Abstract
Epigenetic modifications, characterized by changes in gene expression without altering the DNA sequence, play a crucial role in the development and progression of cancer by significantly influencing gene activity and cellular function. This insight has led to the development of a novel class of therapeutic agents, known as epigenetic drugs. These drugs, including histone deacetylase inhibitors, histone acetyltransferase inhibitors, histone methyltransferase inhibitors, and DNA methyltransferase inhibitors, aim to modulate gene expression to curb cancer growth by uniquely altering the epigenetic landscape of cancer cells. Ongoing research and clinical trials are rigorously evaluating the efficacy of these drugs, particularly their ability to improve therapeutic outcomes when used in combination with other treatments. Such combination therapies may more effectively target cancer and potentially overcome the challenge of drug resistance, a significant hurdle in cancer therapy. Additionally, the importance of nutrition, inflammation control, and circadian rhythm regulation in modulating drug responses has been increasingly recognized, highlighting their role as critical modifiers of the epigenetic landscape and thereby influencing the effectiveness of pharmacological interventions and patient outcomes. Epigenetic drugs represent a paradigm shift in cancer treatment, offering targeted therapies that promise a more precise approach to treating a wide spectrum of tumors, potentially with fewer side effects compared to traditional chemotherapy. This progress marks a step towards more personalized and precise interventions, leveraging the unique epigenetic profiles of individual tumors to optimize treatment strategies.
Keywords: epigenetic drugs, histone deacetylase inhibitors, histone acetyltransferase inhibitors, histone methyltransferase inhibitors, DNA methyltransferase inhibitors, clinical trials
Introduction
The history of epigenetics
The history of epigenetics traces back to ancient philosophical inquiries about organism development, with Aristotle proposing early theories about embryogenesis. The term “epigenesis” was later introduced around 1650 (Deichmann, 2016), following William Harvey’s 17th-century work, to describe the progressive development of an organism’s characteristics, challenging the prevailing idea of preformationism. This concept of epigenesis laid the foundation for modern epigenetics, sparking significant debates in the 19th century, particularly in the field of embryology about the role of chemical processes in trait expression (De la Peña and Vargas, 2018).
Conrad Waddington, often hailed as the father of epigenetics, further advanced the concept by defining epigenetics as the complex developmental process that links an organism’s genotype with its phenotype. This definition marked a pivotal shift towards the contemporary understanding of epigenetics (Bird, 2007; Deichmann, 2016).
Throughout the 20th century, the field of epigenetics expanded rapidly, especially with contributions from scientists like Arthur Riggs. Epigenetics came to be recognized as the study of heritable changes in gene expression that do not involve modifications to the DNA sequence itself (Riggs and Xiong, 2004; Felsenfeld, 2014). Significant breakthroughs, such as the identification of transcription markers like methylation and acetylation of histones by Vincent Allfrey and Alfred Misky, and further research linking epigenetic variations to diseases (e.g., cancer, systemic lupus erythematosus, and type II diabetes) by scientists like David Allis, underscored the field’s importance (Rakyan et al., 2011; Deichmann, 2016; Adams and Shao, 2022).
The advent of pharmacoepigenetics, heralded by the U.S. Food and Drug Administration (FDA)’s 2006 approval of azacytidine for treating myelodysplastic syndrome (Kaminskas et al., 2005), opened new avenues for therapeutic interventions. This approval was a significant milestone, establishing the relevance of epigenetic drugs in medical treatment. Currently, the field boasts thousands of drugs and compounds targeting epigenetic factors, with significant numbers receiving FDA approval for treating various conditions. Drugs such as decitabine, vorinostat, romidepsin, and panobinostat are notable examples, known for their roles in inhibiting methyltransferase and deacetylase, and are approved for the treatment of various diseases (Kringel et al., 2021).
The journey from Aristotle’s initial hypotheses to today’s advanced pharmacoepigenetic treatments highlights the remarkable evolution of epigenetic research (Claes et al., 2010). This dynamic progression has led to the development of sophisticated treatment strategies that consider the intricate interplay between genetics and environmental factors. The importance of nutrition, inflammation control, and circadian rhythm regulation in modulating drug responses and enhancing treatment efficacy has gained recognition in recent years (Masri et al., 2015; Nahmias and Androulakis, 2021; Barrero et al., 2022; Tan et al., 2022). These factors have been identified as crucial modifiers of the epigenetic landscape, directly impacting the effectiveness of pharmacological interventions and patient outcomes (Pérez-Villa et al., 2023). This progression underscores the transformative impact of epigenetics in understanding developmental biology and its potential to revolutionize therapeutic approaches, promising more personalized and effective treatment strategies (Felsenfeld, 2014).
Gene expression and epigenetics
Eukaryotic organisms display complex genomic structures, distinctly different from those of prokaryotes, characterized by a well-defined nucleus, the formation of chromatin, and the critical role of histones (Koonin, 2010). This intricate architecture is essential for gene expression and chromosome regulation, either facilitated or restricted by DNA’s structural domains. Chromatin, existing as either tightly packed heterochromatin or less condensed euchromatin, plays a vital role in gene regulation and epigenetic processes. Heterochromatin, in particular, is crucial for organizing large chromosome domains, necessary for proper chromosome segregation (Grewal and Moazed, 2003). Its formation involves a detailed process of histone modifications by silencing complexes, emphasizing the importance of histone tails and the role of non-coding RNAs in the formation of epigenetic domains. The three-dimensional architecture of the genome, influenced by its function, indicates that chromatin’s structural features serve as modulators of genome activity. This dynamic, elucidated through various scientific approaches, highlights the development of structural chromatin features, the diversity and heterogeneity of nuclear architecture, and the evolutionarily conserved traits of genomes, such as plasticity and robustness (Misteli, 2020). Moreover, the complexity of eukaryotic gene regulation, as detailed by the central dogma of molecular biology and further complicated by processes like mRNA maturation, underscores the sophisticated nature of eukaryotic genetic regulation.
In eukaryotes, it is important to note that not all DNA is transcribed into RNA, and not all RNA is translated into proteins. The DNA includes non-coding sequences and gene-flanking regions, while RNA can regulate the expression of other genes (Hernández et al., 1995). The amount of mRNA transcribed can affect gene expression levels, which depend on both the integrity of DNA and the accessibility to transcriptional and translational machinery. DNA damage from mutagens can lead to mutations, while intact genes that are inaccessible to this machinery, or lack recognizable sections, may not be expressed. This variability is part of what is known as the epigenome (Corella and Ordovas, 2017), enabling cells with the same genome to exhibit different phenotypes within an organism.
Gene expression is also influenced by the gene’s location in euchromatin regions, where reduced compaction due to lower electromagnetic charges from DNA and histones allows easier access for transcriptional machinery. Nucleosomes further impact DNA accessibility by organizing the DNA-protein complex into a three-dimensional structure (Shi, 2007).
Epigenetic regulation, which involves modifying proteins and pathways without altering the DNA sequence, is affected by both the organism’s environment and its cellular microenvironment (Bedregal et al., 2010). This regulation includes mechanisms such as gene silencing and activation, and the control of gene promoters and repressors, making epigenetic changes heritable and reversible. Non-Mendelian inheritances, such as prions and non-coding RNAs, contribute to phenotypic expression through epimutations, which mainly alter the three-dimensional conformation of chromatin (Shi, 2007).
Epimutations can change DNA or chromatin charges, or the structure of histone proteins, affecting gene expression. The proximity of a gene to the cellular periphery can also influence its activation (Shi, 2007). These complex changes represent alterations in entire regulatory mechanisms, not just structural components. The epigenetic state of a cell is constantly modified, allowing for optimal function in diverse environments (Horsthemke, 2006). Lastly, epigenetic alterations can disrupt cellular pathways, potentially leading to cancer.
Molecular underpinnings of epigenetic modifications that lead to cancer
Epigenetic regulation encompasses several molecular mechanisms that operate at different biological scales, notably the post-translational modifications of histones, which are central to the regulation of gene expression (Table 1) (Mohtat and Susztak, 2010; Bell and Spector, 2011). These modifications include, but are not limited to, acetylation, methylation, ubiquitination, and phosphorylation, each carrying distinct regulatory functions (Cao et al., 2005; Glozak et al., 2005; Cavagnari, 2012). DNA-level modifications, such as DNA methylation and ATP-dependent chromatin remodeling, also play pivotal roles in altering chromatin architecture, thereby influencing gene accessibility and expression. Additionally, non-coding RNAs, telomere positioning, genomic imprinting, polycomb and trithorax group proteins, prion-like factors, X-chromosome inactivation, and sumoylation processes contribute to the complex landscape of epigenetic regulation (Blackburn, 2001; Shi, 2007; Bracken and Helin, 2009; Cotton et al., 2015).
TABLE 1.
Landscape of epigenetic mechanisms involved in cancer.
| Mechanism | Description | Cancer types | Reference |
|---|---|---|---|
| DNA methylation | Addition of methyl groups to DNA, leading to gene silencing | Colorectal, breast, lung | Jones and Baylin (2007) |
| Histone acetylation | Addition of acetyl groups to histones, usually associated with gene activation | Leukemia, glioblastoma | Glozak et al. (2005) |
| Histone methylation | Methylation of histones, affecting gene expression depending on the side and state of methylation | Prostate, lymphoma | Greer and Shi (2012) |
| Non-coding RNAs | RNA molecules that regulate gene expression, including microRNAs and long non-coding RNA | Liver, breast, pancreas | Esteller (2011) |
| Chromatin remodeling | Reorganization of chromatin, affecting DNA accessibility and gene expression | Melanoma, sarcoma | Wilson and Roberts (2011) |
| RNA interference | Small RNA molecules inhibit gene expression or translation, impacting cancer genes | Ovarian, kidney | Bader (2012) |
| Histone phosphorylation | Addition of phosphate groups to histones, involved in DNA repair and chromosome condensation | Breast, colorectal | Rossetto et al. (2012) |
| Histone ubiquitination | Addition of ubiquitin to histones, involved in DNA repair and gene expression regulation | Colorectal, prostate | Cao et al. (2005) |
| DNA hydroxymethylation | Conversion of methylated DNA to hydroxymethylated DNA, associated with gene activation | Melanoma, gliomas, liver | Lian et al. (2012) |
| Polycomb repression | Polycomb group proteins repress gene expression through histone modifications | Breast, endometrial | Bracken and Helin (2009) |
| Nucleosome positioning | Arrangement of nucleosomes along DNA, influencing gene expression by accessibility | Leukemia, melanoma | Voigt et al. (2013) |
| Histone variants | Incorporation of histone variants into nucleosomes, affecting chromatin structure and gene expression | Glioblastoma, sarcoma | Talbert and Henikoff (2010) |
| Telomere positioning | Influence of telomere structure and location on gene expression | Lung, bladder | Blackburn (2001) |
| Sumoylation | Addition of SUMO proteins to target proteins, influencing stability and activity | Breast, thyroid | Seeler and Dejean (2003) |
| Enhancer RNAs | Non-coding RNAs transcribed from enhancer regions that regulate gene expression | Prostate, colorectal | Li et al. (2013) |
| DNA demethylation | Removal of methyl groups from DNA, often leading to gene activation | Colorectal, gastric | Wu and Zhang (2010) |
| Chromatin accessibility | Degree to which DNA is exposed and accessible to transcription factors | Lymphoma, leukemia | Thurman et al. (2012) |
| X-chromosome inactivation | Inactivation of one of the X chromosomes in females | Breast, ovarian | Cotton et al. (2015) |
| Genomic imprinting | Parent-specific gene expression due to epigenetic marks | Ovarian, testicular | Ferguson-Smith (2011) |
Histone PTMs are dynamic and multifaceted, capable of occurring concurrently and influencing the interactions between histones and DNA. Such modifications can lead to chromatin compaction and gene silencing or chromatin relaxation, enhancing transcriptional activity (Zhao and Shilatifard, 2019). The specificity of these modifications, including their type and location, underlines the intricate control of gene expression and phenotypic outcomes (Cavagnari, 2012).
