Abstract
Sixteen long-range crosslinks are induced in Escherichia coli 16S rRNA by far-UV irradiation. Crosslinking patterns in two other organisms, Bacillus subtilis and Thermus aquaticus, were investigated to determine if the number and location of crosslinks in E.coli occur because of unusually photoreactive nucleotides at particular locations in the rRNA sequence. Thirteen long-range crosslinks in B.subtilis and 15 long-range crosslinks in T.aquaticus were detected by gel electrophoresis and 10 crosslinks in each organism were identified completely by reverse transcription analysis. Of the 10 identified crosslinks in B.subtilis, eight correspond exactly to E.coli crosslinks and two crosslinks are formed close to sites of crosslinks in E.coli. Of the 10 identified crosslinks in T.aquaticus, five correspond exactly to E.coli crosslinks, three are formed close to E.coli crosslinking sites, one crosslink corresponds to a UV laser irradiation-induced crosslink in E.coli and the last is not seen in E.coli. The overall similarity of crosslink positions in the three organisms suggests that the crosslinks arise from tertiary interactions that are highly conserved but with differences in detail in some regions.
INTRODUCTION
Intramolecular crosslinks in 16S rRNA provide useful information about proximity of specific nucleotides in the rRNA tertiary structure. This information is particularly important for crosslinks located in regions functionally important for translation. Escherichia coli 16S rRNA has been investigated previously to determine the location of UV-induced crosslinks (1–5). The occurrence of a crosslink must depend on a combination of factors, including the inherent photoreactivity of the nucleotides that are joined, the correct distance and geometrical arrangement of the nucleotides and the molecular dynamics at the site of the crosslink. Thus, the appearance of a particular number of UV crosslinks in the E.coli 16S rRNA might not be reproduced in other organisms due to sequence or structural differences.
Comparative sequence analysis has demonstrated the universal conservation of a core secondary structure in the rRNAs (6). Within the central part of the 16S rRNA secondary structure there is maintenance of the arrangement and lengths of the base paired regions even though there are compensating changes in sequence (7). In addition, there is universal conservation of some sequence intervals in 16S rRNA. This implies that the elements containing these sequences are conserved as a requirement for proper function in translation (8). What has not been demonstrated yet is the extent to which the 16S rRNA tertiary structures are identical between different organisms. An example pertaining to the connection between RNA sequence and structure is found in RNase P RNA. Crosslinking analysis of E.coli and Bacillus subtilis RNase P RNAs has shown that even with two sequences and secondary structures that are dissimilar, tertiary structure contacts and crosslinks between these contacts (9) and the essential features of the function of the RNA are conserved. Within the bacterial kingdom the 16S rRNA sequence and secondary structure varies to a much lesser degree than that of RNase P RNA, but the ribosome still functions in a wide environmental range.
Sixteen UV-induced intramolecular RNA×RNA crosslinks have been identified in the E.coli 16S rRNA within the 30S subunit and 70S ribosome (4,5). This study repeats that analysis using ribosomes from B.subtilis and Thermus aquaticus and the distribution of crosslinks between these organisms is compared. Bacillus subtilis is a Gram-positive organism representing the Bacillus genus and has 78% 16S rRNA sequence identity to E.coli. Reconstitution of 30S subunits can be done with Bacillus stearathermophilus small subunit proteins and E.coli 16S rRNA, indicating the compatibility of the ribosomal proteins and RNA (10,11). Thermus thermophilus ribosomal subunits have been extensively studied using electron microscopy (12) and most recently the crystal structures of empty T.thermophilus 30S subunits have been solved at 5.5 (13) and 6.7 Å resolution (14) and the 70S ribosome·tRNA·mRNA complex at 7.8 Å resolution (15) has been determined. The T.aquaticus 16S rRNA sequence is 71% sequence identical to E.coli and closely resembles that of T.thermophilus, providing an example of a thermophilic organism whose ribosomes function at elevated temperatures.
The method used here for determining UV-induced crosslinks involves a step to separate and isolate crosslinked molecules by gel electrophoresis. Crosslinked nucleotides must be at least 50 nt apart in the primary sequence to be separated by this method, so all of the crosslinks are long range. Crosslinks have been classified as being of the secondary or tertiary type depending upon whether they occur between nucleotides that were already known to be close to each other by virtue of the secondary structure. However, UV-induced crosslinks do not normally occur between nucleotides within base paired regions (4), presumably due to the lack of favorable stacking between bases from opposite strands or efficient quenching of the activated state, so secondary structure crosslinks are still an indication of unusual geometry in that region. The tertiary structure crosslinks join nucleotides distant in the secondary structure, establishing evidence of the global 3-dimensional fold of the rRNA. If the crosslinking patterns are very dissimilar between organisms, this could be due to tertiary structure differences, primary sequence context effects or both and it would be necessary to examine the sequences involved at the sites or perform additional mutational experiments to determine the reasons for the different patterns. Differences in the secondary structure should not contribute to crosslinking differences since the present secondary structure has been rigorously secured by comparative sequence analysis (8). On the other hand, if the crosslink patterns are very similar, it will be evidence of conservation of the tertiary structure. In addition, analysis of crosslinks in organisms other than E.coli should give more evidence about the participation of purine and pyrimidine nucleotides in the photochemical processes. The data from these experiments indicate that while a number of secondary and tertiary crosslinks are conserved in the three organisms assayed, several crosslinks vary in position, perhaps due to local structural variations of unknown significance. Also, a large number of purines are involved in crosslinking, suggesting that purines, in the environment of a large, folded RNA, are more photoreactive than previous experiments have suggested.
