Abstract
Saccharomyces cerevisiae contains three genes that encode members of the histone H2A gene family. The last of these to be discovered, HTZ1 (also known as HTA3), encodes a member of the highly conserved H2A.Z class of histones. Little is known about how its in vivo function compares with that of the better studied genes (HTA1 and HTA2) encoding the two major H2As. We show here that, while the HTZ1 gene encoding H2A.Z is not essential in budding yeast, its disruption results in slow growth and formamide sensitivity. Using plasmid shuffle experiments, we show that the major H2A genes cannot provide the function of HTZ1 and the HTZ1 gene cannot provide the essential function of the genes encoding the major H2As. We also demonstrate for the first time that H2A.Z genes are functionally conserved by showing that the gene encoding the H2A.Z variant of the ciliated protozoan Tetrahymena thermophila is able to rescue the phenotypes associated with disruption of the yeast HTZ1 gene. Thus, the functions of H2A.Z are distinct from those of the major H2As and are highly conserved.
INTRODUCTION
In eukaryotes, many genes, including those encoding histones, occur as members of multigene families. Except for very recent gene duplications, in which inactivating mutations have not yet had time to accumulate, this implies that the multiple copies of a gene have some important function subject to positive evolutionary selection. There are three possible explanations for the existence of multigene families. In the first case, referred to as dosage repetition, genes are repeated because, in some physiological state, an organism needs more gene product than can be provided by a single gene. In the second case, referred to as regulatory repetition, multiple genes are required for regulatory reasons. For example, if a gene expressed in two different physiological or developmental conditions is duplicated, each copy may lose sequences required for expression in one or the other condition or gain new regulatory sequences independently of the other gene. Finally, in the third case, referred to as variant repetition, the coding region of one of a pair of duplicated genes may evolve a new function which, like that of the original gene, also confers a selectable advantage.
Examples of each type of gene repetition appear to occur among histone genes. The large number of tandemly repeated clusters of histone genes found in many organisms (1,2) almost certainly reflects dosage repetition. A likely example of regulatory repetition has been described in sea urchins in which different H4 genes are expressed at different stages of development, but the switch does not involve any change in the sequence of the H4 proteins (3).
The most interesting type of histone gene repetition is variant repetition, because it implies that there are functional differences between nucleosomes containing different histone isotypes. However, variant repetition is the most difficult to demonstrate. It requires demonstration that the non-identical coding sequences of two members of a histone gene family placed under the control of the same regulatory sequences have biologically significant differences in function (i.e. different phenotypes). This difficulty is best illustrated by a recent study of the H3 gene family in Tetrahymena (4). Tetrahymena contains two H3 proteins differing at 16 residues, a remarkable difference for copies of such a highly conserved protein in the same organism. Nonetheless, disrupting the gene encoding the divergent, constitutively expressed H3, which is the only H3 expressed in non-growing cells, was without detectable phenotype and resulted in the expression of the other, normally replication-dependent H3 gene. Thus, in spite of the unusually large number of amino acid differences between these two H3s, it appears that it is their expression pattern, not their primary sequence that distinguishes them.
We previously reported the presence of a gene in Saccharomyces cerevisiae encoding a third member of the HTA gene family encoding the H2A class of histones (5). This gene (referred to as HTA3 or HTZ1) encodes a member of the H2A.Z (also referred to as H2A.F or H2A.F/Z) class of H2A variants, a highly conserved type of histone found in diverse eukaryotes, including vertebrates, invertebrates, protozoa and fungi. Phylogenetic analyses indicate that the H2AZs diverged from the major H2As early in eukaryotic evolution and are at least as evolutionarily conserved as the major H2As (6). The gene encoding H2A.Z has been shown either to be essential (ciliates and flies) (7,8) or to be required for normal growth (fission yeast) (9). These properties suggest that H2A.Z has an important function that is distinct from that of the major H2As. However, only a single study has actually demonstrated that the H2A.Z protein itself appears to have a function that is distinct from that of the major H2A (10). A small deletion in the Drosophila H2AvD gene encoding H2A.Z is lethal and can be rescued by transformation with the H2AvD gene. Null mutants in H2AvD fail to develop through pupation. By replacing regions unique to H2AvD with residues in the homologous position from the major H2A, essential elements unique to H2AvD were mapped to histone fold regions involved in docking the H2A/H2B dimer to the H3/H4 tetramer (11). However, because the assay was a developmental defect, these studies leave unanswered the question of whether the two types of H2A protein are each sufficient for cell survival. In addition, since reciprocal replacements were not done, it is not clear if the H2A.Z variant could perform the function of the major H2A. Such a demonstration is difficult in Drosophila inasmuch as it would require placing multiple copies of the H2A.Z coding region under the regulatory sequences of the highly repeated, major H2A genes.
