Abstract
In most eukaryotic organisms the U2 small nuclear RNA (snRNA) gene is transcribed by RNA polymerase II to generate a primary transcript with a 5′ terminal 7-methylguanosine cap structure. Following nuclear export, the U2 snRNA is assembled into a core ribonucleoprotein particle (RNP). This involves binding a set of proteins that are shared by spliceosomal snRNPs to the highly conserved Sm site. Prior to nuclear import, the snRNA-(guanosine-N2)-methyltransferase appears to interact with the core RNP and hypermethylates the cap structure to 2,2,7-trimethylguanosine (m3G). In the protist parasite Trypanosoma brucei, U-snRNAs are complexed with a set of common proteins that are analogous to eukaryotic Sm antigens but do not have a highly conserved Sm sequence motif, and most U-snRNAs are synthesised by RNA polymerase III. Here, we examined the determinants for m3G cap formation in T.brucei by expressing mutant U2 snRNAs in vivo and assaying trimethylation and RNP assembly by immunoprecipitation. Surprisingly, these studies revealed that the Sm-analogous region is not required either for binding of the common proteins or for cap trimethylation. Furthermore, except for the first 24 nt which are part of the U2 promoter, the U2 coding region could be substituted or deleted without affecting cap trimethylation.
INTRODUCTION
The U1, U2, U4/U6 and U5 small nuclear RNAs (snRNAs) are essential for splicing of nuclear pre-mRNA (reviewed in 1–3). In most eukaryotes, all except U6 snRNA are synthesised by RNA polymerase II to produce primary transcripts with a 7-methylguanosine (m7G) cap at their 5′ end and short extensions at the 3′ end (reviewed in 4,5). The precursors are exported from the nucleus to the cytoplasm where they assemble into a stable core ribonucleoprotein particle (RNP) by binding the common proteins, referred to as Sm antigens, at the highly conserved Sm site [5′-PuA(U)nGPu-3′] (6). The 3′ extensions of the pre-snRNAs are removed and, as an essential maturation step, the m7G cap is hypermethylated to 2,2,7-trimethylguanosine (m3G). In Xenopus oocytes it has been shown that formation of the m3G cap is dependent on the Sm site and coincides with binding of the common proteins (7). No other sequence determinants for cap trimethylation were found in spliceosomal U-snRNAs: even changing the 5′-terminal sequences of human U1 or Xenopus U2 snRNAs had no detectable effect on cap trimethylation (7,8). By developing an in vitro trimethylation assay for human U1 snRNA, Plessel et al. (9) provided evidence that the snRNA-(guanosine-N2)-methyltransferase is a non-snRNP protein localised in the cytoplasm and that it recognises the U1 RNP by interacting with the common proteins, presumably by binding to the B/B′ core proteins. These findings were supported by studying the snRNP core assembly pathway: the core U1 RNP was hypermethylated in vitro, whereas a subcore U1 RNP lacking the B/B′ proteins was not (10). Following methylation in the cytoplasm, the U-snRNAs are reimported into the nucleus, where they may recruit additional snRNP-specific proteins. Nuclear import is dependent on a bipartite signal composed of the common proteins (11) and the m3G cap (12,13). In addition, nuclear reimport of the U2 snRNA requires processed 3′ ends (14).
A complete set of spliceosomal U-snRNAs has been identified in trypanosomatids (15–19) and site-directed degradation has shown that U2 and U4/U6 snRNAs are essential components of the mRNA splicing apparatus (20). Similar to what is known in other organisms, U2 and U4 snRNAs contain a trimethylated cap structure (15,16). However, unlike other eukaryotic U2 snRNA genes, the Trypanosoma brucei homologue is synthesised by RNA polymerase III and expression is regulated by both extragenic elements and intragenic sequences located within the first 24 nt of the coding region (21). Little is known about the maturation pathway used by trypanosome U-snRNAs including the determinants for m3G cap formation. Inspection of all available U-snRNA sequences revealed that there is no highly conserved sequence motif analogous to the Sm site. Nevertheless, U2 and U4 snRNAs bind a set of common proteins (22), but in vitro reconstitution experiments combined with methylation/interference analysis revealed that the Sm-analogous region is not required for common protein binding to the U2 snRNA. Instead, the assembly of a core RNP was dependent on sequences located in stem–loop IV (23,24). In the present investigation we carried out a detailed mutational analysis of T.brucei U2 snRNA to localise the determinants for cap trimethylation. The strategy we employed was to express mutant U2 snRNA genes in vivo and to examine m3G cap formation and RNP assembly by immunoprecipitation. Our results showed that binding of common proteins is not required for U2 cap trimethylation and that U2 snRNA sequences, excluding the 5′ terminal 24 nt of the promoter, do not harbour determinants for m3G cap formation.