MATERIALS AND METHODS
Preparation of ribosomes and UV crosslinking procedures
Frozen E.coli [MRE 600 (1/2 log) cells] were obtained from the Cell Culture Fermentation Facility, University of Alabama, Birmingham, AL. Bacillus subtilis (strain Marburg, ATCC 6051) and T.aquaticus (strain RF 4738, ATCC 25104) 70S ribosomes and ribosomal subunits were prepared according to Makhno et al. (16). Bacillus subtilis cells were grown in LB medium to an OD of 1 at 600 nm. Thermus aquaticus cells were grown in Castenholz TYE medium (ATCC culture medium 461) to an OD of 1 at 600 nm. Ribosomal subunits were purified and finally dissolved in activation buffer (20 mM Tris–HCl, pH 7.5, 20 mM MgCl2, 200 mM NH4Cl, 4 mM β-mercaptoethanol) in all instances. To minimize rRNA degradation, T.aquaticus 70S ribosomes were salt washed but not disassociated and reassociated. Escherichia coli and B.subtilis 70S ribosomes were prepared by reassociating equimolar amounts of 30S and 50S subunits and were free of mRNA or tRNA. Irradiation of the ribosomes and RNA purification were done as previously described (4). Agarose gels (1%) were used to isolate 16S rRNA before 5′-end-labeling with [γ-32P]ATP by T4 polynucleotide kinase or 3′-labeling by ligation to [5′-32P]pCp.
Crosslinked 16S rRNA was separated by gel electrophoresis on gels made with 3.6% acrylamide:bis-acrylamide (70:1), 8.3 M urea and BTBE buffer [30 mM bis(2-hydroxyethyl)imino tris(hydroxymethyl)methane, 30 mM boric acid, 2.5 mM EDTA, pH 6.8] as previously described (4). For analysis of crosslinking sites, the location of the bands containing uncrosslinked and crosslinked 16S rRNA were detected with a phosphorimager and bands were cut out and eluted by ultracentrifugation through cushions containing 2 M CsCl, 0.2 M EDTA, pH 7.4, for 12 h at 40 000 r.p.m. (17). RNA pellets were redissolved in 250 µl H2O, phenol extracted and re-precipitated before further analysis.
Determination of crosslinked site identity and frequency
The crosslinking sites in separated 16S rRNA were found by primer extension analysis using DNA primers complementary to regions throughout the respective sequences of 16S rRNA (4). For B.subtilis and T.aquaticus 11 primers corresponding to the same regions as analyzed in E.coli were synthesized.
Primary sequences and secondary structures
Primary sequences for E.coli, B.subtilis and T.aquaticus and secondary structures for E.coli and B.subtilis were taken from the Ribosomal Database Project II (RDP) (http://www.cme.msu.edu/rdp/ ) (18). The sequence and secondary structure diagram for T.aquaticus was modified from the secondary structure diagram of T.thermophilus, also taken from the RDP.
RESULTS
Bacillus subtilis 16S rRNA was chosen in this experiment for the degree of sequence similarity with E.coli 16S rRNA and the environmental similarity in which the ribosomes of these two organisms function. Bacillus subtilis is a Gram-positive organism with a 16S rRNA 1550 nt long, compared to 1542 nt in E.coli. It has a G+C content (55%) that is similar to that of E.coli (54%) and the two organisms are 78% 16S rRNA sequence identical. Alternatively, T.aquaticus 16S rRNA was chosen because it is an extreme thermophile, whose ribosomal structure is stable in vivo at temperatures that are completely inimical to E.coli ribosome function. Thermus aquaticus 16S rRNA has a higher G+C content (63%), as expected for functionality at higher temperatures, is 71% sequence identical to E.coli and is shorter (at 1470 nt) than that of E.coli. The 16S rRNA sequence identity between B.subtilis and T.aquaticus is 72%.
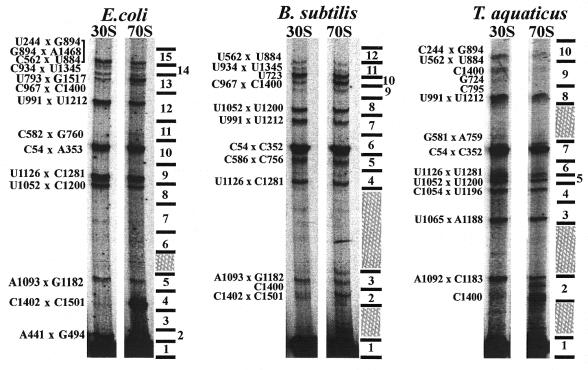
UV irradiation was done on 30S subunits and 70S ribosomes to induce crosslinks in 16S rRNA. Crosslinked 16S rRNA molecules containing loops ranging in size from ∼50 to 600 nt can be separated by gel electrophoresis. Molecules with larger loops are not separated from one another by this method and molecules with smaller loops are not separated from linear RNA. The presence of UV-induced crosslinks in the 16S rRNA is demonstrated by gel electrophoresis for the three organisms (Fig. 1). Similarities in the patterns of the crosslinked bands can be readily seen in the comparison of E.coli and B.subtilis 16S rRNAs, but the pattern of bands is different in T.aquaticus. Sixteen bands in the E.coli pattern were characterized previously. Thirteen and 15 bands are seen in the samples for B.subtilis and T.aquaticus, respectively.