We describe here an experimental analysis of the function of H2A.Z in S.cerevisiae showing that the HTZ1 gene is not essential but its disruption results in slow growth and conditional lethality in formamide-containing medium. These functions can be rescued with plasmids containing the gene encoding H2A.Z from either yeast or from the ciliated protozoan Tetrahymena thermophila. We also show that the major and minor histone H2A sequence variants are functionally distinct, i.e. the minor H2A.Z variant cannot supply major H2A function and vice versa. Coupled with earlier studies showing that the genes encoding the Tetrahymena major H2As can provide the essential function of the yeast major H2A genes (12), these results demonstrate that the functions unique to each type of H2A are highly conserved.
MATERIALS AND METHODS
Yeast strains
The diploid yeast strain used to generate the ΔHTZ1 mutant (JJY1) was EJ72 (gal– trp1 leu2 ura3-52 his4) (M.-H.Kuo, PhD thesis, University of Rochester, 1995). The strains used for the plasmid shuffle experiments were XLY1 (MATα his3Δ200 lys2-801a ade2-101° ura3-52 GAL suc2 hta1 hta2 <pJC102>) and XLY2 (MATa his3Δ200 tyr1 ade2-101° ura3-52 GAL suc2 hta1 hta2 <pJC102>) (12). The yHTZ1 gene was also knocked out in XLY2 using pJJ2.1r and the resulting strain designated JJY100 (MATa his3Δ200 tyr1 ade2-101° ura3-52 GAL suc2 hta1 hta2 hta3 <pJJ90>).
Plasmid construction
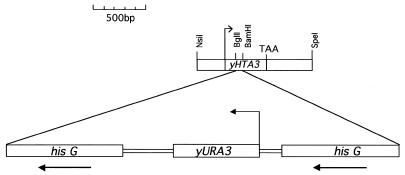
To clone the yHTZ1 gene, a 2.3 kb yHTZ1 fragment was amplified with primers yH2A.Z-1 (CGGAATTCTTTCTCCCCTTCTCTTAC) and yH2A.Z-2 (CCCGGAATTCCTTGCTTTTAGTCCTCTT), digested with NsiI and SpeI and the resulting 1.2 kb fragment was ligated to the PstI and SpeI sites of pBSIISK– to make pJJ1. A longer 1.6 kb yHTZ1 fragment was amplified from cosmid pEOA215 (13), which contains a 29 kb fragment from chromosome XV that includes yHTZ1, using oligonucleotides yH2A.Z-1 and yH2A.Z-20 (GGTATGCTCGAGTTGCAACGCACAAAGCTT). This PCR product was blunt end ligated to the SmaI sites of both pBSISK+ and pRS313 (14), a HIS3 CEN6 ARSH4 plasmid, to make pJJ42 and pJJ43, respectively. To make the yHTZ1 gene disruption construct (pJJ2.1r), pJJ1 was digested at BamHI and BglII sites located within the yHTZ1 gene and ligated to a 3.8 kb BamHI fragment from pDG82 (12) containing the yURA3 gene flanked by 1.1 kb direct repeats of the bacterial hisG sequence (15; Fig. 1). To express the 1.2 kb yHTZ1 fragment in XLY1 and XLY2 yeast strains, the insert from pJJ1 was excised using EcoRI and SpeI and ligated to the EcoRI and SpeI sites of pRS313, to make pJJ3. To express the 1.2 kb yHTZ1 fragment in yeast strain EJ72 and its ΔHTZ1 derivative (JJY1), the insert was excised from pJJ1 at KpnI and XbaI sites and ligated to the KpnI and XbaI sites of YCplac33 (16), a URA3, CEN4, ARS1 plasmid, to make pJJ11. To express the 1.6 kb yHTZ1 fragment in JJY1, the insert from pJJ42 was excised at KpnI and SacI sites and ligated to the KpnI and SacI sites of YCplac33, to make pJJ44. To place the yHTZ1 gene under yHTA1 