MATERIALS AND METHODS
DNA oligonucleotides and plasmid construction
The following DNA oligonucleotides were used: U2x, 5′-CCCTCGAGGGTTAGCTAAAT-3′; U2k, 5′-GCTCTAGACCGTCGCGCTCCATCCGGAC-3′; 7SB, 5′-CGTCACCGGCCTGACCCGTT-3′; stl4-5, 5′-CCCTCGAGGGTCCCGCGTTCTTCCG-3′; stl4-3, 5′-GTTCGCGACCGTCGCGCTCC- ATCCG-3′; SL-5, 5′-GCCCTCGAGGGAAGGTGGGGTCGGATGACCTCCACTCTTTCG-3′; SL-3, 5′-CGAAAGAGTGGAGGTCATCCGACCCC-3′; Tloop-5, 5′-TCGAGCGCTAGCTTCGGCTAGCGTTCG-3′; Tloop-3, 5′-CGAACGCTAGCCGAAGCTAGCGC-3′. Construct TbU2-Xtag contained the 641 bp genomic fragment of the T.brucei gambiense U2 snRNA gene from positions –398 to +243 relative to the transcription start site with the sequence 5′-CCCTCGAGGGTAA-3′ inserted after position +24 (21). Constructs TbU2-Hustl4, TbU2-Hust4, TbU2-Hul4 and TbU2-ssrinv were made by transferring the mutated regions of the corresponding pT7-TbU2 constructs described in Günzl et al. (24) to the genomic clone TbU2-Xtag by standard PCR technology. For generation of TbU2-5′half, gene positions +84 to +150 were removed from TbU2-Xtag with restriction enzymes StyI and NruI. The following constructs were generated by replacing the XhoI–NruI fragment of TbU2-Xtag with corresponding DNA fragments. The fragment of TbU2-stl4 was obtained by PCR with oligonucleotides stl4-5 and stl4-3, and TbU2-Xtag as template. TbU2-dstm4 was constructed from TbU2-stl4 by changing nucleotides 130–143 to 5′-ACATCTACATAATC-3′ by PCR and TbU2-dstm4-rv was derived from TbU2-dstm4 by changing nucleotides 105–117 to 5′-GATTATGTGATGT-3′. For TbU2-SL3′stl, the sequence comprising the 3′-terminal stem–loop of the SL RNA was PCR-amplified with oligonucleotides SL-5 and SL-3. Finally, the DNA fragment for TbU2-Tloop was obtained by hybridising oligonucleotides Tloop-5 and Tloop-3. Expected lengths for mature mutant U2 RNAs derived from these constructs were 162 nt for TbU2-Hul4 RNA, 161 nt for TbU2-Xtag and TbU2-ssrinv RNAs, 160 nt for TbU2-Hustl4 RNA, 159 nt for TbU2-Hust4 RNA, 96 nt for TbU2-5′half, 84 nt for TbU2-stl4, TbU2-dstm4, and TbU2-dstm-rv RNAs, 63 nt for TbU2-SL3′stl RNA and 52 nt for TbU2-Tloop RNA.
A second set of constructs was made for the generation of stable cell lines. For construct TbU2-Xtag-Neo, TbU2-ssrinv-Neo, TbU2-Hust4-Neo, TbU2-stl4-Neo and TbU2-SL3′stl-Neo, the neomycin resistance gene under the control of a procyclin gene promoter and 3′ splice acceptor region, and flanked by the β–α tubulin intergenic region, was excised from plasmid BNsp-Neo-T (25) by restriction digest and inserted into the PvuII site of TbU2-Xtag. In all these constructs, the neomycin resistance gene and U2 genes are arranged head-to-tail with the 3′ end of the neomycin resistance gene unit being separated by 213 bp from the 5′ end of the U2 gene unit.
Trypanosome culture and extract preparation
Cultures of wild-type and stably transfected procyclic form T.brucei brucei strain 427 were cultured as described previously (26). For the cell extract preparation, a 200 ml cell culture was grown to a density of ∼1 × 107 cells per ml. Cells were harvested at 4°C, washed twice with 5 ml of ice-cold wash solution (20 mM Tris–HCl, pH 7.4, 100 mM NaCl, 3 mM MgCl2, 1 mM EDTA) and once with 5 ml of ice-cold E-buffer (150 mM sucrose, 20 mM potassium l-glutamate, 3 mM MgCl2, 20 mM HEPES–KOH, pH 7.7, 2 mM dithiothreitol, 10 µg/ml leupeptin). Cells were resuspended in 1 ml of E-buffer and broken with a Branson B12 sonifier by applying two 10 s bursts at 50% Branson power (75 W). Cell fragments were pelleted at 21 000 g for 10 min at 4°C, and the supernatant was aliquoted, shock-frozen in liquid nitrogen and stored at –70°C.
DNA transfection and RNA isolation
Transient transfections and total RNA preparations were carried out as described previously (21) except that transfected cells were cultured for 16–18 h before RNA was prepared. In preliminary experiments, we found that electroporation had a negative effect on U2 snRNA cap trimethylation. Four hours after transfection, TbU2-Xtag RNA caps were not as efficiently trimethylated as those from endogenous U2 RNA. However, when transfected cells were incubated for 16–18 h, this effect was negligible (data not shown). RNA pellets from a 10 ml cell culture were finally resuspended in 50 µl distilled water. Stable transfections were achieved by electroporating 50 µg of the neomycin resistance gene-containing constructs, which were linearised with MluI within the β–α tubulin intergenic region. TbU2-ssrinv-Neo had to be linearised with BstXI because the mutation generated an additional MluI site inside the U2 coding region. Stably transfected cells were selected with Geneticin (G-418) until the culture was resistant to 100 µg/ml of the antibiotic.
