Abstract
In this study we have characterized a positive regulatory region located in the first intron of the α-fetoprotein (AFP) gene. We show that the enhancer activity of the region depends on a 44 bp sequence centered on a CACCC motif. The sequence is the target of the two zinc fingers transcription factors BKLF and YY1. The introduction of a mutation destroying the CACCC box impairs the binding of BKLF but improves that of YY1. Moreover, the mutated sequence behaves as a negative control element, suggesting that BKLF behaves as a positive factor and that YY1 is a negative one. We also demonstrate the existence of a novel, tissue-specific AFP mRNA isoform present in the yolk sac and fetal liver which initiates from an alternative promoter located ∼100 bp downstream of the enhancer element. The transcriptional start site controlled by this new promoter (called P2), was mapped to 66 bp downstream of a TATA box. A putative AUG translation site in-frame with exon 2 of the classical gene was found 295 bp downstream of the transcription start site. Like the traditional AFP promoter (P1), the P2 promoter is active in the yolk sac and fetal liver. Embryonic stem cells with an AFP knock-in gene containing either the P2 promoter or deleted for it were isolated and comparative analysis of embryonic bodies derived from these cells suggests that the P2 promoter contributes to early expression of the AFP gene.
INTRODUCTION
The albumin gene family comprises four known genes expressed specifically in the liver and encoding serum albumin (ALB), α-fetoprotein (AFP), α-albumin (ALF) and vitamin D-binding protein (Gc, group-specific component) (1–5). These related genes produce specialized products characterized by specific patterns of expression. The molecular basis of the differential gene expression is only partially explained. Two recent reviews provide a general overview of AFP regulation (6,7). The AFP gene is an attractive gene for studying tissue-specific and developmental transcription regulation. Indeed, this gene is transcribed at high levels in the visceral endoderm, the yolk sac and fetal hepatocytes and at low levels in the fetal gut. Expression is shut off at birth, so that the normal adult liver produces 103- to 104-fold less AFP mRNA than the fetal liver (8–10). The AFP gene can be reactivated in the adult during liver regeneration or in the case of hepatocarcinomas or teratomas. The AFP gene has a large and complex transcription control region. cis-acting control domains that govern AFP expression have been identified by transfection assays (11–15) and by insertion of transgenes into mice (16,17). Three upstream enhancer regions have been defined. They are several hundred nucleotides long and are active in most tissues, although at various levels (18–21). The AFP promoter, covering a region of ∼250 bp, is regulated by tissue-specific activators such as HNF1 (22), C/EBP (23), Nkx2.8 (24) and FTF (25), as well as by ubiquitous factors like NF1 (26). Promoter activity is limited to tissues producing AFP, indicating that this region contributes to tissue-specific expression (12,15,27,28). In addition, it has been suggested that a region in the first intron of AFP participates in the control of gene activity (13); indeed, a 146 bp sequence located in the first intron stimulates transcription of a reporter gene preceded by the AFP promoter. The purpose of this work was to characterize this regulatory region. We report that it contains two regulatory elements: an enhancer able to act on the upstream promoter and an alternative promoter active in the yolk sac and fetal liver.
MATERIALS AND METHODS
Gel retardation analysis
Nuclei of HepG2 human hepatoma cells (29) were isolated from sub-confluent cultures as previously described (13) and nuclear extracts were prepared according to a procedure as described before (30). DNA probes were prepared by 3′-end-labeling with [α-32P]dATP and Klenow enzyme. Binding reactions were performed on ice in a final volume of 25 µl [10 mM HEPES–KOH, pH 7.9, 30 mM KCl, 12% glycerol, 5 mM MgCl2, 0.5 mM dithiothreitol (DTT), 0.1 mM EDTA]. Aliquots of 0.5–1 µg HepG2 nuclear extract were incubated with 1 µg poly(dI–dC) and 32P-end-labeled DNA (30 pmol) for 20 min on ice. In competition experiments the competitor was pre-incubated with nuclear extract and poly(dI–dC) for 20 min on ice before incubation with the probe. A Sp1 target sequence (5′-ATTCGATCGGGGCGGGGCGAGC-3′), a Sp1 mutated sequence (5′-ATTCGATCGGTTCGGGGCGAGC-3′) and a YY1 target sequence (5′-CGCTCCGCGGCCATCTTGGCGGCTGGT-3′) were used as competitors. For DNA binding assays in which antibodies were included, antibodies were added to the binding mixtures and incubated for 30 min on ice before addition of radiolabeled oligonucleotide. Anti-BKLF was a gift from M. Crossley (31). Polyclonal rabbit antiserum against the YY1 protein was purchased from Biotechnology Inc. (Santa Cruz, CA). The samples were loaded on a 5% non-denaturing acrylamide gel buffered in 1× TBE (89 mM Tris–borate, 89 mM boric acid, 2 mM EDTA). The complexes were separated at room temperature and, after running, the gels were dried and autoradiographed.
Purification of YY1
Two strategies were used. The first one was classical chromatography. Aliquots of 5 mg HepG2 nuclear extract were prepared and fractioned by ion exchange chromatography on an FPLC system (Hi trap Q; Amersham Pharmacia Biotech, UK). The fractions containing C3 activity were monitored by band shift assay using the 44-mut bp probe. DNA affinity chromatography of pooled active fractions was performed on a SMART system with a NHS-activated Superose column (Amersham Pharmacia Biotech, UK) coupled with a multimer of 44-mut bp sequence as described before (32) and according to the manufacturer’s instructions. Proteins were eluted with band shift buffer containing increasing amounts of KCl. Protein fractions were TCA precipitated, washed with acetone, resuspended in sample buffer and incubated in boiling water for 3 min prior to analysis by SDS–PAGE. The second strategy was based on preparative band shifts in a Prep Cell system (model 491; Bio-Rad, Hercules, CA). Aliquots of 120 µg HepG2 nuclear extract were incubated with 32P-labeled 44-mut bp probe (100 000 c.p.m.) under the band shift assay conditions. The binding reaction was loaded on the Prep Cell column containing a 5% non-denaturing acrylamide gel in 1× TBE. The fractions collected were monitored for radioactivity and the labeled fractions were subjected to a band shift assay and to SDS–PAGE. The gels were silver stained with a PlusOne Silver Staining Kit from Amersham Pharmacia Biotech.