The interplay between histone modifications and chromatin structure is further complicated by cis and trans effects, with cis effects modifying chromatin at the level of internucleosomal contacts, and trans effects arising from the association of various proteins with the chromatin (Esteller, 2007; Talbert and Henikoff, 2010; Cavagnari, 2012; Greer and Shi, 2012). This complexity is augmented by the enzymatic systems responsible for adding or removing histone PTMs, which also target non-histone proteins, thereby extending the scope of epigenetic regulation (Shi, 2007; Voigt et al., 2013).
Among the myriad of histone PTMs, acetylation and methylation are particularly noteworthy. Acetylation, typically associated with transcriptional activation, is mediated by histone acetyltransferases (HATs) and reversed by histone deacetylases (HDACs) (Glozak et al., 2005; Shi, 2007; Gujral et al., 2020). Methylation, however, exhibits a dual role in gene regulation, dependent on the specific amino acids modified and their methylation state, adding layers to the regulatory network (Zhang et al., 2023).
Histone phosphorylation and ubiquitination are additional modifications with significant roles in gene expression and chromatin dynamics. Phosphorylation, often linked to chromatin condensation, is mediated by specific kinases (Shi, 2007; Rossetto et al., 2012), whereas co-activator-mediated ubiquitination is definitively associated with gene activation (Talukdar and Chatterji, 2023). Other modifications, such as glycosylation and sumoylation, although less understood, are recognized for their contribution to chromatin structure and function (Seeler and Dejean, 2003; Ramírez et al., 2009).
DNA methylation, a key epigenetic mark, occurs predominantly at CpG sites and plays a critical role in gene silencing and chromatin remodeling (Jones and Baylin, 2007; Zhang et al., 2023). The intricate balance between methylation, demethylation, hydroxymethylation is crucial for cellular function and is tightly regulated by DNA methyltransferases and other chromatin-associated proteins (Wu and Zhang, 2010; Lian et al., 2012; Dhar et al., 2021).
In the context of cancer epigenetics, this field has made significant strides in understanding how DNA methylation and histone modifications contribute to tumorigenesis (Baylin and Jones, 2016). This review highlights the importance of hypermethylation in silencing tumor suppressor genes, CpG islands, and individual genes, which has implications for the early detection and classification of cancer. Tumor suppressor proteins such as INK4A, INK4B, and APC, along with enzymes and cell adhesion proteins like GSTP1, MGMT, and CDH1, have been studied extensively in cancers of the prostate, liver, stomach, colon, and breast. These studies reveal tissue-specific methylation patterns, underscoring the complexity of epigenetic regulation in different cell types (Esteller, 2002; Gonzalo et al., 2008; Bouyahya et al., 2022).
Microsatellite instability, another focus of cancer epigenetics research, is closely linked to colorectal cancer and Lynch syndrome (Gusev, 2019). It arises from length changes in microsatellites, increasing susceptibility to replication errors and leading to genetic alterations that drive cancer progression (Toledo González and Cruz-Bustillo Clarens, 2005).
To identify methylated sites within the genome, researchers employ a variety of techniques, including methylation-sensitive enzymes, chemical conversion of methylated cytosines using sodium bisulfite, and immunological methods that capture DNA-methyl cytosine complexes (Frommer et al., 1992). These approaches are pivotal in mapping the epigenetic landscape of cancer cells (Weber et al., 2005).
Epigenetic therapies, particularly those targeting DNA methylation like azacitidine, aim to reactivate silenced genes, offering new strategies for cancer treatment. These drugs modify the epigenetic landscape, potentially reversing the gene silencing that contributes to cancer development (Issa and Kantarjian, 2009; Gailhouste et al., 2018).
Chromatin remodeling, a key aspect of epigenetic regulation, involves both covalent histone modifications and ATP hydrolysis mechanisms (Zhou et al., 2016). This remodeling facilitates access to transcriptional machinery by displacing nucleosomes, leading to nucleosomal sliding or eviction and consequent changes in gene expression. The SWI/SNF complex is notable for its role in these processes, highlighting the dynamic interplay between chromatin structure and gene regulation (Schwabish and Struhl, 2007; Wilson and Roberts, 2011; Cavagnari, 2012).
RNA molecules, smaller than mRNA, play crucial roles in regulating gene expression by managing DNA exposure for transcription and translation. The regulation of gene expression by microRNAs (miRNAs), small interfering RNAs (siRNAs), enhancer RNAs (eRNAs), and other non-coding RNAs is vital for cellular processes like differentiation, immune response, and cell proliferation (Bader, 2012; Li et al., 2013; Lam et al., 2015). These RNAs contribute to cancer development and progression by altering the transcriptome, offering potential targets for therapeutic intervention (Esteller, 2011; Gusev, 2019).
In summary, the study of cancer epigenetics encompasses the investigation of DNA methylation patterns, histone modifications, chromatin remodeling, and RNA-mediated gene regulation. These components interplay to control gene expression, with aberrations in these processes contributing to the onset and progression of cancer. Understanding these mechanisms provides valuable insights into cancer biology and opens avenues for novel diagnostic and therapeutic strategies.
Immune system and epigenetics
The immune system in vertebrates is a complex and finely tuned entity, governed not only by signaling pathways but also by a network of epigenetic mechanisms (Busslinger and Tarakhovsky, 2014). These epigenetic regulations are essential for the immune system’s ability to defend against various pathogens, illustrating the system’s sensitivity to both internal and external stimuli. The activity of immune cells, such as macrophages, and their components like toll-like receptors (TLRs), is subject to precise epigenetic control (Fitzgerald and Kagan, 2020). For example, the regulation of inflammatory cytokine genes in macrophages is mediated by histone modifications, particularly the induction of histone lysine acetylation, which is crucial for cytokine gene transcription (Obata et al., 2015).
Bromodomain and extraterminal (BET) proteins, which bind to acetylated lysines, play a significant role in modulating cytokine gene expression (Obata et al., 2015). Studies have shown that synthetic compounds like I-BET can inhibit the expression of inflammatory cytokines by disrupting the interaction between BET proteins and acetylated histones, thereby suppressing the immune response to lipopolysaccharides in macrophages (Nicodeme et al., 2010; Guo et al., 2019; Wang et al., 2021).
Histone deacetylase enzymes, specifically class IV HDAC, are also pivotal in the immune response, targeting histones near the interleukin (IL) 10 gene to compact chromatin and inhibit transcription. This suppression of IL-10, an anti-inflammatory cytokine, highlights the role of epigenetic regulation in modulating immune responses (Villagra et al., 2009).
Methyltransferases further influence the immune landscape, particularly in macrophages. Ash1l, an enzyme that methylates lysine 4 on histone H3 (H3K4), enhances the expression of the A20 gene (Xia et al., 2013), which plays a role in modulating inflammatory responses by inhibiting certain signaling pathways. This methylation leads to the suppression of NF-kB and MAPK signaling pathways, reducing the production of IL-6, an inflammatory cytokine (Xia et al., 2013).
Another H3K4 methyltransferase, Wpp7, is crucial for the antimicrobial response in macrophages (Li et al., 2022). The absence of Wpp7 leads to reduced expression of glycosylphosphatidylinositol (GPI)-anchored proteins, affecting the macrophages’ response to microbial stimuli due to the lack of CD14, a GPI-anchored protein necessary for effective microbial recognition (Austenaa et al., 2012).
This intricate network of epigenetic regulation, from histone modifications to the activity of specific enzymes, underscores the delicate balance maintained in the immune system. Disruptions in this balance can lead to immunological disorders, such as inflammation, demonstrating the critical role of epigenetic controls in immune regulation (Xu et al., 2023).
Before cancer treatment
The role of epigenetic signatures in the pharmacoepigenetic treatments of cancer
Cancer is characterized by complex genomic and epigenomic alterations that drive its development and progression. Among these, epigenomic changes, including DNA methylation, histone modifications, and the reprogramming of non-coding RNAs, play a pivotal role due to their reversible nature, making them attractive targets for therapeutic intervention (Lu et al., 2020). DNA methylation, the most extensively studied epigenetic modification, often occurs early in cancer and influences gene expression by altering the methylation status of oncogenes or tumor suppressor genes (Diaz-Lagares et al., 2016). This can lead to the dysregulation of non-coding RNAs, further impacting the regulation of mRNA targets and potentially contributing to oncogenesis (Nishiyama and Nakanishi, 2021; Tonmoy et al., 2022).
Histone modifications encompass a wide range of changes, such as acetylation, methylation, and phosphorylation, which affect chromatin structure and subsequently the accessibility of transcription factors to DNA (Kouzarides, 2007). These modifications not only regulate gene expression directly but also influence the activity of non-coding RNAs involved in post-transcriptional gene regulation (Ferro et al., 2017).
The concept of the cancer epigenome encompasses the global aberrant epigenetic marks found across various tumor types, a phenomenon known as epigenetic instability. This hallmark of cancer is integral to developing diagnostic and prognostic epigenetic signatures, as seen in the hypermethylation of genes in prostate cancer, altered CpG island methylation in breast cancer associated with poor outcomes, and the silencing of tumor suppressor genes like CDO1 across multiple cancer types due to DNA methylation (Brait et al., 2012; Batra et al., 2021).
Specific epigenetic patterns, such as DNA hypermethylation in lung adenocarcinoma related to smoking history and changes in histone modification levels in colorectal cancer (Gezer et al., 2015; Bakulski et al., 2019), highlight the heterogeneity and complexity of cancer epigenetics (Brait et al., 2012; Diaz-Lagares et al., 2016). These findings underpin the development of diagnostic tools and pharmaco-epigenetic therapies, exemplified by FDA-approved colorectal cancer screening tests that assess DNA methylation (Koch et al., 2018; Batra et al., 2021).
However, the high heterogeneity among tumors, coupled with environmental influences like smoking and aging-related epigenetic changes, presents significant challenges in identifying cancer-specific epigenetic landscapes and therapeutic targets. Distinguishing cancer-related epigenetic modifications from those associated with aging is crucial for advancing precision medicine and optimizing pharmaco-epigenetic approaches to cancer treatment. This differentiation is essential in developing targeted therapies and enhancing the efficacy of epigenetic-based interventions in cancer care (Pérez et al., 2021).