Figure 1.
Separation and identification of crosslinks in 16S rRNA of E.coli, B.subtilis and T.aquaticus by gel electrophoresis. The crosslinked 16S rRNA from each organism is labeled above the corresponding set of crosslinks. Experiments performed with 30S subunits or 70S ribosomes are indicated above each lane. Crosslink identifications are labeled to the left of each set of lanes according to the E.coli secondary structure numbering. The corresponding B.subtilis and T.aquaticus numberings are listed in Table 1. Bands containing orphan stops in the reverse transcription primer extension analysis, indicating nucleotides without an identified crosslink partner, are labeled. Numbered divisions to the right of each set of lanes correspond to sections of preparative electrophoresis gels analyzed by reverse transcription, as shown in the numbered lanes in Figures 2 and 3. Areas of the gels indicated by crosshatching were not used for RNA isolation.
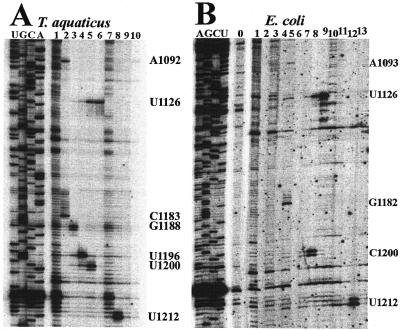
RNA was isolated from polyacrylamide gel bands as indicated in Figure 1 and reverse transcription experiments were done to identify the crosslinked nucleotides in each of the bands. DNA primers for sites throughout the rRNA sequence of each organism were used for overlapping reads of the RNA. A crosslinked nucleotide inhibits the insertion of a complementary nucleotide and causes an interruption in extension of the complementary DNA strand. A single reverse transcription stop at a specific nucleotide is frequently seen and this behavior has been determined previously to indicate a crosslink on the 5′ adjacent nucleotide for UV- (19,20), psoralen- (21) and s4U-induced (22) crosslinks. In other cases, a doublet stop is present because the structure of the crosslink allows pairing of a complementary nucleotide on the crosslinked nucleotide. In this case, the 5′ nucleotide of the doublet is the crosslinked nucleotide (21). One example of the reverse transcription of crosslinks in the interval nt 1075–1215 (E.coli numbering) is shown in Figure 2 for T.aquaticus and E.coli. Comparison of the reverse transcription pattern from RNA in a particular band to that from RNA in other bands (containing other crosslinks) or RNA in the irradiated but uncrosslinked band (fraction 1 in all three samples) allows the identification of specific reverse transcription stops attributable to the crosslink of that particular band. Reverse transcription stops due to other types of UV-induced changes and post-transcriptionally modified nucleotides will be seen in all of the samples, so they will not be mistaken for long-range crosslinking sites. Additional criteria for crosslink assignment are that a crosslinking site is seen with different primers to rule out reverse transcription priming artifacts and that the mobility of the RNA species should be consistent with the loop size predicted by the participating nucleotides.
Figure 2.
Reverse transcription analysis of the pattern of crosslinks in 30S ribosomal subunits of T.aquaticus and E.coli. The primer extension analysis corresponding to nt 1060–1196 of T.aquaticus 16S rRNA is shown in (A). Primer extension analysis corresponding to E.coli nt 1075–1215 is shown in (B). The crosslinked nucleotides are indicated to the right of each panel and are numbered according to the E.coli secondary structure. In both (A) and (B) primer extension stops 1 nt before the labeled crosslinked nucleotide. Reverse transcription samples in the left part of each panel were prepared with total unirradiated 16S rRNA with ddTTP, ddCTP, ddGTP or ddATP or without dideoxynucleotide (lane 0 in B). Lanes numbered 1–10 on the right of (A) and 1–13 on the right of (B) were prepared with irradiated 16S rRNA from 30S ribosomes isolated from crosslinked 16S rRNA preparative samples of either T.aquaticus or E.coli, as indicated in Figure 1.
Reverse transcription performed with RNA from all of the visible crosslinked bands shown in Figure 1 resulted in the complete identification of 10 crosslinks each in B.subtilis and T.aquaticus. In the remaining bands in B.subtilis and T.aquaticus only one side of the crosslink (orphan stops) could be determined by primer extension, perhaps due to masking of crosslinked nucleotide stops by natural stops in the reverse transcription pattern, masking due the presence of post-transcriptional RNA modifications or the inability of AMV reverse transcriptase to arrest at some types of crosslinks.