Plasmids
The plasmid p(–1023 to +33)-CAT has been described previously (pHAF-CAT) (12); it contains a segment of the mouse AFP gene including the promoter (from –1023 to +33) cloned into pBLCAT2 (33). p(–1023 to +33)-KSE-CAT is derived from p(–1023 to +33)-CAT by insertion, at the HindIII restriction site, of a polylinker containing KpnI, SacII and EcoRV restriction sites and flanked by HindIII restriction sites. p(–1023 to +33)-(+347 to +390)-CAT was constructed in two steps. First, an oligonucleotide containing a 44 bp segment (from +347 to +390) (Fig. 3A) flanked by SalI (5′) and XbaI (3′) restriction sites was inserted into the pBSIISK+ (Stratagene, La Jolla, CA) polylinker, opened by SalI and XbaI. Subsequently, the KpnI–SacII fragment of the modified pBSIISK+ vector was inserted into the polylinker of p(–1023 to +33)-KSE-CAT.
Figure 3.
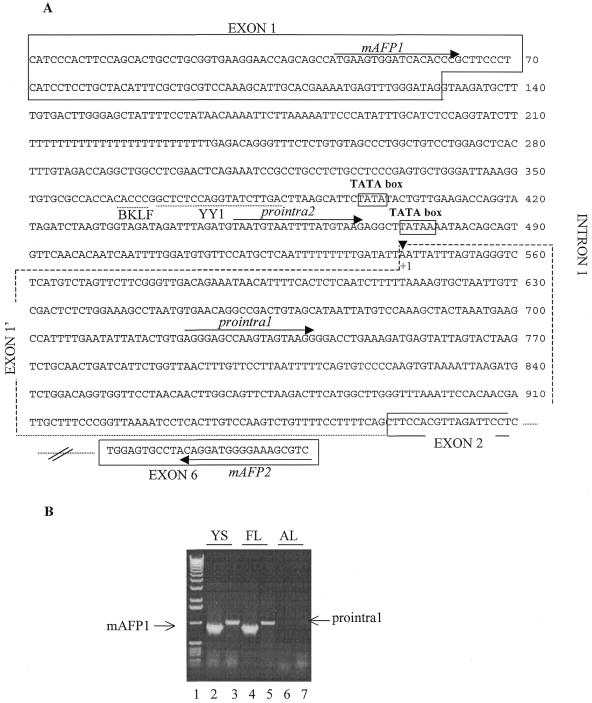
Identification of a novel AFP transcript. (A) Part of the mouse AFP gene sequence. The sequences spanned by exons 1, 1′, 2 and 6 are boxed (by a dashed line for exon 1′). The binding sites for the BKLF and YY1 transcription factors (TFSEARCH program from Kyoto University) are indicated by a dashed line. The primers used in RT–PCR experiments are indicated by arrows. (B) RT–PCR analysis of the AFP transcripts in the yolk sac (YS), the fetal liver (FL) and the adult liver (AL). The primers were used in the following combinations: mAFP1 (exon 1-specific) and mAFP2 (exon 6-specific) (lanes 2, 4 and 6); prointra1 (intron 1-specific) and mAFP2 (lanes 3, 5 and 7). The sizes of the fragments are 838 bp in the case of mAFP1 and 992 bp in the case of prointra1. Lane 1 contained a 1 kb DNA ladder.
The plasmid containing two copies of the 44 bp AFP sequence was also obtained in two steps. First the modified pBSIISK+ described above was digested either by ScaI (site in pBSIISK+ at 2526 bp) and XbaI (site in the polylinker) or by ScaI and SalI (site in the polylinker). The two blunted fragments containing the 44 bp AFP sequence were ligated. The fragment with two copies of the 44 bp AFP sequence was rescued from this plasmid by KpnI and SacII digestion and subsequently inserted at the KpnI and SacII sites of p(–1023 to +33)-KSE-CAT.
The plasmid containing four copies of 44 bp AFP sequence was obtained as follows. Plasmid pBSIISK+ containing two copies of the 44 bp AFP sequence was digested either by ScaI (site in pBSIISK+ at 2526 bp) and XbaI (site in the polylinker) or by ScaI and XhoI (site in the polylinker). The two blunted fragments, each of which contained two copies of the 44 bp AFP sequence, were ligated. The fragment with four copies of AFP sequence rescued by KpnI and SacII digestion was inserted at the KpnI and SacII sites of p(–1023 to +33)-KSE-CAT. The same cloning strategy was used to generate the corresponding constructs containing the mutated sequence (44-mut bp, Fig. 3A).
pSV2-LUC was constructed as follows. pSV2-CAT (34) was digested with PvuII and HindIII and the resulting 323 bp fragment, containing the SV40 early promoter, was inserted in pXp2 (35) between the SmaI and HindIII sites, upstream of the luciferase coding sequence.
pAFP K.O-1 consists of two recombination arms separated by a reporter-selective cassette. A 16 kb genomic fragment of the mouse Afp gene was isolated from a 129 λ library using an Afp promoter fragment as a probe. The genomic insert was subcloned in pKIL-PCR2 (36). The 5′-arm (2.5 kb) was generated by PCR using the following primers: N-MerI (5′-AGAGCGGCCGCGGAAGTGACAAAGCAGAACC-3′) complementary to the MerI sequence of AFP enhancer I (18); X-exon 1 (5′-AGACTCGAGGGATGAGGGAAGCGGGTGTG-3′). The 3′-arm was subcloned from the λ clone into the pBSIISK+ vector (Stratagene, La Jolla, CA). The 5′ recombination arm was introduced upstream of the 3′ recombination arm. The IRES lacZ/neo reporter-selective cassette was introduced between these recombination arms. The thymidine kinase gene from Herpes virus, inserted into the targeting vector, was used to select for homologous recombinants.
pAFP K.O-2 was constructed as for pAFP K.O-1 except that the 5′-arm (3.4 kb) was generated by PCR using N-MerI and X-exon 2 (5′-AGATTGCACCTTCGACTTTC-3′).