regulation, pJJ90, a derivative of pXL90 (12) lacking the ClaI site within the polylinker, was initially isolated from a dam– Escherichia coli strain, digested with ClaI to eliminate the yHTA1 coding region and religated to make pJJ91. The yHTZ1 coding region was then amplified from pJJ1 with primers yH2A.Z-6 (GGGAGAATCGATGGAAATGGGAAAGAAAA), flanking the 3′ coding region, and yH2A.Z-7 (GCCGCACGTATCGATAACTTAACATAATG, initiation codon in bold), flanking the 5′ coding region. This PCR product was digested with ClaI (site underlined above) and ligated to the ClaI site of pJJ91, to make pJJ4. To express this yHTA1–yHTZ1 gene fusion in JJY1, the 2.2 kb insert from pJJ4 was excised at KpnI and SpeI sites and ligated to the KpnI and SpeI sites of YCplac33, to make pJJ10. To place the yHTZ1 gene under yGAL1 regulation, the yHTZ1 coding and 3′ flanking sequences were amplified with primers yH2A.Z-5 (CGGGGTACCTCAGGAAAAGCTCATGGAGG), flanking the 5′ coding region, and yH2A.Z-1, flanking the 3′ coding region. This PCR product was digested with KpnI and EcoRI and ligated to the KpnI and EcoRI sites of pYES2, a yeast URA3 2µ plasmid containing the yGAL1 promoter, to make pJJ15. To express this GAL1–HTZ1 fusion in yeast XLY1 and XLY2 strains, the insert from pJJ15 was excised with AflIII and NaeI, filled in with Klenow and ligated to the SmaI site of pRS313, to make pJJ6. To place the yHTA1 gene under yHTZ1 regulation, 775 bp of the yHTZ1 promoter was initially amplified using primers yH2A.Z-18 (TGTTAAGTTATCGATACGTGCGGCTAT) and yH2A.Z-20 (GGTATGCTCGAGTTGCAACGCACAAAGCTT). This PCR product was digested with ClaI and XhoI and cloned into the ClaI and XhoI sites of pRS313, to make pJJ39. A 780 bp fragment containing the yHTA1 coding region and 3′ flank was excised from pJJ90 at ClaI and SacII sites and then ligated to the ClaI and SacII sites of pJJ39, to make pJJ40. To express this yHTZ1–yHTA1 fusion in JJY1, the insert from pJJ40 was excised at KpnI and SacII sites and ligated to YCplac33 at the KpnI and SacII sites to make pJJ41. To express the yHTZ1–yHTA1 gene fusion from a high copy plasmid, the insert from pJJ40 was excised at XhoI and SacII sites and ligated to pRS423, a HIS3 2µ plasmid, at the XhoI and SacII sites, to make pJJ47. To express the three genes encoding the two Tetrahymena major H2As and H2A.Z in XLY1 and XLY2 yeast strains, tHTA1, tHTA2 and tHTZ1 were placed under yHTA1 regulation to make pXL87, pXL88 and pXL89, respectively. To express these gene fusions in yeast strain JJY1, the inserts from pXL87, pXL88 and pXL89 were excised with KpnI and XbaI and ligated to the KpnI and XbaI sites of YCplac33 to make pJJ87, pJJ88 and pJJ89, respectively. To express the yHTA1 gene under its own regulation in JJY1, a 1.2 kb fragment containing the yHTA1 gene was initially excised from pJJ90 at EcoRI and SacII sites and ligated to the EcoRI and SacII sites of pBSIISK, to make pJJ21. The insert from pJJ21 was then excised at BssHII sites, filled in with Klenow and ligated to the SmaI site of YCplac33, to make pJJ46.
