Abstract
Nucleotide excision repair plays a crucial role in removing many types of DNA adducts formed by UV light and chemical carcinogens. We have examined the interactions of Escherichia coli UvrABC nuclease proteins with three site-specific C8 guanine adducts formed by the carcinogens 2-aminofluorene (AF), N-acetyl-2-acetylaminofluorene (AAF) and 1-nitropyrene (1-NP) in a 50mer oligonucleotide. Similar to the AF and AAF adducts, the 1-NP-induced DNA adduct contains an aminopyrene (AP) moiety covalently linked to the C8 position of guanine. The dissociation constants for UvrA binding to AF–, AAF– and AP–DNA adducts, determined by gel mobility shift assay, are 33 ± 9, 8 ± 2 and 23 ± 9 nM, respectively, indicating that the AAF adduct is recognized much more efficiently than the other two. Incision by UvrABC nuclease showed that AAF–DNA was cleaved ∼2-fold more efficiently than AF– or AP–DNA (AAF > AF ≈ AP), even though AP has the largest molecular size in this group. However, an opened DNA structure of six bases around the adduct increased the incision efficiency for AF–DNA (but not for AP–DNA), making it equivalent to that for AAF–DNA. These results are consistent with a model in which DNA damage recognition by the E.coli nucleotide excision repair system consists of two sequential steps. It includes recognition of helical distortion in duplex DNA followed by recognition of the type of nucleotide chemical modification in a single-stranded region. The difference in incision efficiency between AF– and AAF–DNA adducts in normal DNA sequence, therefore, is a consequence of their difference in inducing structural distortions in DNA. The results of this study are discussed in the light of NMR solution structures of these DNA adducts.
INTRODUCTION
Aromatic amines, such as N-acetyl-2-aminofluorene (AAF), and ubiquitous environmental pollutants, like 1-nitropyrene (1-NP), are well-known chemical carcinogens (1–6). These chemicals are metabolically activated in cells and react with cellular DNA to form covalent adducts predominantly at the C8 position of dG residues. AAF forms two major DNA adducts, deacetylated C8-AF-dG and C8-AAF-dG (Fig. 1A). Similarly, the partially reduced derivative of 1-NP formed during metabolism reacts with DNA to form a major type of adduct, dG-C8-1-aminopyrene (C8-AP-dG, Fig. 1A) (7–9). Failure to remove these adducts could substantially increase the potential for genetic mutations, which may ultimately lead to tumorigenesis. Both the C8-AF-dG and C8-AAF-dG adducts are mutagenic in many bacterial and mammalian systems, although the types of mutations induced by these two adducts are different (5,6,10,11). Similarly, 1-NP, the most common nitroaromatic chemical in many environmental samples, is mutagenic in both bacterial and mammalian cells (12–14). It is also tumorigenic in experimental animals (15–16).
Figure 1.
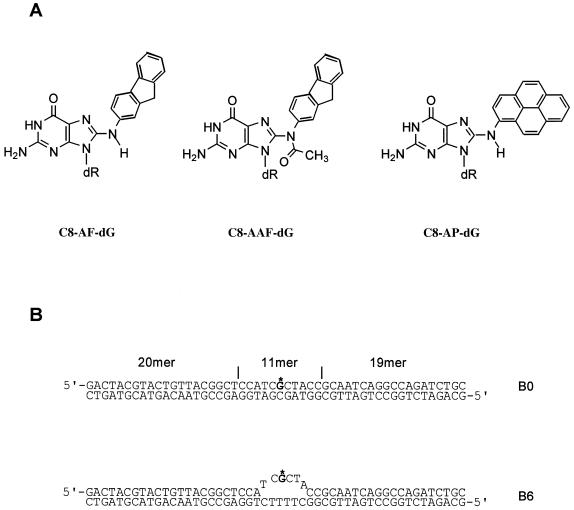
DNA adducts and substrates used in the present study. (A) Chemical structures of 2-aminofluorene (AF)-, N-acetyl-2-aminofluorene (AAF)- and 1-nitropyrene-derived 1-aminopyrene (AP)-C8-dG DNA adducts. (B) Substrates constructed for this study.
