Abstract
Spontaneous recombination between direct repeats at the adenine phosphoribosyltransferase (APRT) locus in ERCC1-deficient cells generates a high frequency of rearrangements that are dependent on the process of homologous recombination, suggesting that rearrangements are formed by misprocessing of recombination intermediates. Given the specificity of the structure-specific Ercc1/Xpf endonuclease, two potential recombination intermediates are substrates for misprocessing in ERCC1– cells: heteroduplex loops and heteroduplex intermediates with non-homologous 3′ tails. To investigate the roles of each, we constructed repeats that would yield no heteroduplex loops during spontaneous recombination or that would yield two non-homologous 3′ tails after treatment with the rare-cutting endonuclease I-SceI. Our results indicate that misprocessing of heteroduplex loops is not the major source of recombination-dependent rearrangements in ERCC1-deficient cells. Our results also suggest that the Ercc1/Xpf endonuclease is required for efficient removal of non-homologous 3′ tails, like its Rad1/Rad10 counterpart in yeast. Thus, it is likely that misprocessing of non-homologous 3′ tails is the primary source of recombination-dependent rearrangements in mammalian cells. We also find an unexpected effect of ERCC1 deficiency on I-SceI-stimulated rearrangements, which are not dependent on homologous recombination, suggesting that the ERCC1 gene product may play a role in generating the rearrangements that arise after I-SceI-induced double-strand breaks.
INTRODUCTION
Nucleotide excision repair (NER) is one of several DNA repair pathways for correcting abnormal DNA structures arising from DNA damage, replication errors or recombination processes (1–3). Bulky DNA lesions such as UV-induced pyrimidine dimers are preferentially removed by NER, which incises a single strand on either side of the lesion and removes it as part of a 24–32 nt fragment (4,5). The resulting gap is filled in by DNA synthesis and ligation (1). Damage recognition and strand incision in mammalian cells require several NER-specific proteins, including two structure-specific endonucleases: the dimeric Ercc1/Xpf endonuclease, which makes the 5′ incision, and the Xpg endonuclease, which makes the 3′ incision. Although these endonucleases efficiently excise bulky adducts, they also remove abasic sites, methylated bases and mismatches, albeit at lower efficiencies, suggesting a partial overlap in specificity with base excision repair and mismatch repair (6).
In addition to their specificity for abnormal bases, purified Ercc1/Xpf and Xpg endonucleases can cleave stem–loop and bubble substrates as well as substrates with flaps, splayed arms or protruding single strands (7–11). Some of these alternative DNA structures can arise naturally during recombination between homologous, but non-identical DNA sequences. In Saccharomyces cerevisiae the Rad1/Rad10 endonuclease, the yeast homolog of the Ercc1/Xpf endonuclease, participates in the removal of non-homologous 3′-ends in the single-strand annealing (SSA) pathway for homologous recombination (12–16).
Previous studies in hamster and mouse cells indicated that deficiency of ERCC1 does not substantially alter the frequencies of extrachromosomal (17) or chromosomal homologous recombination (18,19). In hamster cells, however, ERCC1 deficiency unexpectedly resulted in a high frequency of rearrangements at the adenine phosphoribosyltransferase (APRT) locus, which occurred during homologous recombination between direct repeats (19). When one of the direct repeats was removed so that the possibility for homologous recombination was eliminated, no rearrangements were formed. Given the specificity of the Ercc1/Xpf endonuclease and the structure of the repeated sequences, recombination-dependent rearrangements likely formed by misprocessing of intermediates in homologous recombination: either large heteroduplex loops (800 bp) formed between the non-identical tandem copies of the APRT gene or heteroduplex intermediates that carried non-homologous 3′ tails (19).
To determine whether misprocessing of heteroduplex loops in the absence of the Ercc1/Xpf endonuclease was responsible for recombination-dependent rearrangements, we constructed identical, tandemly duplicated APRT genes in ERCC1+ and ERCC1– hamster cells lines. Heteroduplexes formed between these repeats would carry at most a single base pair mismatch at the site of the marker mutation used to detect recombination events. Comparable chromosomal substrates that yield no non-homologous 3′ tails cannot be constructed, making it difficult to directly demonstrate a role for the Ercc1/Xpf endonuclease in processing non-homologous 3′ tails in spontaneous chromosomal recombination. Thus, we sought to show that the endonuclease is required for removal of non-homologous tails during double-strand-break (DSB) stimulated chromosomal recombination. Although ERCC1 deficiency in previous studies did not affect overall frequencies of homologous recombination, recombination substrates that would have required removal of non-homologous tails were not explicitly tested (17–19). Again, we constructed the same tandem duplication in ERCC1+ and ERCC1– cell lines. One of the duplicated genes carried the recognition site for the I-SceI endonuclease in an 800 bp stretch of non-homologous DNA. I-SceI-induced cleavage in the middle of the non-homologous DNA would require removal of two non-homologous tails in order to generate the crossover (pop-out) products that are characteristic of the SSA pathway for homologous recombination.
Our results indicate that misprocessing of heteroduplex loops is not the major source of recombination-dependent rearrangements during spontaneous recombination in ERCC1– cells. ERCC1– cells are, however, substantially defective in generating crossovers in response to I-SceI-induced DSBs. Thus, it is likely that the Ercc1/Xpf endonuclease is required for processing non-homologous 3′ tails and in the absence of a functional endonuclease recombination intermediates with 3′ tails are misprocessed to form rearrangements.