Figure 1.
yHTZ1 gene disruption construct. yHTZ1 was disrupted by inserting a gene blaster cassette into the body of the yHTZ1 gene at BamHI and BglII sites.
Other procedures
All DNA cloning, sequencing, PCR amplification and site-directed mutagenesis were done essentially as described previously (17). Yeast transformations were done by the lithium acetate method as described (18,19).
RESULTS
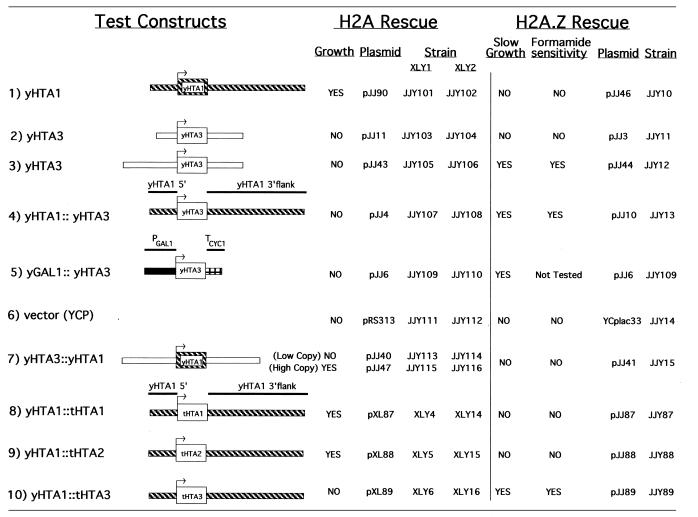
To elucidate the function of histone H2A.Z, we disrupted the HTZ1 gene of S.cerevisiae and assayed for its effect. A ‘gene blaster’ cassette (Fig. 1) containing the URA3 gene (15) was used to disrupt one of the chromosomal HTZ1 genes in diploid yeast strain EJ72 (kindly provided by Dr Min-Hao Kuo), which is auxotrophic for uracil. Four heterozygous HTZ1 knockout transformants were obtained and plated onto medium containing potassium acetate to induce the formation of tetrads containing haploid spores (20). Sixty-three tetrads were dissected and plated onto YPD medium. All spores from each tetrad grew on YPD at 30°C with a 2:2 segregation pattern of small (ΔHTZ1) versus large (wild-type) colonies, indicating that while the HTZ1 gene is not essential, its disruption confers a slow growth phenotype. This slow growth effect was more pronounced at 20°C, at which temperature the ΔHTZ1 mutant grew 4- to 5-fold slower than its isogenic wild-type strain (Table 1). At 37°C the ΔHTZ1 and wild-type strains grew similarly. Slow growth was rescued at both 20 and 30°C by a plasmid (pJJ44) containing yHTZ1 and 700 bp of upstream sequences, demonstrating that this phenotype was due to the absence of H2A.Z (Table 1). A plasmid (pJJ3) containing yHTZ1 and 300 bp of 5′ sequence failed to rescue, suggesting the presence of an upstream activating sequence (UAS) 300–700 bp upstream of the coding region (Table 2, constructs 2 and 3).
