeTS) tool.12 The preprocessing steps included co-registering MRI scans to a template atlas, correcting for bias field distortion, and skull stripping. FeTS was also employed for segmenting the tumors, and the resulting tumor masks were employed as conditions for training the diffusion models. Subsequently, the skull-stripped images were cropped using the bounding box of the non-zero area. The z-score normalization was also applied to all non-zero voxels before feeding the data to the generative model training process.
2.2. Generative models
The skull-stripped images and the corresponding tumor masks were adjusted for size by randomly scaling and zero-padding. This process aimed to create volumes of 128 × 128 × 64 voxels, representing the x, y, and z dimensions of the 3D MR dataset. In addition, flipping was also used as an additional augmentation technique for training both autoencoder and diffusion models. The autoencoder model consists of an encoder, a decoder, and a discriminator network.2 The encoder reduces the dimensions of input images from (4 × 128 × 128 × 64) to a lower-dimensional latent space of (8 × 32 × 32 × 16). The decoder then reconstructs the MRI image from this compressed latent representation. Kullback-Leibler (KL) regularization was employed to slightly enforce the learned latent vectors towards a standard normal distribution.2 We introduced the Structural Similarity Index (SSIM) loss,5 a metric quantifying structural similarity between two images, to ensure the preservation of overall structure and perceptual details in the reconstructed MRI images. The autoencoder model was trained using a combination of perceptual loss,6 pixel-level L1 loss, and SSIM loss.
After achieving satisfactory results with the autoencoder model in generating high-quality reconstructed MRI images, we proceeded to train the diffusion model (DM). The DM consists of two main processes: the forward diffusion process, which adds noise to the latent-space features using a discrete-time diffusion schedule13 with 1000 steps, and the backward process of denoising using the UNet backbone.4 Our study focused on using the DM to learn conditional data distributions in the latent space. To achieve this, the conditional brain tumor mask first underwent a conditional module that downsampled it to match the latent space dimension (32 x 32 x 16). The resulting downsampled mask was then concatenated with the latent space features. To guide the learning process with respect to the conditional mask, cross-attention modules were integrated into the UNet backbone of the DM. These modules facilitated the learning of class-dependent data distribution based on the semantic information provided by the conditional mask.
