Abstract
Scorpion primers can be used to detect PCR products in homogeneous solution. Their structure promotes a unimolecular probing mechanism. We compare their performance with that of the same probe sequence forced to act in a bimolecular manner. The data suggest that Scorpions indeed probe by a unimolecular mechanism which is faster and more efficient than the bimolecular mechanism. This mechanism is not dependent on enzymatic cleavage of the probe. A direct comparison between Scorpions, TaqMan and Molecular Beacons on a Roche LightCycler indicates that Scorpions perform better, particularly under fast cycling conditions. Development of a cystic fibrosis mutation detection assay shows that Scorpion primers are selective enough to detect single base mutations and give good sensitivity in all cases. Simultaneous detection of both normal and mutant alleles in a single reaction is possible by combining two Scorpions in a multiplex reaction. Such favourable properties of Scorpion primers should make the technology ideal in numerous applications.
INTRODUCTION
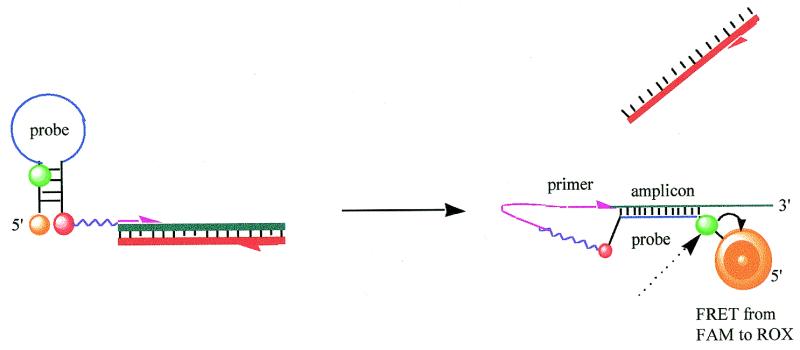
Scorpion primers are used in a fluorescence based method for the specific detection of PCR products (1,2). A Scorpion (Fig. 1) consists of a specific probe sequence that is held in a hairpin loop configuration by complementary stem sequences on the 5′ and 3′ sides of the probe. The fluorophore attached to the 5′-end is quenched by a moiety (normally methyl red) joined to the 3′-end of the loop. The hairpin loop is linked to the 5′-end of a primer via a PCR stopper. After extension of the primer during PCR amplification, the specific probe sequence is able to bind to its complement within the same strand of DNA. This hybridisation event opens the hairpin loop so that fluorescence is no longer quenched and an increase in signal is observed. The PCR stopper prevents read-through, which could lead to opening of the hairpin loop in the absence of the specific target sequence. Such read-through would lead to the detection of non-specific PCR products, e.g. primer dimers or mispriming events.
Figure 1.

Scorpion probing mechanism. Step 1: initial denaturation of target and Scorpion stem sequence. Step 2: annealing of Scorpion primer to target. Step 3: extension of Scorpion primer produces double-stranded DNA. Step 4: denaturation of double-stranded DNA produced in step 3. This gives a single-stranded target molecule with the Scorpion primer attached. Step 5: on cooling, the Scorpion probe sequence binds to its target in an intramolecular manner. This is favoured over the intermolecular binding of the complementary target strand.
Unlike Molecular Beacons (3–5) and TaqMan probes (6,7) which can be used as alternatives, the Scorpion system does not require a separate probe. In the TaqMan assay an oligonucleotide probe is labelled at one end with a fluorophore and at the other end with a fluorescence quencher. When the probe binds to the target site in a PCR product the 5′–3′ exonuclease activity of Taq DNA polymerase cleaves it between the fluorophore and quencher, thereby producing an increase in fluorescence. Molecular Beacons consist of a probe flanked by a hairpin loop that holds a fluorophore and quencher in close proximity until specific binding of the probe to its target opens out the structure, producing a fluorescent signal. Although it is believed that Scorpions work in a unimolecular manner (1) this has not been proved and it is possible that they could also act as Molecular Beacons, probing by an inter- rather than intra-molecular manner. The mechanism of Scorpion action is investigated and compared to the bimolecular probing of Molecular Beacons and TaqMan.
Having compared the efficiency of Scorpion primers with Molecular Beacons and TaqMan, we investigate the use of Scorpion primers for allelic discrimination. With the near completion of the human genome project a large number of multi-allelic SNPs (8) will be characterised and used for a variety of applications such as genetic mapping. Scorpions can be adapted to mutation or allelic discrimination by monitoring the fluorescence at a temperature where the probe has dissociated from a target with a mismatch but remains bound to a complementary target. This is different to the allelic discrimination by Scorpions described by Whitcombe et al. (1) as they used the ARMS system, whereby the primer is sited over the polymorphic site rather than the probe. We chose cystic fibrosis mutation analysis as a test assay. Cystic fibrosis is an autosomal recessive disease that is estimated to occur in one in 2000 live births and is carried by 5% of the population (9). It is caused by mutations in a gene on chromosome 7 (ABCC7) that produces the cystic fibrosis transmembrane conductance regulator protein. These mutations affect the transport of chloride ions across the apical membrane of epithelial cells, producing a number of clinical symptoms (10). The most common mutation is ΔF508, a 3 bp deletion found on ~70% of all cystic fibrosis chromosomes, causing severe clinical symptoms (11). However, a number of other mutations have been found, which give a range of symptoms that differ in severity (12). Various methods have been employed for detecting ABCC7 mutations. Direct sequencing of the regions containing the mutations can determine presence or absence of mutations (12) but it is costly and time consuming, particularly if all known mutations sites have to be assayed. Other methods include allele specific PCR (13), single-strand conformation polymorphism (14), allele specific oligonucleotide hybridisation (12) and TaqMan probes (PE Biosystems).