The plasmids were purified on Qiagen columns (Qiagen GmbH, Hilden, Germany).
Transfections and enzymatic assays
HepG2 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum. Twenty-four hours before transfection the cells (2 × 106) were seeded in a 100 mm culture dish. Transfections were performed using the calcium phosphate method with 5 µg of test plasmids containing the CAT reporter gene and with 0.5 µg of pSV2-LUC, used as an internal control (27). The culture medium was removed 6 h after transfection and replaced by fresh medium. Forty-eight hours after transfection the cells were harvested for CAT activity assays (33). The luciferase assay was performed as described before (37), in a volume of 50 µl of cell extract. The values of CAT activities were normalized against those of luciferase.
RT–PCR
The primer mAFP2 (5′-ACGCTTTCCCCATCCTGTAG-3′) was used to generate a first strand cDNA from 1 mg of total RNA, with a First Strand cDNA Synthesis kit (Roche Diagnostics, Brussels, Belgium) in a volume of 20 µl. In all experiments total RNA was extracted with the RNA Insta-Pure System (Eurogentec Bel SA, Seraing, Belgium). The reverse transcription mix was incubated at 42°C for 1 h, after annealing the primer at 25°C for 10 min. Reactions were stopped by heating at 90°C for 3 min. This first strand cDNA was amplified by PCR in 100 µl of reaction mixture using an Expand High Fidelity kit (Roche Diagnostics) and different forward specific primers [mAFP1 (5′-ATGAAGTGGAGCGCATCCAT-3′), prointra1 (5′-GAGGGAGCCAAGTAGTAAGG-3′) and prointra2 (5′-TGTAATTTTATGTAAGA-3′)] in a GeneAmp PCR System 9600 thermocycler (35 cycles of 30 s at 94°C, 30 s at 52°C and 1 min at 72°C) (Applied Biosystems Inc., Foster City, CA). The reverse primer was chosen in exon 6 to avoid misinterpretation of results due to possible contaminating genomic DNA. Aliquots of 20 µl of each reaction product were run on 1% agarose gels. Ethidium bromide-stained PCR bands were visualized under UV and in all cases a PCR product of the expected size was sequenced with a model ABI Prism 310 Genetic Analyzer and the Big Dye terminator cycle sequencing protocol (Applied Biosystems Inc.).
Primer extension analysis
Aliquots of 10 µg of total RNA were hybridized with 200 000 c.p.m. of a 32P-labeled oligonucleotide complementary to the sequence covering positions +591 to +575 (Fig. 1A), in 15 µl of hybridization buffer (10 mM Tris–HCl pH 8.3, 150 mM KCl, 1 mM EDTA) at 42°C for 16 h. The primer reaction was started by addition of 30 µl of reverse transcriptase reaction buffer (50 mM Tris–HCl, pH 8.5, 30 mM KCl, 8 mM MgCl2, 1 mM DTT, 1 mM dNTP, 20 U RNase inhibitor) containing 5 U avian myeloblastosis virus reverse transcriptase (AMV; Roche Diagnostics). After incubation at 42°C for 60 min, the RNA template was removed by adding 105 µl of RNase mix. After incubation at 37°C for 15 min followed by phenol extraction, the DNA was precipitated with ethanol. The pellets were dissolved in 5 µl of 80% formamide–dye mix and the DNA was separated on a 7% acrylamide–7 M urea gel. Radioactive signals were detected by autoradiography.
Figure 1.
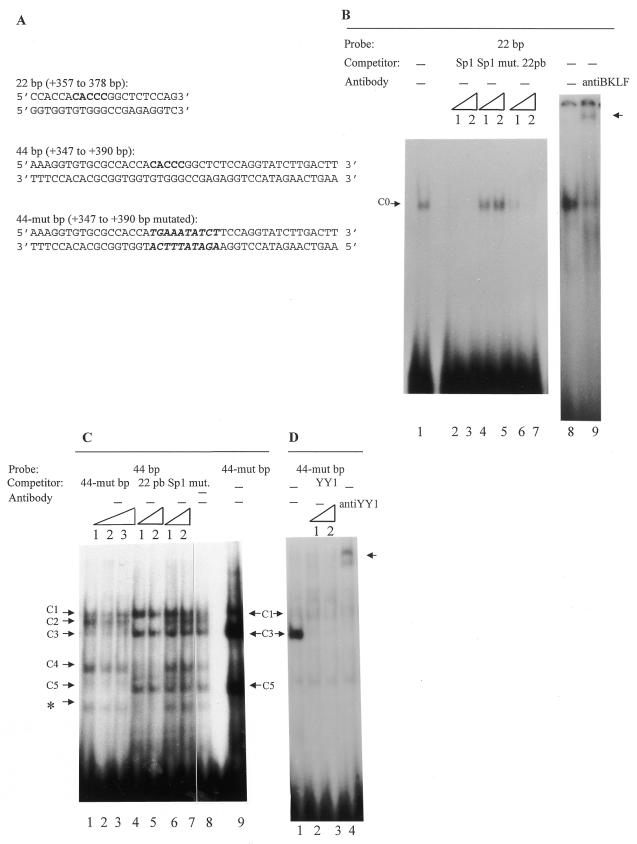
Gel retardation assays with hepatoma cell (HepG2) extracts and different probes. The competition experiments were performed in the presence of increasing amounts of unlabeled double-stranded competitors: 1, 50-fold excess; 2, 100-fold excess; 3, 200-fold excess. – indicates absence of competitor. The probes and the competitors used are indicated above the corresponding lanes. (A) Sequences of the 22 bp, 44 bp and 44-mut bp used. (B) The 22 bp radiolabeled probe (30 pmol/reaction) was incubated in the absence (lanes 1–8) or presence of unlabeled 22 bp sequence (lanes 6 and 7) and of an oligonucleotide corresponding to the Sp1 binding site (lanes 2 and 3) and of a mutated Sp1 binding site (lanes 4 and 5). Lane 9 shows a supershift assay with 1 µl of specific anti-BKLF antibody. The arrow indicates the supershift. (C) Gel retardation assays with the 44 bp probe in the presence of either unlabeled 44-mut bp sequence (lanes 1–3) or unlabeled 22 bp sequence (lanes 4 and 5) or mutated Sp1 binding site (lanes 6 and 7) as non-specific competitor and with 44-mut bp sequence as a probe (lane 9). The asterisk indicates a non-reproducible band. (D) Gel retardation assays with the 44-mut bp probe and purified HepG2 nuclear extract in the presence of either an oligonucleotide corresponding to the YY1 binding site (lanes 2 and 3) or a specific anti-YY1 antibody (lane 4). Similar results were obtained with the 44 bp probe (though the signals were less intense). The arrow indicates the supershift.