Immunoprecipitation and RNA analysis
For m3G-specific immunoprecipitations, 20 µg of mouse anti-m3G monoclonal antibody (generously provided by Adrian Krainer, Cold Spring Harbor Laboratory, NY) and 20 µg of rabbit anti-mouse IgG1 antibody were bound to 500 µl of a 1:1 slurry of protein A–Sepharose beads and NET-150 buffer [50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.05% Nonidet P-40 (v/v)]. For a single immunoprecipitation reaction, beads and bound antibodies from 50 µl of this solution were washed three times with NET-150 buffer. The bead pellet was then resuspended in 170 µl NET-150 buffer and mixed with 30 µl of total RNA prepared from transiently transfected cells. After antigen–antibody binding for 2 h at 4°C, beads were pelleted and the supernatant was transferred to a new tube and mixed with 200 µl 2× PK buffer [200 mM Tris–HCl, pH 7.5, 25 mM EDTA, 300 mM NaCl, 2% SDS (w/v)]. The bead pellet containing the immunoprecipitate was washed five times in 1 ml of NET-400 buffer (as NET-150, but with 400 mM NaCl) and resuspended in 400 µl of 1× PK buffer containing 10 µg glycogen as carrier. RNA of both supernatant and immunoprecipitate was extracted with phenol/chloroform and precipitated with ethanol. Finally, the RNA was separated on 6% polyacrylamide/50% urea gels, blotted onto nylon membrane and detected by northern hybridisation. Each membrane was successively probed with 5′ end-labelled oligonucleotides U2x, U2k and 7SB to detect U2 snRNA from transfected genes, endogenous U2 snRNA and endogenous 7SL RNA, respectively. Northern blot signals were quantified by the E.A.S.Y Win32 imaging system (Herolab). Anti-m3G immunoprecipitation efficiency varied between 50 and 90% and was found to be the same in transfected and non-transfected cells (data not shown).
For immunoprecipitation of U2 core RNPs, IgG antibodies of the polyclonal anti-CP antiserum (27) were bound to protein A–Sepharose beads at a 1:1 ratio of antiserum and bead volume. The beads were washed three times with NET-150 buffer and equilibrated in RNP binding buffer (20 mM HEPES–KOH, pH 7.7, 100 mM KCl, 0.2 mM EDTA, 20% glycerol). For binding of U2 RNP to polyclonal antibodies, 50 µl of beads were mixed with 300 µl RNP binding buffer and 50 µl of cell extract, and rotated for 6 h at 4°C. For the last 10 min, the KCl concentration of the binding reaction was raised to 400 mM. Subsequently, bead pellet and supernatant were separated, and processed as described above. The immunoprecipitation efficiency of these reactions varied between 30 and 60%.
RESULTS
Cap trimethylation of U2 snRNA does not depend on the Sm-analogous region or stem–loop IV sequences
As detailed above, previous experiments (24) revealed that sequences in U2 RNA loop IV (Fig. 1) are important for core snRNP formation, whereas the sequence of the Sm-analogous region is dispensable for common protein binding. Since in higher eukaryotes Sm sequences are critical determinants for cap trimethylation by binding the common proteins (9,10), these results suggested that in T.brucei U2 snRNA loop IV sequences might play a role similar to that of the Sm binding site. To address this possibility, we assayed cap trimethylation of mutant U2 snRNAs after transient DNA transfection of procyclic T.brucei cells. In a first set of experiments, we tested a series of previously generated human–trypanosome sequence substitutions (24), in which stem–loop IV (Hustl4), stem IV (Hust4) or loop IV (Hul4) of the trypanosome U2 RNA was replaced by the corresponding human sequences (Fig. 2A). In addition, we constructed a mutant U2 gene in which the single-stranded region adjacent to stem–loop IIb, namely the Sm-analogous region, was inverted (ssrinv). Mutations were introduced in a U2 snRNA gene containing a tag at position 24 of the coding region, termed Xtag, which we previously showed contains all the sequences necessary for efficient and accurate expression in vivo (21). The resulting constructs were transfected by electroporation into procyclic T.brucei cells and after 16–18 h total RNA was prepared and immunoprecipitated with an anti-m3G monoclonal antibody. Immunoprecipitates and supernatants were analysed by northern hybridisation using as a probe a 5′ end-labelled oligonucleotide complementary to the U2-specific tag (Fig. 2B, upper panels). To control for immunoprecipitation specificity and for quality of RNA isolation, the same blot was subsequently rehybridised with oligonucleotide probes complementary to endogenous U2 snRNA or to 7SL RNA which both are constitutively expressed. Under our experimental conditions ∼50% of endogenous U2 snRNA was immunoprecipitated by anti-m3G antibodies (Fig. 2B, middle panels), whereas 7SL RNA, lacking a m3G cap structure, was not detected in immunoprecipitates (Fig. 2B, lower panels). As shown in Figure 2B, tagged wild-type U2 snRNA (lanes Xtag) and RNAs mutated in stem IV (lanes Hust4) or in the Sm-analogous region (lanes ssrinv) were immunoprecipitated with approximately the same efficiency as endogenous U2 snRNA indicating that they all possessed a trimethylated guanosine cap structure (summary in Table 1). These results correlated with our previous reconstitution studies, since these RNAs formed stable core RNPs in vitro (24).
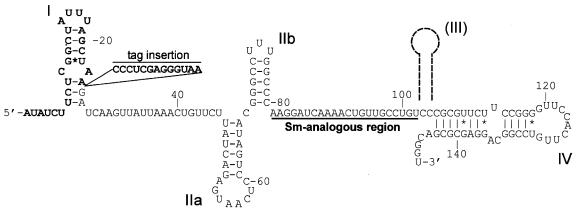
Figure 1.
Secondary structure of T.brucei U2 snRNA. The sequence and secondary structure of the U2 snRNA from T.brucei gambiense are shown (28,29). Stem–loop structures are named according to Ares and Igel (30). Stem–loop III is not present in trypanosome U2 snRNAs, but its position in vertebrate U2 RNA is indicated (broken line). The 24 nt of the 5′ terminus representing the essential intragenic promoter element are indicated in bold letters, the Sm-analogous region is underlined and numbers refer to nucleotide positions. G/U base pairings are indicated by asterisks.