In both prokaryotic and eukaryotic cells, nucleotide excision repair (NER) is capable of repairing DNA damaged by a large variety of chemicals, albeit with varying efficiencies (17–21). The UvrABC nuclease system which initiates NER in Escherichia coli represents a paradigm in understanding the general mechanism of DNA damage recognition and incision (17). This model system has been widely used to study the interactions of NER with the DNA damage induced by various chemical and physical agents. We have recently postulated that DNA damage recognition by E.coli NER may be achieved through a sequential two-step mechanism in which the adduct-induced disruption of Watson–Crick DNA structure is recognized at the initial step while the type of modification of the nucleotide is recognized at the following step upon strand opening (Y.Zou and N.E.Geacintov, submitted for publication). A better understanding of how a specific DNA adduct is recognized and repaired by NER may be achieved by analyzing each step of repair in conjunction with structural information on the adduct. Specifically, the NMR solution structures of C8-AF-dG, C8-AAF-dG and C8-AP-dG adducts in DNA duplexes showed that both C8-AAF-dG and C8-AP-dG adopt a conformation in which the aminofluorene and aminopyrene rings are intercalated into the DNA helix and stack with the neighboring base pairs with the modified guanine displaced into the major groove (22–26). In contrast, the C8-AF-dG DNA adduct remains in equilibrium between AF-intercalated and AF-external (in the major groove) conformers with the external one as the predominant form (27–31). However, the interconversion is dependent on the DNA sequences neighboring the adduct. It is conceivable that these structural and conformational differences may result in the different biological effects. It has been reported that the UvrABC nuclease recognizes and cleaves both the C8-AF-dG and C8-AAF-dG DNA adducts in a similar manner when the adducts are randomly introduced into DNA (32,33). However, the relative efficiency of UvrABC incision of these two adducts varied dramatically depending on the DNA sequence surrounding the adducts (33). This is most likely attributable to the effects of sequence on the structures of AF– and AAF–DNA adducts. More studies need to be performed to address the structure–function issues.
Recent advances in defining the solution conformations of the AF–, AAF– and AP–DNA adducts by NMR spectroscopy enable us to probe the structure–function relationship of NER of these DNA adducts. In the present study we have systematically examined and compared the interactions of the UvrABC nuclease system with a DNA duplex containing a single site-specifically placed C8-AF-dG, C8-AAF-dG or C8-AP-dG adduct. This group of adducts includes a pair having a similar conformation but with different chemical modifications (C8-AAF-dG and C8-AP-dG) and another having an identical aromatic ring system but with different conformations (C8-AF-dG and C8-AAF-dG). Our study provides a better understanding of the relationship between the structural characteristics of the DNA adducts and their recognition and the efficiency of incision.
MATERIALS AND METHODS
Protein purification
UvrA was purified from E.coli strain MH1 ΔUvrA containing the overproducing plasmid pSST10 (graciously supplied by L. Grossman, Johns Hopkins University), in which the uvrA gene is under the control of the heat-inducible PL promoter. UvrB was purified in one step through a chitin column from E.coli strain XL-1 Blue transformed with the overexpressing plasmid pUTG97 containing the uvrB gene under control of the IPTG-induced Ptac promoter as described previously (34). UvrC was overproduced from E.coli C41(DE3) cells (35) harboring plasmid pUTG98 containing the PCR-amplified uvrC gene, which was subcloned via NdeI and KpnI restriction sites into vector pTYB1 (IMPACT T7 system; New England Biolabs). The UvrC protein was also purified on a chitin column in one step following the same procedures as described previously for UvrB (34) except that 500 mM rather than 100 mM NaCl was used in the cleavage and elution buffers.
DNA substrate construction
The 11mers d(CCATCG*CTACC) modified at G* with the C8 guanine adduct of AF, AAF or AP were synthesized, purified and characterized as described previously (22,29,36). The 50 bp oligonucleotides containing a single AF, AAF or AP were constructed as described previously (34,37). Briefly, the phosphorylated 11mers (30 pmol) were ligated with stoichiometric quantities of 20mer and phosphorylated 19mer, using T4 DNA ligase in the presence of a 55mer template strand containing the complementary sequence of 50 bases in a 30 µl solution containing 50 mM Tris–HCl, pH 7.8, 10 mM MgCl2, 10 mM DTT, 1 mM ATP and 50 µg/ml BSA. The ligation was carried out at 16°C for 12 h. After ligation, the products were purified and then re-annealed with various 50mer template strands to make appropriate substrates, as shown in Figure 1. The annealed substrates were purified on a non-denaturing 8% polyacrylamide gel.