RNase protection assay
A PCR fragment from intron 1 (positions +245 to +741) was cloned using a Topo-XL cloning kit (Invitrogen, Carlsbad, CA). The vector was linearized with BglII and a 32P-labeled RNA probe was produced using T7 RNA polymerase. The reaction mixture contained 1 µg DNA, 40 mM Tris–HCl pH 7.5, 6 mM MgCl2, 2 mM spermidine, 100 µM UTP, 0.5 mM GTP, CTP and ATP, 10 mM DTT, 20 U RNase inhibitor (Roche Diagnostics), 50 µCi [α-32P]UTP (3000 Ci/mmol; NEN/Dupont) and 10 U T7 RNA polymerase (Roche Diagnostics) in 10 µl. Labeled RNA was purified on an acrylamide–urea gel and eluted from the gel slice in 0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA and 0.1% SDS at 37°C for 16 h. Total RNA (50 µg) from mouse tissues was hybridized with 30 000 c.p.m. of labeled RNA probe in 30 µl of hybridization buffer (40 mM PIPES pH 6.4, 80% formamide, 0.4 M NaCl, 1 mM EDTA) at 45°C for 16 h after heat denaturation at 90°C for 5 min. Reaction mixtures were placed at room temperature and 350 µl of RNase mix [10 mM Tris–HCl pH 7.5, 5 mM EDTA, 300 mM NaCl, 3.5 µg RNase A, 25 U RNase T1 (Roche Diagnostics)] was added. Samples were incubated at 30°C for 30 min before addition of 10 µl of 20% SDS and 50 µg proteinase K and continuation of incubation at 37°C for 15 min before phenol/chloroform extraction and ethanol precipitation with 10 µg carrier tRNA. Dried samples were resuspended in 80% formamide–dye mix and run on 7% acrylamide–7 M urea gels, which were dried and exposed to autoradiograph film.
Recombinant embryonic stem (ES) cells
The targeting vectors pAFP K.O-1 and pAFP K.O-2 were linearized with NotI and electroporated into E14 ES cells. Correct targeted clones were identified by Southern blot analysis using an external probe from the AFP 5′-region. ES cells were cultured as described before (38) except that Glasgow’s modified Eagle’s medium (GMEM) was used. Embryoid bodies (EBs) were produced according to the method of Robertson (39) and stained as described before (40).
RESULTS
Analysis of DNA–protein interactions
In a previous study a 142 bp region containing a methylable cytosine located in the first intron of the AFP gene was shown, by transfection experiments in HepG2 AFP-producing hepatoma cells, to participate in the control of gene activity (13). This cytosine is the last base of a CACCC box which itself is a putative binding site for transcription factors of the Sp/XKLF zinc finger family (for reviews see 41,42). Band shift assays were performed with nuclear proteins extracted from AFP-producing hepatoma cells (HepG2) and a 22 bp oligonucleotide centered on this CACCC box which covers the segment from +357 to +378 bp (Fig. 1A). A single complex was obtained (Fig. 1B, lane 1, C0). The reaction product was specific since this shift was eliminated by competition with the homologous unlabeled probe (Fig. 1B, lanes 6 and 7). To determine whether this complex was due to Sp/XKLF family factors, new gel mobility shift assays were performed using HepG2 nuclear extracts and, as a competitor, an oligonucleotide corresponding to the most common Sp1 binding site (GC box, see Materials and Methods). The C0 shift was specifically abolished by this oligonucleotide (Fig. 1B, lanes 2 and 3). This shift thus appears to result from binding of a factor(s) belonging to the Sp/XKLF family. Among these factors, BKLF (for basic Krüppel-like factor; also called KLF3) (31) binds the CACCC motif and is known to be abundant in the mouse yolk sac and fetal liver, i.e. in AFP-expressing tissues. An antibody directed against BKLF was added to the gel shift assay reactions; under these conditions, complex C0 was strongly inhibited and a supershift was clearly visible (Fig. 1B, lane 9). This result shows that BKLF binds the 22 bp segment and is (at least partially) responsible for the C0 shift. A longer sequence centered on the 22 bp oligonucleotide (and thus on the CACCC motif) covering the segment from +347 to +390 bp (Fig. 1A, 44 bp) was also used in band shift assays. Five DNA–protein complexes were obtained with this 44 bp probe (Fig. 1C, lane 8, C1–C5). The C2 and C4 shifts were abolished when the 22 bp sequence was used as competitor, strongly suggesting that these shifts contain BKLF as the main component. Moreover, an oligonucleotide mutated in the CACCC motif (44-mut bp, 10 bp including the CACCC box were mutated; see Fig. 1A) was unable to compete for formation of the C2 and C4 complexes (Fig. 1C, lanes 1–3) and, when the mutated sequence was used as probe, the C2 and C4 complexes were not formed while formation of the C3 and C5 shifts was improved (Fig. 1C, lane 9). The later observation suggests that BKLF competes with the factor(s) generating the C3 and C5 shifts. In order to identify this factor(s), HepG2 nuclear extracts were partially purified either by ion exchange chromatography followed by affinity chromatography (FPLC) or by preparative gel shifts. With the purified extracts and the 44 bp or 44-mut bp probe, the C3 complex was still produced, and even reinforced with the 44-mut bp probe. The same extracts showed a major protein of 68 kDa on polyacrylamide gel electrophoresis (data not shown). This protein must be a zinc finger protein because the C3 shift disappeared in the presence of the specific zinc chelator 1,10-phenathroline. The 44 bp and 44-mut bp sequences contain a potential binding site for the multifunctional zinc finger protein YY1, which has a molecular weight of 68 kDa (43–46). To test the role of YY1 in formation of the C3 complex, the 44-mut bp sequence was used as probe and incubated with purified HepG2 nuclear extracts (in this case only the C3 complex was observed) and either an unlabeled oligonucleotide containing a YY1 binding site (Fig. 1D, lanes 2 and 3) or an antibody specific to YY1 (Fig. 1D, lane 4). The C3 complex disappeared under these conditions and a supershift was observed in the presence of the antibody. These results thus show that the 347–390 bp sequence possesses binding sites for BKLF and YY1 and suggest that in the presence of BKLF, binding of YY1 to its site is reduced.