MATERIALS AND METHODS
Vectors
Vector pJLM1, which was used to create cell lines JAM1-41 (ERCC1+) and JAM1-77a (ERCC1–), was constructed by removing the GPT gene from pGS73 (20). Vector pAK45, which was used to generate cell lines AK784 (ERCC1+) and GSAK1 (ERCC1–), was constructed by inserting an 800 bp non-homologous DNA segment into the SalI and NotI sites in pGS101, a vector identical to pGS100 (20) except that the plasmid backbone is reversed in orientation. Recombinant PCR was used to amplify a non-homologous segment from intron 2 (nt 14928–15730) of the hypoxanthine-guanine phosphoribosyl transferase (HPRT) gene and to position an I-SceI recognition site in the center of the fragment. The I-SceI expression vector, pCMV-I-SceI, contains the structural gene for the I-SceI endonuclease driven by the cytomegalovirus (CMV) promoter (21). Transfection control vectors were pCMV-β-galactosidase (22), pEGFP-N1 (Clontech, Palo Alto, CA) and pcDNA4/Myc-His A (Invitrogen, Carlsbad, CA).
Cell construction
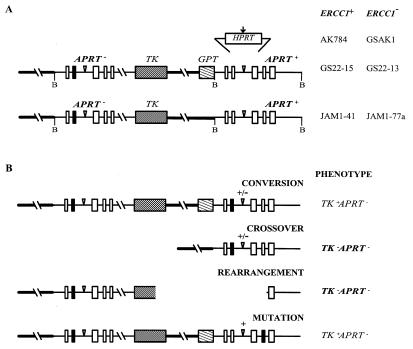
Vectors pJML1 and pAK45 were inserted into the APRT gene in RMP41 (ERCC1+) and RMP41-77 (ERCC1–) cells using FLP/FRT site-specific recombination (19,20). The recombinant cell lines, JAM1-41 (ERCC1+), JAM1-77a (ERCC1–), AK784 (ERCC1+) and GSAK1 (ERCC1–), were shown by Southern blotting and PCR analysis to have the molecular structures indicated in Figure 1A.
Figure 1.
Molecular structures of the substrates at the APRT locus in ERCC1+ and ERCC1– cell lines and of the products isolated in various selections. (A) The inserted HPRT sequence is shown above its site of insertion into the second intron of the downstream, functional APRT gene (the five exons of APRT are shown as boxes). The vertical arrow indicates the location of the I-SceI recognition site in the HPRT insert. Inverted triangles indicate the positions of the FRT recognition sequences. The upstream copy of APRT is non-functional by virtue of a truncated exon 5 and a single base substitution in exon 2 (filled box). The upstream and downstream copies share 6.8 kb of homology; 4.5 kb upstream of the APRT gene (thick line) and 2.3 kb of homology within the gene itself. Cleavage sites for the restriction enzyme BamHI (B), which was used in Southern analyses, are indicated. The hybridization probe corresponds to the downstream BamHI fragment that encompasses the APRT gene, but included no inserted sequences. (B) Types of possible APRT– products. Only TK–APRT– products (bold) were analyzed in these experiments; they were identified based on Southern patterns after BamHI cleavage and PCR analysis. Crossovers yielded a single BamHI cleavage fragment of 4.0 kb for JAM1-41 and JAM1-77a and a single BamHI cleavage fragment of 4.0 or 4.8 kb, depending on whether the insert was lost or retained, for AK784 and GSAK1 (+/– indicates presence or absence of the insert). Rearrangements gave a Southern pattern that did not correspond to crossovers (or any other class of products).
Recombination experiments
Parallel cultures of JAM1-41 and JAM1-77a were grown from initial populations of 100 cells. Cultures were selected for TK–APRT– colonies by growth in medium containing 0.3 µM 1-(2-deoxy-2-fluro-β-d)-arabinofuranosyl-5-iodouracil and 0.4 mM 8-azaadenine (23). A single colony was picked from each parallel culture to ensure that all analyzed colonies arose independently.
Uncut pCMV-I-SceI or pCMV-β-galactosidase was transfected into GSAK1 and AK784 cells at 0.25 µg/well in 24-well plates, using the cationic lipid vesicle preparation Lipofectamine as recommended by the manufacturer (Gibco BRL, Rockville, MD). After transfection, cells were grown for 5–7 days in non-selective medium to allow expression of the recombinant phenotypes. Cells were then replated in medium containing 0.3 µM FIAU to select for TK– cells, in 0.4 mM 8-azaadenine for APRT– cells or in medium containing both drugs for TK–APRT– cells (23). To ensure that independent I-SceI-induced events were characterized, only one colony was picked from each transfection. Rates of recombination were measured by fluctuation analysis (19).
Transfection efficiency in ERCC1+ and ERCC1– cells was measured by gene expression and by stable transformation using plasmid markers. Transfection of pEGFP-N1 followed by FACS analysis after 18 h indicated that 68 ± 7% of AK784 (ERCC1+) cells and 52 ± 2% of GSAK1 (ERCC1–) cells were fluorescent (average of four transfections). Transfection of pcDNA4/Myc-His A and selection for zeocin resistance indicated that 0.50 ± 0.09% of AK784 (ERCC1+) and 0.36 ± 0.04% of GSAK1 (ERCC1–) cells were stably transformed (average of five transfections).
DNA analysis
One colony from each parallel culture and drug selection was expanded in non-selective medium, DNA was isolated and the structure of the APRT locus was determined by Southern blotting, using the 4.0 kb BamHI fragment of the APRT gene as a hybridization probe, and by PCR analysis (23). Primer sequences are available on request.