We have used Scorpion primers to detect five common ABCC7 mutations, ΔF508 (11–13), N1303K (15), W1282X (12), G542X (12) and G551D (12,16) (Table 1). We have also adapted the technology for multiplex PCR on the Roche LightCycler. This instrument is extremely fast and is capable of producing results in <30 min.
Table 1. ABCC7 loci and GenBank accession nos.
| Loci | GenBank accession no. | Mutation site | Base change | Probe–target mismatch |
|---|---|---|---|---|
| ΔF508 |
M55115 |
436–438 |
CTT del |
– |
| N1303K |
M55128 |
329 |
C→G |
C–C |
| W1282X |
M55127 |
395 |
G→A |
C–A |
| G551D |
M55116 |
362 |
G→A |
C–A |
| G542X | M55116 | 334 | G→T | C–T |
Mutation sites are numbered according to the GenBank base numbering.
MATERIALS AND METHODS
Human genomic DNA samples were obtained from Coriell and were stored in 1% bovine serum albumin (BSA) at a concentration of 5 ng/µl. Sequence data for the ABCC7 loci were obtained from GenBank (17) (see Table 1 for loci, accession nos and mismatches). Primer sequences were designed close to the mutation site using Oligo 4.0. software (National Biosciences Inc., Plymouth, MN) to give amplicons of ~100–200 bases. The Scorpion probe sequence was attached to the primer closer to the mutation site except for G90 (Table 2). All unlabelled primers and Scorpions were synthesised by Oswel Research Products Ltd and purified by reversed-phase HPLC (1). Scorpion folding was modelled using the DNA mfold programme (http://www.ibc.wustl.edu/∼zuker ) and thermodynamic parameters of Santalucia (18).
Table 2. Scorpion and primer sequences.
| Oligo name | Code | Oligo sequence |
|---|---|---|
| MTHFR forward primer | BPF | 5′-CTGACCTGAAGCACTTGAAGG-3′ |
| MTHFR reverse primer | BPR | 5′-ATGTCGGTGCATGCCTTCAC-3′ |
| MTHFR Molecular Beacon | MMB | 5′-FAM GCGAGTGCGGGAGCCGATTTCTCGC MR-3′ |
| MTHFR TaqMan | MT | 5′-FAM TGCGGGAGCCGATTT TAMRA-3′ |
| MTHFR Scorpion | MS | 5′-FAM CCCGCGGAAATCGGCTCCCGCACCGCGGG MR HEG CTGACCTGAAGCACTTGAAGG-3′ |
| ΔF508 normal Scorpion | 508S | 5′-FAM CCGCGCAAACACCAAAGATGATATTTTCTGCGCGG MR HEG AGTTTTCCTGGATTATGCCT-3′ |
| ΔF508 mutant Scorpion | 508M | 5′-ROX CCGC(F)GCAAACACCAATGATATTTTCTGCAGCGG MR HEG AGTTTTCCTGGATTATGCCT-3′ |
| ΔF508 reverse primer | 508R | 5′-TTGGGTAGTGTGAAGGGTTC-3′ |
| ΔF508 forward primer | 508F | 5′-AGTTTTCCTGGATTATGCCT-3′ |
| Hybrid Scorpion | HS | 5′-FAM CCGCGCAAACACCAAAGATGATATTTTCTGCGCGG MR HEG CTTGGAGAAGGTGGAATCAC-3′ |
| N1303K Scorpion | N13S | 5′-FAM CCCGCGCGGAACATTTAGAAAAAACTTGGATCCCGCGCGGG MR HEG TTTCTTGATCACTCCACTGTTC-3′ |
| N1303K reverse primer | N13R | 5′-CATACTTTCTTCTTCTTTTCTTT-3′ |
| W1282X Scorpion | W12S | 5′-FAM CCCGCGCCTTTCCTCCACTGTTGCGCGCGGG MR HEG ATGGTGTGTCTTGGGATTCA-3′ |
| W1282X reverse primer | W12R | 5′-GGCTAAGTCCTTTTGCTCAC-3′ |
| G551D Scorpion | 551S | 5′-FAM CCCGCGCCTCGTTGACCTCCACTCGCGCGGG MR HEG CTTGGAGAAGGTGGAATCAC-3′ |
| G551D reverse primer | 551R | 5′-AAATGCTTGCTAGACCAATA-3′ |
| G551D forward primer | 551F | 5′-CTTGGAGAAGGTGGAATCAC-3′ |
| G551D-DIST Scorpion (90 bases between 3′-end of Scorpion primer and 5′-end of probe target) | G90 | 5′-FAM CCCGCGCCTCGTTGACCTCCACTCGCGCGGG MR HEG CAGATTGAGCATACTAAAAG-3′ |
| G542X Scorpion | G542S | 5′-FAM CCGCGCACCTTCTCCAAGAACTAGCGCGG MR HEG CCAAGTTTGCAGAGAAAGAC-3′ |
| G542X reverse primer | G542R | 5′-AAATGCTTGCTAGACCAATA-3′ |
MR, methyl red attached to the 1 position of deoxyribose by a hexyl linker; HEG, hexethylene glycol; FAM, fluorescein attached to the oligonucleotide by a hexyl linker; F, internal FAM attached to the 5 position of deoxyuridine by an 11 atom spacer.