Analysis of the regulatory role of the 44 bp intronic sequence
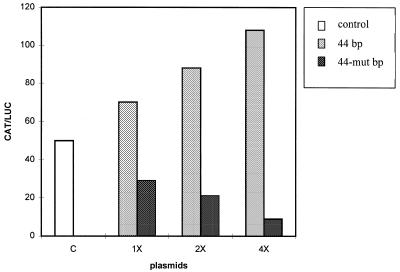
To determine whether the region containing the binding sites for BKLF and YY1 is involved in the activity of the 142 bp intronic region (13), the 44 bp and 44-mut bp sequences were inserted as monomers or as multimers (two or four) in chimeric constructs between the AFP promoter region (–1023 to +33) and the CAT reporter gene (Fig. 1A and Materials and Methods). The different constructs were transfected into HepG2 cells and CAT activities were compared. The results are summarized in Figure 2. They show that the constructs containing the 44 bp sequence are more active than the constructs lacking this sequence or containing the 44-mut bp sequence. Furthermore, their respective activities increase with the number of copies of the 44 bp sequence. The 44 bp sequence thus behaves as an enhancer, acting on the upstream promoter. Moreover, the constructs bearing the 44-mut bp sequence are less active than the constructs lacking the 44 bp sequence, suggesting that the mutation transforms the 44 bp sequence into a negative element. Since the mutated sequence no longer binds BKLF and is a better target for YY1, these results strongly suggest that BKLF and YY1 control the activity of the 44 bp sequence oppositely, BKLF stimulating its activity and YY1 repressing it.
Figure 2.
Activity of chimeric constructs used to test the effect of the 44 bp sequence containing the CACCC motif and the mutated sequence. One, two or four copies of each sequence was cloned in a vector containing the AFP promoter (–1023 to +33) and the CAT reporter gene (control plasmid, c) (plasmids 1×, 2× and 4×, respectively). The constructs were transfected into HepG2 cells with pSV2-LUC used as an internal control. The CAT activities were normalized against those of luciferase. The values shown are the means of six experiments, done with independent plasmid preparations. The standard deviations were <20% of the mean values.
Identification of a variant AFP mRNA
Computer analysis of the intron 1 sequence not only revealed the presence of putative binding sites for transcription factors but also indicated the presence of two TATA boxes. One or two alternative promoters could thus exist in the region determined to be the first intron of the AFP gene. In order to test this hypothesis, RT–PCR amplification was done on RNA preparations using forward primers complementary to sequences of the first intron (prointra1 located downstream of the second TATA box and prointra2 located between the two TATA boxes; see Fig. 3A). For the first strand cDNA synthesis another primer, mAFP2, complementary to a sequence of exon 6, was used. Finally, a primer corresponding to a sequence in the first exon, mAFP1, was used as a control to amplify the cDNA complementary to the traditional mRNA originally described (47). With the mAFP1 and mAFP2 primers and using RNA preparations from mouse yolk sac and fetal and adult liver, an 838 bp fragment was amplified in the yolk sac and fetal liver cases (Fig. 3B, lanes 2 and 4). In addition, a cDNA of 992 bp was amplified from the yolk sac and fetal liver with prointra1 and mAFP2 (Fig. 3B, lane 3, yolk sac, and lane 5, fetal liver) but no amplification was obtained using the prointra2 and mAFP2 primers, indicating the existence of a transcript which covers the prointra1 sequence (downstream of the second TATA box) but not that of prointra2 (upstream of the second TATA box). No cDNA was produced from the adult liver with any primer pair (Fig. 3B, lanes 6 and 7). The sequence of the 992 bp RT–PCR product was determined and revealed that this cDNA corresponded to the expected one: this cDNA starts at the beginning of prointra1, reaches and includes exon 2 and ends in exon 6, thereby defining an alternative exon 1 (exon 1′, Fig. 3A). A novel transcription start site is thus active in this region, which we will call P2, designating the traditional AFP promoter P1. We will continue to use the base numbering of Tilghman (47) and in this context the P2 promoter is thus located between position +470 (corresponding to the end of the prointra2 primer) and +722 (corresponding to the beginning of the prointra1 primer). The prointra1 and mAFP2 primers were also used for RT–PCR amplification with RNA preparations from murine AFP-producing hepatoma cells (BWTG3) and the corresponding human primers were used with RNA preparations from human AFP-producing hepatoma cells (HepG2). In both cases no amplification product was obtained.
The AFP mRNAs controlled by promoters P1 and P2 thus co-exist in the murine yolk sac and fetal liver. Both are absent in the murine adult liver and only one (the P1 mRNA) is present in hepatoma cells, suggesting that the two AFP transcripts are subject to different regulation mechanisms.