Gel mobility shift assays
Binding of the UvrA protein to the DNA substrates was determined by gel mobility shift assay. Typically, the substrate (2 nM) was incubated with UvrA at varying concentrations as indicated at 37°C for 15 min in 20 µl of UvrABC buffer (50 mM Tris–HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM DTT) in the presence of 1 mM ATP. After incubation, 2 µl of 80% (v/v) glycerol was added and the mixture was immediately loaded onto a 3.5% native polyacrylamide gel in TBE running buffer and electrophoresed at room temperature. After quantification of the radioactivity of the corresponding bands in the gel, the data were analyzed as described previously to obtain the dissociation constants (38).
Incision assays
The 5′-terminally labeled DNA substrates (2 nM) were incised by UvrABC (UvrA, 10 nM, UvrB, 250 nM and UvrC, 50 nM) in UvrABC buffer (1 mM ATP) at 37°C for 0, 5, 10 and 30 min. The Uvr subunits were diluted and pre-mixed into storage buffer before mixing with DNA. The reactions were terminated by adding EDTA (20 mM) or heating to 90°C for 3 min. The samples were denatured with formamide and heated to 90°C for 5 min and then quick chilled on ice. The digested products were analyzed by electrophoresis on a 12% polyacrylamide sequencing gel under denaturing conditions with TBE buffer.
RESULTS
Binding of UvrA to AF–, AAF– and AP–DNA adducts
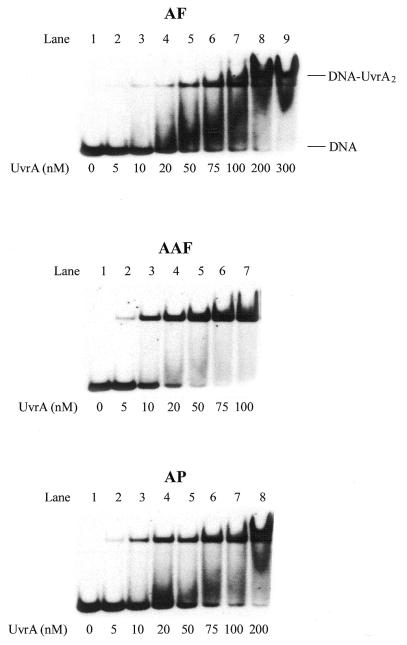
Figure 1A shows the structure of the three C8 guanine adducts and Figure 1B shows the 50mer substrates used in this work. In E.coli NER UvrA plays a crucial role in initial damage recognition. To understand the structural basis of recognition and incision of C8-AF-dG, C8-AAF-dG and C8-AP-dG adducts by UvrABC, a gel mobility shift assay was performed to examine binding of these DNA adducts with increasing concentration of UvrA. As shown in Figure 2, incubation of the substrates with UvrA produced a complex as determined from the slower mobility of a band containing 32P-radiolabeled DNA, which is consistent with the formation of a UvrA2–DNA complex. The faster running band represents the substrate free of proteins. Titration of the binding with varying UvrA concentrations generated binding isotherms, which were then used for determination of the dissociation constants (38), as listed in Table 1. In general, the order of binding affinity was AAF >> AP > AF, which implies that the AAF adduct distorts the DNA helix most. The solution NMR studies are in agreement with this conclusion (22–24).
Figure 2.
Binding of UvrA to AF–, AAF– and AP–DNA substrates. UvrA at the concentrations specified was incubated at 37°C for 10 min with 1 nM AF–, AAF– or AP–DNA substrates (B0) in UvrABC buffer and then analyzed on a 4% polyacrylamide native gel in a gel mobility shift assay.
Table 1. Equilibrium dissociation constants for binding of DNA adducts by UvrA at 37°Ca.
| Adducts | Kd (nM) |
|---|---|
| AF | 33 ± 9 |
| AAF | 8 ± 2 |
| AP | 23 ± 9 |
aThe data represent the means ± SD of at least three independent experiments. The dissociation constants were determined for UvrA dimers unless otherwise indicated.