During cancer treatment
Histone deacetylase inhibitors
Histone deacetylase inhibitors are essential chemotherapeutic agents used in cancer therapy, serving as both cytotoxic and cytostatic drugs (Ferrarelli, 2016; Eckschlager et al., 2017) (Figure 1). The FDA has approved HDAC inhibitors such as vorinostat, romidepsin, belinostat, and panobinostat (Table 2). Additionally, HDAC inhibitors like chidamine, valproic acid, entinostat, tacedinaline, quisinostat, and resminostat are currently under investigation (Mann et al., 2007; Piekarz et al., 2009; San-Miguel et al., 2014; O’Connor et al., 2015; Mendez et al., 2019; Ochoa et al., 2023). These inhibitors function by blocking the removal of acetyl groups from histones, thereby preserving an open chromatin structure essential for enhancing gene expression (Gonzalo et al., 2008; Stimson et al., 2009). Such an open chromatin structure is critical for inhibiting tumor cell activity and repressing gene expression involved in processes like angiogenesis, cell cycle disruption, immunity, and cell survival (Gonzalo et al., 2008).
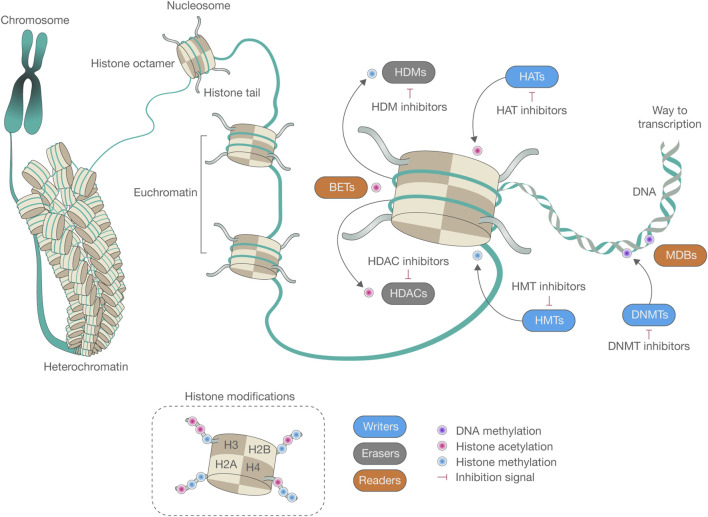
FIGURE 1.
The epigenetic machinery plays an essential role in shaping the conformation of chromatin and regulating genome functionality. DNA is intricately packed and wound around a core composed of histone octamers, thus forming nucleosomes, the fundamental structural units of chromatin. This sophisticated network of epigenetic modifications, encompassing DNA methylation and histone modifications, profoundly impacts the structure of chromatin and the functionality of the genome. Central to epigenetics are enzymes that serve three primary roles: adding (writers), recognizing (readers), and removing (erasers) epigenetic marks on DNA or histone tails. DNA methylation, primarily carried out by DNA methyltransferases (DNMTs), can be reversed by ten-eleven translocation enzymes (TETs) or can diminish progressively over successive cell divisions. Among histone modifications, acetylation and methylation have been extensively studied. The equilibrium of histone methylation is controlled by the opposing activities of histone methyltransferases (HMTs) and histone demethylases (HDMs). In a similar vein, histone acetylation levels are modulated by the concerted efforts of histone acetyltransferases (HATs) and histone deacetylases (HDACs), which add or remove acetyl groups from lysine residues on the histone tails, respectively. This “epigenetic code” is interpreted by specific reader or effector proteins that selectively bind to certain types of modifications. For instance, methyl-CpG-binding domain (MBDs) proteins bind to methylated DNA, whereas bromodomain and extraterminal domain proteins (BETs) recognize acetylated lysines. These epigenetic modifications play a critical role in altering chromatin conformation, leading to either the transcriptional silencing or activation of genes, often through the recruitment of additional proteins to these sites.
TABLE 2.
FDA-approved epigenetic inhibitors.
| Inhibitor | Drug | First approval | Target | Cancer treatment | Relevant clinical trial | Reference |
|---|---|---|---|---|---|---|
| HDAC | Vorinostat | 2006 | HDAC 1, 2, 3, and 6 | Cutaneous T-cell lymphoma | NCT00091559 | Mann et al. (2007) |
| HDAC | Romidepsin | 2009 | HDAC 1, 2, 4, and 6 | Cutaneous T-cell lymphoma, peripheral T-cell lymphoma | NCT00106431 | Piekarz et al. (2009) |
| HDAC | Belinostat | 2014 | HDAC 1, 2, 3, and 10 | Peripheral T-cell lymphoma | NCT00106431 | O’Connor et al. (2015) |
| HDAC | Panobinostat | 2015 | HDAC 1, 2, 3, 6, and 10 | Multiple myeloma (in combination with bortezomib and dexamethasone) | NCT01023308 | San-Miguel et al. (2014) |
| HMT | Tazemetostat | 2020 | EZH2 | Epitheloid sarcoma, follicular lymphoma | NCT01897571 | Italiano et al. (2018) |
| DNMT | Azacitidine | 2004 | DNMT | Myelodysplastic syndromes, acute myeloid leukemia, chronic myelomonocytic leukemia | NCT00071799 | Fenaux et al. (2010) |
| DNMT | Decitabine | 2006 | DNMT | Myelodysplastic syndromes, acute myeloid leukemia | NCT00313586 | Kantarjian et al. (2006) |
As epigenetic modulators, HDAC inhibitors alter the expression of histone and non-histone proteins without changing the DNA sequence itself. They help restore normal cell differentiation and apoptotic functions by maintaining histone acetylation, modifying chromatin structure, and facilitating access for transcription factors (Bolden et al., 2006; Mayr et al., 2021).
Histone acetylation, promoted by HAT/KAT enzymes, leads to changes in gene expression and chromatin structure. Conversely, HDAC enzymes act as transcriptional repressors by removing these acetyl groups. Such histone deacetylation results in transcriptional silencing, either through increased charge density in histone N-terminal groups or chromatin compaction, thus reducing transcription accessibility (Gallinari et al., 2007; Seto and Yoshida, 2014).
HDACs are targeted by several classes of inhibitors, including hydroxamic acids, cyclic peptides, aliphatic fatty acids, benzamides, epoxy ketones, and hybrids, categorized by their chemical structures and action mechanisms (Hess-Stumpp et al., 2007; Jain and Zain, 2011; Ghasemi, 2020; Lu et al., 2020). These inhibitors, affecting the 18 known HDACs through zinc-dependent or NAD + -dependent mechanisms, offer therapeutic potential by inducing apoptosis, cell cycle arrest, and modifying non-coding RNA expression (Bose et al., 2014; Suraweera et al., 2018). They boost the acetylation of genes that regulate the cell cycle and apoptosis, like p53, thereby inducing apoptosis through various pathways (Kulka et al., 2020).
Nonetheless, the clinical application of HDAC inhibitors encounters challenges such as drug resistance, often due to the overexpression of proteins like CDC25A in tumor cells, and side effects like nausea, vomiting, headache, and fluid and electrolyte imbalances (Liu et al., 2020; Wang et al., 2023). Addressing these issues requires patients to adhere to medical advice specifically tailored to their treatment plan. Comprehending the molecular mechanisms of HDAC inhibitors, including their effects on chromatin structure and gene expression, is vital for refining cancer treatment strategies and overcoming resistance and adverse effects (Matore et al., 2022).
Histone acetyltransferase inhibitors
Histone acetyltransferases play a crucial role in regulating gene expression by acetylating histones, thereby making DNA more accessible for transcription (Sterner and Berger, 2000). Instead of inhibiting, they enhance the acetyltransferase activity on lysine residues of histones, a process essential for transcriptional activation associated with euchromatin. This activity is vital for suppressing tumor cell growth and inhibiting cellular mitosis (Dalvoy Vasudevarao et al., 2012; Sabnis, 2021). However, developing inhibitors that target HATs has been challenging, with most potential inhibitors still in the experimental or preclinical stages.
HAT inhibitors aim to target specific human enzymes such as CREBBP, linked to acute myeloid leukemia, as well as CDY1 and CDY2, important for erythropoiesis and spermatogenesis, and CLOCK, crucial for regulating circadian rhythms (Mullighan et al., 2011; Pérez-Villa et al., 2023). The significance of HAT inhibitors spans various enzymes implicated in cancers, including hematological neoplasms like diffuse large B-cell lymphoma, non-Hodgkin lymphoma, acute lymphocytic leukemia, and solid tumors such as colorectal, breast, and prostate cancer (Wu et al., 2020). Noteworthy inhibitors under investigation include anacardic acid, C646, curcumin, garnicol, and MB-3.
The exploration of HAT inhibitors has raised concerns about potential side effects, which might include issues typical of chemotherapeutic agents, such as cytotoxicity and off-target effects (Manzo et al., 2009). Concerns about chemotherapy resistance, potentially driven by the activation of multidrug resistance genes and increased growth factors, also underscore the need for careful therapeutic design and monitoring.
HATs are bisubstrate enzymes that catalyze the transfer of an acetyl group from the cofactor acetyl coenzyme A (Ac-CoA) to a lysine substrate on histones, playing a pivotal role in gene regulation. They function through mechanisms including a random ternary complex, an obligatory ordered ternary complex requiring a general base like glutamic acid, and a Ping-Pong mechanism, wherein Ac-CoA binds first, transferring the acetyl group to an enzyme’s amino acid before CoA binds to the substrate (Wapenaar and Dekker, 2016).
Research continues to focus on developing small molecule HAT inhibitors as potential therapeutic agents. Efforts include designing HAT mimics, utilizing natural products, and conducting virtual screenings to identify high-performance HAT inhibitors. Examples such as KAT2B, KAT3B, KAT5, and KAT5 ESA1 are promising due to their selectivity but face challenges including low metabolic stability and lack of cellular permeability (Poziello et al., 2021). Natural product-based HAT inhibitors, such as garcinol, show potential in preclinical models for inhibiting cancer cell growth, highlighting the ongoing challenge of balancing the biological activity of HAT inhibitors with their drug applicability for treating diseases like cancer (Kopytko et al., 2021).
Histone methyltransferase inhibitors
Histone methyltransferase (HMT) inhibitors are emerging as significant agents in cancer therapy, targeting enzymes responsible for methylating lysine or arginine residues on histones, especially on proteins H3 and H4 (Figure 1). This methylation plays a critical role in epigenetic regulation of gene expression, including silencing tumor suppressor genes in cancer cells. By inhibiting HMTs, these drugs aim to correct aberrant methylation patterns, potentially reactivating genes that can suppress tumor growth and affect processes such as cell replication, differentiation, apoptosis, angiogenesis, and senescence (Kim and Bae, 2011).
Understanding chromatin structure is fundamental. Every cell in the human body contains identical DNA, packaged with histones into chromatin. The nucleosome, comprising an octameric core of histones (two copies each of H2A, H2B, H3, and H4) wrapped around a segment of DNA 145–147 base pairs in length, forms the basic unit of chromatin (Simon and Lange, 2008).
Histones regulate gene expression through various epigenetic mechanisms, including methylation, which occurs on lysine and arginine residues and involves enzymes such as G9a. G9a, a histone methyltransferase, is responsible for the monomethylation and dimethylation of histone H3 lysine 9 (H3K9), influencing chromatin structure and gene expression (Robertson, 2001; Liu et al., 2015). Methylation, particularly by G9a, plays a part in coordinating gene regulation along with DNA methyltransferases and demethylases.