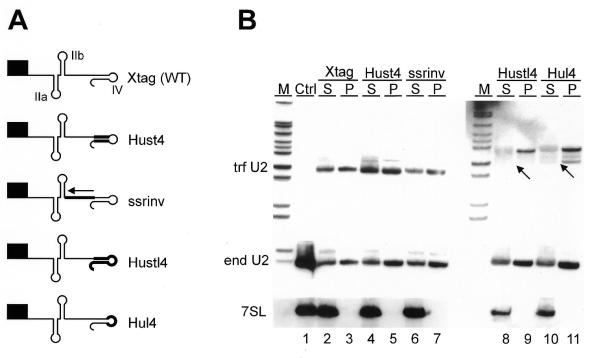
Figure 2.
Substitution of stem–loop IV or of the Sm-analogous region does not affect cap trimethylation of trypanosome U2 RNA. (A) Schematic outline of Xtag, Hust4, ssrinv, Hustl4 and Hul4 U2 RNAs. The filled box represents the 5′ terminal promoter nucleotides and the tag sequence, which are part of all mutant U2 RNAs analysed in this study. Xtag represents the tagged wild-type control (WT) and the stem–loop structures are denoted as in Figure 1. Thick lines represent mutated regions. The arrow indicates the inverted sequence of the Sm-analogous region in ssrinv RNA. (B) Northern blot analysis of anti-m3G immunoprecipitates. Total RNA from transiently transfected cells was immunoprecipitated with a monoclonal anti-m3G antibody. RNA from supernatants (S) and immunoprecipitates (P) was separated on 6% polyacrylamide/50% urea gels, blotted onto nylon membrane and hybridised sequentially to 5′ end-labelled oligonucleotide probes complementary to the tag of U2 RNAs expressed from transfected genes (trf U2), to endogenous U2 snRNA sequences (end U2), and to 7SL RNA (7SL). As a hybridisation control, RNA from non-transfected cells was analysed (Ctrl). Arrows point to the expected length of mature transcripts. Lanes 8 to 11 in panel trf U2 were exposed approximately three times longer than lanes 1 to 7. M, marker (MspI-digested pBR322).
Table 1. Summary of mutant U2 RNA expression and immunoprecipitation analyses.
| RNA | m3G cap | core RNP | mature 3′ end | stability |
|---|---|---|---|---|
| Xtag | + | + | + | + |
| Hust4 | + | + | + | + |
| ssrinv | + | + | + | + |
| Hustl4 | + | n.d. | – | – |
| Hul4 | + | n.d. | – | – |
| 5′half, | m+/e– | n.d. | – | + |
| 3′half* | + | + | + | + |
| stl4 | + | m+/e– | +/– | + |
| stl4-dstm | + | n.d. | – | – |
| stl4-dstm-rv | + | n.d. | – | + |
| SL3′stl | + | – | – | + |
| Tloop | + | n.d. | – | – |
Efficiencies of m3G cap formation (m3G cap), core RNP formation (core RNP) and mature 3′ end formation (mature 3′ end) of mutant U2 RNAs were determined by a combination of immunoprecipitation and northern blot analysis. In addition, expression levels of these RNAs, which in this particular case represent a measure of RNA stability, were analysed in transient assays (stability). Results are compared with those of TbU2-Xtag and presented as + (>90% of TbU2-Xtag), +/– (20–90%) and – (<20%). It is stated if mature (m) and 3′ extended (e) RNAs exhibited different results. n.d., not determined. *, data not shown.
Most of the Hustl4 and Hul4 RNAs were ∼12 nt longer than the size expected for mature U2 snRNAs, which instead were barely detectable (indicated by arrows in Fig. 2B, lanes 8–11). Primer extension analysis revealed that the longer transcripts have the correct 5′ end (data not shown), implying that these RNAs carry 3′ extensions. Both the mature transcripts and the 3′ extended forms were efficiently immunoprecipitated with anti-m3G antibodies demonstrating that both RNA species were trimethylated.
One further observation was made. Expression levels of Hustl4 and Hul4 RNAs were between 5- and 10-fold lower than that of the other RNAs tested (Fig. 2B, note the 3-fold longer exposure time of lanes 8–11 in comparison to lanes 1–7, and data not shown). Since stem–loop IV sequences do not contribute to transcription efficiency of the U2 snRNA gene (21), these lower expression levels are most likely due to a change in RNA stability caused by substituting stem–loop IV (summary in Table 1).
In conclusion, the sequence of stem–loop IV and of the Sm-analogous region was not relevant for U2 snRNA cap trimethylation. Since stem–loop IV and loop IV mutant RNAs did not bind common proteins in vitro (24), these results suggested that common protein binding is not a prerequisite for cap trimethylation in T.brucei. The longer U2 transcripts detected in our analysis are likely to represent termination products at a stretch of T residues 12 bp downstream of the U2 RNA mature 3′ end, as it was observed in our in vitro transcription system (31; A.Günzl, unpublished results). The accumulation of these longer U2 RNAs as observed in stem–loop IV and loop IV substitutions suggested that these mutations impaired 3′ end processing.
Are there U2 RNA sequences critical for cap trimethylation?