UvrABC incision of AF–, AAF– and AP–DNA adducts
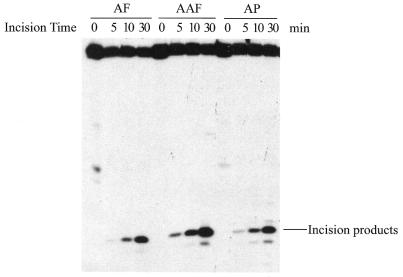
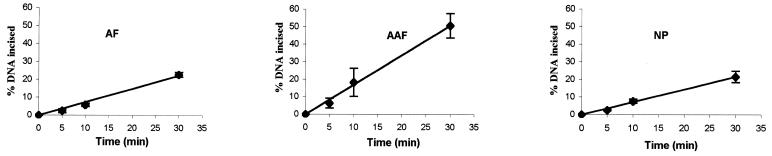
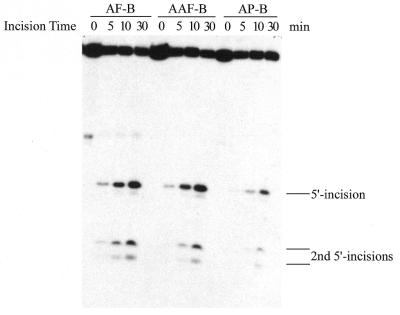
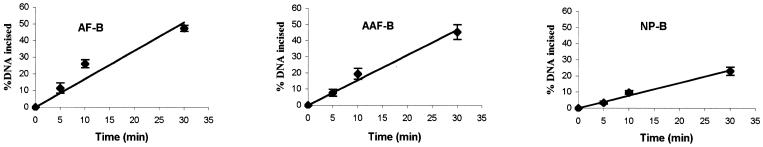
As shown in Figure 3A, the 50 bp DNA substrate containing a single C8-AF-dG, C8-AAF-dG or C8-AP-dG adduct (B0 in Fig. 1B) was incised by UvrABC nuclease in a kinetic assay. These substrates were labeled at the 5′-end of the modified strand. The results indicated that the repair proteins recognized and cleaved all three types of adducts but with varying efficiencies, and in each case the 5′ incision occurred at the eighth phosphate 5′ to the modified guanine. At least three independent experiments were performed to determine the rate of incision (Fig. 3B). The initial rate was determined by a linear least squares fit of the data collected over the incision period. Table 2 shows the incision efficiency for these substrates, which follows the order AAF > AF ≈ AP. The AF adduct was incised much more slowly than the AAF–DNA adduct, while the AP and AF adducts showed similar incision efficiencies.
Figure 3.
Incision of AF–, AAF– and AP–DNA adducts by UvrABC nuclease. (A) The 5′-terminally labeled DNA substrates (B0, 3 nM) containing an AF, AAF or AP adduct were incubated with UvrABC (10 nM UvrA, 250 nM UvrB and 100 nM UvrC) in UvrABC buffer at 37°C for the period indicated. The incision products were then analyzed on a 12% polyacrylamide sequencing gel. (B) Incision kinetics of the DNA substrates incised by UvrABC. Data represent the means ± SD of at least three independent experiments.
Table 2. Initial rate of UvrABC incisions of DNA substratesa.
| Substrate | Initial incision rate (fmol/min) | ||
|---|---|---|---|
| AF | AAF | AP | |
| B0 | 0.22 ± 0.02 | 0.51 ± 0.09 | 0.22 ± 0.03 |
| B6 | 0.51 ± 0.03 | 0.47 ± 0.05 | 0.24 ± 0.03 |
aThe data represent the means ± SD of three to six independent experiments.