All PCR reactions were carried out on a Roche LightCycler. Ten microlitre volumes were used, which included 0.5 µM unlabelled primers and Scorpions [0.4 µM primers and probes for methylenetetrahydrofolate reductase (MTHFR) tests], 200 µM dNTPs, 4 mM magnesium chloride, 250 ng/µl BSA and 0.5 U of Thermoprimeplus Taq polymerase with the manufacturer’s buffer IV (Abgene). For comparison between Scorpions, TaqMan and Molecular Beacons we used 0.5 U of GoldStar Taq polymerase with the manufacturer’s buffer (Eurogentec). GoldStar and Thermoprimeplus are similar enzymes and the change was made due to availability of enzyme rather than difference in activity. To test the susceptibility of Scorpions to 5′-exonuclease degradation we used a 5′–3′ exonuclease deficient enzyme (DNAmp Ltd) that worked under the same reaction conditions as GoldStar. For those reactions requiring SYBR Gold, 1 µl was added from a 1/1000 dilution of the stock. Human genomic DNA (5 ng) was added to each reaction. This was replaced with sterile water for the negative controls. Cycling conditions were an initial denaturation at 95°C for 15 s (3 min for GoldStar), 100 cycles of 95°C for 0 s, annealing temperature for 0 s and monitoring for 3 s (see Table 3 for annealing and monitoring temperatures). A single fluorescence measurement was made for each cycle during the monitoring step. The slow cycling conditions for the MTHFR experiments described in Results were 95°C for 30 s, 58°C for 60 s and 72°C for 30 s. The amplifications reached a plateau before the hundredth cycle in all reactions described in this paper. Melt curves and cooling steps were carried out as described in the LightCycler manual (19). The fluorescence monitoring temperature was initially found from the mfold programme as the temperature at which the Scorpion remained bound to the normal amplicon but had dissociated from the mutant. However, the melt curve carried out at the end of each PCR reaction gave a more accurate measure of the temperature that should be used for the best possible mutation discrimination. Monitoring temperatures were adjusted accordingly. Fluorescence gains (used to set sensitivity of detection) employed for the LightCycler were F1–5 and F2–15 when multiplexing was carried out and F1–2 for singleplex reactions.
Table 3. Annealing and fluorescence monitoring temperatures used for each locus.
| Loci/test | Annealing temperature (°C) | Monitoring temperature (°C) |
|---|---|---|
| ΔF508 |
48 |
51 |
| N1303K |
44 |
61 |
| W1282X |
49 |
53 |
| G551D |
47 |
55 |
| G551D-DIST |
46 |
55 |
| G542X |
48 |
53 |
| Unimolecular versus bimolecular test |
47 |
51 |
| Distance constraints |
|
|
| G90 Scorpion |
46 |
55 |
| G551DS Scorpion |
47 |
55 |
| MTHFR | 58 | 58 |
RESULTS AND DISCUSSION
Unimolecular versus bimolecular mechanism
In order to test the probing mechanism we made a hybrid Scorpion (Table 2, HS). This consisted of the primer from one locus attached to the probe of another locus. We used the cystic fibrosis loci ΔF508 and G551D (12). The probe sequence for ΔF508 was attached to the G551D forwards primer (Table 2, 551F). Several experiments were then carried out within the same LightCycler run. All samples were run in duplicate with negative controls for each test. The tests were as follows.
(i) HS, G551D unlabelled lower primer (Table 2, 551R) and two unlabelled ΔF508 primers without a probe sequence attached (Table 2, 508F and 508R), were added to the PCR reactions (Fig. 2, test 1). In this case the probe is unable to bind to the extension of the Scorpion primer, which contains the G551D rather than ΔF508 target site. However, 508F and 508R will give a PCR product to which the probe can bind in a bimolecular manner. Therefore the Scorpion can only act as a Molecular Beacon although it will have the G551D PCR product attached.
Figure 2.
Unimolecular versus bimolecular test 1. Unimolecular probing is not possible as the amplicon attached to the Scorpion primer does not contain the probe target site. All probing must be bimolecular with the probe having the 551 amplicon as a tail. Unimolecular versus bimolecular test 2. Probing is bimolecular as in test 1 but extension of the Scorpion primer is not possible as the unlabelled primer is not present. Unimolecular versus bimolecular test 3. Unimolecular probing by a scorpion primer. The key is the same as for Figure 1.
(ii) As above except 551R was excluded. Again the Scorpion can only act as a Molecular Beacon but without the G551D PCR product tail attached (Fig. 2, test 2).
(iii) The standard ΔF508 Scorpion (Table 2, 508S) was run to compare the above bimolecular reactions with a reaction that can act in a unimolecular manner (Fig. 2, test 3).