RESULTS
Rearrangements in ERCC1+ and ERCC1– cells
The tandem duplications in JAM1-41 (ERCC1+) and JAM1-77a (ERCC1–) share 6.8 kb of homology that is identical except for a single nucleotide difference, the APRT-inactivating mutation in exon 2 of the upstream copy (Fig. 1A). Recombination-dependent rearrangements were originally demonstrated in GS22-13 (ERCC1–) cells, which carry an identical tandem duplication except for the presence of the GPT gene in the downstream copy (Fig. 1A; 19). In those studies selection for TK–APRT– cells yielded about equal numbers of crossovers and rearrangements (Fig. 1B and Table 1). In the absence of the GPT gene, heteroduplexes that form during homologous recombination would have at most a single base pair mismatch, which is repaired inefficiently by NER (6). Thus, if misprocessing of heteroduplex loops was the basis for recombination-dependent deletion formation in ERCC1– cells, rearrangements should be rare in JAM1-77a cells.
Table 1. Analysis of TK–APRT– colonies.
| Cell line | TK–APRT– coloniesb | ||||
|---|---|---|---|---|---|
| – I-SceI | + I-SceI | ||||
| C | R | C | R | ||
| GS22-15a | ERCC1+ | 72 | 0 | ||
| JAM1-41 | ERCC1+ | 49 | 0 | ||
| AK784 | ERCC1+ | 16 | 0 | 15 | 8 |
| GS22-13a | ERCC1– | 24 | 23 | ||
| JAM1-77a | ERCC1– | 13 | 19 | ||
| GSAK1 | ERCC1– | 3 | 17 | 10 | 12 |
aData are from Sargent et al. (19). GS22-13 and GS22-15 have tandem duplications identical to AK784 and GSAK1 except that they do not carry the 800 bp segment of non-homology or the I-SceI site in the second intron of APRT.
bColonies were classified as crossovers (C) or rearrangements (R) based on Southern blotting and PCR analyses.
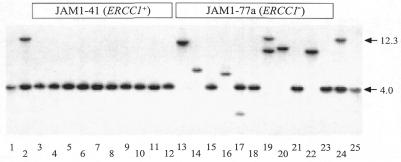
Independent TK–APRT– colonies from JAM1-41 and JAM1-77a cells were analyzed by Southern blotting (Fig. 2). Crossovers were expected to have a single band at 4.0 kb, whereas rearrangements yield an unpredictable pattern. Rearrangements were not detected among the colonies derived from ERCC1+ JAM1-41 cells, however, they were as common among the colonies derived from ERCC1– JAM1-77a cells as they were in the original ERCC1– GS22-13 cells, which carried the GPT gene (Fig. 2 and Table 1). Most of the rearrangements had a single novel band by Southern blotting, consistent with a simple deletion encompassing portions of the APRT and TK genes. Two of the 19 rearrangements, however, had two bands, suggesting that more complex rearrangements, such as translocations or insertions, can arise during spontaneous recombination in ERCC1– cells. The high fraction of rearrangements among TK–APRT– progeny arising from identical tandem duplications indicates that misprocessing of large heteroduplex loops cannot be the major cause of recombination-dependent rearrangements in ERCC1– cells.
Figure 2.
Southern blot analysis of DNA from TK–APRT– colonies isolated from JAM1-41 (ERCC1+) and JAM1-77a (ERCC1–) cells. The structure of the tandem duplication along with the locations of BamHI sites is shown in Figure 1A. The downstream copy of APRT in the parental structure gives rise to a 4.0 kb fragment. Crossover products retain only the downstream copy of the APRT gene. The hybridization probe corresponds to the 4.0 kb BamHI fragment of the APRT gene. Lanes 1 and 25 show DNA from a control cell line that has a single copy of the APRT gene and gives a band at 4.0 kb; lanes 2 and 24 show DNA from the parent cell line, which gives rise to bands at 4.0 and 12.3 kb. Lanes 3–12 are TK–APRT– colonies from JAM1-41; lanes 13–23 are TK–APRT– colonies from JAM1-77a.
Experimental design to detect the role of Ercc1p in removal of non-homologous tails
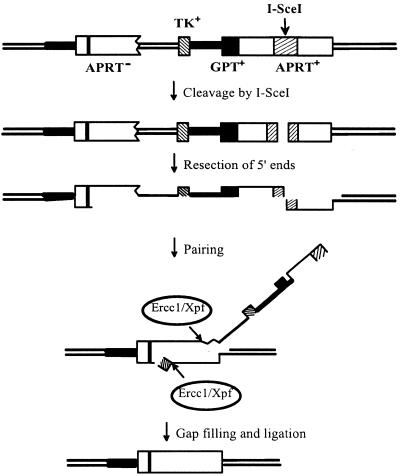
The mechanism of spontaneous homologous recombination on chromosomes is unknown, but is unlikely to be the same as that induced by endonuclease-mediated DSBs (24). Nevertheless, we can test the requirement for Ercc1p in removal of non-homologous 3′ tails by introducing DSBs and assaying for formation of crossovers (pop-outs) between directly repeated sequences by the SSA pathway of homologous recombination (15). In this pathway a 5′→3′ exonuclease removes one strand from each side of a DSB until homologous single-stranded regions are exposed, which can then pair. For this paired structure to be resolved, non-homologous single-stranded tails must be removed so that the gapped intermediate can be filled in by priming DNA synthesis from the 3′-ends (Fig. 3). In yeast such 3′ protruding non-homologous tails are removed by the Rad1/Rad10 endonuclease and in its absence DSB-induced crossover formation is substantially reduced (15). If the Ercc1/Xpf endonuclease carries out the analogous function in mammalian cells, then crossovers should be reduced in ERCC1– cells relative to ERCC1+ cells.