The majority of the crosslinks in B.subtilis and T.aquaticus involve the same nucleotides, or nearly the same nucleotide, compared to E.coli when aligned with the E.coli 16S rRNA sequence and secondary structure (Table 1). For all of these sites the alignment is unambiguous and E.coli numbering will be used to describe all of the positions. Eight of the 10 crosslinks identified in B.subtilis involve crosslinked nucleotide pairs that are in exactly the same positions as in E.coli. In the first exception, crosslink C586×C756 in B.subtilis, the 5′- and the 3′-ends of the crosslink shift 5 and 3 nt, respectively, when compared to the equivalent E.coli crosslink (Fig. 3). This moves the location of the crosslink to a position a short distance away in the base paired stem from the related crosslink in E.coli. In the second exception in B.subtilis, crosslink C54×C352, the downstream participating nucleotide is at 352 rather than at 353, as seen in E.coli.
Table 1. Identification of crosslinks found in 16S rRNA in E.coli, B.subtilis and T.aquaticus ribosomes.
| Crosslink | Escherichia colia | Bacillus subtilisa | Thermus aquaticusa |
|---|---|---|---|
| 1 | C54×A353 | C54×C352 (55×358) | C54×C352 (55×348) |
| 2 | U244×G894 | C244×G894 (240–872) | |
| 3 | A441×G494 | ||
| 4 | U562×U884 | U562×U884 (568×892) | U562×U884 (546×862) |
| 5 | C582×G760 | C586×C756 (592×763) | G581×A759 (565×743) |
| 6 | G894×A1468 | ||
| 7 | C934×U1345 | U934×U1345 (944×1352) | |
| 8 | C967×C1400 | C967×C1400 (975×1407) | |
| 9 | U991×U1212 | U991×U1212 (999×1219) | U991×U1212 (969×1194) |
| 10 | U1052×C1200 | U1052×U1200 (1060×1206) | U1052×U1200 (1035×1181) |
| 11 | A1093×G1182 | A1093×G1182 (1101×1189) | A1092×C1183 (1075×1165) |
| 12 | U1126×C1281 | U1126×C1281 (1134×1288) | U1126×U1281 (1109×1263) |
| 13 | C1402×C1501 | C1402×C1501 (1409×1510) | |
| 14 | C1400×C1501 | ||
| 15 | C1397×U1495 | ||
| 16 | U793×G1517 | ||
| 17 | C1054×A1196b | C1054×U1196 (1037×1178) | |
| 18 | U1065×A1188 (1048×1170) | ||
| 19c | G724×? (708×?) | ||
| 20c | C795×? (779×?) | ||
| 21c | C1397×? (1378×?)d | ||
| 22c | C1400×? (1407×?) | C1400×? (1381×?)d | |
| 23c | U723×? (729×?) |
aNumbering is according to the primary sequence of E.coli, with the numbering in B.subtilis and T.aquaticus in parentheses. Underlined crosslinks are at identical locations in the respective organisms.
bCrosslink produced in E.coli with UV laser irradiation (T.Shapkina, S.Franzen and P.Wollenzien, unpublished data).
cCrosslinks are partially identified in bands where only one band-specific reverse transcription stop was identified.
dCrosslink observed at 70 but not at 4°C.
Figure 3.
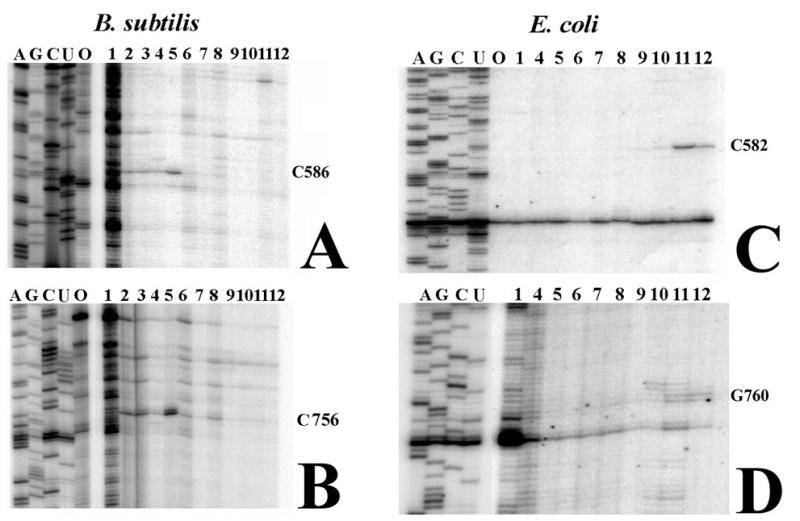
Reverse transcription analysis of crosslink 5 in 30S ribosomal subunits of B.subtilis and E.coli. The primer extension analysis corresponding to nt 564–616 of B.subtilis 16S rRNA is shown in (A). Primer extension analysis corresponding to nt 740–787 is shown in (B). In both (A) and (B) lane 5 shows a primer extension stop 1 nt before the crosslinked nucleotide. The crosslinked nucleotides are indicated to the right of each panel. Primer extension analysis of E.coli 16S rRNA nt 554–612 is shown in (C). Primer extension analysis of nt 740–780 is shown in (D). In (C) and (D) lane 11 contains the primer extension stop corresponding to each end of the crosslink. Numbering is according to the E.coli secondary structure. Reverse transcription samples on the left of each panel were prepared with total unirradiated 16S rRNA with ddTTP, ddCTP, ddGTP or ddATP or without dideoxynucleotide (lane 0). Lanes numbered 1–12 on the right part of (A) and (B) and 1–12 on the right of (C) and (D) were prepared with irradiated 16S rRNA from 30S ribosomes isolated from crosslinked 16S rRNA preparative samples of B.subtilis or E.coli, as indicated in Figure 1.