During damage recognition by E.coli NER two modification characteristics of a DNA adduct are believed to be recognized in two separate steps (Y.Zou and N.E.Geacintov, submitted for publication). First, adduct-induced structural distortion of the DNA double helix is identified by UvrA2/UvrA2B. After first recognition, a local DNA strand opening occurs, which then allows discrimination based on the type of DNA base modification at the second step. So the second step depends on success in the first step of recognition. In order to understand the mechanism involved in recognition and thus incision of C8-AF-dG-, C8-AAF-dG- and C8-AP-dG-containing DNA, bubble substrates containing a six-base mismatched region around the adduct were constructed (B6, Fig. 1B). The pre-existing strand separation in the substrates mimics the strand opening during NER and allows the initial recognition step to be skipped. Incision of these substrates by UvrABC nuclease was conducted in the same way and under the same conditions as in Figure 3 and the results are shown in Figure 4A and B and Table 2. The second 5′ incisions of the bubble substrates at the fifteenth and sixteenth phosphates 5′ to the adduct in Figure 4A are directly correlated and coupled with the normal 5′ incision at the eighth phosphate (39,40; Y.Zou and N.E.Geacintov, submitted for publication). Therefore, to determine incision efficiency, the incision products from both the normal and second 5′ incisions have been combined. In comparison with the full duplex substrates (B0), the incision efficiency for the C8-AF-dG-containing bubble substrate increased to the same level as that for C8-AAF-dG DNA. In contrast, however, the incision of C8-AAF-dG and C8-AP-dG DNA bubble substrates remained at the same level as the normal duplex substrate B0 (Table 2). These results strongly suggest that the differential incision between AF– and AAF–DNA is most likely due to the difference in the extent of DNA helical distortion they induce, which may be recognized at the first step. A comparison of incisions between bubbled C8-AAF-dG and C8-AP-dG DNA substrates seems to indicate that the former causes a more severe helix distortion than the latter in terms of damage recognition, as the repair proteins incised the C8-AAF-dG DNA bubble substrate more efficiently.
Figure 4.
Incision of AF–, AAF– and AP–DNA bubble substrates by UvrABC. (A) 5′-Terminally labeled DNA bubble substrates (B6, 3 nM) containing an AF, AAF or AP adduct were incubated with UvrABC (10 nM UvrA, 250 nM UvrB and 100 nM UvrC) in UvrABC buffer at 37°C for the period indicated. The incision products were then analyzed on a 12% polyacrylamide sequencing gel. (B) Incision kinetics of the DNA substrates incised by UvrABC. Data represent the means ± SD of at least three independent experiments.
DISCUSSION
One of the major challenges in molecular toxicology is to define the structural and chemical basis for mutagenesis and carcinogenesis induced by lesions in DNA. DNA repair has been recognized as the crucial cellular pathway that corrects DNA damage and avoids genetic alterations. Therefore, it is important to understand the structure–function relationship of DNA damage recognition and repair. In the present study we have investigated and compared the interactions of UvrABC nuclease with the C8 guanine adducts of AF, AAF and AP. Elucidation of the structures of these adducts by NMR spectroscopy (22–31) provides important information required for understanding the differential incision of these adducts and the mechanism governing recognition and repair.
Rather than residing in the major groove, the AAF and AP moieties of C8-AF-dG and C8-AP-dG intercalate into the DNA helix and stack with the neighboring base pairs. In contrast, the AF adduct remains in an equilibrium in which the major conformer maintains Watson–Crick hydrogen bonding with the AF ring positioned externally in the major groove of a minimally perturbed B-DNA helix (25,27–31). Specific binding of UvrA2 to DNA adducts is believed to be important in the initial step of damage recognition, although during normal repair in a cellular environment the UvrA2B protein complex is actually involved. Since UvrA is a molecular matchmaker, direct measurement of UvrA2B binding would seem to be difficult. The equilibrium dissociation constants presented in Table 1 indicate that AAF–DNA has a much higher affinity (4- to 5-fold) for UvrA2 than AF–DNA. The incision results for these substrates (B0), summarized in Table 1, also follow the same trend. In contrast to the striking conformational differences between the AF and AAF adducts, dG residues in both the AAF– and AP–DNA adducts are in the syn orientation and the aromatic ring systems are intercalated into the DNA. Nevertheless, the AP adduct actually has a much lower UvrA binding affinity. Our finding that AAF–DNA is subject to more efficient incision than AP–DNA suggests that some critical structural features may be recognized at the second step of recognition. The behavior of the adduct in a local single-stranded region probably plays a major role in incision by the UvrBC proteins.