Since none of the substitution mutations tested so far affected cap trimethylation of the U2 snRNA, we next generated more severe mutations by deleting portions of the U2 snRNA (Fig. 3A and Table 1). However, it was not possible to assess the importance of the first 24 nt of the coding region, since this sequence is part of the U2 snRNA gene promoter (21). Thus, all subsequent constructs contained the first 24 bp of the coding region followed by the 13 nt tag. Removing sequences in the 5′ half of the molecule from nucleotide positions 25–83 resulted in transcripts of the predicted length and they were capped with m3G (construct 3′half, see Table 1). In contrast, if sequences downstream of stem–loop IIb (positions 84–148) were deleted (construct 5′half; Fig. 3A), >90% of the RNA expressed from this construct carried a 3′ extension, as assessed by primer extension analysis, and only a small amount of the RNA had the expected size (Fig. 3B, lane 3). Following immunoprecipitation with anti-m3G antibodies, a minor fraction of the extended form was detected in the pellet, whereas the mature form was trimethylated as efficiently as wild-type U2 snRNA (lane 4). Thus, it appeared from these results that the 3′ half of the U2 snRNA is involved in m3G cap formation, as well as in 3′ end processing of pre-U2 RNA.
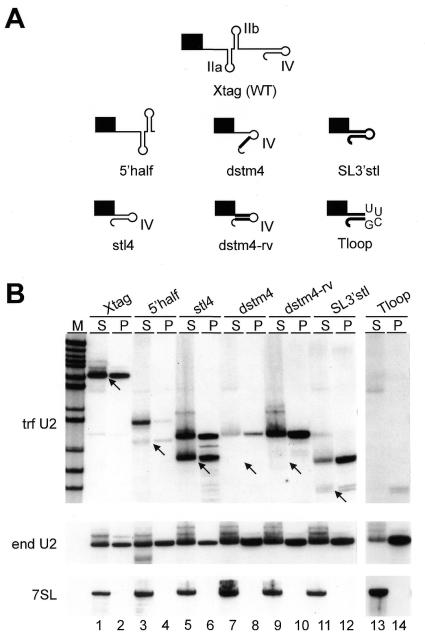
Figure 3.
Anti-m3G immunoprecipitation analysis of U2 RNA deletion mutants. (A) Schematic outline of the tagged, wild-type U2 Xtag RNA (Xtag) and mutant U2 RNAs 5′half, stl4, dstm4, dstm4-rv, SL3′stl and Tloop. Non-U2 sequences are represented by thick lines. (B) Northern blot of RNA obtained from supernatants (S) and precipitates (P) after immunoprecipitation of m3G-capped RNA from transiently transfected cells. The same blot was probed for the tagged U2 snRNA (trf U2), the endogenous U2 snRNA (end U2), and 7SL RNA (7SL). Arrows point to the expected lengths of mature transcripts. M, marker (MspI-digested pBR322).
To investigate this further, a minimal construct (stl4) was made in which sequences from position 25 to 100, including the Sm-analogous region, were deleted, generating a construct where the first 24 nt and the tag were fused to stem–loop IV (Fig. 3A). Northern blot analysis showed that this RNA is efficiently expressed resulting in two distinct RNA species (Fig. 3B, lane 5): one of the correct size and one with a 3′ extension of ∼12 nt. Both RNA species were immunoprecipitated with anti-m3G antibodies as efficiently as the tagged or endogenous U2 snRNA (lanes 5 and 6), demonstrating that stem–loop IV is sufficient for formation of the m3G cap structure. However, in contrast to the results with the 5′half construct, where only a minor proportion of the 3′ extended RNA carried the m3G cap structure, the 3′ extended form of stl4 RNA was efficiently trimethylated (compare lanes 4 and 6).
To examine the importance of stem IV on cap trimethylation, the structure of the stem was disrupted in stl4 by replacing the bottom strand with an unrelated sequence (dstm4, Fig. 3A). Next, the top strand in dstm4 was mutated to restore a stem structure (dstm4-rv, Fig. 3A). As shown in Fig. 3B (lanes 7–10) both RNAs acquired m3G caps demonstrating that neither the structure nor the sequence of stem IV was essential for cap hypermethylation. The low abundance of dstm4 RNA compared to dstm4-rv RNA suggested that the structure, but not the sequence of stem IV, plays a role in RNA stability. Finally, both RNAs occurred exclusively with 3′ extensions implying that stem IV nucleotides are involved in 3′ end processing.
Taken together, our results so far suggested that sequences from nucleotide position 25 to the very 3′ end of the U2 snRNA are not essential for the formation of the trimethylated guanosine cap structure. To confirm this, we fused the first 24 nt of the U2 snRNA and the tag sequence to the 3′ terminal stem–loop sequence of the m7G–capped SL RNA (SL3′stl) and to an artificial stem with a tetraloop 5′-UUCG-3′ (Tloop). The expression level of TbU2-SL3′stl was comparable to that of the tagged wild-type U2 snRNA gene (TbU2-Xtag) and exclusively transcripts with a 3′ extension were detected (Fig. 3B, lane 11). On the other hand, the artificial stem–loop (Tloop) was only weakly expressed as a 3′ extended form, and mature transcripts were below the detection level of our assay (lane 13). Nevertheless, both SL3′stl (lane 12) and Tloop (lane 14) RNAs were efficiently immunoprecipitated with anti-m3G antibodies, demonstrating that U2 snRNA sequences downstream of position 25 are not required for m3G cap formation.
In an attempt to identify the role of the 24 5′ terminal nt of the U2 RNA in formation of a correct cap structure we made U2/U6 hybrid gene constructs, because the U6 snRNA does not possess a guanosine cap. First, we replaced the intragenic promoter element of a U2 gene construct with the corresponding U6 snRNA gene element which comprised the 41 5′ terminal nt of the U6 snRNA gene (32). The resulting U6-U2 hybrid RNA was expressed and did not carry a m3G cap (data not shown). Hence, the U2 5′ terminus is essential for the formation of a correct cap structure. In a reciprocal experiment, we introduced this sequence into a U6 gene construct thereby replacing the U6 intragenic promoter element. However, in the context of the U6 gene, the U2 5′ terminus was not able to direct m3G cap formation (data not shown).