Five of the 10 crosslinked nucleotide pairs identified in T.aquaticus correspond exactly to crosslinked nucleotide pairs in E.coli. Three other T.aquaticus crosslinks are located in similar positions as crosslinks in E.coli, but have either one or both crosslinking sites located up to several nucleotides adjacent to the nucleotide identified in E.coli. The 3′-end of the crosslink C54×C352 is 1 nt in the 5′ direction (placing it in the same location as in B.subtilis). Both ends of the G581×A759 crosslink in T.aquaticus are displaced 1 nt in the 5′ direction compared to the corresponding E.coli crosslink. Both the 5′- and 3′-ends of the T.aquaticus crosslink A1092×C1183 are 1 nt 5′, compared to the equivalent E.coli crosslink (Fig. 2). Two crosslinks seen in domain III (Fig. 2) in T.aquaticus 16S rRNA, C1054×U1196 (crosslink 17) and U1065×A1188 (crosslink 18), do not have corresponding crosslinks identified in E.coli in this experiment.
Several orphan stops were identified in both B.subtilis and T.aquaticus irradiation experiments. These are strong primer extension stops that occur in specific RNA fractions for which no partner can be found. RNA from two fractions, one in the upper half of the gel containing U723 (E.coli numbering) and one in the lower half containing C1400 (E.coli numbering), were seen in B.subtilis experiments. Three orphan stops were seen in the crosslinks from separated T.aquaticus 16S rRNA. These were C1400, G724 and C795 (E.coli numbering), respectively, and are indicated in Figure 1.
To determine if elevated temperature would alter the pattern of crosslinking of T.aquaticus 16S rRNA, the crosslinking experiment was repeated on T.aquaticus 30S subunits and 70S ribosomes at 70°C (Fig. 4). Because of RNA fragmentation, this crosslinking experiment could not be done at temperatures >70°C and could not be done with reassociated subunits (12,23). The frequency of crosslinking and locations of nucleotide participants determined by reverse transcription analysis (results not shown) were unchanged for the majority of the crosslinks, with the exception of two bands seen in the 70S ribosome sample irradiated at 70°C. These are noted by arrows in Figure 4. Primer extension analysis (results not shown) indicates that these bands contained stops at C1397 and C1400 (E.coli numbering). It has not been possible to identify second primer extension stops in the RNA for these fractions that would indicate partner crosslinking sites. Further, the 70S ribosomes used in the analysis may contain residual tRNA or mRNA and these may participate in crosslinks to the rRNA in this region (24; J.W.Noah and P.Wollenzien, unpublished observation). However, the crosslinks involving T.aquaticus C1397 and C1400 did not occur at the lower temperature in the same ribosome preparation and this indicates differences in the structure of the decoding region of T.aquaticus 16S rRNA that are temperature dependent.
Figure 4.
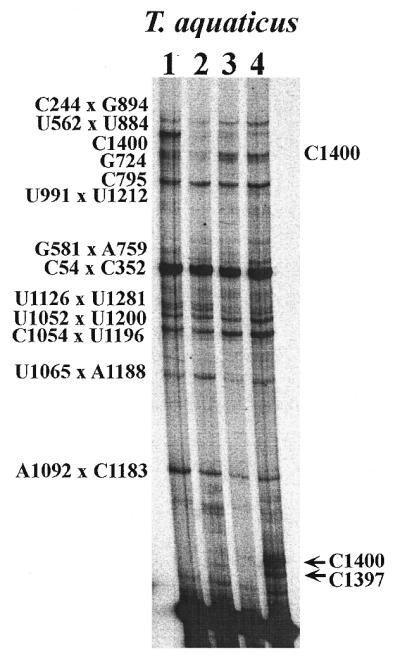
Comparison of crosslinks seen in T.aquaticus 16S rRNA from 30S and 70S ribosomes at 4 and 70°C. Lanes 1–4 show the crosslinks identified in each sample, according to the crosslinking conditions. Lane 1 shows the crosslinks seen in 30S subunits at 4°C; lane 2 shows the crosslinks seen in 70S ribosomes at 4°C; lane 3 show 30S subunits at 70°C; lane 4 shows 70S ribosomes crosslinked at 70°C. Crosslinks are identified to the left of the panel. The two arrows at the bottom right of the panel indicate two orphan reverse transcription stops identified in this region of the gel only in the 70S ribosome sample crosslinked at 70°C.
DISCUSSION
The pattern of UV-induced crosslinking in E.coli 16S rRNA indicates that a limited number of positions in the folded RNA exist where the arrangement allows crosslinking between nucleotides distant in the primary sequence. That result is repeated in B.subtilis and T.aquaticus, because 13 and 15 crosslinks are detected in the 16S rRNA in those organisms, respectively. Eight of the 10 completely identified crosslinks in B.subtilis are identical to those seen in E.coli and the remaining two are similar. Five of the 10 completely identified crosslinks in T.aquaticus are identical and three more are similar to those seen in E.coli. Further, of the 18 completely identified crosslinks in these organisms, 11 are common (identical or similar) in at least two organisms. Of these, seven are secondary structure crosslinks and four are tertiary structure crosslinks. This gives a lower estimate for the similarity of the crosslink patterns because three of the E.coli crosslinks either were made under conditions different from those described here or were determined by techniques that supplement reverse transcription analysis. Also, five other crosslinks in B.subtilis and T.aquaticus are incompletely determined, but the partial information for them again suggests similarity between the two organisms. Thus the similarity in the location of these indicates a tertiary structure that is highly conserved overall.