Figure 3.
Generation of a crossover recombinant by the SSA pathway of homologous recombination after cleavage of the APRT tandem duplication by I-SceI. The positions of the GPT and TK genes are indicated. The duplicated APRT genes are shown as open boxes; the vertical bar in the upstream APRT gene represents the inactivating exon 2 mutation; the hatched segment in the downstream APRT gene represents the 800 bp segment of non-homologous DNA derived from the HPRT gene. Thick lines represent a 4.5 kb portion of chromosomal DNA (not shown to scale) that is also duplicated in the tandem duplication at the APRT locus.
To ensure a uniform population of SSA heteroduplex intermediates with two non-homologous tails, we inserted into the downstream copy of APRT an 800 bp segment of non-homologous HPRT DNA carrying a centrally located recognition sequence for the rare-cutting endonuclease I-SceI. By expressing the I-SceI endonuclease in cells, homologous recombination can be initiated at a specific site such that heteroduplex intermediates formed by SSA will carry two non-homologous tails (Fig. 3).
Spontaneous recombination in ERCC1+ and ERCC1– cells
To assess whether the 800 bp HPRT insert had any significant effect on the spontaneous rates at which the TK–, TK–APRT– and APRT– phenotypes were generated (predominantly by homologous recombination), we measured the rates by fluctuation analysis. The rates in AK784 (ERCC1+) and GSAK1 (ERCC1–) cells were similar to previous measurements in GS22-15 (ERCC1+) and GS22-13 (ERCC1–) cells, indicating that the 800 bp HPRT segment had no substantial influence on spontaneous recombination at the APRT locus (Table 2). As noted previously, there is a small (2- to 4-fold) increase in rates for all phenotypes in ERCC1– cells (19).
Table 2. Rates at which selectable phenotypes are generated in ERCC1+ and ERCC1– cells.
| Selection | Rates (×10–7) | |||
|---|---|---|---|---|
| ERCC1+ | ERCC1– | |||
| GS22-15a | AK784b | GS22-13a | GSAK1b | |
| TK–APRT– | 1.4 ± 0.5 | 1.1 | 6 ± 3 | 1.8 |
| TK– | 15 ± 3 | 17 | 48 ± 23 | 62 |
| APRT– | 14 ± 3 | 6.9 | 31 ± 25 | 42 |
aData for GS22-13 and GS22-15 are from Sargent et al. (19). GS22-13 and GS22-15 have tandem duplications identical to AK784 and GSAK1 except that they do not carry the 800 bp segment of non-homology or an I-SceI site in the second intron of APRT.
bData for AK784 and GSAK1 are from a single 12 pot fluctuation experiment analyzed to determine recombination rates, as described previously (23,24).
I-SceI-stimulated crossovers in ERCC1+ and ERCC1– cells
To determine whether Ercc1p is required for removal of non-homologous tails during homologous recombination, we measured the ability of I-SceI endonuclease to stimulate crossovers in AK784 (ERCC1+) and GSAK1 (ERCC1–) cells. The I-SceI expression vector, pCMV-I-SceI, or a control plasmid was transfected into AK784 and GSAK1 cells and TK–APRT– cells were selected. I-SceI expression in AK784 (ERCC1+) cells stimulated TK–APRT– colony formation >1000-fold, whereas its expression in GSAK1 (ERCC1–) cells stimulated TK–APRT– colony formation an average of only 14-fold (Table 3). Because the background frequency of TK–APRT– cells is higher in GSAK1 (ERCC1–) cells, reflecting their higher rates of formation (Table 2), it is more appropriate to compare the stimulated levels, which were ∼20-fold higher in AK784 (ERCC1+) cells than in GSAK1 (ERCC1–) cells. These results are consistent with Ercc1p playing a role in removal of non-homologous tails during homologous recombination (Fig. 3).
Table 3. I-SceI-stimulation of TK–APRT– colony formation in ERCC1+ and ERCC1– cell populations.
| Cell line | DNA | TK–APRT– cells (×10–6)a | ||
|---|---|---|---|---|
| Experiment 1 | Experiment 2 | |||
| AK784 | ERCC1+ | pCMV-β-galactosidase | <2 | 3.9 ± 3.8 |
| AK784 | ERCC1+ | pCMV-I-SceI | 1100 ± 340 | 4100 ± 2600 |
| GSAK1 | ERCC1– | pCMV-β-galactosidase | 11 ± 1.9 | 8.1 ± 4.3 |
| GSAK1 | ERCC1– | pCMV-I-SceI | 62 ± 28 | 200 ± 130 |
aFor experiment 1 the frequencies of TK–APRT– cells are an average of results from four transfected wells. For experiment 2 the frequencies for pCMV-β-galactosidase transfections are an average of results from four transfected wells, whereas the frequencies for pCMV-I-SceI transfections are averaged from 20 transfected wells. Standard deviations are indicated.
Molecular analysis of TK–APRT– cells derived from ERCC1+ and ERCC1– cell lines
Although I-SceI-induced DSBs stimulate homologous recombination, they also stimulate non-homologous (illegitimate) recombination (24,25). To measure the proportions of the frequencies in Table 3 that were due to homologous and non-homologous recombination, we analyzed independent TK–APRT– colonies from untreated and I-SceI-treated AK784 (ERCC1+) and GSAK1 (ERCC1–) cells by Southern blotting, analogous to that shown in Figure 2 (data not shown). The TK–APRT– colonies from untreated AK784 (ERCC1+) cells were all homologous recombinants, whereas most of the TK–APRT– colonies from untreated GSAK1 (ERCC1–) cells were rearrangements (Table 1). These results parallel those for ERCC1+ and ERCC1– cells that lack the 800 bp HPRT insert or the GPT gene (Table 1).