Identification of the transcription start site controlled by the P2 promoter
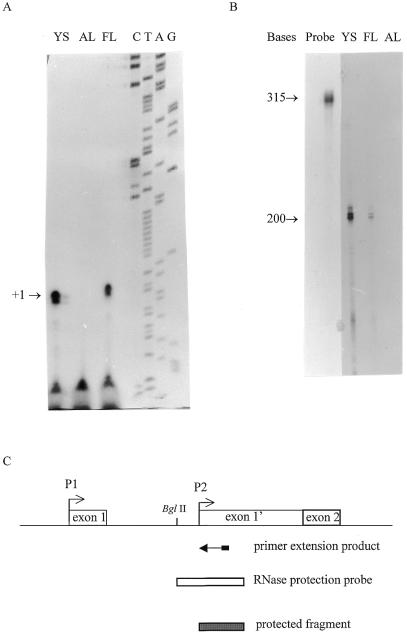
The transcriptional start site controlled by the P2 promoter was identified by primer extension and RNase protection experiments carried out on total RNA purified from murine yolk sac and fetal liver. Using a reverse primer corresponding to the sequence located between +591 and +575 bp, a major transcription start site was identified at position +541 (Fig. 4A), ∼66 bp downstream of the second TATAA box. The RNA probe (see Fig. 4C and Materials and Methods) was hybridized to total RNA and subjected to digestion by RNase T1 and RNase A in order to remove unhybridized RNA. A protected fragment of ∼200 bp was detected (Fig. 4B). This size corresponds to that of a segment extending from the prointra1 primer 5′-end to the position of the start site (+541) identified by the above primer extension experiment. The signal observed after RNase protection was stronger with yolk sac RNA than with fetal liver RNA, suggesting that the P2 promoter is more active in the yolk sac. Sequence analysis of the region downstream of the P2 promoter revealed a putative AUG translation start site (at +838 bp) in exon 1′ which is in-frame with the traditional exon 2.
Figure 4.
Identification of the alternative intronic transcriptional start site. (A) Autoradiograph showing the results of primer extension analysis of the 5′-end of the P2 mRNA using an intron-specific end-labeled DNA primer. Samples were RNA from yolk sac (YS), adult liver (AL) and fetal liver (FL). The arrow indicates the +1 position corresponding to +541 bp by comparison with the sequence of intron 1 with the intron-specific primer used in this experiment (lanes C, T, A and G). (B) Autoradiograph showing the results of an RNase protection assay with a labeled antisense RNA probe extending from the BglII site to the prointra1 primer used in RT–PCR (Fig. 1A). The probe was hybridized to yolk sac (YS), fetal liver (FL) and adult liver (AL) RNA. The numbers on the left indicate the sizes of the protected fragments obtained by RNase protection assay and with the RNA probe. (C) Graphic illustration of the position of the P2 intronic promoter, as determined by primer extension studies and RNase protection analysis.
Activity of the AFP promoters during the early steps of gene transcription activation
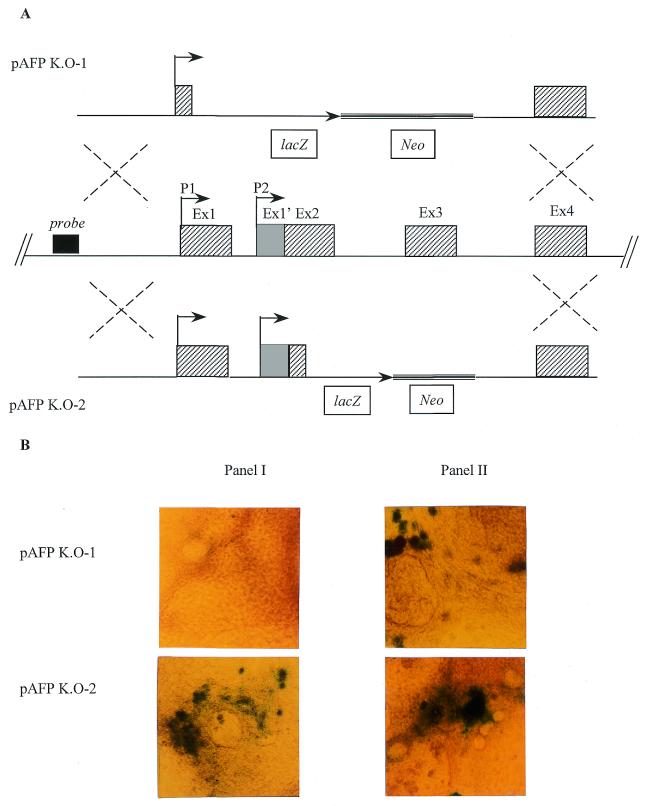
It has been suggested that EBs may serve as an in vitro system to study the early steps of AFP activation (48). We used the potential of aggregated ES cells to differentiate in vitro in EBs (49) to assess the role of the P2 promoter in turning on AFP transcription. Homologous recombination in ES cells was used to produce cells with a knock-in AFP gene in which the bacterial LacZ gene replaces part of the gene: either the sequence from +72 to +2773 bp (pAFP K.O-1) or that from +973 to +2773 bp (pAFP K.O-2) (see Fig. 5). The pAFP-KO.2 chimeric gene contains the P2 promoter described above (at +541 bp) whereas the pAFP-KO.1 chimeric gene does not. Previous studies with EBs showed that AFP expression starts a few days before the appearance of autonomous beating cells (48). In our study we observed that during EB differentiation, expression of β-galactosidase was detected 5 days earlier with cells containing the pAFP K.O-2 transgene than with those containing the pAFP K.O-1 vector and that β-galactosidase expression by EBs containing the pAFP K.O-2 vector was observed a few days before the appearance of autonomous beating cells, just like AFP expression (48; Fig. 5B). These results suggest that the P2 promoter is responsible for earlier expression of the AFP gene.
Figure 5.
(A) Schematic representation of the strategy used to introduce the lacZ reporter gene under control of the AFP promoter region. The structures of the targeting vectors, pAFP K.O-1 (top) and pAFP K.O-2 (bottom), and of the AFP genomic locus (middle) are shown. The AFP exons are represented by shaded boxes. Homologous recombination resulted in replacement of the 5′-region of the AFP intragenic sequence (exon 1, except the first 73 bp, exons 2 and 3 in pAFP K.O-1 or exons 2, except the first 13 bp, and exon 3 in pAFP K.O-2) by an IRES lacZ/neo cassette. The 5′-probe used to screen for homologous recombination is represented by a black box. (B) Photograph of EBs containing either pAFP K.O-1 (top) or pAFP K.O-2 (bottom) after lacZ staining. The EBs in panel I were stained at the time of appearance of autonomous beating cells. The EBs in panel II were stained 5 days later.