The results point to the utility of UV-induced crosslinking for these types of analyses. In spite of significant RNA sequence differences, many similarities in the RNA tertiary structure are readily revealed. This suggests that tertiary interactions specifically poise certain nucleotide pairs for relatively efficient UV-induced covalent bond formation. Importantly, these interactions seem to be specific, limited in number and the crosslinking efficiencies are good enough to allow their use in monitoring conformational changes.
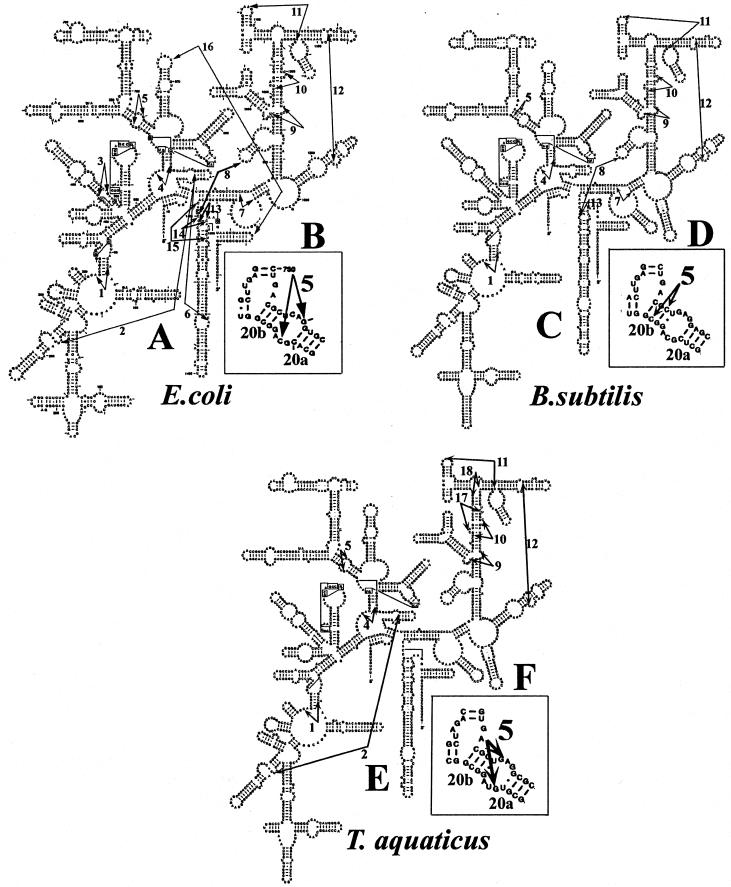
At the same time that similarities are seen, there are also some differences in the exact location of several crosslinks that indicate that certain regions exhibit variations in the details of the tertiary structure. In B.subtilis and T.aquaticus C54×C352 (crosslink 1) differs from a crosslink found in the same region of E.coli by 1 nt. In a second exception the B.subtilis C586×C756 and the T.aquaticus G581×A759 crosslinks (crosslink 5) correspond to E.coli crosslink C582×G760, but each of the versions occurs in a slightly different location in base paired regions 20a and 20b (see Fig. 5). The 20a/20b RNA region is found in the platform region in the 30S subunit (13; M.A.Dolan, P.Babin and P.Wollenzien, unpublished observations). The platform region in the 30S subunit in E.coli undergoes structural changes during subunit association (25) and IF-3 binding (24,26–28). These UV crosslinking results indicate that a slightly different tertiary arrangement may be present in this region in the three organisms, even in empty non-working ribosomes. In the last example of a crosslink difference, the T.aquaticus crosslink A1092×C1183, both nucleotides are 5′ in the sequence compared to the E.coli version of the same crosslink.
Figure 5.
Crosslinks mapped onto the secondary structure of 16S rRNA in E.coli, B.subtilis and T.aquaticus. (A, C and E) The complete secondary structure of the respective 16S rRNAs. (B) A detailed view of nt 750–764 and 577–589 in E.coli, which is the region containing crosslink 5 (C582×G760). The corresponding regions in B.subtilis and T.aquaticus, shown in (D) and (F), indicate the change in position of crosslink 5. Base paired regions 20a and 20b are indicated in panels (B), (D) and (F). Nucleotide×nucleotide crosslinks are indicated with large arrows, numbered 1–18. Numbered crosslinks correspond to those listed in Table 1.
Two crosslinks found in the decoding region of E.coli, C967×C1400 and C1402×C1501, have also been identified in B.subtilis. However, these crosslinks were not unambiguously identified in T.aquaticus. A primer extension stop is seen at the nucleotide that corresponds to E.coli C1400 in T.aquaticus samples isolated from the region of the preparative gel that should contain crosslinks between nucleotides distant in the primary sequence. However, a stop that would correspond to E.coli position C967 is not seen. This may be due to an intense natural stop in the 967 region in T.aquaticus in the primer extension analysis that masks adjacent nucleotides (data not shown), making crosslink identification difficult. The crosslink equivalent to E.coli C1402×C1501 does not occur in T.aquaticus at 4°C. However, crosslinks involving the decoding region were seen in T.aquaticus at 70°C (Fig. 4), so the absence may be related to structure dynamics. Alternatively, the decoding region in the 16S rRNA is heavily post-transcriptionally modified and these may affect photochemistry.