Table 1. Doubling times of the wild-type and ΔHTZ1 strains in liquid culture at various temperatures.
| Strain | Plasmid | Doubling time (min) | ||
|---|---|---|---|---|
| 20°C | 30°C | 37°C | ||
| ΔH2A.F/Z | None | 1666 ± 10 | 226 ± 11 | 278 |
| Wild-type | None | 459 ± 13 | 143 ± 16 | 280 |
| ΔH2A.F/Z | YCP-yHTA3 | 447 ± 9 | 144 ± 11 | 275 |
Table 2. Summary of the plasmid shuffle and mutant phenotype rescue experiments.
Each construct was tested for its ability to replace yHTA1 and to rescue the mutant phenotypes of the ΔHTZ1 mutant.
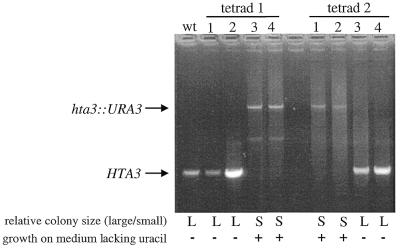
To determine if the 2:2 pattern of slow growth reflected segregation of the disrupted HTZ1 gene, the gene was amplified from the genomic DNA of eight spores (four from each of two dissected tetrads). PCR products from amplifications using primers that flanked the site of the disruption were sized by agarose gel electrophoresis. The spores that exhibited slow growth also contained the disrupted yHTZ1 gene (Fig. 2). A haploid strain from one of these ΔHTZ1 spores (JJY1) was used in subsequent experiments.
Figure 2.
The 2:2 segregation pattern of the yHTZ1 disruption correlates with the 2:2 segregation pattern of slow growth in haploid spores. L, large colony with wild-type growth; S, small colony with slow growth.
To assay for other phenotypes associated with the knockout, the ΔHTZ1 mutant (JJY1) was tested for its ability to grow on medium supplemented with a number of different chemicals shown to affect the growth of yeast mutants (17). Both mutant and wild-type strains were tested for growth at 15, 30 and 37°C on medium in which glucose was replaced with either galactose, maltose, glycerol, ethanol, potassium acetate or sucrose. Little or no difference was observed between mutant and wild-type strains. Mutant and wild-type strains were also tested for growth on medium supplemented with chloramphenicol, caffeine, sorbitol, cycloheximide or formamide, as well as acidic medium. Only formamide (18) selectively inhibited growth of the ΔHTZ1 mutant compared to its isogenic wild-type strain. To characterize this formamide sensitivity further, mutant and wild-type strains were tested on YPD plates containing from 1.5 to 4% formamide. The growth difference between the ΔHTZ1 mutant and wild-type strains increased with increasing amounts of formamide. At a concentration of 4% the ΔHTZ1 mutant exhibited little or no growth. This formamide sensitivity was rescued by an HTZ1-containing plasmid, indicating that the phenotype was indeed due to the absence of histone H2A.Z (Table 2). Two independently derived HTZ1 knockout strains (derivatives of W303 and S288C, kindly provided by Drs M. Mitchell Smith and Maria S. Santisteban, University of Virginia) were also formamide sensitive. Thus, this phenotype is likely to be a consistent feature of the absence of H2A.Z in S.cerevisiae.
To address whether the yeast H2A.Z variant could serve as the sole H2A variant in S.cerevisiae, we attempted to replace the yHTA1 gene (encoding yH2A.1) with yHTZ1 in a plasmid shuffle experiment. The major H2A is essential in yeast and cells can survive with either one of the genes (HTA1 or HTA2) encoding a major H2A (19). The yeast strains used in this experiment, XLY1 and XLY2 (12), contain disrupted chromosomal HTA1 and HTA2 genes, with HTA1 function supplied on the URA3, ARS1, CEN3, pJC102 plasmid (21). We initially tested the yHTZ1 gene regulated by its own promoter on a HIS3 test plasmid (pJJ43) using a plasmid (pJJ90) containing the wild-type yHTA1 gene as a positive control and the plasmid vector (pRS313) as a negative control. The XLY1 and XLY2 strains transformed with either the minimal or the extended promoter yHTZ1 construct (pJJ11 or pJJ43) were plated without histidine and uracil to yield transformants with two plasmids (the test plasmid and the yHTA1 plasmid). To obtain cells without the yHTA1 plasmid, colonies were streaked onto medium containing the drug 5-FOA, which kills cells expressing URA3. We obtained colonies on the positive control plate but not on either of the experimental plates, suggesting that yHTZ1, regulated by its own flanking sequences, is unable to replace yHTA1 (Table 2, constructs 2 and 3).