DISCUSSION
In a previous study we suggested that a region located in the first intron of the mouse AFP gene and containing a methylable cytosine at +347 bp behaves as an enhancer (13). We report here that this cytosine is included in a CACCC box and that a 44 bp segment centered on this motif binds nuclear factors. One of these factors is BKLF, a transcription factor which belongs to the Sp/XKLF family and binds the CACCC motif with high affinity (31). Another factor is YY1, a multifunctional zinc finger protein that activates or represses gene transcription in a promoter context-dependent manner and is capable of acting as an initiator of transcription (46). In transfection experiments, the 44 bp segment stimulates the activity of the upstream AFP promoter but a mutation destroying the CACCC motif eliminates the binding of BKLF and abrogates this stimulatory activity. Furthermore, the mutated element, on the one hand, acquires an inhibitory activity and, on the other hand, binds YY1 more efficiently. These observations strongly suggest that BKLF acts as a positive factor whereas YY1 acts as an inhibitor and that the binding of YY1 is hindered by BKLF.
The first intron of the mouse AFP gene also contains two TATA boxes, downstream of the BKLF and YY1 binding sites, suggesting that this region could thus behave as a promoter. In recent years several examples of alternative promoters that direct the transcription of multiple RNA transcripts from a single gene have been described (see for example 50–54). We thus tested whether an alternative promoter exists in the AFP gene and using RT–PCR we could demonstrate the existence of a previously undetected mRNA isoform. The combination of primer extension analysis and protection assay has revealed that transcription initiates 66 bp downstream of a TATA sequence located at position +475 (according to the numbering of ref. 47). These results thus show that an alternative promoter is located in the region known as intron 1 (promoter P2 at +541). We showed that production of the novel transcript is, as for the traditional one, regulated in a tissue-specific and developmental manner. It is present in the yolk sac, less abundant in the fetal liver and absent in the adult liver. The use of these two alternative promoters will result in the production of at least two mRNA isoforms that differ in their 5′-untranslated region. A putative AUG codon was found 297 bp downstream of the new transcriptional start site and is in-frame with the traditional exon 2, located at +838 bp. The novel variant mRNA must thus contain a relatively long 5′-untranslated region and the N-terminal region of the corresponding protein could be different from that of the traditional AFP, at least before processing. Different 5′ mRNA sequences can lead to variations in translation efficiencies of the mRNA, different cellular distributions of the proteins and finally to proteins with different functions.
From a study of transgenic mice generated with a transgene lacking the AFP exonic and intronic sequences, it has been concluded that proper transcriptional control of the AFP gene does not require intragenic sequences (17). However, this analysis started at day 16.5, i.e. with a relatively long delay after the AFP gene had been switched on. We have shown that EBs carrying a knock-in gene containing both the P1 and P2 promoters produce transcript which is turned on 5 days before that of EBs carrying a knock-in gene containing the P1 promoter only. The P2 promoter thus probably contributes to earlier gene expression. Knock-in mice carrying the knock-in genes with either the P1 promoter alone or with both the P1 and P2 promoters will be interesting to analyze in this respect.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr M. Crossley for the BKLF antibody. This work was supported by the Association contre le Cancer and the CGER-Assurances. S.S. was supported by a FRIA fellowship and a Televie grant (FNRS, Belgium). C.S. is a Research Director of the FNRS (Belgium).
REFERENCES
- 1.Gibbs P.E., Zielinski,R., Boyd,C. and Dugaiczyk,A. (1987) Biochemistry, 26, 1332–1343. [DOI] [PubMed] [Google Scholar]
- 2.Minghetti P.P., Ruffner,D.E., Kuang,W.J., Dennison,O.E., Hawkins,J.W., Beattie,W.G. and Dugaiczyk,A. (1986) J. Biol. Chem., 261, 6747–6757. [PubMed] [Google Scholar]
- 3.Nishio H., Heiskanen,M., Palotie,A., Belange,L. and Dugaiczyk,A. (1996) J. Mol. Biol., 259, 113–119. [DOI] [PubMed] [Google Scholar]
- 4.Ray K., Wang,X., Zhao,M. and Cooke,N.E. (1991) J. Biol. Chem., 266, 6221–6229. [PubMed] [Google Scholar]
- 5.Witke W.F., Gibbs,P.E., Zielinski,R., Yang,F., Bowman,B.H. and Dugaiczyk,A. (1993) Genomics, 16, 751–754. [DOI] [PubMed] [Google Scholar]
- 6.Chen H., Egan,J.O. and Chiu,J.-F. (1997) Crit. Rev. Eukaryot. Gene Expr., 7, 11–41. [DOI] [PubMed] [Google Scholar]
- 7.Lazarevich N.L. (2000) Biokhimiya, 65, 139–158. [PubMed] [Google Scholar]
- 8.Belayew A. and Tilghman,S.M. (1982) Mol. Cell. Biol., 2, 1427–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruoslahti E. and Sepällä,M. (1979) Adv. Cancer Res., 29, 276–336. [DOI] [PubMed] [Google Scholar]
- 10.Sell S. and Becker,F. (1978) J. Natl Cancer Inst., 60, 19–26. [DOI] [PubMed] [Google Scholar]