TK–APRT– colonies isolated from AK784 (ERCC1+) and GSAK1 (ERCC1–) cells after treatment with I-SceI were a mixture of crossovers and rearrangements (Table 1). All crossovers had lost the HPRT insert and the GPT gene (data not shown), as expected for the SSA pathway (Fig. 3). TK–APRT– rearrangements isolated from AK784 (ERCC1+) and GSAK1 (ERCC1–) cells after treatment with I-SceI resembled those previously characterized (24). Southern blotting and PCR analyses showed that three of eight rearrangements from AK784 (ERCC1+) cells and six of 12 from GSAK1 (ERCC1–) cells were simple deletions that included the I-SceI site and the TK gene (data not shown). The more complex rearrangements have not been further characterized.
The specific effect of I-SceI expression on the frequency of crossovers can be estimated by multiplying the average of the TK–APRT– frequencies (Table 3) by the proportion of crossovers among the TK–APRT– products (Table 1), as shown in Table 4. This calculation indicates that I-SceI expression stimulated crossover formation ∼800-fold in AK784 (ERCC1+) cells versus ∼40-fold in GSAK1 (ERCC1–) cells. Comparison of the stimulated frequencies indicates that crossovers were stimulated some 30-fold less in GSAK1 (ERCC1–) cells than in AK784 (ERCC1+) cells (Table 4). A similar calculation indicates that I-SceI expression stimulates rearrangements several thousand fold in AK784 cells versus ∼10-fold in GSAK1 cells. Comparison of the stimulated frequencies indicates that rearrangements were stimulated some 10-fold less in GSAK1 (ERCC1–) cells versus AK784 (ERCC1+) cells (Table 4).
Table 4. Frequencies of crossovers and rearrangements in ERCC1+ and ERCC1– cells with and without I-SceI treatment.
| Cell line | Treatment | TK–APRT– cells (×10–6)a | Crossovers | Rearrangements | |||
|---|---|---|---|---|---|---|---|
| Colonies | Frequency (×10–6)b | Colonies | Frequency (×10–6)b | ||||
| AK784 | ERCC1+ | – I-SceI | 2 | 16/16 | 2 | 0/16 | <0.1 |
| AK784 | ERCC1+ | + I-SceI | 2600 ± 1500 | 15/23 | 1700 ± 1070 | 8/23 | 900 ± 600 |
| GSAK1 | ERCC1– | – I-SceI | 9.6 ± 1.5 | 3/20 | 1.4 ± 0.8 | 17/20 | 8.2 ± 2.4 |
| GSAK1 | ERCC1– | + I-SceI | 130 ± 69 | 10/22 | 59 ± 36 | 12/22 | 71 ± 43 |
aValues are the means of the two experiments shown in Table 3. Standard errors of the means are indicated.
bValues represent the frequency of TK–APRT– cells multiplied by the proportion of crossovers or rearrangements among the colonies analyzed. Standard rules for propagation of error were used to combine the standard error of the mean for TK–APRT– cells with the counting errors associated with the number of colonies counted in each class; the propagated errors are indicated.
DISCUSSION
Recombination-dependent rearrangements in the absence of Ercc1p were first defined in studies at the APRT locus in Chinese hamster cells and attributed to loss of function of the Ercc1/Xpf endonuclease (19). General genome instability in the absence of a significant effect on homologous recombination was also described in ERCC1– mouse cells (18). Drosophila that are mutant in the mei-9 gene, which encodes an XPF homolog, have what may be analogous defects, exhibiting an increased incidence of chromosomal instability (26–28). Homozygous ERCC1 knockout mice exhibit severe runting and a greatly reduced lifespan (29,30), effects that are unlikely to be due to a deficiency in NER, since XPA and XPC knockout mice show only an elevated susceptibility to UV-induced carcinogenesis, like their human XP counterparts (31–33). These studies point to a role for the Ercc1/Xpf endonuclease in genome stability.
In yeast the RAD10 gene, the counterpart of the mammalian ERCC1 gene, encodes one component of the Rad1/Rad10 endonuclease, which functions in NER and in the removal of non-homologous 3′ tails in homologous recombination (15). Recombination-dependent rearrangements, however, have not been observed in experiments that used RAD1- or RAD10-defective S.cerevisiae (14,34–36). Unlike the loss of function assays used here, which can detect both homologous and non-homologous products, those studies used gain of function assays that specifically select for homologous recombinants. Even if loss of function assays had been used in S.cerevisiae, they may not have detected rearrangements because non-homologous recombination is rare in S.cerevisiae except in RAD52-defective cells or in cells where no homolog is available for repair by homologous recombination (15,37). Because non-homologous recombination is very active in mammalian cells (38), the consequences of ERCC1 deficiency may be fundamentally different than in S.cerevisiae.
Purified Ercc1/Xpf endonuclease can cleave two types of artificial substrate, duplexes containing single-stranded loops and duplexes with 3′ tails, that are analogs of structures that can arise as heteroduplex intermediates in homologous recombination (7,8,10). Misprocessing of such intermediates in the absence of Ercc1/Xpf endonuclease could reasonably give rise to rearrangements. In our original study, which identified recombination-dependent rearrangement formation in ERCC1– cells, both structures were potential intermediates in the recombination events that generated rearrangements (19). Formation of heteroduplex loops during spontaneous recombination at the APRT locus was inferred from the formation of crossovers with conversion (inclusion of the GPT gene in the crossover), which constituted 25% (17/68) of crossover events in ERCC1+ cells (23). The potential relevance of heteroduplex loops to the rearrangement process was indicated by a decrease in this class of events to 4% (1/24) in ERCC1– cells (19).