Tazemetostat (an EZH2 inhibitor) has received FDA approval for treating epithelioid sarcoma and certain types of follicular lymphoma, marking a significant achievement in the clinical use of HMT inhibitors (Table 2) (Italiano et al., 2018). Other inhibitors in development include GSK2816126 for EZH2, pinometostat for DOT1L, GSK3326595 and JNJ-64619178 for PRMTs, and GSK2879552 and iadademstat for LSD1. Tazemetostat specifically targets EZH2, which is implicated in suppressing tumor genes and epithelial-mesenchymal transition (EMT)-related genes such as p16INK4a and e-cadherin (Baell et al., 2018).
However, HMT inhibitors can cause side effects, including nausea and vomiting, with some patients experiencing symptoms akin to tumor lysis syndrome. Therefore, patients often require close monitoring, especially during initial treatment cycles. Resistance to HMT inhibitors can arise from mutations in the tumor’s genomic sequence, emphasizing the need to understand the molecular mechanisms of action to optimize cancer treatment and manage resistance (Wang et al., 2023).
Histones undergo modifications such as acetylation and methylation. Acetylation neutralizes the positive charge on lysine residues, reducing histone-DNA interactions and leading to more relaxed chromatin conducive to gene expression (Liu et al., 2015). The effect of methylation on chromatin and gene expression depends on the specific residue methylated. These epigenetic modifications are crucial targets for drug development, as demonstrated by the approval of DNA methyltransferase and histone deacetylase inhibitors (Pócza et al., 2016).
In the epigenetic landscape, enzymes like G9a are termed “writers” for adding chemical groups, “readers” recognize these modifications, and “erasers” remove them (Subramaniam et al., 2014). Targeting G9a and other HMTs is a promising strategy for cancer therapy, underscoring the significance of these “writers” in maintaining malignant phenotypes and regulating gene expression (Ghoshal and Bai, 2007; Morera et al., 2016).
DNA methyltransferase inhibitors
DNA methyltransferase (DNMT) inhibitors are increasingly recognized as essential agents in cancer therapy, specifically targeting enzymes responsible for methylating cytosine residues within CpG dinucleotides in DNA (Jin and Robertson, 2013). This methylation is fundamental to the epigenetic regulation of gene expression, including the silencing of tumor suppressor genes in cancer cells. By inhibiting DNMTs, these drugs aim to correct aberrant methylation patterns, potentially leading to the reactivation of genes that can suppress tumor growth and impact key processes such as cell replication, differentiation, apoptosis, angiogenesis, and senescence (Kim and Bae, 2011).
Histones play a critical role in regulating gene expression through various epigenetic mechanisms, notably methylation at cytosine residues, primarily carried out by DNMT enzymes such as DNMT1, DNMT3A, and DNMT3B (Apuri and Sokol, 2016). This methylation usually leads to gene silencing, playing a key role in coordinating gene regulation alongside DNA methyltransferases and demethylases.
FDA-approved DNMT inhibitors, including azacitidine and decitabine, are designated for treating myelodysplastic syndromes, acute myeloid leukemia, and chronic myelomonocytic leukemia (Table 2) (Kantarjian et al., 2006; Fenaux et al., 2010). Functioning as nucleoside analogs, these inhibitors integrate into DNA, leading to DNA hypomethylation (Giri and Aittokallio, 2019). At therapeutic doses, they primarily exhibit a cytostatic effect, enabling the reactivation of previously silenced genes (Traynor et al., 2023). Additionally, compounds like guadecitabine and CC-486 (oral azacitidine) are being evaluated in clinical trials for various conditions but have not yet been approved by the FDA (Ochoa et al., 2023).
However, repeated use of DNMT inhibitors can result in drug resistance, characterized by mutations and gene overexpression that enhance tumor cell survival against treatment (Laranjeira et al., 2023). DNMT inhibitors can cause side effects such as nausea, vomiting, dehydration, headache, anorexia, and myelosuppression (anemia, neutropenia, thrombocytopenia), necessitating close monitoring, particularly during the initial treatment cycles (Chen et al., 2023).
Current research focuses on developing non-nucleoside DNMT inhibitors with minimal off-target effects and enhanced specificity. Experimental compounds, including non-nucleoside analogs, are under investigation for their ability to more selectively block DNMT activity and reactivate tumor suppressor genes (Traynor et al., 2023).
In cancer treatment, DNMT inhibitors have demonstrated potential across various cancers, including bladder and gastric cancer, by inducing the overexpression of tumor suppressor genes, inhibiting tumor cell growth, and promoting apoptosis (Norollahi et al., 2019; Juárez-Mercado et al., 2020). These outcomes underscore the significant potential of DNMT inhibitors in clinical trials and experimental models. The DNMTs, as a conserved family of cytosine methyltransferases, are crucial for epigenetic regulation. Targeting these enzymes offers a strategic approach to developing cancer therapies, aiming to integrate such treatments into comprehensive cancer management strategies. As the field evolves, continued research on DNMT inhibitors is critical for advancing cancer treatment strategies.
Combinational therapy
Combinational therapy marks a significant evolution in medical treatments by employing two or more therapeutic agents together, an approach that has been shown to be more effective than using a single agent alone (Bayat Mokhtari et al., 2017). This strategy is particularly beneficial in targeting proliferating cells, a common characteristic of cancer, but it also poses the risk of harming healthy cells alongside cancerous ones. Historically, combinational therapies have been instrumental in managing diseases with a high social impact, such as cardiovascular, metabolic, infectious, and autoimmune disorders. A prime example of this is the POMP regimen, introduced in 1965 for acute leukemia, which includes methotrexate, 6-mercaptopurine, vincristine, and prednisone, and has significantly improved remission rates in pediatric patients (Bayat Mokhtari et al., 2017).
In the context of cancer, combinational therapy often involves pairing standard chemotherapy or radiotherapy with a variety of drugs to address the complex nature of tumors, their compensatory mechanisms, and their rapid progression. This multi-targeted approach, through the sequential use of therapeutic agents, aims to reduce tumor growth, limit metastasis, and promote cell death in mitotically active cells, thus minimizing the risk of drug resistance (Ascierto and Marincola, 2011).
A newer strategy, restrictive combinations, is under clinical investigation. It focuses on optimizing drug dosing and scheduling to protect healthy cells while targeting cancer cells more effectively. For example, low doses of doxorubicin can induce a cell cycle arrest in healthy cells, shielding them from the cytotoxic effects of subsequent treatments like Taxol, which specifically targets cancer cells for mitotic arrest (Blagosklonny, 2008). The goal is to enhance the therapeutic impact by adding drugs in a manner that synergistically boosts their effectiveness (Bayat Mokhtari et al., 2017).
Drug repurposing is another innovative approach, where medications initially intended for other conditions are used in cancer treatment (Mokhtari et al., 2013). Acetazolamide, traditionally used for glaucoma, epilepsy, and altitude sickness, is one such drug being explored for cancer therapy. This is based on the observation that cancer cells frequently have high levels of carbonic anhydrase activity, contributing to their malignant properties. By inhibiting this enzyme, it's hoped that anticancer benefits can be achieved (Islam et al., 2016). Repurposing offers the advantages of using drugs with established safety profiles and potentially reducing treatment costs, making it a promising avenue in cancer therapy.
After cancer treatment
Epigenetic biomarkers in post-treatment evaluation
In the evolving landscape of cancer treatment and post-treatment evaluation, the role of epigenetic biomarkers is gaining prominence. The use of these biomarkers provides a practical method for assessing treatment efficacy and identifying potential risks that might emerge after treatment. This review explores the advancements and significance of various epigenetic biomarkers in different cancer types.
Among these biomarkers, DOK7, known as downstream of kinase, plays a pivotal role in the growth, migration, and invasion of cancer cells (Heyn et al., 2013). Intensive research has been conducted to understand its involvement in signaling pathways such as PI3K, PTEN, AKT. Clinical trials have shed light on how the overexpression of DOK7 can inhibit the activation of p-AKT and amplify the expression of PTEN, crucial for suppressing tumors associated with its overexpression. Specifically, in the context of breast cancer control, DOK7 is proposed as a powerful biomarker for forecasting the future presence of cancer and gauging its remission well in advance (Yue et al., 2021).
The field of bladder cancer detection also illustrates the potential of biomarkers like GDF15, TMEFF2, and VIM. These have shown a notable capability to detect the cellular absence of cancer and predict recurrence. Derived from extensive studies involving samples from healthy individuals and patients with renal and prostate cancer, these biomarkers have achieved remarkable sensitivity and specificity in both tissue and urine samples (Costa et al., 2010). This approach, being non-invasive, early, precise, and cost-effective, offers a viable option for the early detection of low-grade tumors, compared to traditional methods such as urine cytology and cystoscopy (Costa et al., 2010).
In colorectal carcinoma, the diagnostic landscape is still predominantly guided by the classic tumor-node-metastasis (TNM) methodology, especially in stage II where surgical interventions are common. However, these procedures carry a significant risk of tumor recurrence and fatal disease progression. To enhance accuracy and predictability, newer alternatives have been explored, involving specific biomarkers like carcinoembryonic antigen and carbohydrate antigen 19–9 (CA19-9), though the latter has faced criticism for its relative lack of specificity. Advances in technology such as microarray analysis, genomic screening, and sequencing have paved the way for more specific biomarkers. These biomarkers provide heightened precision in detecting microsatellite instability and microRNA instability (Luo et al., 2021). Biomarkers like KRAS, associated with viral oncogenesis, APC, linked to adenomatous polyposis coli, and TP53, coding for the tumor protein p53, have significantly contributed to the understanding of CRC pathogenesis and show promise in improving risk stratification and personalizing therapeutic approaches (Luo et al., 2021).
Finally, the epigenetic biomarker PHLPP2 has emerged as a significant indicator in pancreatic cancer. Research indicates that increased expression of PHLPP2 correlates with enhanced survival in patients with pancreatic adenocarcinoma. Studies have shown that vitamin C can elevate levels of 5-hydroxymethylcytosine in the promoter region of the PHLPP2 tumor suppressor gene, suggesting that VC-mediated DNA demethylation may positively regulate its expression. The epigenetic modulation of PHLPP2 could thus be pivotal in predicting recurrence and aiding in the treatment of pancreatic cancer (Chen et al., 2020).
In summary, the exploration of pharmacogenomic and epigenetic biomarkers in post-treatment evaluation opens new avenues for personalized medicine in cancer treatment, offering insights into patient prognosis, the efficacy of therapy, and potential future strategies in disease management based on the ethnicity (Paz-y-Miño et al., 2010; Paz-Y-Miño et al., 2015; López-Cortés et al., 2017b; 2020; López-Cortés et al., 2018; Yumiceba et al., 2020; Salas-Hernández et al., 2023).