Next, we addressed whether differences in the way the HTA genes are regulated might be responsible for the failure of yHTZ1 to replace yHTA1. Because H2A.Z is expressed at a much lower level than the major H2As in yeast (M.M.Smith and M.S.Santisteban, personal communication), we tested whether the highly inducible yGAL1 promoter could elevate yHTZ1 expression to a level sufficient to replace yHTA1. The yHTZ1 coding region fused to the yGAL1 promoter was unable to replace yHTA1 in XLY1 and XLY2 strains grown in galactose (Table 2, construct 5), suggesting that the failure of yHTZ1 to replace yHTA1 was not due to differences in expression levels between the two genes.
In S.cerevisiae, as in other eukaryotes, the difference in the level of expression of the H2A.Z variant between S phase and other phases of the cell cycle is less than that of the major H2As, suggesting that expression of this conserved variant is only partially replication dependent (9,22–24). To rule out the possibility that differences in the pattern of gene expression during the cell cycle might be responsible for the inability of yHTZ1 to replace yHTA1, we tested whether the H2A.Z variant could supply the major H2A function when regulated under control of the yHTA1 flanking sequences. A plasmid, pJJ4, was constructed in which the 5′ and 3′ flanking sequences of yHTZ1 were replaced with those of yHTA1. Again, yHTZ1 failed to replace yHTA1 (Table 2, construct 4). The yHTA1 coding region flanked by these same sequences rescued the major HTA deficiency (Table 2, construct 1). These results suggest that structural differences between H2A.1 and H2A.Z rather than differences in their expression are responsible for the inability of yHTZ1 to supply yHTA1 function.
To ensure that the chimeric constructs containing yHTZ1 were functional, each was tested for its ability to rescue phenotypes associated with the yHTZ1 knockout. The gene fusion containing the yHTA1 flanking sequences and the yHTZ1 coding sequences was tested in the yHTZ1 knockout strain described above and found to rescue both the slow growth and formamide-sensitive phenotypes (Table 2, construct 4). Testing the GAL1–yHTZ1 fusion required knocking out the yHTZ1 gene in another yeast strain, as JJY1 does not grow on galactose. The yHTZ1 gene was knocked out in XLY2 to make JJY100, lacking all three chromosomal HTA genes, with yHTA1 supplied on a plasmid (pJJ90). The GAL1–yHTZ1 fusion rescued the slow growth of colonies on plates, demonstrating that this construct also expresses a functional yHTZ1 (Table 2, construct 5).
Although yHTZ1 was unable to supply yHTA1 function, the possibility remained that yHTA1 could supply yHTZ1 function. To address this, yHTA1, under the control of its own flanking sequences or those of yHTZ1, was tested for its ability to rescue the slow growth and formamide-sensitive phenotypes in JJY1 (Table 2, constructs 1 and 7). Neither construct was able to rescue the mutant phenotypes associated with the yHTZ1 disruption, supporting the conclusion that these genes have distinct functions. To confirm that the gene fusion was expressing functional yHTA1 using the yHTZ1 flanking sequences, this construct was tested for its ability to replace yHTA1 in XLY1 and XLY2. Although it failed to replace yHTA1 when expressed from a low copy yeast centromeric plasmid (YCP) vector, it successfully replaced yHTA1 when expressed from a high copy episomal plasmid (YEP) vector (Table 2, construct 7). In addition to demonstrating that yHTA1 can be expressed by the yHTZ1 flanking sequences, this result is also consistent with a lower level of expression of the variant gene than the major gene.