To investigate the role of heteroduplex loops, we constructed a tandem duplication at APRT in JAM1-77a (ERCC1–) cells in which the repeated segments were identical except for the single base difference used to detect recombination. In these cells 19/32 spontaneous TK–APRT– colonies carried a rearranged APRT locus, which is similar to the frequency of rearrangements (23/47) observed originally in cell line GS22-13 (ERCC1–), which had the potential to form an 800 bp heteroduplex loop (Table 1). Although misprocessing of heteroduplex loops, when present, may contribute to rearrangements, these results rule out their misprocessing as the major source of rearrangements in ERCC1– cells.
By analogy with the above experiments, it would be most informative to test the possible role of misprocessing of non-homologous 3′ tails by using a substrate that formed a heteroduplex intermediate lacking 3′ tails. Construction of such a chromosomal substrate is not possible. Heteroduplex intermediates formed by the SSA pathway after a DSB would consist of a variety of structures, some with one non-homologous tail (for breaks in regions of homology) and some with two non-homologous tails (for breaks in regions of non-homology). Only in the special case where the direct repeats abut and the break occurs at that junction would the heteroduplex intermediate formed by SSA not carry a 3′ tail. The expedient of placing an I-SceI recognition site at that junction is not an option in mammalian cells because I-SceI cleavage generates rearrangements even in ERCC1+ cells (24).
In the absence of a direct test, we sought to demonstrate a requirement for the Ercc1/Xpf endonuclease in removal of non-homologous 3′ tails in mammalian cells, as demonstrated for the Rad1/Rad10 endonuclease in yeast (12,13,15,16). By placing the I-SceI recognition sequence in the middle of an 800 bp segment of non-homologous DNA in one copy of the APRT gene we were able to initiate the SSA pathway at a site of non-homology, generating a heteroduplex intermediate with two non-homologous 3′ tails. Spontaneous recombination with this substrate occurred at similar rates in ERCC1+ and ERCC1– cell lines (Table 2). Expression of I-SceI in ERCC1– cells, however, was some 30-fold less effective at generating crossovers than it was in ERCC1+ cells (Table 4). These results mirror those in yeast. Spontaneous pop-out recombination between direct repeats longer than 1 kb in yeast defective for RAD1 or RAD10 occurred at roughly equal rates (14,34–36), whereas DSB-induced pop-outs were reduced 20- to 30-fold (12,13).
Two trivial explanations for the decrease in I-SceI-stimulated crossovers, decreased transfection efficiency and increased toxicity of breaks, seem unlikely. The measured transfection efficiencies in AK784 (ERCC1+) and GSAK1 (ERCC1–) cells differed only slightly (Materials and Methods). Increased toxicity of DNA breaks in ERCC1– cells could reduce detected crossovers if the stimulated population were preferentially killed. This is unlikely because ERCC1+ and ERCC1– cells have been shown to be equally resistant to X-irradiation, which kills by introducing DSBs (39,40), and electroporation of restriction enzymes into ERCC1+ and ERCC1– cells gave the same killing curve (G.M.Adair, unpublished results).
We conclude, therefore, that Ercc1p, most likely as a component of the Ercc1/Xpf endonuclease, is required for efficient removal of non-homologous 3′ tails during homologous recombination between chromosomal repeats. We recently demonstrated a similar requirement in plasmid-by-chromosome targeted recombination at the APRT locus in CHO cells (41). Plasmids with long non-homologous tails (271 and 670 nt) were some 20-fold less effective at generating diagnostic classes of targeted recombinants in ERCC1– cells than were plasmids with short non-homologous tails (11 and 18 nt). In contrast, in ERCC1+ cells plasmids with long or short tails yielded equivalent results. Thus the inability to remove long non-homologous tails in ERCC1– cells is likely to be the principal source of recombination-dependent rearrangement formation in mammalian cells.
Although I-SceI-induced crossovers in ERCC1– cells were some 30-fold less than in ERCC1+ cells, they were still stimulated ∼45-fold above uninduced levels (Table 4). This result implies that there are other mechanisms for removing non-homologous tails that do not depend on Ercc1p. This background stimulation is unlikely to be due to any residual Ercc1 activity because previous characterization of the ERCC1-targeted knockout in the parents of these cells revealed no ERCC1 transcript by northern analysis, no Ercc1 protein by western analysis and full mitomycin C sensitivity (19,42). Similar low level stimulation has been observed in RAD1- and RAD10-defective yeast, but the mechanism is undefined (12,13,15).
In the absence of I-SceI treatment, rearrangements were at least 80-fold more frequent in ERCC1– cells than in ERCC1+ cells (Table 4), confirming our original observations (19). The spectrum of spontaneous TK–APRT– rearrangements generated in ERCC1– cells is distinctive. Nearly all give rise to a single BamHI fragment upon Southern analysis, as expected for deletions extending from APRT to TK (Fig. 1B) and confirmed by PCR analysis and sequencing (19). Of the 59 spontaneous rearrangements indicated in Table 1, however, five generated more complex Southern patterns, inconsistent with simple deletions. This distribution of 90% deletions and 10% other rearrangements is quite different from that observed after I-SceI cleavage, which generated only three deletions out of eight rearrangements in ERCC1+ cells and six out of 12 in ERCC1– cells, consistent with previous characterization of I-SceI-induced rearrangements in ERCC1+ cells, where 13 out of 23 rearrangements were deletions (24). These distinctive spectra suggest that there may be two pathways for generating the rearrangements observed in our experiments: a recombination-dependent, spontaneous pathway and a DSB-induced pathway, which is not dependent on homologous recombination (24).