Optimizing pharmacoepigenetic interventions: the role of nutrition, inflammation control, and circadian rhythm regulation
Optimizing pharmacoepigenetic interventions requires a nuanced understanding of the roles played by nutrition, inflammation control, and circadian rhythm regulation (Masri et al., 2015; Barrero et al., 2022; Tan et al., 2022). Chronic inflammation is a well-documented root cause of various diseases, including a significant proportion (40%–60%) of carcinoma cases (Zitvogel et al., 2017). This inflammation can stem from multiple sources, such as dietary metabolic syndromes like high cholesterol (LDL), which are known to trigger chronic inflammation leading to the formation of atheroma plaques in blood vessels. Conversely, certain dietary components, like omega-3 fatty acids, have been shown to possess anti-inflammatory properties, highlighting the importance of diet in managing inflammation and its related epigenetic effects (Pisaniello et al., 2021).
Inflammatory dietary agents can exacerbate conditions in certain individuals. For instance, gluten can cause abdominal and/or colorectal inflammation in celiac patients, while lactose may lead to inflammation in the large intestine of lactose-intolerant individuals (Prodhan et al., 2022; Iversen and Sollid, 2023). Additionally, meat and its by-products, containing heme iron, aromatic amino acids, and LDL, contribute to chronic inflammation through the formation of the trimethylamine N-oxide byproduct and cytokine cascade activation (Janeiro et al., 2018).
Diet choices play a crucial role in modulating inflammation and, by extension, cancer risk (Soldati et al., 2018). For instance, plant-based diets, rich in anti-inflammatory agents and antioxidants, have been associated with a reduced risk of developing colorectal cancer and cardiovascular diseases. This association is attributed to their ability to mitigate inflammation and influence epigenetic markers related to disease progression, including high-sensitivity C-reactive protein (hsCRP), IL-18, interleukin-1 receptor antagonist (IL-1Ra), intercellular adhesion molecule 1 (ICAM-1), adiponectin, omentin-1, and resistin (Chovert, 2013; Menzel et al., 2020b). However, the specific impact of these diets on inflammatory biomarkers and epigenetic modifications requires further research to fully understand their potential in preventing disease and enhancing pharmacoepigenetic treatments (Menzel et al., 2020a).
Nutrition’s dual role as both a risk factor and a protective agent against epigenetic changes underscores its significance in disease outcomes. Nutrients such as folate (B9 or M) and polyphenols can influence the methylation of oncogenes and tumor suppressor genes, affecting cancer risk and progression (López-Cortés et al., 2013; 2015; 2017a; Bishop and Ferguson, 2015). Additionally, isothiocyanates in colorectal cancer demonstrate the ability to suppress DNA methyltransferase expression, acting as oncogene repressors and influencing cellular proliferation (Gupta et al., 2019). In breast cancer, they appear to reduce aggressiveness by making estrogen-receptor interactions more responsive to tumor proliferation inhibitors (Hernando-Requejo et al., 2019). This intricate interplay between diet, inflammation, and epigenetics is crucial for accurate disease prediction, diagnostics, and treatment.
Moreover, the circadian rhythm, often referred to as the body’s biological clock, plays a crucial role in regulating various cellular functions essential in cancer progression (Liu et al., 2023). It orchestrates key bodily activities such as sleep patterns, hormonal fluctuations, body temperature regulation, and metabolic processes (Pérez-Villa et al., 2023). Disruptions in this rhythm have been closely linked to an increased risk of developing cancer, a connection supported by epidemiological research (Kaakour et al., 2023). Studies have shown that disturbances in the circadian rhythm significantly raise the risk for various types of cancer, including breast, colon, prostate, and skin cancers (Zhou et al., 2022). Therefore, adopting healthy lifestyle habits, such as regular sleep routines, minimal nighttime light exposure, and a balanced diet, can significantly reduce cancer risk and support the effectiveness of pharmacoepigenetic interventions (Zhou et al., 2022).
In summary, the optimization of pharmacoepigenetic treatments necessitates a holistic approach that incorporates nutritional guidance, inflammation control, and circadian rhythm regulation. This approach not only promises to enhance the efficacy of treatments but also to prevent disease onset and progression through lifestyle modifications and dietary interventions.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Latin American Society of Pharmacogenomics and Personalized Medicine (SOLFAGEM). Publication of this article was funded by Universidad de Las Américas from Quito, Ecuador.
Author contributions
BO-P: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. SR-O: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. DR-S: Conceptualization, Data curation, Formal Analysis, Methodology, Writing–original draft, Writing–review and editing. JM-G: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. MF: Data curation, Formal Analysis, Methodology, Writing–original draft, Writing–review and editing, Conceptualization. SE-F: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. AA-C: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. PE-E: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. MR-M: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. GE-G: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. DG-M: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. AJ-A: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. MA: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. AL-C: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing–original draft, Writing–review and editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Adams D. E., Shao W.-H. (2022). Epigenetic alterations in immune cells of systemic lupus erythematosus and therapeutic implications. Cells 11, 506. 10.3390/cells11030506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apuri S., Sokol L. (2016). An overview of investigational Histone deacetylase inhibitors (HDACis) for the treatment of non-Hodgkin’s lymphoma. Expert Opin. Investig. Drugs 25, 687–696. 10.1517/13543784.2016.1164140 [DOI] [PubMed] [Google Scholar]
- Ascierto P. A., Marincola F. M. (2011). Combination therapy: the next opportunity and challenge of medicine. J. Transl. Med. 9, 115. 10.1186/1479-5876-9-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austenaa L., Barozzi I., Chronowska A., Termanini A., Ostuni R., Prosperini E., et al. (2012). The histone methyltransferase Wbp7 controls macrophage function through GPI glycolipid anchor synthesis. Immunity 36, 572–585. 10.1016/j.immuni.2012.02.016 [DOI] [PubMed] [Google Scholar]
- Bader A. G. (2012). miR-34 - a microRNA replacement therapy is headed to the clinic. Front. Genet. 3, 120. 10.3389/fgene.2012.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell J. B., Leaver D. J., Hermans S. J., Kelly G. L., Brennan M. S., Downer N. L., et al. (2018). Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 560, 253–257. 10.1038/s41586-018-0387-5 [DOI] [PubMed] [Google Scholar]
- Bakulski K. M., Dou J., Lin N., London S. J., Colacino J. A. (2019). DNA methylation signature of smoking in lung cancer is enriched for exposure signatures in newborn and adult blood. Sci. Rep. 9, 4576. 10.1038/s41598-019-40963-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero M. J., Cejas P., Long H. W., Ramirez de Molina A. (2022). Nutritional epigenetics in cancer. Adv. Nutr. 13, 1748–1761. 10.1093/advances/nmac039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra R. N., Lifshitz A., Vidakovic A. T., Chin S.-F., Sati-Batra A., Sammut S.-J., et al. (2021). DNA methylation landscapes of 1538 breast cancers reveal a replication-linked clock, epigenomic instability and cis-regulation. Nat. Commun. 12, 5406. 10.1038/s41467-021-25661-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayat Mokhtari R., Homayouni T. S., Baluch N., Morgatskaya E., Kumar S., Das B., et al. (2017). Combination therapy in combating cancer. Oncotarget 8, 38022–38043. 10.18632/oncotarget.16723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin S. B., Jones P. A. (2016). Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol. 8, a019505. 10.1101/cshperspect.a019505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedregal P., Shand B., Santos M. J., Ventura-Juncá P. (2010). Contribution of epigenetics to understand human development. Rev. Méd. Chile 138, 366–372. 10.4067/S0034-98872010000300018 [DOI] [PubMed] [Google Scholar]
- Bell J. T., Spector T. D. (2011). A twin approach to unraveling epigenetics. Trends Genet. 27, 116–125. 10.1016/j.tig.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. (2007). Perceptions of epigenetics. Nature 447, 396–398. 10.1038/nature05913 [DOI] [PubMed] [Google Scholar]
- Bishop K. S., Ferguson L. R. (2015). The interaction between epigenetics, nutrition and the development of cancer. Nutrients 7, 922–947. 10.3390/nu7020922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H. (2001). Switching and signaling at the telomere. Cell 106, 661–673. 10.1016/s0092-8674(01)00492-5 [DOI] [PubMed] [Google Scholar]
- Blagosklonny M. V. (2008). “Targeting the absence” and therapeutic engineering for cancer therapy. Cell Cycle 7, 1307–1312. 10.4161/cc.7.10.6250 [DOI] [PubMed] [Google Scholar]
- Bolden J. E., Peart M. J., Johnstone R. W. (2006). Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 5, 769–784. 10.1038/nrd2133 [DOI] [PubMed] [Google Scholar]
- Bose P., Dai Y., Grant S. (2014). Histone deacetylase inhibitor (HDACI) mechanisms of action: emerging insights. Pharmacol. Ther. 143, 323–336. 10.1016/j.pharmthera.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyahya A., Mechchate H., Oumeslakht L., Zeouk I., Aboulaghras S., Balahbib A., et al. (2022). The role of epigenetic modifications in human cancers and the use of natural compounds as epidrugs: mechanistic pathways and pharmacodynamic actions. Biomolecules 12, 367. 10.3390/biom12030367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken A. P., Helin K. (2009). Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer 9, 773–784. 10.1038/nrc2736 [DOI] [PubMed] [Google Scholar]
- Brait M., Ling S., Nagpal J. K., Chang X., Park H. L., Lee J., et al. (2012). Cysteine dioxygenase 1 is a tumor suppressor gene silenced by promoter methylation in multiple human cancers. PLoS ONE 7, e44951. 10.1371/journal.pone.0044951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger M., Tarakhovsky A. (2014). Epigenetic control of immunity. Cold Spring Harb. Perspect. Biol. 6, a019307. 10.1101/cshperspect.a019307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Tsukada Y.-I., Zhang Y. (2005). Role of bmi-1 and Ring1A in H2A ubiquitylation and hox gene silencing. Mol. Cell 20, 845–854. 10.1016/j.molcel.2005.12.002 [DOI] [PubMed] [Google Scholar]
- Cavagnari B. M. (2012). Regulation of gene expression: how do epigenetic mechanisms work. Arch. Argent. Pediatr. 110, 132–136. 10.5546/aap.2012.132 [DOI] [PubMed] [Google Scholar]