In terms of the photochemistry, the crosslinking is apparently less sensitive to base identity than would be predicted from DNA experiments. Theoretical and experimental work has shown that the lone electrons in the excited S1 or T1 states are localized around the C(5)=C(6) double bond in pyrimidines (29–31), which explains in part the photoreactivity of pyrimidines. Purine crosslinks have also been characterized, although to a much lesser extent. The quantum yields for purine photoreactivity in short DNA oligomers are almost 10-fold lower than that of pyrimidines (32) and purine crosslinks are generally in the minority, as would be statistically expected in a mixed purine/pyrimidine system (33).
There are three instances that illustrate that purines as well as pyrimidines are efficient participants in UV-induced crosslinking in 16S rRNAs. The first is the C54×A353 crosslink in E.coli, which involves a C×A crosslink. However, the crosslink (using E.coli numbering) in B.subtilis and T.aquaticus is C54×C352 and involves a C×C crosslink (see Table 1). Thus, in E.coli the UV-induced crosslink involves a purine residue, even though the adjacent nucleotide is C352 and C352 is involved in the crosslink in B.subtilis and T.aquaticus. The second example is in the crosslink found in the 582×760 region. There are three different versions of the crosslink found here (all E.coli numbering): C582×G760 (E.coli), C586×C756 (B.subtilis) and G581×A759 (T.aquaticus). Thus three different versions of crosslinks between the interacting strands in this region are seen even though the sequences in the three organisms are nearly identical and the pattern does not depend on the involvement of a pyrimidine nucleotide in the crosslink. Lastly, identical crosslinks at A1093×G1182 are found in E.coli and B.subtilis, but the crosslink in T.aquaticus is instead A1092×C1183 (E.coli numbering).
The summary for the 16S rRNA crosslinks in these organisms is 24 pyrimidine×pyrimidine, seven pyrimidine×purine and five purine×purine crosslinks. These numbers suggest a strong preference for pyrimidine×pyrimidine crosslinks, but sufficient examples of purine-containing crosslinks exist to suggest that the influencing factors of the orientation of the involved nucleotides, the degree of dynamic energy associated with those nucleotides and the surrounding electronic environment are important (34). This indicates that in the context of ribosome structure, purine residues may be arranged in interactions that change their electronic features, increase the efficiency of internucleotide reactions or decrease the rate of loss of excitation energy through energy transfer. In a number of instances in other RNAs and ribonucleoproteins, purines have been found as UV crosslink participants. These include specific and high yield G×U products in potato spindle tuber viroid RNA and HeLa 5S rRNA (35), a G×U minor product after irradiation of pre-tRNA and RNase P (36), a specific and high yield A×A product in the Tetrahymena group I ribozyme (37), specific and high yield G×U and A×U products in the hairpin ribozyme and dimerized hairpin ribozyme (38), a specific G×C product in U6 snRNA (39) and a specific G×A product in annealed U2 snRNA/U6 snRNA (40). Thus there are other examples in RNAs in which purine-containing photoproducts are specifically produced.
Acknowledgments
ACKNOWLEDGEMENTS
Our thanks are due to Dr James Brown (North Carolina State University) for supplying the strain of B.subtilis used in these experiments and for his advice and comments. This work was supported by National Institutes of Health Grant GM43237 and by a GAANN Fellowship to J.W.N.
REFERENCES
- 1.Atmadja J. and Brimacombe,R. (1985) Nucleic Acids Res., 13, 6919–6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmadja J., Stiege,W., Zobawa,M., Greur,B., Osswald,M. and Brimacombe,R. (1986) Nucleic Acids Res., 14, 659–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doring T., Greuer,B. and Brimacombe,R. (1992) Nucleic Acids Res., 20, 1593–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilms C., Noah,J.W., Zhong,D. and Wollenzien,P. (1997) RNA, 3, 602–612. [PMC free article] [PubMed] [Google Scholar]
- 5.Noah J. and Wollenzien,P. (1998) Biochemistry, 37, 15442–15448. [DOI] [PubMed] [Google Scholar]
- 6.Gutell R., Larsen,N. and Woese,C. (1994) Microbiol. Rev., 58, 10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noller H.F. (1993) In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World, Cold Spring Harbor Monograph Series no. 24. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 8.Gutell R.R. (1993) In Nierhaus,K.H., Franceschi,F. and Subramanian,A.R. (eds), The Translation Apparatus. Plenum Press, New York, NY, pp. 477–488.
- 9.Chen J.L., Nolan,J.M., Harris,M.E. and Pace,N.R. (1998) EMBO J., 17, 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nomura M. and Held,W.A. (1974) In Nomura,M., Tissieres,A. and Lengyel,P. (eds), Ribosomes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 193–223.