The formation of rearrangements via the recombination-dependent, spontaneous pathway is clearly stimulated in the absence of Ercc1p, as shown here and previously (19). In surprising contrast, formation of rearrangements via the recombination-independent, DSB-induced pathway is decreased some 10-fold in ERCC1– cells relative to ERCC1+ cells (Table 4). This result suggests that Ercc1p may normally participate in formation of these rearrangements, which inactivate both the APRT gene and the TK gene. Inactivation by simple deletion would require removal of 6.5 kb, the distance from the I-SceI site in the APRT gene to the TK gene.
Three studies in yeast bear on this issue but do not entirely clarify the role of the Rad1/Rad10 endonuclease in non-homologous recombination. In one study, rad1 (endonuclease-deficient) strains were unaffected in their ability to repair HO endonuclease-induced chromosomal DSBs by non-homologous end joining (NHEJ) (37). Most repair events, however, were additions or deletions of <50 nt around the site of the break and the effect of rad1 mutations on large deletions was not examined (37). In a second study, repair of HO-induced DSBs in plasmids occasionally generated deletions (>2 kb) in wild-type yeast cells, but these products were absent in identical experiments in rad1 cells, suggesting that generation of such deletions depends on the RAD1 gene product (43). Finally, expression of the restriction enzyme EcoRI in a Δhdf1 (NHEJ-deficient) haploid strain caused a much greater loss of viability than its expression in a Δrad52 (recombination-deficient) haploid strain, which had the same survival kinetics as a wild-type haploid strain (44). EcoRI expression in a Δrad1 haploid strain caused a loss of viability that was intermediate between these two, suggesting that a subset of EcoRI-induced DSBs may be processed for NHEJ by the Rad1/Rad10 endonuclease under these conditions (44).
The unexpected effect of Ercc1p on TK–APRT– rearrangements after I-SceI treatment may have been revealed in our experiments by the requirement for simultaneous inactivation of both the APRT and TK genes, which are separated by several kilobases in our constructs. The mechanism for deletion of such a length of DNA is undefined, but has been variously suggested to arise by crossing a nick during resection of the 5′→3′ strand, by the action of a 3′→5′ exonuclease or by the action of a single-strand endonuclease on the protruding 3′ single strand. In vitro purified Ercc1/Xpf endonuclease will cleave a protruding 3′ strand from duplex DNA (8). Such an activity of Ercc1/Xpf endonuclease on broken, resected DNA ends in cells could account for the higher frequency of TK–APRT– rearrangements detected in ERCC1+ cells than in ERCC1– cells. In a normal cell it may be that the periodic action of Ercc1/Xpf endonuclease on protruding 3′ single strands is required to continually regenerate termini appropriate for NHEJ, thereby giving homologous and non-homologous recombination multiple chances to repair an otherwise lethal chromosome break. Additional experiments will be needed to define the role of ERCC1 in the rearrangements described here.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Fung Chan and Kathleen Marburger for technical support and Dr Karen Vasquez for critical discussions. This investigation was supported by NIH grants to J.H.W. (GM38219), R.S.N. (CA36361) and G.M.A. (CA28711), by Pilot Project support from NIEHS Center grant ES07784 to G.M.A. and by Department of Defense Breast Cancer Research Program grant DAMD17-97-1-7283 to J.H.W. B.D.P. was supported by a training grant from the NIH (T32 EY07102).
REFERENCES
- 1.Friedberg E.C., Walker,G.C. and Seide,W. (1995) DNA Repair and Mutagenesis. American Society for Microbiology, Washington, DC.
- 2.Sancar A. (1995) Annu. Rev. Genet., 29, 69–105. [DOI] [PubMed] [Google Scholar]
- 3.Weeda G., Hoeijmakers,J.H. and Bootsma,D. (1993) Bioessays, 15, 249–258. [DOI] [PubMed] [Google Scholar]
- 4.Huang J.C., Svoboda,D.L., Reardon,J.T. and Sancar,A. (1992) Proc. Natl Acad. Sci. USA, 89, 3664–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moggs J.G., Yarema,K.J., Essigmann,J.M. and Wood,R.D. (1996) J. Biol. Chem., 271, 7177–7186. [DOI] [PubMed] [Google Scholar]
- 6.Huang J.C., Hsu,D.S., Kazantsev,A. and Sancar,A. (1994) Proc. Natl Acad. Sci. USA, 91, 12213–12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bessho T., Sancar,A., Thompson,L.H. and Thelen,M.P. (1997) J. Biol. Chem., 272, 3833–3837. [DOI] [PubMed] [Google Scholar]
- 8.de Laat W.L., Appeldoorn,E., Jaspers,N.G.J. and Hoeijmakers,J.H.J. (1998) J. Biol. Chem., 273, 7835–7842. [DOI] [PubMed] [Google Scholar]