- Chen J., Zuo Z., Gao Y., Yao X., Guan P., Wang Y., et al. (2023). Aberrant JAK-STAT signaling-mediated chromatin remodeling impairs the sensitivity of NK/T-cell lymphoma to chidamide. Clin. Epigenetics 15, 19. 10.1186/s13148-023-01436-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Song H., Luo Z., Cui H., Zheng W., Liu Y., et al. (2020). PHLPP2 is a novel biomarker and epigenetic target for the treatment of vitamin C in pancreatic cancer. Int. J. Oncol. 56, 1294–1303. 10.3892/ijo.2020.5001 [DOI] [PubMed] [Google Scholar]
- Chovert A. M. (2013). Medicina ortomolecular. [Google Scholar]
- Claes B., Buysschaert I., Lambrechts D. (2010). Pharmaco-epigenomics: discovering therapeutic approaches and biomarkers for cancer therapy. Heredity 105, 152–160. 10.1038/hdy.2010.42 [DOI] [PubMed] [Google Scholar]
- Corella D., Ordovas J. M. (2017). Basic concepts in molecular biology related to genetics and epigenetics. Rev. Española Cardiol. 70, 744–753. 10.1016/j.rec.2017.05.011 [DOI] [PubMed] [Google Scholar]
- Costa V. L., Henrique R., Danielsen S. A., Duarte-Pereira S., Eknaes M., Skotheim R. I., et al. (2010). Three epigenetic biomarkers, GDF15, TMEFF2, and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin. Cancer Res. 16, 5842–5851. 10.1158/1078-0432.CCR-10-1312 [DOI] [PubMed] [Google Scholar]
- Cotton A. M., Price E. M., Jones M. J., Balaton B. P., Kobor M. S., Brown C. J. (2015). Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet. 24, 1528–1539. 10.1093/hmg/ddu564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvoy Vasudevarao M., Dhanasekaran K., Selvi R. B., Kundu T. K. (2012). Inhibition of acetyltransferase alters different histone modifications: probed by small molecule inhibitor plumbagin. J. Biochem. 152, 453–462. 10.1093/jb/mvs093 [DOI] [PubMed] [Google Scholar]
- Deichmann U. (2016). Epigenetics: the origins and evolution of a fashionable topic. Dev. Biol. 416, 249–254. 10.1016/j.ydbio.2016.06.005 [DOI] [PubMed] [Google Scholar]
- De la Peña C., Vargas V. M. L. (2018). De la genética a la epigenética: La herencia que no está en los genes. [Google Scholar]
- Dhar G. A., Saha S., Mitra P., Nag Chaudhuri R. (2021). DNA methylation and regulation of gene expression: guardian of our health. Nucl. (Calcutta) 64, 259–270. 10.1007/s13237-021-00367-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Lagares A., Mendez-Gonzalez J., Hervas D., Saigi M., Pajares M. J., Garcia D., et al. (2016). A novel epigenetic signature for early diagnosis in lung cancer. Clin. Cancer Res. 22, 3361–3371. 10.1158/1078-0432.CCR-15-2346 [DOI] [PubMed] [Google Scholar]
- Eckschlager T., Plch J., Stiborova M., Hrabeta J. (2017). Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. 18, 1414. 10.3390/ijms18071414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. (2002). CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene 21, 5427–5440. 10.1038/sj.onc.1205600 [DOI] [PubMed] [Google Scholar]
- Esteller M. (2007). Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 8, 286–298. 10.1038/nrg2005 [DOI] [PubMed] [Google Scholar]
- Esteller M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. (2014). A brief history of epigenetics. Cold Spring Harb. Perspect. Biol. 6, a018200. 10.1101/cshperspect.a018200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenaux P., Mufti G. J., Hellström-Lindberg E., Santini V., Gattermann N., Germing U., et al. (2010). Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. J. Clin. Oncol. 28, 562–569. 10.1200/JCO.2009.23.8329 [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith A. C. (2011). Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 12, 565–575. 10.1038/nrg3032 [DOI] [PubMed] [Google Scholar]
- Ferrarelli L. K. (2016). HDAC inhibitors in solid tumors and blood cancers. Sci. Signal. 9, ec216. 10.1126/scisignal.aaj2316 [DOI] [Google Scholar]
- Ferro M., Ungaro P., Cimmino A., Lucarelli G., Busetto G. M., Cantiello F., et al. (2017). Epigenetic signature: a new player as predictor of clinically significant prostate cancer (pca) in patients on active surveillance (AS). Int. J. Mol. Sci. 18, 1146. 10.3390/ijms18061146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K. A., Kagan J. C. (2020). Toll-like receptors and the control of immunity. Cell 180, 1044–1066. 10.1016/j.cell.2020.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M., McDonald L. E., Millar D. S., Collis C. M., Watt F., Grigg G. W., et al. (1992). A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U. S. A. 89, 1827–1831. 10.1073/pnas.89.5.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailhouste L., Liew L. C., Hatada I., Nakagama H., Ochiya T. (2018). Epigenetic reprogramming using 5-azacytidine promotes an anti-cancer response in pancreatic adenocarcinoma cells. Cell Death Dis. 9, 468. 10.1038/s41419-018-0487-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinari P., Di Marco S., Jones P., Pallaoro M., Steinkühler C. (2007). HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res. 17, 195–211. 10.1038/sj.cr.7310149 [DOI] [PubMed] [Google Scholar]
- Gezer U., Yörüker E. E., Keskin M., Kulle C. B., Dharuman Y., Holdenrieder S. (2015). Histone methylation marks on circulating nucleosomes as novel blood-based biomarker in colorectal cancer. Cancer. Int. J. Mol. Sci. 16, 29654–29662. 10.3390/ijms161226180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemi S. (2020). Cancer’s epigenetic drugs: where are they in the cancer medicines? Pharmacogenomics J. 20, 367–379. 10.1038/s41397-019-0138-5 [DOI] [PubMed] [Google Scholar]
- Ghoshal K., Bai S. (2007). DNA methyltransferases as targets for cancer therapy. Drugs Today 43, 395–422. 10.1358/dot.2007.43.6.1062666 [DOI] [PubMed] [Google Scholar]
- Giri A. K., Aittokallio T. (2019). DNMT inhibitors increase methylation in the cancer genome. Front. Pharmacol. 10, 385. 10.3389/fphar.2019.00385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak M. A., Sengupta N., Zhang X., Seto E. (2005). Acetylation and deacetylation of non-histone proteins. Gene 363, 15–23. 10.1016/j.gene.2005.09.010 [DOI] [PubMed] [Google Scholar]
- Gonzalo V., Castellví-Bel S., Balaguer F., Pellisé M., Ocaña T., Castells A. (2008). Epigenetics of cancer. Hepatol 31, 37–45. 10.1157/13114573 [DOI] [PubMed] [Google Scholar]
- Greer E. L., Shi Y. (2012). Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357. 10.1038/nrg3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S. I. S., Moazed D. (2003). Heterochromatin and epigenetic control of gene expression. Science 301, 798–802. 10.1126/science.1086887 [DOI] [PubMed] [Google Scholar]
- Gujral P., Mahajan V., Lissaman A. C., Ponnampalam A. P. (2020). Histone acetylation and the role of histone deacetylases in normal cyclic endometrium. Reprod. Biol. Endocrinol. 18, 84. 10.1186/s12958-020-00637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N.-H., Zheng J.-F., Zi F.-M., Cheng J. (2019). I-BET151 suppresses osteoclast formation and inflammatory cytokines secretion by targetting BRD4 in multiple myeloma. Biosci. Rep. 39. 10.1042/BSR20181245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Bhatt L. K., Momin M. (2019). Potent antitumor activity of Laccaic acid and Phenethyl isothiocyanate combination in colorectal cancer via dual inhibition of DNA methyltransferase-1 and Histone deacetylase-1. Toxicol. Appl. Pharmacol. 377, 114631. 10.1016/j.taap.2019.114631 [DOI] [PubMed] [Google Scholar]
- Gusev Y. (2019). MicroRNA profiling in cancer: a bioinformatics perspective. [DOI] [PubMed] [Google Scholar]
- Hernández A., Martín Vasallo P., Torres A., Salido E. (1995). Análisis del RNA: Estudio de la expresión genética. Nefrología. [Google Scholar]
- Hernando-Requejo O., García de Quinto H., Rubio Rodríguez M. a. C. (2019). Nutrition as an epigenetic factor in develops of cancer. Nutr. Hosp. 36, 53–57. 10.20960/nh.02810 [DOI] [PubMed] [Google Scholar]
- Hess-Stumpp H., Bracker T. U., Henderson D., Politz O. (2007). MS-275, a potent orally available inhibitor of histone deacetylases--the development of an anticancer agent. Int. J. Biochem. Cell Biol. 39, 1388–1405. 10.1016/j.biocel.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Heyn H., Carmona F. J., Gomez A., Ferreira H. J., Bell J. T., Sayols S., et al. (2013). DNA methylation profiling in breast cancer discordant identical twins identifies DOK7 as novel epigenetic biomarker. Carcinogenesis 34, 102–108. 10.1093/carcin/bgs321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsthemke B. (2006). Epimutations in human disease. Curr. Top. Microbiol. Immunol. 310, 45–59. 10.1007/3-540-31181-5_4 [DOI] [PubMed] [Google Scholar]
- Islam S. S., Mokhtari R. B., Akbari P., Hatina J., Yeger H., Farhat W. A. (2016). Simultaneous targeting of bladder tumor growth, survival, and epithelial-to-mesenchymal transition with a novel therapeutic combination of acetazolamide (AZ) and sulforaphane (SFN). Target. Oncol. 11, 209–227. 10.1007/s11523-015-0386-5 [DOI] [PubMed] [Google Scholar]
- Issa J.-P. J., Kantarjian H. M. (2009). Targeting DNA methylation. Clin. Cancer Res. 15, 3938–3946. 10.1158/1078-0432.CCR-08-2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A., Soria J.-C., Toulmonde M., Michot J.-M., Lucchesi C., Varga A., et al. (2018). Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 19, 649–659. 10.1016/S1470-2045(18)30145-1 [DOI] [PubMed] [Google Scholar]
- Iversen R., Sollid L. M. (2023). The immunobiology and pathogenesis of celiac disease. Annu. Rev. Pathol. 18, 47–70. 10.1146/annurev-pathmechdis-031521-032634 [DOI] [PubMed] [Google Scholar]
- Jain S., Zain J. (2011). Romidepsin in the treatment of cutaneous T-cell lymphoma. J. Blood Med. 2, 37–47. 10.2147/JBM.S9649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeiro M. H., Ramírez M. J., Milagro F. I., Martínez J. A., Solas M. (2018). Implication of trimethylamine N-oxide (TMAO) in disease: potential biomarker or new therapeutic target. Nutrients 10, 1398. 10.3390/nu10101398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B., Robertson K. D. (2013). DNA methyltransferases, DNA damage repair, and cancer. Adv. Exp. Med. Biol. 754, 3–29. 10.1007/978-1-4419-9967-2_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A., Baylin S. B. (2007). The epigenomics of cancer. Cell 128, 683–692. 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juárez-Mercado K. E., Prieto-Martínez F. D., Sánchez-Cruz N., Peña-Castillo A., Prada-Gracia D., Medina-Franco J. L. (2020). Expanding the structural diversity of DNA methyltransferase inhibitors. Pharm. (Basel) 14, 17. 10.3390/ph14010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakour D., Fortin B., Masri S., Rezazadeh A. (2023). Circadian clock dysregulation and prostate cancer: a molecular and clinical overview. Clin. Med. Insights Oncol. 17, 11795549231211521. 10.1177/11795549231211521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminskas E., Farrell A., Abraham S., Baird A., Hsieh L.-S., Lee S.-L., et al. (2005). Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin. Cancer Res. 11, 3604–3608. 10.1158/1078-0432.CCR-04-2135 [DOI] [PubMed] [Google Scholar]
- Kantarjian H., Issa J.-P. J., Rosenfeld C. S., Bennett J. M., Albitar M., DiPersio J., et al. (2006). Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer 106, 1794–1803. 10.1002/cncr.21792 [DOI] [PubMed] [Google Scholar]