As in the case of UvrA binding, AAF–DNA served as the best substrate for UvrABC incision, while the AF– and AP–DNA adducts were incised less efficiently. The differential incision between AF– and AAF–DNA may be attributed to larger helical distortion or disruption of the Watson–Crick DNA structure in the latter, which is probably easier for the UvrA2B protein complex to recognize. The adducts appear to be discriminated primarily at the first step of recognition. To further clarify this aspect, the same adducts were constructed in a DNA sequence with a six base mismatched region surrounding the damaged site (Fig. 1B). We used these bubble substrates to separate the two steps of damage recognition by UvrABC and to determine the origin of the differential incisions. We believe that the second level of recognition is dependent on the type of chemical modification rather than helix destabilization (35; Y.Zou and N.E.Geacintov, submitted for publication). The strand separation may permit UvrB or UvrBC to fully access the modified nucleotide. The data presented here indicate that strand opening results in a significant enhancement of incision for the C8-AF-dG adduct but not for C8-AAF-dG (Fig. 3 and Table 2). Both the AF– and AAF–DNA adducts when placed in the bubble were incised with equal efficiency. These results, when compared with those for the normal DNA substrate (B0) (Fig. 3 and Table 2), strongly suggest that the AF– and AAF–DNA adducts are equally recognized in the second step and discriminated only during initial recognition due to their differences in helical distortion. Similar to AAF–DNA, opening of the DNA duplex around the C8-AP-dG site has only a marginal effect on incision efficiency, consistent with the fact that C8-AP-dG is an intercalating adduct in which the induced helical distortion may be relatively more efficiently recognized at the initial step than chemical modification at the second step. This suggests that differential incision between AAF– and AP–DNA is due to differences in chemical structure, since both adducts prefer to assume a conformation such that the polycyclic aromatic moiety is intercalated with the DNA bases. Therefore, they are discriminated primarily during the second step of recognition in which the aminofluorene ring may better fit a pocket of UvrB than the aminopyrene ring. It is conceivable that the 3-dimensional features of AAF, which include an sp3 hybridized C9, are more recognizable to the Uvr system than the completely planar ring system of AP.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs R. Stephen Lloyd and Ben Van Houten for their generous support and encouragement. We are grateful to Dr David Konkel for critical reading of this manuscript. This study was supported by NIEHS grants ES09127 and ES00318 (to A.K.B.) and ES07955 (to R.S.L.) and an NIEHS Center grant ES06676 (to R.S.L.).
REFERENCES
- 1.Rosenkranz H.S., McCoy,E.C., Sanders,D.R., Butler,M., Kiriazides,D.K. and Mermelstein,R. (1980) Science, 209, 1039–1043. [DOI] [PubMed] [Google Scholar]
- 2.Tokiwa H. and Ohnishi,Y. (1986) CRC Crit. Rev. Toxicol., 17, 23–60. [DOI] [PubMed] [Google Scholar]
- 3. IARC (1989) IARC Monographs on the Evaluation of Carcinogenic Risk to Humans, Vol. 39. IARC, Lyon.
- 4.Kriek E. (1992) J. Cancer Res. Clin. Oncol., 118, 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heflich R.H. and Neft,R.E. (1994) Mutat. Res., 318, 73–114. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann G.R. and Fuchs,R.P.P. (1997) Chem. Res. Toxicol., 10, 347–359. [DOI] [PubMed] [Google Scholar]
- 7.Howard P.C., Beland,F.A. and Cerniglia,C.E. (1983) Carcinogenesis, 4, 985–990. [DOI] [PubMed] [Google Scholar]
- 8.Howard P.C., Heflich,R.H., Evans,F.E. and Beland,F.A. (1983) Cancer Res., 43, 2052–2058. [PubMed] [Google Scholar]