- 9.Evans E., Fellows,J., Coffer,A. and Wood,R.D. (1997) EMBO J., 16, 625–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsunaga T., Park,C.H., Bessho,T., Mu,D. and Sancar,A. (1996) J. Biol. Chem., 271, 11047–11050. [DOI] [PubMed] [Google Scholar]
- 11.O’Donovan A., Davies,A.A., Moggs,J.G., West,S.C. and Wood,R.D. (1994) Nature, 371, 432–435. [DOI] [PubMed] [Google Scholar]
- 12.Fishman-Lobell J. and Haber,J.E. (1992) Science, 258, 480–484. [DOI] [PubMed] [Google Scholar]
- 13.Ivanov E.L. and Haber,J.E. (1995) Mol. Cell. Biol., 15, 2245–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein H.L. (1988) Genetics, 120, 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paques F. and Haber,J.E. (1999) Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prado F. and Aguilera,A. (1995) Genetics, 139, 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nairn R.S., Adair,G.M., Christmann,C.B. and Humphrey,R.M. (1991) Mol. Carcinog., 4, 519–526. [DOI] [PubMed] [Google Scholar]
- 18.Melton D., Ketchen,A.M., Nu,F., Bonatti-Abbondandolo,S., Abbondandolo,A., Squires,S. and Johnson,R. (1998) J. Cell Sci., 111, 395–404. [DOI] [PubMed] [Google Scholar]
- 19.Sargent R.G., Rolig,R.L., Kilburn,A.E., Adair,G.M., Wilson,J.H. and Nairn,R.S. (1997) Proc. Natl Acad. Sci. USA, 94, 13122–13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merrihew R.V., Sargent,R.G. and Wilson,J.H. (1995) Somat. Cell Mol. Genet., 21, 299–307. [DOI] [PubMed] [Google Scholar]
- 21.Rouet P., Smih,F. and Jasin,M. (1994) Proc. Natl Acad. Sci. USA, 91, 6064–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacGregor G.R. and Caskey,C.T. (1989) Nucleic Acids Res., 17, 2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sargent R.G., Merrihew,R.V., Nairn,R., Adair,G., Meuth,M. and Wilson,J.H. (1996) Nucleic Acids Res., 24, 746–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sargent R.G., Brenneman,M. and Wilson,J.H. (1997) Mol. Cell. Biol., 17, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouet P., Smih,F. and Jasin,M. (1994) Mol. Cell. Biol., 14, 8096–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker B.S., Carpenter,A.T.C. and Ripoll,R. (1978) Genetics, 90, 531–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker B.S., Gatti,M., Carpenter,A.T., Pimpinelli,S. and Smith,D.A. (1980) Basic Life Sci., 15, 189–208. [DOI] [PubMed] [Google Scholar]
- 28.Gatti M. (1979) Proc. Natl Acad. Sci. USA, 76, 1377–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McWhir J., Selfridge,J., Harrison,D.J., Squires,S. and Melton,D.W. (1993) Nature Genet., 5, 217–224. [DOI] [PubMed] [Google Scholar]
- 30.Weeda G., Donker,I., de Wit,J., Morreau,H., Janssens,R., Vissers,C.J., Nigg,A., van Steeg,H., Bootsma,D. and Hoeijmakers,J.H.J. (1997) Curr. Biol., 7, 427–439. [DOI] [PubMed] [Google Scholar]
- 31.de Vries A., van Oostrom,C.T., Hofhuis,F.M., Dortant,P.M., Berg,R.J., de Gruijl,F.R., Wester,P.W., van Kreijl,C.F., Capel,P.J., van Steeg,H. and Verbeek,S.J. (1995) Nature, 377, 169–173. [DOI] [PubMed] [Google Scholar]
- 32.Nakane H., Takeuchi,S., Yuba,S., Saijo,M., Nakatsu,Y., Murai,H., Nakatsuru,Y., Ishikawa,T., Hirota,S., Kitamura,Y., Kato,Y., Tsunoda,Y., Miyauchi,H., Horio,T., Tokunaga,T., Matsunaga,T., Nikaido,O., Nishimune,Y., Okada,Y. and Tanaka,K. (1995) Nature, 377, 165–168. [DOI] [PubMed] [Google Scholar]
- 33.Sands A.T., Abuin,A., Sanchez,A., Conti,C.J. and Bradley,A. (1995) Nature, 377, 162–165. [DOI] [PubMed] [Google Scholar]
- 34.Schiestl R.H. and Prakash,S. (1990) Mol. Cell. Biol., 10, 2485–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang K.N. and Symington,L.S. (1994) Mol. Cell. Biol., 14, 6039–6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas B.J. and Rothstein,R. (1989) Genetics, 123, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore J.K. and Haber,J.E. (1996) Mol. Cell. Biol., 16, 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth D.B. and Wilson,J.H. (1988) In Kucherlapati,R. and Smith,G.R. (eds), Genetic Recombination. American Society for Microbiology, Washington, DC, pp. 621–653.
- 39.Thompson L.H., Rubin,J.S., Cleaver,J.E., Whitmore,G.F. and Brookman,K. (1980) Somat. Cell Genet., 6, 391–405. [DOI] [PubMed] [Google Scholar]
- 40.Collins A.R. (1993) Mutat. Res., 293, 99–118. [DOI] [PubMed] [Google Scholar]
- 41.Adair G.M., Rolig,R.L., Moore-Faver,D., Zabelshansky,M., Wilson,J.H. and Nairn,R.S. (2000) EMBO J., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rolig R.L., Layher,S.K., Santi,B., Adair,G.M., Gu,F., Rainbow,A.J. and Nairn,R.S. (1997) Mutagenesis, 12, 277–283. [DOI] [PubMed] [Google Scholar]
- 43.Colaiacovo M.P., Paques,F. and Haber,J.E. (1999) Genetics, 151, 1409–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis L.K., Westmoreland,J.W. and Resnick,M.A. (1999) Genetics, 152, 1513–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]