- Kim H.-J., Bae S.-C. (2011). Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs. Am. J. Transl. Res. 3, 166–179. [PMC free article] [PubMed] [Google Scholar]
- Koch A., Joosten S. C., Feng Z., de Ruijter T. C., Draht M. X., Melotte V., et al. (2018). Analysis of DNA methylation in cancer: location revisited. Nat. Rev. Clin. Oncol. 15, 459–466. 10.1038/s41571-018-0004-4 [DOI] [PubMed] [Google Scholar]
- Koonin E. V. (2010). The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 11, 209. 10.1186/gb-2010-11-5-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopytko P., Piotrowska K., Janisiak J., Tarnowski M. (2021). Garcinol-A natural histone acetyltransferase inhibitor and new anti-cancer epigenetic drug. Int. J. Mol. Sci. 22, 2828. 10.3390/ijms22062828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128, 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Kringel D., Malkusch S., Lötsch J. (2021). Drugs and epigenetic molecular functions. A pharmacological data scientometric analysis. Int. J. Mol. Sci. 22, 7250. 10.3390/ijms22147250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulka L. A. M., Fangmann P.-V., Panfilova D., Olzscha H. (2020). Impact of HDAC inhibitors on protein quality control systems: consequences for precision medicine in malignant disease. Front. Cell Dev. Biol. 8, 425. 10.3389/fcell.2020.00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J. K. W., Chow M. Y. T., Zhang Y., Leung S. W. S. (2015). siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids 4, e252. 10.1038/mtna.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laranjeira A. B. A., Hollingshead M. G., Nguyen D., Kinders R. J., Doroshow J. H., Yang S. X. (2023). DNA damage, demethylation and anticancer activity of DNA methyltransferase (DNMT) inhibitors. Sci. Rep. 13, 5964. 10.1038/s41598-023-32509-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A. Y., et al. (2013). Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520. 10.1038/nature12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ye Y., Peng K., Zeng Z., Chen L., Zeng Y. (2022). Histones: the critical players in innate immunity. Front. Immunol. 13, 1030610. 10.3389/fimmu.2022.1030610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian C. G., Xu Y., Ceol C., Wu F., Larson A., Dresser K., et al. (2012). Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150, 1135–1146. 10.1016/j.cell.2012.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liu Y., Hai R., Liao W., Luo X. (2023). The role of circadian clocks in cancer: mechanisms and clinical implications. Genes Dis. 10, 1279–1290. 10.1016/j.gendis.2022.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Zheng M., Lu R., Du J., Zhao Q., Li Z., et al. (2020). The role of CDC25C in cell cycle regulation and clinical cancer therapy: a systematic review. Cancer Cell Int. 20, 213. 10.1186/s12935-020-01304-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Kimball S., Liu H., Holowatyj A., Yang Z.-Q. (2015). Genetic alterations of histone lysine methyltransferases and their significance in breast cancer. Oncotarget 6, 2466–2482. 10.18632/oncotarget.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Cortés A., Cabrera-Andrade A., Salazar-Ruales C., Zambrano A. K., Guerrero S., Guevara P., et al. (2017a). Genotyping the high altitude mestizo Ecuadorian population affected with prostate cancer. Biomed. Res. Int. 2017, 3507671. 10.1155/2017/3507671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Cortés A., Echeverría C., Oña-Cisneros F., Sánchez M. E., Herrera C., Cabrera-Andrade A., et al. (2015). Breast cancer risk associated with gene expression and genotype polymorphisms of the folate-metabolizing MTHFR gene: a case-control study in a high altitude Ecuadorian mestizo population. Tumour Biol. 36, 6451–6461. 10.1007/s13277-015-3335-0 [DOI] [PubMed] [Google Scholar]
- López-Cortés A., Guerrero S., Redal M. A., Alvarado A. T., Quiñones L. A. (2017b). State of art of cancer pharmacogenomics in Latin american populations. Int. J. Mol. Sci. 18, 639. 10.3390/ijms18060639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Cortés A., Jaramillo-Koupermann G., Muñoz M. J., Cabrera A., Echeverría C., Rosales F., et al. (2013). Genetic polymorphisms in MTHFR (C677T, A1298C), MTR (A2756G) and MTRR (A66G) genes associated with pathological characteristics of prostate cancer in the Ecuadorian population. Am. J. Med. Sci. 346, 447–454. 10.1097/MAJ.0b013e3182882578 [DOI] [PubMed] [Google Scholar]
- López-Cortés A., Leone P. E., Freire-Paspuel B., Arcos-Villacís N., Guevara-Ramírez P., Rosales F., et al. (2018). Mutational analysis of oncogenic AKT1 gene associated with breast cancer risk in the high altitude Ecuadorian mestizo population. Biomed. Res. Int. 2018, 7463832. 10.1155/2018/7463832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Cortés A., Paz-Y-Miño C., Guerrero S., Jaramillo-Koupermann G., León Cáceres Á., Intriago-Baldeón D. P., et al. (2020). Pharmacogenomics, biomarker network, and allele frequencies in colorectal cancer. Pharmacogenomics J. 20, 136–158. 10.1038/s41397-019-0102-4 [DOI] [PubMed] [Google Scholar]
- Lu Y., Chan Y.-T., Tan H.-Y., Li S., Wang N., Feng Y. (2020). Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy. Mol. Cancer 19, 79. 10.1186/s12943-020-01197-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X.-J., Zhao Q., Liu J., Zheng J.-B., Qiu M.-Z., Ju H.-Q., et al. (2021). Novel genetic and epigenetic biomarkers of prognostic and predictive significance in stage II/III colorectal cancer. Mol. Ther. 29, 587–596. 10.1016/j.ymthe.2020.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann B. S., Johnson J. R., He K., Sridhara R., Abraham S., Booth B. P., et al. (2007). Vorinostat for treatment of cutaneous manifestations of advanced primary cutaneous T-cell lymphoma. Clin. Cancer Res. 13, 2318–2322. 10.1158/1078-0432.CCR-06-2672 [DOI] [PubMed] [Google Scholar]
- Manzo F., Tambaro F. P., Mai A., Altucci L. (2009). Histone acetyltransferase inhibitors and preclinical studies. Expert Opin. Ther. Pat. 19, 761–774. 10.1517/13543770902895727 [DOI] [PubMed] [Google Scholar]
- Masri S., Kinouchi K., Sassone-Corsi P. (2015). Circadian clocks, epigenetics, and cancer. Curr. Opin. Oncol. 27, 50–56. 10.1097/CCO.0000000000000153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matore B. W., Banjare P., Guria T., Roy P. P., Singh J. (2022). Oxadiazole derivatives: histone deacetylase inhibitors in anticancer therapy and drug discovery. Eur. J. Med. Chem. Rep. 5, 100058. 10.1016/j.ejmcr.2022.100058 [DOI] [Google Scholar]
- Mayr C., Kiesslich T., Erber S., Bekric D., Dobias H., Beyreis M., et al. (2021). HDAC screening identifies the HDAC class I inhibitor romidepsin as a promising epigenetic drug for biliary tract cancer. Cancers (Basel) 13, 3862. 10.3390/cancers13153862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez D., Gaulton A., Bento A. P., Chambers J., De Veij M., Félix E., et al. (2019). ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 47, D930–D940. 10.1093/nar/gky1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel J., Biemann R., Longree A., Isermann B., Mai K., Schulze M. B., et al. (2020a). Associations of a vegan diet with inflammatory biomarkers. Sci. Rep. 10, 1933. 10.1038/s41598-020-58875-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel J., Jabakhanji A., Biemann R., Mai K., Abraham K., Weikert C. (2020b). Systematic review and meta-analysis of the associations of vegan and vegetarian diets with inflammatory biomarkers. Sci. Rep. 10, 21736. 10.1038/s41598-020-78426-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. (2020). The self-organizing genome: principles of genome architecture and function. Cell 183, 28–45. 10.1016/j.cell.2020.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohtat D., Susztak K. (2010). Fine tuning gene expression: the epigenome. Semin. Nephrol. 30, 468–476. 10.1016/j.semnephrol.2010.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokhtari R. B., Kumar S., Islam S. S., Yazdanpanah M., Adeli K., Cutz E., et al. (2013). Combination of carbonic anhydrase inhibitor, acetazolamide, and sulforaphane, reduces the viability and growth of bronchial carcinoid cell lines. BMC Cancer 13, 378. 10.1186/1471-2407-13-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morera L., Lübbert M., Jung M. (2016). Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin. Epigenetics 8, 57. 10.1186/s13148-016-0223-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan C. G., Zhang J., Kasper L. H., Lerach S., Payne-Turner D., Phillips L. A., et al. (2011). CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 471, 235–239. 10.1038/nature09727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahmias Y., Androulakis I. P. (2021). Circadian effects of drug responses. Annu. Rev. Biomed. Eng. 23, 203–224. 10.1146/annurev-bioeng-082120-034725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E., Jeffrey K. L., Schaefer U., Beinke S., Dewell S., Chung C.-W., et al. (2010). Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123. 10.1038/nature09589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A., Nakanishi M. (2021). Navigating the DNA methylation landscape of cancer. Trends Genet. 37, 1012–1027. 10.1016/j.tig.2021.05.002 [DOI] [PubMed] [Google Scholar]
- Norollahi S. E., Mansour-Ghanaei F., Joukar F., Ghadarjani S., Mojtahedi K., Gharaei Nejad K., et al. (2019). Therapeutic approach of Cancer stem cells (CSCs) in gastric adenocarcinoma; DNA methyltransferases enzymes in cancer targeted therapy. Biomed. Pharmacother. 115, 108958. 10.1016/j.biopha.2019.108958 [DOI] [PubMed] [Google Scholar]
- Obata Y., Furusawa Y., Hase K. (2015). Epigenetic modifications of the immune system in health and disease. Immunol. Cell Biol. 93, 226–232. 10.1038/icb.2014.114 [DOI] [PubMed] [Google Scholar]
- Ochoa D., Hercules A., Carmona M., Suveges D., Baker J., Malangone C., et al. (2023). Open Targets Platform: supporting systematic drug-target identification and prioritisation. Nucleic Acids Res. 51, D1302–D1310. 10.1093/nar/gkaa1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor O. A., Horwitz S., Masszi T., Van Hoof A., Brown P., Doorduijn J., et al. (2015). Belinostat in patients with relapsed or refractory peripheral T-cell lymphoma: results of the pivotal phase II BELIEF (CLN-19) study. J. Clin. Oncol. 33, 2492–2499. 10.1200/JCO.2014.59.2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-y-Miño C., López-Cortés A., Muñoz M. J., Cabrera A., Castro B., Sánchez M. E. (2010). Incidence of the L858R and G719S mutations of the epidermal growth factor receptor oncogene in an Ecuadorian population with lung cancer. Cancer Genet. cytogenet. 196, 201–203. 10.1016/j.cancergencyto.2009.10.007 [DOI] [PubMed] [Google Scholar]
- Paz-Y-Miño C. A., García-Cárdenas J. M., López-Cortés A., Salazar C., Serrano M., Leone P. E. (2015). Positive association of the cathepsin D ala224val gene polymorphism with the risk of alzheimer’s disease. Am. J. Med. Sci. 350, 296–301. 10.1097/MAJ.0000000000000555 [DOI] [PubMed] [Google Scholar]