3. RESULTS
3.1. Autoencoder quality
We first evaluated the quality of the images reconstructed through the autoencoder model. Figure 1 depicts the original and reconstructed images achieved via the autoencoder. We observe that the autoencoder effectively preserved details during the image reconstruction process.
Figure 1.
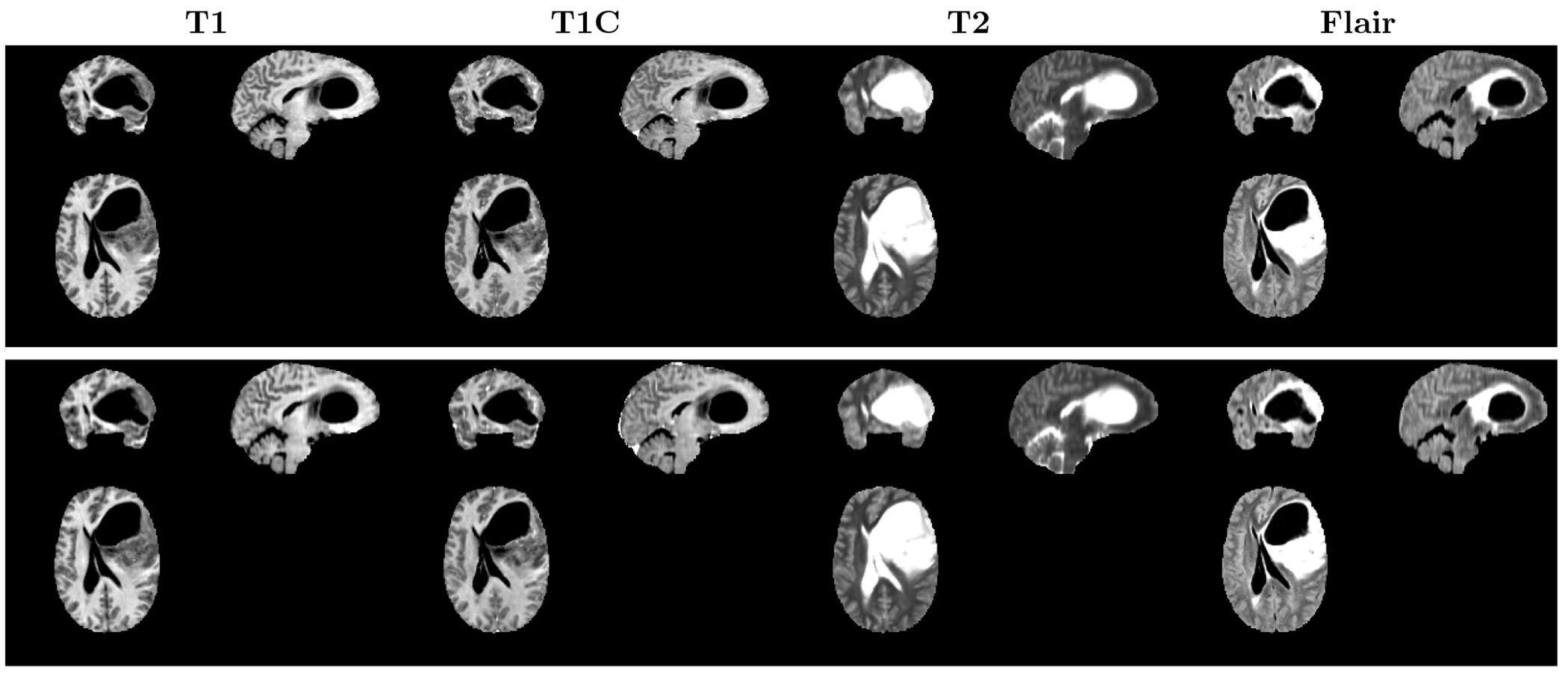
Comparison of the original and reconstructed images obtained by the autoencoder. The top row depicts the original images, while the bottom row shows the images that have been reconstructed by the autoencoder.
Table 1 shows the qualitative evaluation outcomes of the reconstructed images obtained by the autoencoder. To assess image quality and similarity between the original and reconstructed images, we employed metrics including Peak Signal to Noise Ratio (PSNR), Structural Similarity Index (SSIM), Mean Absolute Error (MAE), and Perceptual Loss. These metrics were calculated using a set of 50 test images. The quantitative analysis underscores that the autoencoder generated high-quality reconstructed images, even when subjected to a 32-fold compression factor.
Table 1.
Qualitative evaluation results of the reconstructed images obtained by the autoencoder.
| MRI Sequence | PSNR ↑ | SSIM ↑ | MAE ↓ | Perceptual loss ↓ |
|---|---|---|---|---|
| T1 | 30.9 | 0.84 | 0.069 | 0.046 |
| T1C | 34.8 | 0.96 | 0.037 | 0.051 |
| T2 | 32.5 | 0.94 | 0.049 | 0.040 |
| FLAIR | 32.9 | 0.91 | 0.046 | 0.062 |
3.2. Image Generation with 3D Multi-contrast Latent Diffusion
Figure 2 shows two examples of the synthetic images generated by our condition 3D LDMs. The conditional tumor masks were denoted by the transparent red area overlaid on the T1 sequence. The LDMs effectively generate brain tumor MRI samples, aligning the tumor locations with the provided masks. The generated samples also exhibit diversity despite having only a single mask as input. Figure 3 presents 6 samples of T1C MRI sequence, generated by the LDMs employing an identical mask.
Figure 2.