- 11.Godbout R., Ingram,R.S. and Tilghman,S.M. (1986) Mol. Cell. Biol., 6, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molné M., Houart,C., Szpirer,J. and Szpirer,C. (1989) Nucleic Acids Res., 17, 3447–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Opdecamp K., Rivière,M., Molné,M., Szpirer,J. and Szpirer,C. (1992) Nucleic Acids Res., 20, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott R.W. and Tilghman,S.M. (1983) Mol. Cell. Biol., 3, 1295–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Widen S.G. and Papaconstantinou,J. (1986) Proc. Natl Acad. Sci. USA, 83, 8196–8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krumlauf R., Hammer,R.E., Tilghman,S.M. and Brinster,R.L. (1985) Mol. Cell. Biol., 5, 1639–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spear B.T. (1994) Mol. Cell. Biol., 14, 6497–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godbout R., Ingram,R.S. and Tilghman,S.M. (1988) Mol. Cell. Biol., 8, 1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groupp E.R., Crawford,N. and Locker,J. (1994) J. Biol. Chem., 269, 22178–22187. [PubMed] [Google Scholar]
- 20.Hammer R.E., Krumlauf,R., Camper,S.A., Brinster,R.L. and Tilghman,S.M. (1987) Science, 235, 53–58. [DOI] [PubMed] [Google Scholar]
- 21.Ramesh T.M., Ellis,A.W. and Spear,B.T. (1995) Mol. Cell. Biol., 15, 4947–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feuerman M.H., Godbout,R., Ingram,R.S. and Tilghman,S.M. (1989) Mol. Cell. Biol., 9, 4204–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D.E., Hoyt,P.R. and Papaconstantinou,J. (1990) J. Biol. Chem., 265, 3382–3391. [PubMed] [Google Scholar]
- 24.Apergis G.A., Crawford,N., Ghosh,D., Steppan,C.M., Vorachek,W.R., Wen,P. and Locker,J. (1998) J. Biol. Chem., 273, 2917–2925. [DOI] [PubMed] [Google Scholar]
- 25.Galerneau L., Pare,J.-F., Allard,D., Hamel,D., Levesque,L., Tugwood,J.D., Green,S. and Belanger,L. (1996) Mol. Cell. Biol., 16, 3853–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernier D., Thomassin,H., Allard,D., Guertin,M., Hamel,D., Blaquiere,M., Beauchemin,M., LaRue,H., Estable-Puig,M. and Belanger,L. (1993) Mol. Cell. Biol., 13, 1619–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henriette M.-F., Gabant,P., Dreze,P-L., Szpirer,C. and Szpirer,J. (1997) Folia Biol., 43, 5–13. [PubMed] [Google Scholar]
- 28.Scott R.W., Vogt,T.F., Croke,M.E. and Tilghman,S.M. (1984) Nature, 310, 562–567. [DOI] [PubMed] [Google Scholar]
- 29.Knowles B.B., Howe,C.C. and Aden,D.P. (1980) Science, 209, 497–499. [DOI] [PubMed] [Google Scholar]
- 30.Cereghini S., Raymondjean,M., Carranca,A.G., Herbomel,P. and Yaniv,M. (1987) Cell, 50, 627–638. [DOI] [PubMed] [Google Scholar]
- 31.Crossley M., Whielaw,E., Perkins,A., Williams,G., Fujiwara,Y. and Orkin,S.H. (1996) Mol. Cell. Biol., 16, 1695–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadonega J.T. and Tjian,R. (1986) Proc. Natl Acad. Sci. USA, 83, 5889–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luckow B. and Schütz,G. (1987) Nucleic Acids Res., 15, 5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorman C.M., Moffat,L.F. and Howard,B.H. (1982) Mol. Cell. Biol., 2, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nordeen S.K. (1988) Biotechniques, 6, 454–457. [PubMed] [Google Scholar]
- 36.Gabant P., Dreze,P-L., Van Reeth,T., Szpirer,J. and Szpirer,C. (1997) Biotechniques, 23, 938–941. [DOI] [PubMed] [Google Scholar]
- 37.de Wet J.R., Wood,K.V., Deluca,M., Helsinki,D.R. and Subramani,S. (1987) Mol. Cell. Biol., 7, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith A.G. (1991) J. Tissue Cult. Methods, 13, 89–94. [Google Scholar]
- 39.Robertson E.J. (1987) In Robertson,E.J. (ed.), Teratocarcinomas and Embryonic Stem Cells: A Practical Approach. IRL Press, Oxford, UK, pp. 71–151.
- 40.Forrester L.M., Nagy,A., Sam,M., Watt,A., Stevenson,L., Bernstein,A.L., Joyner,A.L. and Wurst,W. (1996) Proc. Natl Acad. Sci. USA, 93, 1677–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philipsen S. and Suske,G. (1999) Nucleic Acids Res., 27, 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner J. and Crossley,M. (1999) Trends Biochem. Sci., 24, 236–240. [DOI] [PubMed] [Google Scholar]
- 43.Hariharan N., Kelley,D.E. and Perry,R.P. (1991) Proc. Natl Acad. Sci. USA, 88, 9799–9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park D. and Atchinson,M.L. (1991) Proc. Natl Acad. Sci. USA, 88, 9804–9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi Y., Seto,E., Chang,L.-S. and Shenk,T. (1991) Cell, 67, 377–388. [DOI] [PubMed] [Google Scholar]
- 46.Shrivastava A. and Calame,K. (1994) Nucleic Acids Res., 22, 5151–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eiferman A., Young,P.R., Scott,R.W. and Tilghman,S.M. (1981) Nature, 294, 24–31. [Google Scholar]
- 48.Abe K., Niwa,H., Takiguchi,M., Mori,M., Abé,S.-I., Abe,K. and Yamamura,K.-I. (1996) Exp. Cell Res., 229, 27–34. [DOI] [PubMed] [Google Scholar]
- 49.Keller G.M. (1995) Curr. Opin. Cell Biol., 7, 862–869. [DOI] [PubMed] [Google Scholar]
- 50.Ayoubi T.A.Y. and Van De Ven,W.J.M. (1996) FASEB J., 10, 453–460. [PubMed] [Google Scholar]
- 51.Ben-Nehiah Y., Bernards,A., Paskind,M., Daley,G.Q. and Baltimore,D. (1986) Cell, 44, 577–586. [DOI] [PubMed] [Google Scholar]
- 52.Fautsch M.P., Vrabel,A., Subramaniam,M., Hefferen,T.E., Spelsberg,T.C. and Wieben,E.D. (1998) Genomics, 51, 408–416. [DOI] [PubMed] [Google Scholar]
- 53.Jackson P. and Baltimore,D. (1989) EMBO J., 8, 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shtivelman E., Lifshitz,B., Gale,R.P., Roe,B.A. and Canaani,E. (1986) Cell, 47, 277–284. [DOI] [PubMed] [Google Scholar]