- Pérez R. F., Tejedor J. R., Santamarina-Ojeda P., Martínez V. L., Urdinguio R. G., Villamañán L., et al. (2021). Conservation of aging and cancer epigenetic signatures across human and mouse. Mol. Biol. Evol. 38, 3415–3435. 10.1093/molbev/msab112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Villa A., Echeverría-Garcés G., Ramos-Medina M. J., Prathap L., Martínez-López M., Ramírez-Sánchez D., et al. (2023). Integrated multi-omics analysis reveals the molecular interplay between circadian clocks and cancer pathogenesis. Sci. Rep. 13, 14198. 10.1038/s41598-023-39401-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarz R. L., Frye R., Turner M., Wright J. J., Allen S. L., Kirschbaum M. H., et al. (2009). Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J. Clin. Oncol. 27, 5410–5417. 10.1200/JCO.2008.21.6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisaniello A. D., Psaltis P. J., King P. M., Liu G., Gibson R. A., Tan J. T., et al. (2021). Omega-3 fatty acids ameliorate vascular inflammation: a rationale for their atheroprotective effects. Atherosclerosis 324, 27–37. 10.1016/j.atherosclerosis.2021.03.003 [DOI] [PubMed] [Google Scholar]
- Pócza T., Krenács T., Turányi E., Csáthy J., Jakab Z., Hauser P. (2016). High expression of DNA methyltransferases in primary human medulloblastoma. Folia Neuropathol. 54, 105–113. 10.5114/fn.2016.60365 [DOI] [PubMed] [Google Scholar]
- Poziello A., Nebbioso A., Stunnenberg H. G., Martens J. H. A., Carafa V., Altucci L. (2021). Recent insights into Histone Acetyltransferase-1: biological function and involvement in pathogenesis. Epigenetics 16, 838–850. 10.1080/15592294.2020.1827723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodhan U. K., Milan A. M., Shrestha A., Vickers M. H., Cameron-Smith D., Barnett M. P. G. (2022). Circulatory amino acid responses to milk consumption in dairy and lactose intolerant individuals. Eur. J. Clin. Nutr. 76, 1415–1422. 10.1038/s41430-022-01119-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan V. K., Down T. A., Balding D. J., Beck S. (2011). Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 12, 529–541. 10.1038/nrg3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez L. A., Zayas R. R., González S. H. (2009). Los procesos de glucosilación no enzimática. Archivo Médico de. [Google Scholar]
- Riggs A. D., Xiong Z. (2004). Methylation and epigenetic fidelity. Proc. Natl. Acad. Sci. U. S. A. 101, 4–5. 10.1073/pnas.0307781100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K. D. (2001). DNA methylation, methyltransferases, and cancer. Oncogene 20, 3139–3155. 10.1038/sj.onc.1204341 [DOI] [PubMed] [Google Scholar]
- Rossetto D., Avvakumov N., Côté J. (2012). Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics 7, 1098–1108. 10.4161/epi.21975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabnis R. W. (2021). Novel histone acetyltransferase (HAT) inhibitors for treating diseases. ACS Med. Chem. Lett. 12, 1198–1199. 10.1021/acsmedchemlett.1c00337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Hernández A., Galleguillos M., Carrasco M., López-Cortés A., Redal M. A., Fonseca-Mendoza D., et al. (2023). An updated examination of the perception of barriers for pharmacogenomics implementation and the usefulness of drug/gene pairs in Latin America and the Caribbean. Front. Pharmacol. 14, 1175737. 10.3389/fphar.2023.1175737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Miguel J. F., Hungria V. T. M., Yoon S.-S., Beksac M., Dimopoulos M. A., Elghandour A., et al. (2014). Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: a multicentre, randomised, double-blind phase 3 trial. Lancet Oncol. 15, 1195–1206. 10.1016/S1470-2045(14)70440-1 [DOI] [PubMed] [Google Scholar]
- Schwabish M. A., Struhl K. (2007). The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo . Mol. Cell. Biol. 27, 6987–6995. 10.1128/MCB.00717-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler J.-S., Dejean A. (2003). Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4, 690–699. 10.1038/nrm1200 [DOI] [PubMed] [Google Scholar]
- Seto E., Yoshida M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6, a018713. 10.1101/cshperspect.a018713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. (2007). Taking epigenetics center stage. Cell 128, 639–640. 10.1016/j.cell.2007.02.011 [DOI] [Google Scholar]
- Simon J. A., Lange C. A. (2008). Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 647, 21–29. 10.1016/j.mrfmmm.2008.07.010 [DOI] [PubMed] [Google Scholar]
- Soldati L., Di Renzo L., Jirillo E., Ascierto P. A., Marincola F. M., De Lorenzo A. (2018). The influence of diet on anti-cancer immune responsiveness. J. Transl. Med. 16, 75. 10.1186/s12967-018-1448-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D. E., Berger S. L. (2000). Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64, 435–459. 10.1128/MMBR.64.2.435-459.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimson L., Wood V., Khan O., Fotheringham S., La Thangue N. B. (2009). HDAC inhibitor-based therapies and haematological malignancy. Ann. Oncol. 20, 1293–1302. 10.1093/annonc/mdn792 [DOI] [PubMed] [Google Scholar]
- Subramaniam D., Thombre R., Dhar A., Anant S. (2014). DNA methyltransferases: a novel target for prevention and therapy. Front. Oncol. 4, 80. 10.3389/fonc.2014.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera A., O’Byrne K. J., Richard D. J. (2018). Combination therapy with histone deacetylase inhibitors (hdaci) for the treatment of cancer: achieving the full therapeutic potential of hdaci. Front. Oncol. 8, 92. 10.3389/fonc.2018.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P. B., Henikoff S. (2010). Histone variants--ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11, 264–275. 10.1038/nrm2861 [DOI] [PubMed] [Google Scholar]
- Talukdar P. D., Chatterji U. (2023). Transcriptional co-activators: emerging roles in signaling pathways and potential therapeutic targets for diseases. Signal Transduct. Target. Ther. 8, 427. 10.1038/s41392-023-01651-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. Y. X., Zhang J., Tee W.-W. (2022). Epigenetic regulation of inflammatory signaling and inflammation-induced cancer. Front. Cell Dev. Biol. 10, 931493. 10.3389/fcell.2022.931493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman R. E., Rynes E., Humbert R., Vierstra J., Maurano M. T., Haugen E., et al. (2012). The accessible chromatin landscape of the human genome. Nature 489, 75–82. 10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo González D., Cruz-Bustillo Clarens D. (2005). La inestabilidad en microsatélites: algunos aspectos de su relación con el cáncer colorrectal hereditario no-polipoide. Rev. Cubana Investig. Biomédicas. [Google Scholar]
- Tonmoy M. I. Q., Fariha A., Hami I., Kar K., Reza H. A., Bahadur N. M., et al. (2022). Computational epigenetic landscape analysis reveals association of CACNA1G-AS1, F11-AS1, NNT-AS1, and MSC-AS1 lncRNAs in prostate cancer progression through aberrant methylation. Sci. Rep. 12, 10260. 10.1038/s41598-022-13381-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynor S., Terp M. G., Nielsen A. Y., Guldberg P., Jakobsen M., Pedersen P. G., et al. (2023). DNA methyltransferase inhibition promotes recruitment of myeloid-derived suppressor cells to the tumor microenvironment through induction of tumor cell-intrinsic interleukin-1. Cancer Lett. 552, 215982. 10.1016/j.canlet.2022.215982 [DOI] [PubMed] [Google Scholar]
- Villagra A., Cheng F., Wang H.-W., Suarez I., Glozak M., Maurin M., et al. (2009). The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat. Immunol. 10, 92–100. 10.1038/ni.1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P., Tee W.-W., Reinberg D. (2013). A double take on bivalent promoters. Genes Dev. 27, 1318–1338. 10.1101/gad.219626.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Ma T., Yu B. (2023). Targeting epigenetic regulators to overcome drug resistance in cancers. Signal Transduct. Target. Ther. 8, 69. 10.1038/s41392-023-01341-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Wu R., Tang D., Kang R. (2021). The BET family in immunity and disease. Signal Transduct. Target. Ther. 6, 23. 10.1038/s41392-020-00384-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapenaar H., Dekker F. J. (2016). Histone acetyltransferases: challenges in targeting bi-substrate enzymes. Clin. Epigenetics 8, 59. 10.1186/s13148-016-0225-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M., Davies J. J., Wittig D., Oakeley E. J., Haase M., Lam W. L., et al. (2005). Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37, 853–862. 10.1038/ng1598 [DOI] [PubMed] [Google Scholar]
- Wilson B. G., Roberts C. W. M. (2011). SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492. 10.1038/nrc3068 [DOI] [PubMed] [Google Scholar]
- Wu D., Qiu Y., Jiao Y., Qiu Z., Liu D. (2020). Small molecules targeting hats, hdacs, and brds in cancer therapy. Front. Oncol. 10, 560487. 10.3389/fonc.2020.560487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. C., Zhang Y. (2010). Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 11, 607–620. 10.1038/nrm2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M., Liu J., Wu X., Liu S., Li G., Han C., et al. (2013). Histone methyltransferase Ash1l suppresses interleukin-6 production and inflammatory autoimmune diseases by inducing the ubiquitin-editing enzyme A20. Immunity 39, 470–481. 10.1016/j.immuni.2013.08.016 [DOI] [PubMed] [Google Scholar]
- Xu H., Lin S., Zhou Z., Li D., Zhang X., Yu M., et al. (2023). New genetic and epigenetic insights into the chemokine system: the latest discoveries aiding progression toward precision medicine. Cell. Mol. Immunol. 20, 739–776. 10.1038/s41423-023-01032-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue C., Bai Y., Piao Y., Liu H. (2021). DOK7 inhibits cell proliferation, migration, and invasion of breast cancer via the PI3K/PTEN/AKT pathway. J. Oncol. 2021, 4035257. 10.1155/2021/4035257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumiceba V., López-Cortés A., Pérez-Villa A., Yumiseba I., Guerrero S., García-Cárdenas J. M., et al. (2020). Oncology and pharmacogenomics insights in polycystic ovary syndrome: an integrative analysis. Front. Endocrinol. (Lausanne) 11, 585130. 10.3389/fendo.2020.585130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Xia C., Yang Y., Sun F., Zhang Y., Wang H., et al. (2023). DNA methylation and histone post-translational modifications in atherosclerosis and a novel perspective for epigenetic therapy. Cell Commun. Signal. 21, 344. 10.1186/s12964-023-01298-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Shilatifard A. (2019). Epigenetic modifications of histones in cancer. Genome Biol. 20, 245. 10.1186/s13059-019-1870-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C. Y., Johnson S. L., Gamarra N. I., Narlikar G. J. (2016). Mechanisms of ATP-dependent chromatin remodeling motors. Annu. Rev. Biophys. 45, 153–181. 10.1146/annurev-biophys-051013-022819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Zhang Z., Nice E., Huang C., Zhang W., Tang Y. (2022). Circadian rhythms and cancers: the intrinsic links and therapeutic potentials. J. Hematol. Oncol. 15, 21. 10.1186/s13045-022-01238-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L., Pietrocola F., Kroemer G. (2017). Nutrition, inflammation and cancer. Nat. Immunol. 18, 843–850. 10.1038/ni.3754 [DOI] [PubMed] [Google Scholar]