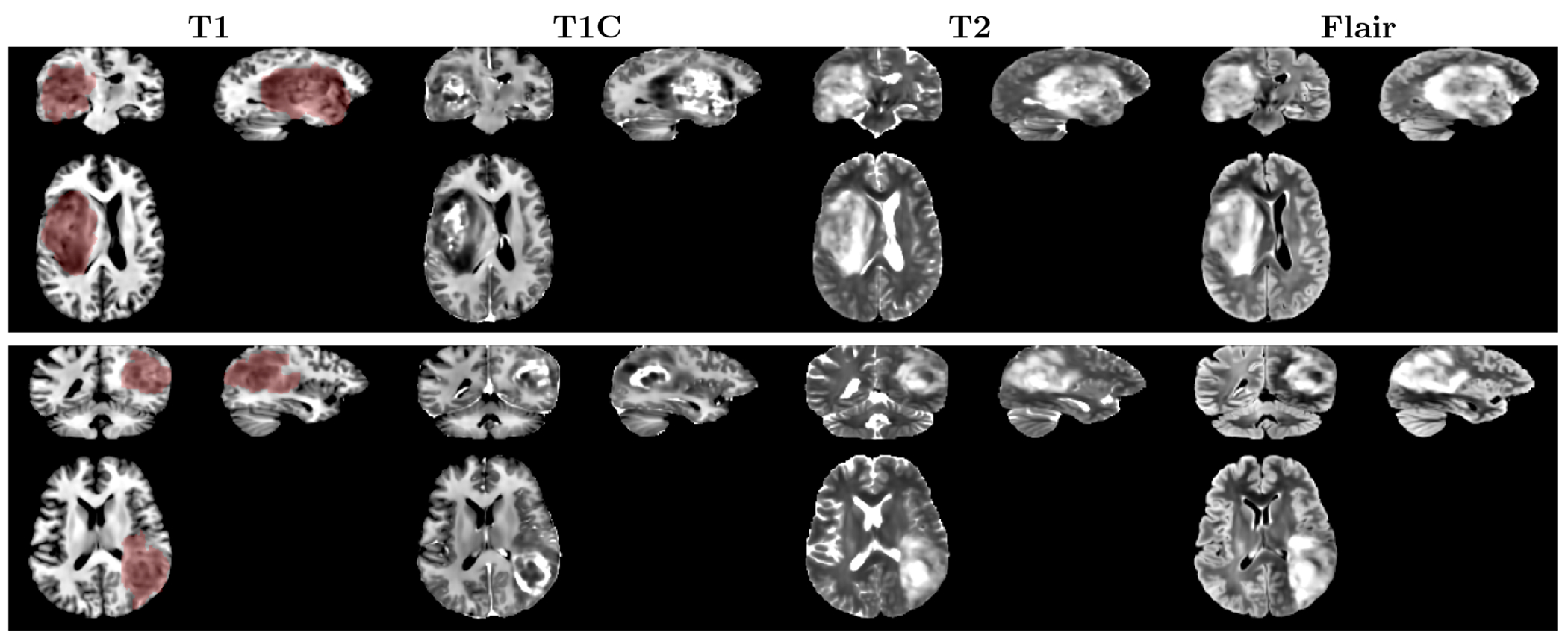
Synthetic images generated by the conditional 3D LDMs. The transparent red area indicates the whole tumor mask used as a condition for the LDMs.
Figure 3.
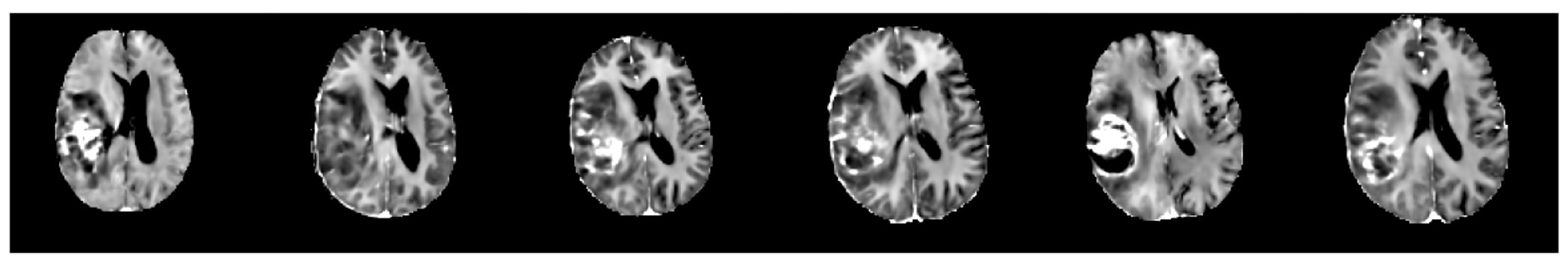
Different T1C samples generated by LDMs using the same whole tumor mask.
To evaluate the realism of the synthetic images, we employed the Fréchet Inception Distance (FID), a widely used metric to estimate the distances between feature representations of synthetic and real images. A smaller FID indicates greater similarity in distribution between the real and generated images. FID was calculated for each MRI sequence using a set of 500 generated samples and 500 real samples. We compared the synthetic image quality generated by two LDMs, each utilizing a UNet backbone with base channel sizes of 256 and 352, respectively. The quantitative results demonstrated that employing the larger UNet enhances the visual fidelity of the generated samples.