SYBR Gold, a DNA intercalator that binds non-specifically to double-stranded DNA (20), showed that all the samples amplified efficiently (results not shown). Therefore, the observed differences in signal in tests 1 to 3 could not be due to differing levels of PCR product. It should be noted that when using carboxy fluorescein (FAM)-labelled probes the fluorescence from the probe cannot be observed when SYBR Gold is added to the reaction because both SYBR Gold and FAM are monitored on the same channel. The SYBR Gold reaction has to be carried out in a separate capillary so is a less valid control than the cases when the Scorpion fluorescence and SYBR Gold can be monitored within the same reaction tube (i.e. if the Scorpion is labelled with a different fluorophore). Reactions without SYBR Gold indicate the level of fluorescence given by the probe (Fig. 3). In tests 1 and 2 the probing gave a level of fluorescence much lower than that of the Scorpion. The Molecular Beacon-like probing also gave different shaped curves to the Scorpion, which increased rapidly before levelling off. Both of these factors suggest that normal Scorpions work by a different mechanism than the bimolecular mechanism of tests 1 and 2. This is evidence for unimolecular probing by Scorpions.
Figure 3.
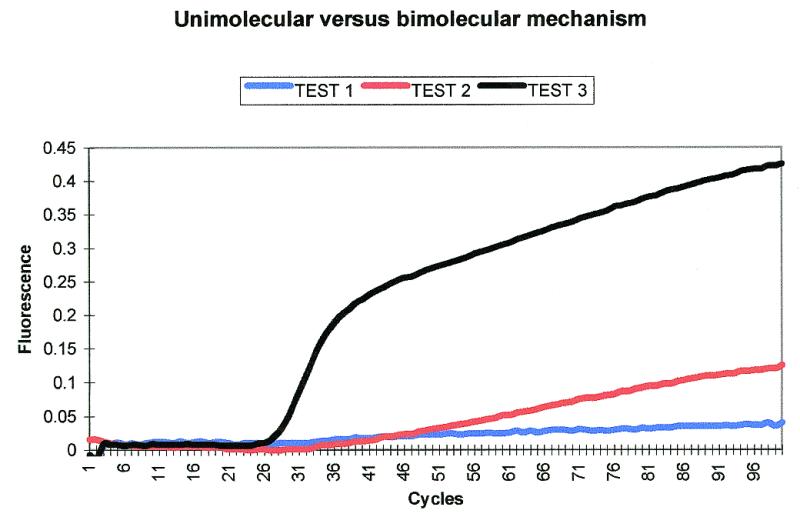
Unimolecular versus bimolecular mechanism test. The level of fluorescence of the Scorpion in test 3 is much higher than the fluorescence seen for the bimolecular probing of tests 1 and 2. Test 3 also has a different shaped curve compared to tests 1 and 2. Fluorescence is shown with a proportional baseline adjustment (19).
Comparison between Scorpions, TaqMan and Molecular Beacons
In order to directly compare the unimolecular probing of Scorpions with the bimolecular probing of Molecular Beacons and TaqMan, the MTHFR Molecular Beacon sequence (Table 2, MMB) and primers BPF and BPR (21) were used. The probe was redesigned as a TaqMan probe (Table 2, MT) by removing the stem sequences and adding FAM to the 5′-end and TAMRA to the 3′-end. The quencher used in the Molecular Beacon was methyl red rather than dabcyl. The probe region of the Scorpion (Table 2, MS) consisted of the reverse complement of the probe sequence used in the above two probe systems. The target region is close to the BPF primer making it preferable to attach the Scorpion probe to this primer rather than BPR. In order to ensure that the Scorpion assay is unimolecular, the probe must bind to the extension of BPF, hence the need to use the reverse complement of the probe sequence. The distance requirements between the Scorpion probe binding site (target site) and the Scorpion primer binding site are discussed later. FAM and methyl red were the fluorophore and quencher used, respectively. The experiments were run under both fast and slow cycling conditions. Under slow cycling conditions the TaqMan and Scorpion give similar results whilst the molecular beacon gives a much weaker signal (Fig. 4A). Under fast cycling conditions the Scorpion gives a much more intense signal than either of the other two probe systems (Fig. 4B). The performance of the Scorpion also improved in terms of signal strength under faster cycling conditions. This may be due to a proportion of the Scorpion probes dissociating from the target when reaching an equilibrium between the bound and disassociated forms under slow cycling conditions. Faster cycling conditions may be under kinetic control. Although the backgrounds of the three assays were different, with the Molecular Beacon being the lowest, followed by TaqMan and then the Scorpion, all three were extremely low, ranging from 2 to 8 fluorescent units. As the baselines did show variation, the arithmetic baseline adjustment was used as recommended in the LightCycler manual (19).
Figure 4.


(A) Comparison between Scorpions, TaqMan and Molecular Beacons under slow cycling conditions. The TaqMan probe and Scorpion perform similarly whilst the Molecular Beacon gives a much lower level of fluorescence. (B) Comparison between Scorpions, TaqMan and Molecular Beacons under fast cycling conditions. The Scorpion performs much better than the other probing systems and also improves on its own performance under slow cycling conditions.