- 9.Stanton C.A., Chow,F.L., Phillips,D.H., Grover,P.L., Garner,R.C. and Martin,C.N. (1985) Carcinogenesis, 6, 535–538. [DOI] [PubMed] [Google Scholar]
- 10.Shibutani S., Suzuki,N. and Grollman,A.P. (1998) Biochemistry, 37, 12034–12041. [DOI] [PubMed] [Google Scholar]
- 11.Mah M.C.-M., Bolt,J., Culp,S.J., Maher,V.M. and McCormick,J.J. (1991) Proc. Natl Acad. Sci. USA, 83, 10193–10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenkranz H.S. and Mermelstein,R. (1983) Mutat. Res., 114, 217–267. [DOI] [PubMed] [Google Scholar]
- 13.Stanton C.A., Garner,R.C. and Martin,C.N. (1988) Carcinogenesis, 9, 1153–1157. [DOI] [PubMed] [Google Scholar]
- 14.Yang J.-L., Maher,V.M. and McCormick,J.J. (1988) Mol. Cell. Biol., 8, 3364–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Bayoumy K., Hecht,S.S., Sacki,T. and Stoner,G.D. (1984) Carcinogenesis, 5, 1449–1452. [DOI] [PubMed] [Google Scholar]
- 16.Hirose M., Lee,M.-S., Wang,C.Y. and King,C.M. (1984) Cancer Res., 44, 1158–1162. [PubMed] [Google Scholar]
- 17.Van Houten B. (1990) Microbiol. Rev., 54, 18–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood R.D. (1996) Annu. Rev. Biochem., 65,135–167. [DOI] [PubMed] [Google Scholar]
- 19.Sancar A. (1996) Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- 20.Lindahl T. and Wood,R.D. (1999) Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- 21.Batty D.P. and Wood,R.D. (2000) Gene, 241, 193–204. [DOI] [PubMed] [Google Scholar]
- 22.O’Handley S.F., Sanford,D.G., Xu,R., Lester,C.C., Hingerty,B.E., Broyde,S. and Krugh,T.R. (1993) Biochemistry, 32, 2481–2497. [DOI] [PubMed] [Google Scholar]
- 23.Milhe C., Dhalluin,C., Fuchs,R.P. and Lefevre,J.F. (1994) Nucleic Acids Res., 22, 4646–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milhe C., Fuchs,R.P. and Lefevre,J.F. (1996) Eur. J. Biochem., 235, 120–127. [DOI] [PubMed] [Google Scholar]
- 25.Cho B.P and Zhou,L. (1999) Biochemistry, 38, 7572–7583. [DOI] [PubMed] [Google Scholar]
- 26.Mao B., Vyas,R.R., Hingerty,B.E., Broyde,S., Basu,A.K. and Patel,D.J. (1996) Biochemistry, 35, 12659–12670. [DOI] [PubMed] [Google Scholar]
- 27.Eckel L.M. and Krugh,T.R. (1994) Biochemistry, 33, 13611–13624. [DOI] [PubMed] [Google Scholar]
- 28.Eckel L.M. and Krugh,T.R. (1994) Nature Struct. Biol., 1, 89–94. [DOI] [PubMed] [Google Scholar]
- 29.Cho B.P., Beland,F.A. and Marques,M.M. (1994) Biochemistry, 33, 1373–1384. [DOI] [PubMed] [Google Scholar]
- 30.Mao B. and Hingerty,B.E., Broyde,S. and Patel,D.J. (1998) Biochemistry, 37, 81–94. [DOI] [PubMed] [Google Scholar]
- 31.Mao B. and Hingerty,B.E., Broyde,S. and Patel,D.J. (1998) Biochemistry, 37, 95–106. [DOI] [PubMed] [Google Scholar]
- 32.Fuchs R.P.P. and Seeberg,E. (1984) EMBO J., 3, 757–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierce J.R., Case,R. and Tang,M. (1989) Biochemistry, 28, 5821–5826. [DOI] [PubMed] [Google Scholar]
- 34.Zou Y. and Van Houten,B. (1999) EMBO J., 18, 4889–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miroux B. and Walker,J.E. (1996) J. Mol. Biol., 260, 289–298. [DOI] [PubMed] [Google Scholar]
- 36.Vyas R.R., Nolan,S.J. and Basu,A.K. (1993) Tetrahedron Lett., 34, 2247–2250. [Google Scholar]
- 37.Zou Y., Liu,T.M., Geacintov,N.E. and Van Houten,B. (1995) Biochemistry, 34, 13582–13593. [DOI] [PubMed] [Google Scholar]
- 38.Zou Y., Bassett,H., Walker,R., Bishop,A., Amin,S., Geacintov,N.E. and Van Houten,B. (1998) J. Mol. Biol., 281, 107–119. [DOI] [PubMed] [Google Scholar]
- 39.Moolenaar G.F., Bazuine,M., van Knippenberg,I.C., Visse,R. and Goosen,N. (1998) J. Biol. Chem., 273, 34896–34903. [DOI] [PubMed] [Google Scholar]
- 40.Gordienko I. and Rupp,W.D. (1998) EMBO J., 17, 626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]