4. CONCLUSIONS
This study presents the development of conditional LDMs to generate 3D multi-contrast brain tumor MRI data. Our models were trained on a dataset consisting of MRI scans of 583 glioma patients. The models effectively produced high-quality multi-contrast MRI samples of brain tumors with the tumor location aligned by the input condition mask. This work has promising implications as it can alleviate the scarcity of brain tumor data and enhance the performance of deep learning models employing brain tumor MRI data.
Table 2.
Qualitative evaluation results of the synthetic images obtained by two LDMs with the UNet base channel sizes of 256 and 352, respectively. For each MRI sequence, FID scores were calculated using a set of 500 synthetic samples and 500 real samples.
| FID ↓ | ||||
|---|---|---|---|---|
| T1 | T1C | T2 | FLAIR | |
| LDMs - 256 | 112.76 | 50.9 | 46.01 | 45.77 |
| LDMs - 352 | 23.01 | 17.14 | 26.43 | 32.42 |
ACKNOWLEDGMENTS
This research was supported by the NIH/NCI U01CA207091 (AJM, JAM) and R01CA260705 (JAM).
Footnotes
DISCLOSURES
The authors declare no conflict of interest.
REFERENCES
- [1].Abdal R, Qin Y, and Wonka P, “Image2StyleGAN: How to Embed Images Into the StyleGAN Latent Space?,” in [Proceedings of the IEEE/CVF International Conference on Computer Vision], 4432–4441 (2019). [Google Scholar]
- [2].Rombach R, Blattmann A, Lorenz D, Esser P, and Ommer B, “High-Resolution Image Synthesis With Latent Diffusion Models,” in [Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition], 10684–10695 (2022). [Google Scholar]
- [3].Dhariwal P and Nichol A, “Diffusion Models Beat GANs on Image Synthesis,” in [Advances in Neural Information Processing Systems], 34, 8780–8794, Curran Associates, Inc. (2021). [Google Scholar]
- [4].Nichol AQ and Dhariwal P, “Improved Denoising Diffusion Probabilistic Models,” in [Proceedings of the 38th International Conference on Machine Learning], 8162–8171, PMLR; (July 2021). [Google Scholar]
- [5].Zhang L, Rao A, and Agrawala M, “Adding Conditional Control to Text-to-Image Diffusion Models,” in [Proceedings of the IEEE/CVF International Conference on Computer Vision], 3836–3847 (2023). [Google Scholar]
- [6].Na Y, Kim K, Ye S-J, Kim H, and Lee J, “Generation of Multi-modal Brain Tumor MRIs with Disentangled Latent Diffusion Model,” in [Medical Imaging with Deep Learning, Short Paper Track], (Apr. 2023). [Google Scholar]
- [7].Khader F, Müller-Franzes G, Tayebi Arasteh S, Han T, Haarburger C, Schulze-Hagen M, Schad P, Engelhardt S, Baeßler B, Foersch S, Stegmaier J, Kuhl C, Nebelung S, Kather JN, and Truhn D, “Denoising diffusion probabilistic models for 3D medical image generation,” Sci Rep 13, 7303 (May 2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pinaya WHL, Tudosiu P-D, Dafflon J, Da Costa PF, Fernandez V, Nachev P, Ourselin S, and Cardoso MJ, “Brain Imaging Generation with Latent Diffusion Models,” in [Deep Generative Models: Second MICCAI Workshop, DGM4MICCAI 2022, Held in Conjunction with MICCAI 2022, Singapore, September 22, 2022, Proceedings], 117–126, Springer-Verlag, Berlin, Heidelberg (Sept. 2022). [Google Scholar]
- [9].Peng W, Adeli E, Bosschieter T, Park SH, Zhao Q, and Pohl KM, “Generating Realistic Brain MRIs via a Conditional Diffusion Probabilistic Model,” (Sept. 2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ceccarelli M, Barthel FP, and et al. , “Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma,” Cell 164, 550–563 (Jan. 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, and Prior F, “The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository,” J Digit Imaging 26, 1045–1057 (Dec. 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pati S, Baid U, Edwards B, Sheller MJ, Foley P, Anthony Reina G, Thakur S, Sako C, Bilello M, Davatzikos C, Martin J, Shah P, Menze B, and Bakas S, “The federated tumor segmentation (FeTS) tool: An open-source solution to further solid tumor research,” Phys Med Biol 67 (Oct. 2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ho J, Jain A, and Abbeel P, “Denoising Diffusion Probabilistic Models,” in [Advances in Neural Information Processing Systems], 33, 6840–6851, Curran Associates, Inc. (2020). [Google Scholar]