We had previously shown that the genes encoding the Tetrahymena major H2As, (tHTA1 and tHTA2) but not the gene encoding Tetrahymena H2A.Z (tHTZ1), could replace yHTA1 when tested in a plasmid shuffle experiment in the XLY1 and XLY2 strains (12). However, at the time these experiments were done it was not known that yeast had a HTZ1 gene. Therefore, we tested whether the Tetrahymena HTA genes could rescue the phenotypes associated with the ΔHTZ1 mutant. The coding regions of the three Tetrahymena H2A genes, flanked by yHTA1 sequences, were tested in JJY1 for their ability to rescue slow growth and formamide sensitivity. Tetrahymena HTZ1 rescued both phenotypes while Tetrahymena HTA1 and HTA2 failed to rescue either phenotype (Table 2, constructs 8–10). These results indicate that H2A.Z performs similar functions in T.thermophila and S.cerevisiae and that these functions are distinct from those of the major H2A.
DISCUSSION
Formal demonstration that two protein encoding sequences in the same multigene family have distinct functions requires rigorous elimination of the possibility that any functional differences between the two genes are due to qualitative or quantitative differences in gene expression. Such criteria are rarely met. In the studies described here we have demonstrated that the sequences encoding the major H2A of yeast or the H2A.Z variant cannot replace one another’s function. This is the case even when they are overexpressed or expressed under the control of the regulatory sequences of the other gene. Therefore, the conclusion seems inescapable that these two types of H2As have reciprocally distinct functions in yeast. We have also shown that these distinct functions are remarkably conserved. Thus, a gene encoding a major H2A of a ciliated protozoan can rescue the function of the major H2A in yeast, but cannot rescue the function of H2A.Z. Conversely, the gene encoding the Tetrahymena H2A.Z can rescue the function of H2A.Z in yeast, but not the function of the major H2A. These results confirm and extend phylogenetic analyses (6) that indicated that these two distinct and conservatively evolving H2As had diverged very early in eukaryotic evolution.
It is common to think of the function of the major H2As in terms of nucleosome structure and to seek specialized functions for conserved histone H2A.Z variants. However, the studies described here argue that (at least) two distinct, highly conserved functions must be sought for the H2A multigene family, in addition to the obvious structural function of participating in a nucleosome. The overall conservation of the structure of the major and the variant H2As argues that both are likely to participate in most of the protein–protein interactions required to form the protein core of the nucleosome (11,25). As shown here, and in Drosophila (10), the major H2A cannot perform all of the functions of the H2A.Z variant. In addition, we have shown here, for the first time, that the H2A.Z variant cannot perform all of the functions of the major H2A.
Despite having been studied in a number of different organisms, the function of histone H2A.Z is unknown. Studies in Tetrahymena strongly suggest a role for H2A.Z in transcription. In Tetrahymena, H2A.Z (formerly called hv1) is localized only in the transcriptionally active macronucleus and not in the transcriptionally inert micronucleus of vegetative cells. However, it does appear in micronuclei during conjugation when the micronucleus becomes transcriptionally active (26). In S.cerevisiae, expression of the HTZ1 gene was disrupted as part of a large-scale gene disruption screen involving 2000 individual genes and found to result in reduced growth in a variety of media (27). Studies have suggested a role for H2A.Z in chromosome segregation during mitosis in fission yeast (9) and during development in Drosophila (10). All of these diverse effects could be secondary manifestations of an effect on transcription.
In summary, we have shown that despite their sequence similarities, the major and minor H2A genes of S.cerevisiae each have at least one important, conserved function that cannot be supplied by the other, even when their regulatory sequences are swapped. Thus, the differences between these two genes must lie within their coding regions and it should now be possible to apply the powerful techniques of yeast genetics to determine these functions.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Min-Hao Kuo for yeast strains and advice on yeast genetics and Lynn Spitz for help with the tetrad dissections. This research was supported by Public Health Service grant GM-21793 from the National Institutes of Health.