Investigation into the susceptibility of Scorpions to 5′-exonuclease degradation
One possibility is that Scorpions act as substrates for the exonuclease activity of Taq DNA polymerase and so function by a TaqMan-like mechanism. To test this we repeated the MTHFR Scorpion and TaqMan reactions using both GoldStar Taq polymerase and a 5′–3′ exonuclease deficient enzyme (DNAmp Ltd). SYBR Gold showed that all the PCR amplifications worked (data not shown). When using GoldStar, under the fast cycling conditions, both the Scorpion and TaqMan probe gave an increase in fluorescence with the Scorpion producing a stronger fluorescent signal (Fig. 5). The TaqMan probe failed to produce a signal when the target was amplified using the 5′–3′ exonuclease deficient enzyme confirming that this enzyme does not have any 5′–3′ exonuclease activity. However, the Scorpion gave a good signal with this enzyme, indicating that Scorpions do not require an enzymatic cleavage process. Further evidence for this has been acquired by running completed Scorpion reactions on an ABI Prism‘ 310 or 377. The small fluorescent fragments expected from enzymatic cleavage are not observed (D.Whitcombe, personal communication). In addition, if the fluorescence is monitored some time after a Scorpion reaction has been completed it will have dropped to background levels but can be restored by heating and cooling (D.Whitcombe, personal communication). In contrast the increase in fluorescence caused by enzymatic cleavage must be irreversible.
Figure 5.

MTHFR amplifications using a Scorpion or TaqMan probe with either GoldStar Taq polymerase or 5′–3′ exonuclease deficient Taq polymerase. The fluorescence increase observed for the Scorpion when using the 5′–3′ exonuclease deficient enzyme indicates that the Scorpion signal is not entirely due to TaqMan-like enzymatic cleavage of the probe. The drop in fluorescence seen for the Scorpion 5′–3′ sample after the plateau phase may be due to the reaction moving towards an equilibrium state between the probe being bound or dissociated. The other samples have not reached the plateau phase of the reaction. Pos, positive sample; Neg, negative control; GS, GoldStar; 5′–3′, the 5′–3′ exonuclease deficient enzyme.
ABCC7 mutation analysis
SYBR Gold results (not shown) indicate all DNAs used, i.e. DNA from a normal individual and a heterozygous individual, amplify with the same efficiency for all loci. This result was reproducible. Results with no SYBR Gold show reproducibly that all Scorpions can discriminate between normal and mutant DNA, with heterozygotes showing approximately half the level of fluorescence of normal individuals (Fig. 6A–E). Homozygous mutant samples were not available for testing. They would be expected to give a negative result with Scorpions designed to be complementary to the normal sequence.
Figure 6.
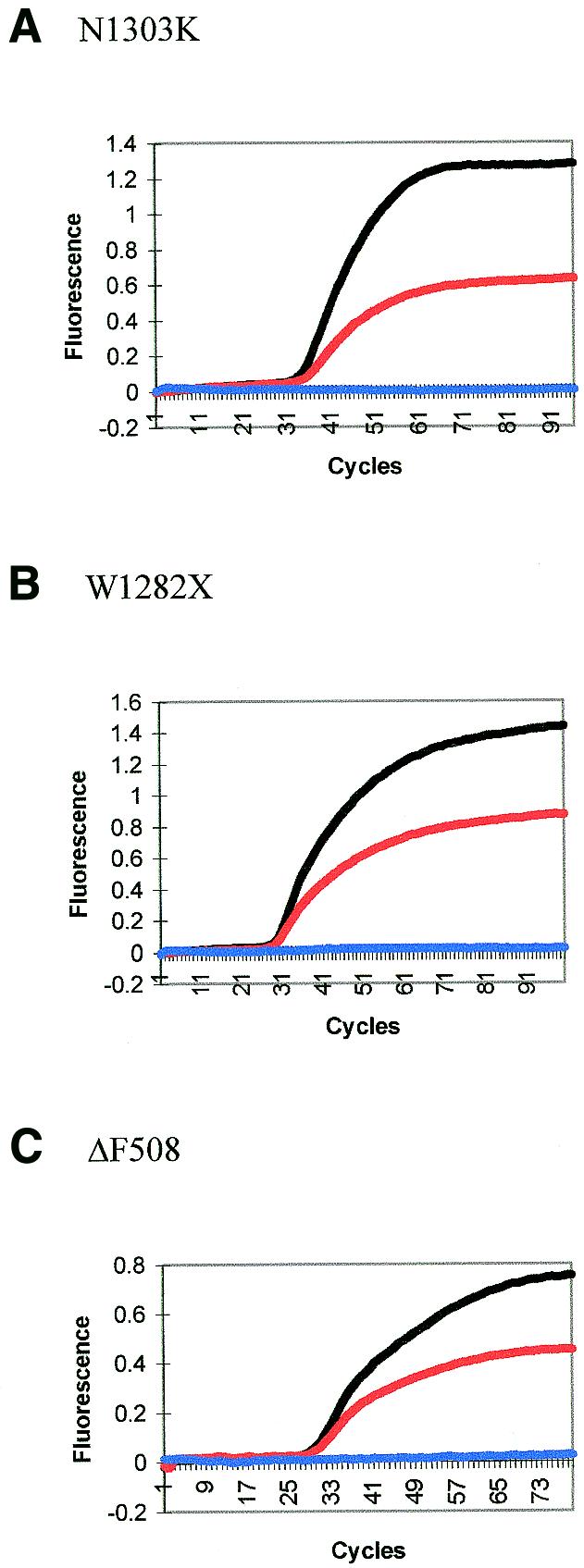

(A–E) Mutation discrimination using Scorpions. Black lines indicate that the sample is from a normal individual, red lines indicate a heterozygous individual and blue lines are negative controls.