- 11.Nomura M. (1987) Cold Spring Harbor Symp. Quant. Biol., 52, 653–663. [DOI] [PubMed] [Google Scholar]
- 12.Harauz G., Letvenuk,L. and Flannigan,D. (1992) Biochim. Biophys. Acta, 1129, 207–214. [DOI] [PubMed] [Google Scholar]
- 13.Clemons W.M.J., May,J.L.C., Wimberly,B.T., McCutcheon,J.P., Capel,M.S. and Ramakrishnan,V. (1999) Nature, 400, 833–840. [DOI] [PubMed] [Google Scholar]
- 14.Tocilj A., Schlunzen,F., Janell,D., Gluhmann,M., Hansen,H.A.S., Harms,J., Bashan,A., Bartels,H., Agmon,I., Franceschi,F. and Yonath,A. (1999) Proc. Natl Acad. Sci. USA, 96, 14252–14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cate J.H., Yusupov,M.M., Yusupova,G.Z., Earnest,T.N. and Noller,H.F. (1999) Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- 16.Makhno V.I., Peshin,N.N., Semenkov,Y.P. and Kirillov,S.V. (1988) Mol. Biol., 22, 528–537. [PubMed] [Google Scholar]
- 17.Wilms C. and Wollenzien,P. (1994) Anal. Biochem., 221, 204–205. [DOI] [PubMed] [Google Scholar]
- 18.Maidak B.L., Cole,J.R., Parker,C.T.,Jr, Garrity,G.M., Larsen,N., Li,B., Lilburn,T.G., McCaughey,M.J., Olsen,G.J., Overbeek,R., Pramanik,S., Schmidt,T.M., Tiedje,J.M. and Woese,C.R. (1999) Nucleic Acids Res., 27, 171–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denman R., Negre,D., Cunningham,P.R., Nurse,K., Colgan,J., Weitzmann,C. and Ofengand,J. (1989) Biochemistry, 28, 1012–1019. [DOI] [PubMed] [Google Scholar]
- 20.Cunningham P.R., Nurse,K., Bakin,A., Weitzmann,C.J., Pflumm,M. and Ofengand,J. (1992) Biochemistry, 31, 12012–12022. [DOI] [PubMed] [Google Scholar]
- 21.Ericson G. and Wollenzien,P. (1988) Anal. Biochem., 174, 215–223. [DOI] [PubMed] [Google Scholar]
- 22.Lemaigre-Debreuil Y., Expert-Bezancon,A. and Favre,A. (1991) Nucleic Acids Res., 19, 3653–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khaitovich R., Mankin,A.S., Green,R., Lancaster,L. and Noller,H.F. (1999) Proc. Natl Acad. Sci. USA, 96, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapkina T.G., Dolan,M.A., Babin,P. and Wollenzien,P. (2000) J. Mol. Biol., 299, 617–630. [DOI] [PubMed] [Google Scholar]
- 25.Merryman C., Moazed,D., McWhirter,J. and Noller,H. (1999) J. Mol. Biol., 285, 97–105. [DOI] [PubMed] [Google Scholar]
- 26.Muralikrishna P. and Wickstrom,E. (1989) Biochemistry, 28, 7505–7510. [DOI] [PubMed] [Google Scholar]
- 27.Moazed D., Samaha,R., Gualerzi,C. and Noller,H. (1995) J. Mol. Biol., 248, 207–210. [DOI] [PubMed] [Google Scholar]
- 28.McCutcheon J.P., Agrawal,R., Philips,S., Grassucci,R., Gerchman,S., Clemons,W.,Jr, Ramakrishnan,V. and Frank,J. (1999) Proc. Natl Acad. Sci. USA, 96, 3401–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shulman R.G. and Rahn,R.O. (1966) J. Chem. Phys., 45, 2940. [DOI] [PubMed] [Google Scholar]
- 30.Snyder L.C., Shulman,R.G. and Neumann,D.B. (1970) J. Chem. Phys., 53, 256. [DOI] [PubMed] [Google Scholar]
- 31.Pullman B. (1968) Photochem. Photobiol., 7, 525. [Google Scholar]
- 32.Smith K.C. (1967) In Silini,G. (ed.), Radiation Research. North-Holland Publishing, Amsterdam, The Netherlands.
- 33.Patrick M.H. and Rahn,R.O. (1976) In Wang,S.Y. (ed.), Photochemistry and Photobiology of Nucleic Acids. Academic Press, New York, NY.
- 34.Gilbert A. and Baggott,J. (1991) Essentials of Molecular Photochemistry. Blackwell Scientific, Cambridge, MA.
- 35.Branch A.D., Benefeld,B.J. and Robertson,H.D. (1985) Proc. Natl Acad. Sci. USA, 82, 6590–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerrier-Takada C., Lumelsky,N. and Altman,S. (1989) Science, 246, 1578–1584. [DOI] [PubMed] [Google Scholar]
- 37.Downs W.D. and Cech,T.R. (1990) Biochemistry 29, 5605–5613. [DOI] [PubMed] [Google Scholar]
- 38.Butcher S.E. and Burke,J.M. (1994) Biochemistry 33, 992–999. [DOI] [PubMed] [Google Scholar]
- 39.Sun J.-S., Valadkhan,S. and Manley,J.L. (1998) RNA, 4, 489–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valadkhan S. and Manley,J.L. (2000) RNA, 6, 206–219. [DOI] [PMC free article] [PubMed] [Google Scholar]