REFERENCES
- 1.Hentschel C.C. and Birnstiel,M.L. (1981) Cell, 25, 301–313. [DOI] [PubMed] [Google Scholar]
- 2.Old R.W. and Woodland,H.R. (1984) Cell, 38, 624–626. [DOI] [PubMed] [Google Scholar]
- 3.Grunstein M., Diamond,K.E., Knoppel,E. and Grunstein,J.E. (1981) Biochemistry, 20, 1216–1223. [DOI] [PubMed] [Google Scholar]
- 4.Yu L.L. and Gorovsky,M.A. (1997) Mol. Cell. Biol., 17, 6303–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson J.D., Falciano,V.T. and Gorovsky,M.A. (1996) Trends Biochem. Sci., 21, 466–467. [DOI] [PubMed] [Google Scholar]
- 6.Thatcher T.H. and Gorovsky,M.A. (1994) Nucleic Acids Res., 22, 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Daal A. and Elgin,S.C.R. (1992) Mol. Biol. Cell, 3, 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X., Li,B. and Gorovsky,M.A. (1996) Mol. Cell. Biol., 16, 4305–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr A.M., Dorrington,S.M., Hindley,J., Phear,G.A., Aves,S.J. and Nurse,P. (1994) Mol. Gen. Genet., 245, 628–635. [DOI] [PubMed] [Google Scholar]
- 10.Clarkson M.J., Wells,J.R.E., Gibson,F., Saint,R. and Tremethick,D.J. (1999) Nature, 399, 694–697. [DOI] [PubMed] [Google Scholar]
- 11.Luger K., Mäder,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 12.Liu X., Bowen,J. and Gorovsky,M.A. (1996) Mol. Cell. Biol., 16, 2878–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tettelin H., Thierry,A., Goffeau,A. and Dujon,B. (1998) Yeast, 14, 601–616. [DOI] [PubMed] [Google Scholar]
- 14.Sikorski R.S. and Hieter,P. (1989) Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alani E., Cao,L. and Kleckner,N. (1987) Genetics, 116, 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gietz R.D. and Sugino,A. (1988) Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- 17.Hampsey M. (1997) Yeast, 13, 1099–1133. [DOI] [PubMed] [Google Scholar]
- 18.Aguilera A. (1994) Genetics, 136, 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolodrubetz D., Rykowski,M.O. and Grunstein,M. (1982) Proc. Natl Acad. Sci. USA, 79, 7814–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherman F. and Hicks,J. (1991) Methods Enzymol., 194, 21–37. [DOI] [PubMed] [Google Scholar]
- 21.Schuster T., Han,M. and Grunstein,M. (1986) Cell, 45, 445–451. [DOI] [PubMed] [Google Scholar]
- 22.Wu R.S., Tsai,S. and Bonner,W.M. (1982) Cell, 31, 367–374. [DOI] [PubMed] [Google Scholar]
- 23.White E.M. and Gorovsky,M.A. (1988) Mol. Cell. Biol., 8, 4780–4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R., Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arents G., Burlingame,R.W., Wang,B.-C., Love,W.E. and Moudrianakis,E.N. (1991) Proc. Natl Acad. Sci. USA, 88, 10148–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stargell L.A., Bowen,J., Dadd,C.A., Dedon,P.C., Davis,M., Cook,R.G., Allis,C.D. and Gorovsky,M.A. (1993) Genes Dev., 7, 2641–2651. [DOI] [PubMed] [Google Scholar]
- 27.Ross-Macdonald P., Coelho,P.S.R., Roemer,T., Agarwal,S., Kumar,A., Jansen,R., Cheung,K.H., Sheehan,A., Symoniatis,D., Umansky,L., Heldtman,M., Nelson,F.K., Iwasaki,H., Hager,K., Gerstein,M., Miller,P., Roeder,G.S. and Snyder,M. (1999) Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]