The G551D locus (Fig. 6E) gave the worst discrimination of all five Scorpions. This was improved by choosing a different Scorpion primer site. The 3′-end of the new Scorpion primer (G90) was 90 bases from the 5′-end of the target sequence as opposed to three bases. Increasing the distance will increase the size of the single-stranded loop produced when the probe binds to the target site (Fig. 1). Although intramolecular kinetics should give rise to faster and more efficient probing than intermolecular kinetics, the effect will be inversely related to the distance between the probe and the target. The two Scorpions (G90 and 551S) were first run with SYBR Gold, using the same reverse primer (Table 2, 551R) (results not shown). This indicated that both primer sets were equally efficient in producing PCR amplification. Without SYBR Gold, although the efficiency of the Scorpion was decreased the mutation discrimination was good (Fig. 7).
Figure 7.

Mutation discrimination using a Scorpion primer with a binding site 90 bases from the probe target. Improved mutation discrimination can be observed compared with that seen in Figure 6E, where the probe target and Scorpion primer binding site are three bases apart. The fluorescence scale has been proportionally adjusted and actual fluorescent increase cannot be compared to Figure 6E in which the scale is also proportionally adjusted.
In order to obtain robust genotypic data at a particular locus it is necessary to reinforce negative results from a probe that detects one allele by obtaining positive results from a probe that detects another. Whilst this could be carried out for the ABCC7 alleles by performing two separate PCR reactions with a mutant and wild-type specific Scorpion, it would be more economic to combine the two reactions in a multiplex. The set-up of the LightCycler (one excitation source at 488 nm and three channels for detecting emission at 520, 640 and 705 nm) determines that multiplexing is a problem for analysis with Scorpion primers, Molecular Beacons and TaqMan probes. Originally this machine was produced for the Roche hybridisation probe technology (22) and although it has three channels for fluorescence detection it only has one excitation source to excite fluorescein (FAM). Hybridisation probes consist of two probes that are complementary to adjacent regions of a specific PCR product. One probe is labelled with a FAM molecule at the 3′-end and the other with an acceptor dye at the 5′-end. This dye has an excitation spectrum that overlaps with the emission spectrum of FAM. When the two probes bind to the target the FAM and acceptor dye are brought into close proximity and fluorescence resonance energy transfer (FRET) (23) occurs. FAM absorbs the energy from the excitation source and transfers it to the acceptor dye. The increase in fluorescence of the acceptor dye is detected in channel 2 or 3. We have incorporated FRET into a Scorpion to allow the use of channel 2 (Fig. 8). The Scorpion is labelled at the 5′-end with carboxy X-rhodamine (ROX) as the acceptor dye. It also has an internal FAM on a modified thymidine several bases into the Scorpion stem [5(6-fluoresceinylcarboxyamidocaproyl) uracil-2′-deoxyriboside, Oswel Research Products Ltd]. This monomer is added as a phosphoramidite during oligonucleotide synthesis. As the internal FAM is attached to a uracil, a corresponding adenine has to be placed in the complementary stem region 3′ to the probe sequence. Experiments showed that FRET is most efficient when the FAM is placed at least four or five bases from the ROX. When the molecules are closer collisional quenching is thought to occur. We designed a FRET Scorpion to detect the mutant form of ΔF508 (Table 2, 508M), for use in a multiplex reaction with the normal Scorpion (508S).
Figure 8.

A FRET Scorpion primer. In the closed form the methyl red quenches the FAM/ROX. After extension of the Scorpion primer the probe binds to the target separating the quencher from the two fluorophores. The FAM absorbs energy from the excitation source and passes it on to the ROX. Excitation of the ROX occurs and an increase in fluorescence is observed.
The two Scorpions were run in singleplex and multiplex reactions with and without SYBR Gold. The SYBR Gold reactions (not shown) indicate that amplification efficiency is not affected by the mutant or normal probe being attached to the Scorpion primer or by having a mixture of two probes in a multiplex reaction. Channel 1 detects 508S. This Scorpion discriminates between the normal (N) and heterozygous (H) samples in both the singleplex (sw) and multiplex (mu) reactions (Fig. 9A). Channel 2 detects 508M. Although the normal sample (N) should give a negative result with this Scorpion, it gives a very weak positive signal in both the multiplex (mu) and singleplex (sm) reactions. The fluorescence was monitored at 54 rather than 51°C to reduce this but we were unable to obtain a completely negative result. However, since the heterozygous sample (H) gives a strong positive signal, this Scorpion is able to discriminate between samples containing no mutant DNA (N) and heterozygous samples (H) (Fig. 9B). Whilst giving the same results as the singleplex reactions (sw and sm), the multiplex reactions (mu) give less intense fluorescent signals (Fig. 9A and B). This is expected as some of the products will be labelled with the wrong Scorpion, i.e. the product with the normal sequence may have extended from a primer with the mutant scorpion probe sequence attached. It is essential to use a colour compensation file when running multiplex reactions with FRET Scorpions. Without this compensation, FAM-labelled Scorpions detected in channel 1 can also be detected in channel 2. Therefore, if a ROX FRET Scorpion reaction is being carried out in the same tube, channel 2 will give a result that is a mixture of crosstalk from the FAM Scorpion and the ROX Scorpion. The singleplex reactions described above showed that when using this file the normal FAM-labelled Scorpion is only detected at a very low level in channel 2 and the baseline remains flat despite changes in fluorescence in channel 1. The same is true of the ROX Scorpion in channel 1.