Cap trimethylation does not require common protein binding
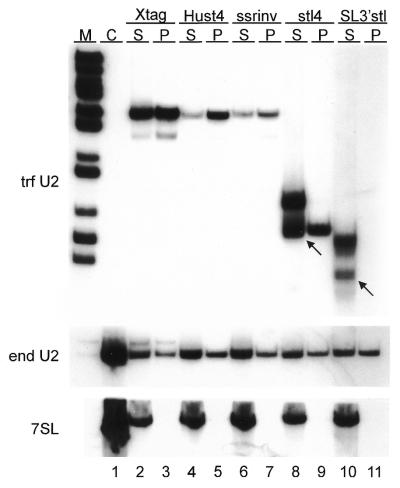
Previous in vitro studies (24) in combination with the results presented here (Fig. 2B) suggest that binding of common proteins to the U2 snRNA is not required for cap trimethylation. To investigate this further, we analysed core RNP formation of mutant U2 snRNAs in stably transfected cell lines. Expression constructs were generated in which the U2 snRNA gene was linked to a neomycin resistance gene under the control of a procyclin gene promoter and flanked by the spacer region between β and α tubulin genes. The constructs were targeted to the β–α tubulin intergenic region and stable cell lines expressing mutant U2 RNAs were selected by resistance to G418. For U2 RNP analysis, total cell extracts were prepared and RNPs were immunoprecipitated with a polyclonal antiserum directed against the T.brucei common proteins (27). RNA preparations of supernatants and immunoprecipitates were then subjected to northern analysis (Fig. 4 and Table 1). Endogenous 7SL RNA, which does not bind common proteins, and endogenous U2 snRNA served as a negative and a positive control, respectively. As shown in Figure 4, tagged wild-type U2 snRNA (lanes Xtag) and RNAs mutated in stem IV (lanes Hust4) or in the Sm-analogous region (lanes ssrinv) were immunoprecipitated with the same efficiency as endogenous U2 snRNA demonstrating that these RNAs were capable of forming a U2 core RNP by binding the common proteins. Consistent with in vitro U2 reconstitution experiments (24), binding of common proteins in vivo does not depend on the sequence of the Sm-analogous region, since this sequence was inverted in TbU2-ssrinv. Similar to what we observed in transient transfections (Fig. 3B, lane 5), analysis of the stable cell line expressing a U2 snRNA truncated to stem–loop IV (stl4) revealed two RNA species: one with the correct 3′ end and the other with a 3′ extension (Fig. 4, lane 8). Only the RNA with the correct 3′ end was in the form of a stable core RNP, whereas the 3′ extended form was not associated with common proteins to any detectable level. Since the longer RNA was efficiently trimethylated (Fig. 3B, lane 6), we conclude that common protein association is not a pre-requisite for cap trimethylation. This result was corroborated with construct SL3′stl, in which the U2 snRNA promoter was fused to the 3′ terminal stem–loop of the SL RNA. Although both the processed and 3′ extended form of this RNA were efficiently trimethylated (Fig. 3B, lane 12), we could not detect binding of common proteins to either RNA species (Fig. 4, lane 11). Furthermore, our results showed that stem–loop IV suffices for stable binding of the common proteins (Fig. 4, lane 9) and indicated that assembly into a core RNP occurs after 3′ end processing (Fig. 4, compare lanes 8 and 9).
Figure 4.
Immunoprecipitation analysis of mutant U2 RNPs with an anti-common protein antiserum. Cell extracts from stably transfected cell lines expressing Xtag, Hust4, ssrinv, stl4 or SL3′stl RNA and from non-transfected cells (Ctrl) were immunoprecipitated with anti-common protein antiserum. RNA prepared from supernatants (S) and precipitates (P) was separated on 6% polyacrylamide/50% urea gels, blotted, and subjected to three successive rounds of northern hybridisation using 5′ end-labelled oligonucleotides complementary to transfected U2 snRNA (trf U2), endogenous U2 snRNA (end U2) and 7SL RNA (7SL). Arrows indicate positions of mature transcripts. M, marker (MspI-digested pBR322).
DISCUSSION
The T.brucei m3G-capped U2 snRNA is exceptional in that it is transcribed by RNA polymerase III and does not contain a classical Sm site. Nevertheless, the U2 snRNA forms a core RNP by binding common proteins, which are present in each spliceosomal snRNP particle. In this study, we have begun a dissection of the maturation pathway of the U2 snRNP by expressing mutant RNAs in procyclic trypanosome cells. In particular, we have searched for determinants in the U2 snRNA essential for m3G cap formation and for common protein binding. The most surprising results were that most of the U2 coding region is dispensable for cap trimethylation and that binding of the common proteins is similarly not required. This is in contrast to m3G formation of the U2 snRNA in higher eukaryotes and yeast, where association of the Sm proteins is essential for cap hypermethylation. The core protein complex probably provides a binding site for the snRNA-(guanosine-N2)-methyltransferase, responsible for converting m7G to m3G.
In the Xenopus oocyte system it was shown that there exists an additional pathway for cap trimethylation which is active in the nucleus and is distinct from that acting on spliceosomal U snRNAs. U3 snRNA is an essential factor in rRNA processing and is retained in the nucleus where it obtains a m3G cap without binding the common proteins (33). In T.brucei, spliceosomal U snRNAs most likely have a cytoplasmic maturation phase like their counterparts in higher eukaryotes (27,34). But presuming that T.brucei m3G caps do not represent nuclear retention signals, it is possible that similar to the Xenopus U3 cap trimethylation, methyl groups are added to the trypanosomal U2 snRNA cap by a nuclear methyltransferase prior to nuclear export, common protein binding and 3′ end maturation. Such a scenario is in agreement with our finding that precursor and mature U2 RNAs possessed m3G caps to the same extent.
Searching for the cap trimethylation determinant in the U2 snRNA, we confined this signal to the 5′ terminal 24 nt of U2. Since the first 24 bp of the U2 snRNA gene coding region represent an essential intragenic promoter element, we could not discriminate whether the determinant resides in the U2 snRNA gene or in the U2 RNA itself. It is possible that the U2 internal promoter element recruits a specific RNA polymerase III transcription complex, which interacts with the methyltransferase. Alternatively, the methyltransferase may directly interact with the 5′ terminal region of the U2 RNA or with a protein bound to this RNA domain. We favour the possibility that determination occurs on the RNA and not on the DNA level since 3′ extended 5′half RNA was the only U2 RNA in our study on which formation of the m3G cap was very inefficient. The corresponding gene contains all necessary transcription signals and lacks unrelated sequences, except for the tag, which did not interfere with cap trimethylation in all other U2 RNAs analysed. Hence, co-transcriptional m3G formation should have occurred on this RNA. On the other end, it is possible that this RNA was not transported to the right compartment or that the RNA folded into an aberrant secondary structure preventing correct cap formation.
Another possibility is that recognition of the U2 cap by the methyltransferase may not require specific U2 sequences at all but may be mediated by the monomethylated guanosine cap itself. In T.brucei, only the SL, which constitutes the 5′ terminus of all mRNAs and the SL RNA, carries a m7G cap. However, the SL possesses an unusual cap 4 structure in which the first four nt are methylated (35). It is therefore possible that the cap 4 structure prevents hypermethylation of the SL cap and that guanosine caps without a cap 4 structure are targets for hypermethylation.
Our previous studies revealed that the sequence determinants for common protein binding reside in stem–loop IV and that the sequence of the Sm-analogous region is dispensable for this process. Previous in vitro reconstitution experiments have led to a model of U2 RNP assembly, in which a yet to be characterised protein initially binds to loop IV before the U2-specific 40 kDa protein and common proteins are recruited to form a core complex (24). Interestingly, in these studies stem–loop IV by itself was only sufficient for binding the 40 kDa protein, but not the common proteins. This and other data suggested that the single-stranded region 5′ to stem–loop IV associates with common proteins in a non-specific manner, possibly by serving as a landing pad rather than a sequence-specific binding site. Support for this view comes from RNase H protection experiments which showed that in U2 core RNPs, the Sm-analogous region is protected (24). Therefore, since the RNA substrate used for in vitro reconstitution contained only 3 nt 5′ to stem–loop IV, it was not sufficient for common protein binding. On the other hand, in vivo expression of U2 snRNA truncated to stem–loop IV was able to bind the common proteins, most likely due to the presence of a 37-nt 5′ extension containing the promoter element and the tag sequence.
Whereas the U2 snRNA gene in higher eukaryotes is transcribed by RNA polymerase II, the T.brucei U2 snRNA is the product of RNA polymerase III. Despite this difference, both our in vitro results (31; A.Günzl, unpublished results) and the present study suggest that, like the U2 snRNA of higher eukaryotes, the trypanosome U2 snRNA primary transcript is synthesised with a 3′ extension of ∼12 nt. Although we have not determined a precise precursor–product relationship, the 3′ extension of trypanosome pre-U2 RNA is most likely removed during U2 RNP maturation. The evolutionary conservation of this process suggests a functional significance of the 3′ extension. As investigated in the human system, processing of pre-U2 snRNA is essential for nuclear reimport (14), and the processing event is directed by internal elements distinct from the processing site (36). More specifically, 3′ end processing depended on the structure of the bottom of stem III and the integrity of stem IV, whereas the sequence of these stem structures were of little significance. The T.brucei U2 snRNA lacks stem–loop III (Fig. 1), but our results showed that 3′ end processing of T.brucei pre-U2 RNA is determined in the same region, namely through sequence and structure of stem–loop IV, indicating that the processing mechanism is evolutionarily conserved. Furthermore, the human pre-U2 RNA is assembled into a core RNP before 3′ end processing (37). In our study, we found that the 3′ extended U2 RNA was not associated with common proteins to any detectable level, whereas transcripts of correct length were in form of a core RNP (see Fig. 4, stl4 RNA, lanes 8 and 9). Hence, in contrast to the human system, pre-U2 RNA 3′ end processing in T.brucei may precede core RNP formation or, alternatively, occurs very rapidly during core RNP formation.
In conclusion, we have shown that the determinants for m3G cap formation on the U2 snRNA are different in the protist parasite T.brucei as compared to higher eukaryotes. Our results imply that in T.brucei the methyltransferase responsible for cap trimethylation interacts with the 5′ end of the U2 snRNA without docking onto the common protein complex. Analysing this interaction in more detail would greatly benefit from the development of an in vitro system for cap trimethylation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Sigrid Hojak for excellent technical assistance. This study received support from National Institutes of Health Grant AI43594 to C.T. A.G. was supported by an EMBO long-term fellowship and grant 371/3-1 of the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Zieve G.W. and Sauterer,R.A. (1990) Crit. Rev. Biochem. Mol. Biol., 25, 1–46. [DOI] [PubMed] [Google Scholar]
- 2.Lührmann R., Kastner,B. and Bach,M. (1990) Biochim. Biophys. Acta, 1087, 265–292. [DOI] [PubMed] [Google Scholar]
- 3.Yu Y.-T., Scharl,E.C., Smith,C.M. and Steitz,J.A. (1999) In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, Vol. 2, pp. 487–524.