Figure 9.

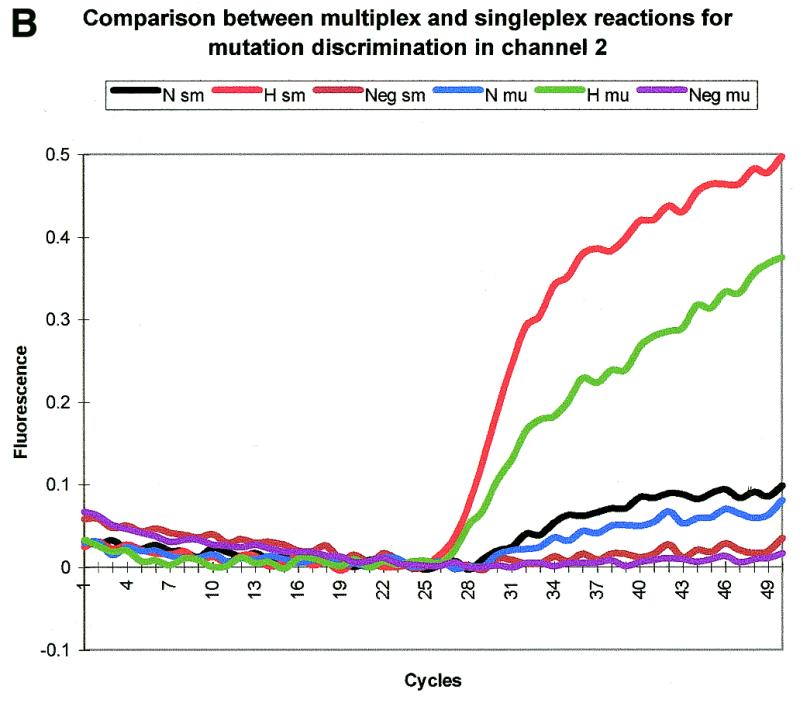
Mutation discrimination with multiplex and singleplex reactions for ΔF508. Both the mutant and normal Scorpions are able to differentiate between samples with or without mutant DNA in the single- and multiplex reactions. N, normal sample; H, heterozygous sample; Neg, negative control; sw, singleplex reaction with the normal Scorpion; sm, singleplex reaction with the mutant Scorpion; mu, multiplex reaction. The baseline is proportionally adjusted in this case as although there is sample–sample variation in baseline this is mainly between samples containing different Scorpions. Importantly there is little variation between samples with the same Scorpion. (A) Fluorescence detection on channel 1. This channel detects the normal Scorpion labelled with FAM. (B) Fluorescence detection on channel 2. This channel detects the mutant Scorpion labelled with ROX and an internal FAM.
CONCLUSION
Our results suggest that whilst under forcing conditions Scorpion probes are able to act in an intermolecular manner (like Molecular Beacons), the primary probing mechanism is intramolecular. Unimolecular probing is kinetically favourable and highly efficient. Covalent attachment of the probe to the target amplicon ensures that each probe has a target in the near vicinity. Further support for the unimolecular mechanism comes from our direct comparison between TaqMan, Molecular Beacons and Scorpions. Scorpions give a strong signal and act much faster than Molecular Beacons or TaqMan probes. Enzymatic cleavage is not required, thereby reducing the time needed for signalling compared to TaqMan probes, which must bind and be cleaved before an increase in fluorescence is observed.
The speed of signalling of Scorpions is highly desirable for the high throughput assays currently being used, e.g. ABCC7 mutation analysis, and is likely to be at the heart of future medical and biological studies. Potential constraints on the design of Scorpions, i.e. distance between probe target and Scorpion primer binding site, are not prohibitive. This makes Scorpions at least as versatile as TaqMan and Molecular Beacons in terms of the range of assays that can be adapted to this technology. For example, although G–T mismatches are relatively stable, with Scorpions it is possible to discriminate between a product that gives a G–T mismatch and one product that is exactly complementary to the probe. The probe can be shortened until properties of the mismatch become significant. When using bimolecular systems, shortening the probe will decrease specificity as there is a greater probability that it will bind to a sequence elsewhere in the genome or to non-specific amplification products. In contrast, the unimolecular probing mechanism of Scorpions gives extra specificity as the primer places the attached probe close to the desired locus. We have successfully designed Scorpion primers with probes of only 15 bases in length (unpublished data).
When performing a sensitive experiment like PCR, using negative results in decision making should be avoided as amplification failures are fairly common, particularly for old or degraded samples. The capacity for multiplexing Scorpion primers for all possible mutations means that negative results will not lead to mistakes in scoring. Currently there are only two possible channels for use with Scorpion primers on the LightCycler. However, Scorpion primers are also adaptable to other platforms like the ABI Prism“ 7700 or the Bio-Rad iCycler, and to end point detection on a fluorescent plate reader. On the ABI 7700 we have observed that using FAM–ROX FRET Scorpions increases the ROX signal (J.Theaker, personal communication). Using a fluorescent plate reader for endpoint colour detection will make the assay cheaper as a normal PCR machine and plate reader are the only instruments required.
In summary, Scorpions could revolutionise mutation screening, making it fast and reliable, whilst not increasing the cost. Scorpions technology can be adapted to high throughput analysis for large-scale screening programmes by using ≥96-well plate formats and kits that are likely to become available in the near future. Alternatively, using equipment such as the LightCycler, small numbers of urgent clinical samples can be assayed at high speed.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank BBSRC and JREI (UK Joint Research Equipment Initiative) for funding for the Roche LightCycler.
REFERENCES
- 1.Whitcombe D., Theaker,J., Guy,S.P., Brown,T. and Little,S. (1999) Nat. Biotechnol., 17, 804–807. [DOI] [PubMed] [Google Scholar]
- 2.Whitcombe D., Kelly,S., Mann,J., Theaker,J., Jones,C. and Little,S. (1999) Am. J. Hum. Genet., 65, 2333. [Google Scholar]
- 3.Tyagi S. and Kramer,F.R. (1996) Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 4.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 5.Kostrikis L.G., Tyagi,S., Mhlanga,M.M., Ho,D.D. and Kramer,F.R. (1998) Science, 279, 1228–1229. [DOI] [PubMed] [Google Scholar]
- 6.Holland P.M., Abramson,R.D., Watson,R. and Gelfand,D.H. (1991) Proc. Natl Acad. Sci. USA, 88, 7276–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo T.H.S., Patel,B.K.C., Smythe,L.D., Norris,M.A., Symonds,M.L. and Dohnt,M.F. (1998) Anal. Biochem., 256, 132–134. [DOI] [PubMed] [Google Scholar]
- 8.Roberts L. (2000) Science, 287, 1898–1899. [DOI] [PubMed] [Google Scholar]
- 9.Rommens J.M., Iannuzzi,M.C., Kerem,B.-S., Drumm,M.L., Melmer,G., Dean,M., Rozmahel,R., Cole,J.L., Kennedy,D., Hidaka,N., Zsiga,M., Buchwald,M., Riordan,J.R., Tsui,L.-C. and Collins,F.S. (1989) Science, 245, 1059–1065. [DOI] [PubMed] [Google Scholar]
- 10.Riordan J.R., Rommens,J.M., Kerem,B.-S., Alon,N., Rozmahel,R., Grzelczak,Z., Zielenski,J., Lok,S., Plavsic,N., Chou,J.-L., Drumm,M.L., Iannuzzi,M.C., Collins,F. and Tsui,L.-C. (1989) Science, 245, 1066–1073. [DOI] [PubMed] [Google Scholar]
- 11.Kerem B.-S., Rommens,J.M., Buchanan,J.A., Markiewicz,D., Cox,T.K., Chakravarti,A., Buckwald,M. and Tsui,L.-C. (1989) Science, 245, 1073–1080. [DOI] [PubMed] [Google Scholar]
- 12.Kerem B.-S., Zielenski,J., Markiewicz,D., Bozon,D., Gazit,E., Yahav,J., Kennedy,D., Riordan,J.R., Collins,F.S., Rommens,J.M. and Tsui,L.-C. (1990) Proc. Natl Acad. Sci. USA, 87, 8447–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ballabio A., Gibbs,R.A. and Caskey,C.T. (1990) Nature, 343, 220. [DOI] [PubMed] [Google Scholar]
- 14.Dean M., White,M.B., Amos,J., Gerrard,B., Stewart,C., Khaw,K.-T. and Leppert,M. (1990) Cell, 61, 863–870. [DOI] [PubMed] [Google Scholar]
- 15.Osborne L., Santis,G., Schwarz,M., Klinger,K., Dork,T., Mcitosh,I., Schwartz,M., Nunes,V., Macek,M., Reiss,J. et al. (1992) Hum. Genet., 89, 653–658. [DOI] [PubMed] [Google Scholar]
- 16.Cutting G.R., Kasch,L.M., Rosenstein,B.J., Zielenski,J., Tsui,L.-C., Antonarakis,S.E. and Kazazian,H.H. (1990) Nature, 346, 366–368. [DOI] [PubMed] [Google Scholar]
- 17.Zielenski J., Rozmahel,R., Bozon,D., Kerem,B., Grzelczak,Z., Riordan,J.R., Rommens,J. and Tsui,L.-C. (1991) Genomics, 10, 214–228. [DOI] [PubMed] [Google Scholar]
- 18.Santalucia J. (1998) Proc. Natl Acad. Sci. USA, 5, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roche Molecular Biochemicals (1999) LightCycler Operator’s Manual Version 3.0. May 1999.
- 20.Tuma R.S., Beaudet,M.P., Jin,X., Jones,L.J., Cheung,C.Y., Yue,S. and Singer,V.L. (1999) Anal. Biochem., 268, 278–288. [DOI] [PubMed] [Google Scholar]
- 21.Giesendorf B.A.J., Vet,J.A.M., Tyagi,S., Mensink,E.J.M.G., Trijbels,F.J.M. and Blom,H.J. (1998) Clin. Chem., 44, 482–486. [PubMed] [Google Scholar]
- 22.Caplin B.E., Rasmussen,R.P., Bernard,P.S. and Wittwer,C.T. (1999) Biochemica, 1, 5–8. [Google Scholar]
- 23.Ju J., Ruan,C., Fuller,C.W., Glazer,A.A. and Mathies,R.A. (1995) Proc. Natl Acad. Sci. USA, 92, 4347–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]