- 4.Dahlberg J.E. and Lund,E. (1988) In Birnstiel,M.L. (ed.), Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer-Verlag, Berlin, pp. 38–70.
- 5.Mattaj I.W. (1988) In Birnstiel,M.L. (ed.), Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer Verlag, Berlin, pp. 100–114.
- 6.Branlant C., Krol,A., Ebel,J.P., Lazar,E., Haendler,B. and Jacob,M. (1982) EMBO J., 1, 1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattaj I.W. (1986) Cell, 46, 905–911. [DOI] [PubMed] [Google Scholar]
- 8.Skuzeski J.M., Lund,E., Murphy,J.T., Steinberg,T.H., Burgess,R.R. and Dahlberg,J.E. (1984) J. Biol. Chem., 259, 8345–8352. [PubMed] [Google Scholar]
- 9.Plessel G., Fischer,U. and Lührmann,R. (1994) Mol. Cell. Biol., 14, 4160–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raker V.A., Plessel,G. and Lührmann,R. (1996) EMBO J., 15, 2256–2269. [PMC free article] [PubMed] [Google Scholar]
- 11.Mattaj I.W. and De Robertis,E.M. (1985) Cell, 40, 111–118. [DOI] [PubMed] [Google Scholar]
- 12.Fischer U. and Lührmann,R. (1990) Science, 249, 786–790. [DOI] [PubMed] [Google Scholar]
- 13.Hamm J., Darzynkiewicz,E., Tahara,S.M. and Mattaj,I.W. (1990) Cell, 62, 569–577. [DOI] [PubMed] [Google Scholar]
- 14.Huang Q. and Pederson,T. (1999) Nucleic Acids Res., 27, 1025–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tschudi C., Richards,F.F. and Ullu,E. (1986) Nucleic Acids Res., 14, 8893–8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mottram J., Perry,K.L., Lizardi,P.M., Lührmann,R., Agabian,N. and Nelson,R.G. (1989) Mol. Cell. Biol., 9, 1212–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dungan J.M., Watkins,K.P. and Agabian,N. (1996) EMBO J., 15, 4016–4029. [PMC free article] [PubMed] [Google Scholar]
- 18.Lücke S., Klöckner,T., Palfi,Z., Boshart,M. and Bindereif,A. (1997) EMBO J., 16, 4433–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnare M.N. and Gray,M.W. (1999) J. Biol. Chem., 274, 23691–23694. [DOI] [PubMed] [Google Scholar]
- 20.Tschudi C. and Ullu,E. (1990) Cell, 61, 459–466. [DOI] [PubMed] [Google Scholar]
- 21.Fantoni A., Dare,A.O. and Tschudi,C. (1994) Mol. Cell. Biol., 14, 2021–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palfi Z., Günzl,A., Cross,M. and Bindereif,A. (1991) Proc. Natl Acad. Sci. USA, 88, 9097–9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Günzl A., Cross,M. and Bindereif,A. (1992) Mol. Cell. Biol., 12, 468–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Günzl A., Cross,M., Palfi,Z. and Bindereif,A. (1993) J. Biol. Chem., 268, 13336–13343. [PubMed] [Google Scholar]
- 25.Lee M.G. and Van der Ploeg,L.H. (1990) Science, 250, 1583–1587. [DOI] [PubMed] [Google Scholar]
- 26.Laufer G., Schaaf,G., Bollgönn,S. and Günzl,A. (1999) Mol. Cell. Biol., 19, 5466–5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palfi Z. and Bindereif,A. (1992) J. Biol. Chem., 267, 20159–20163. [PubMed] [Google Scholar]
- 28.Hartshorne T. and Agabian,N. (1990) Genes Dev., 4, 2121–2131. [DOI] [PubMed] [Google Scholar]
- 29.Tschudi C., Williams,S.P. and Ullu,E. (1990) Gene, 91, 71–78. [DOI] [PubMed] [Google Scholar]
- 30.Ares M. Jr and Igel,A.H. (1990) Genes Dev., 4, 2132–2145. [DOI] [PubMed] [Google Scholar]
- 31.Günzl A., Tschudi,C., Nakaar,V. and Ullu,E. (1995) J. Biol. Chem., 270, 17287–17291. [DOI] [PubMed] [Google Scholar]
- 32.Nakaar V., Günzl,A., Ullu,E. and Tschudi,C. (1997) Mol. Biochem. Parasitol., 88, 13–23. [DOI] [PubMed] [Google Scholar]
- 33.Terns M.P. and Dahlberg,J.E. (1994) Science, 264, 959–961. [DOI] [PubMed] [Google Scholar]
- 34.Bell M. and Bindereif,A. (1999) Nucleic Acids Res., 27, 3986–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bangs J.D., Crain,P.F., Hashizume,T., McCloskey,J.A. and Boothroyd,J.C. (1992) J. Biol. Chem., 267, 9805–9815. [PubMed] [Google Scholar]
- 36.Jacobson M.R., Rhoadhouse,M. and Pederson,T. (1993) Mol. Cell. Biol., 13, 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleinschmidt A.M., Patton,J.R. and Pederson,T. (1989) Nucleic Acids Res., 17, 4817–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]