Abstract
Large-genome bacteriophages (jumbo phages) of the proposed family Chimalliviridae assemble a nucleus-like compartment bounded by a protein shell that protects the replicating phage genome from host-encoded restriction enzymes and DNA-targeting CRISPR-Cas nucleases. While the nuclear shell provides broad protection against host nucleases, it necessitates transport of mRNA out of the nucleus-like compartment for translation by host ribosomes, and transport of specific proteins into the nucleus-like compartment to support DNA replication and mRNA transcription. Here, we identify a conserved phage nuclear shell-associated protein that we term Chimallin C (ChmC), which adopts a nucleic acid-binding fold, binds RNA with high affinity in vitro, and binds phage mRNAs in infected cells. ChmC also forms phase-separated condensates with RNA in vitro. Targeted knockdown of ChmC using mRNA-targeting dCas13d results in accumulation of phage-encoded mRNAs in the phage nucleus, reduces phage protein production, and compromises virion assembly. Taken together, our data show that the conserved ChmC protein plays crucial roles in the viral life cycle, potentially by facilitating phage mRNA translocation through the nuclear shell to promote protein production and virion development.
Graphical Abstract
Graphical Abstract.
Introduction
The continual arms race between bacteria and bacteriophages (phages) has driven the development of myriad immune systems in bacteria, along with an equally complex set of phage-encoded immune countermeasures (1,2). A striking example of these countermeasures is the nucleus-like compartment assembled by a family of large-genome ‘jumbo’ phages (defined as phages with genomes >200 kb in size) with the proposed name Chimalliviridae (chimalliviruses) (3–7). This compartment shields the phages’ replicating genomes from host-encoded defenses including restriction enzymes and DNA-targeting CRISPR-Cas nucleases (8,9). The phage nuclear boundary or shell primarily comprises a single protein, termed Chimallin (ChmA), which assembles into a lattice with pores less than ∼2 nm in size, which can allow the passage of metabolites but restricts the passage of most proteins (7,10). Most chimallivriuses also encode a tubulin homolog called PhuZ, which assembles into dynamic filaments that center and rotate the phage nucleus within the infected cell while also trafficking pro-capsids to the phage nucleus for genome packaging (3–5).
The physical barrier erected by chimalliviruses between their replicating genomes and the host cytoplasm introduces a number of challenges to the phage that mirror challenges faced by eukaryotic cells and their nuclei. In particular, since mRNAs are produced within the phage nucleus but the translation machinery is located in the host cytoplasm, mRNAs must be translocated out of the phage nucleus for translation. The susceptibility of chimalliviruses to RNA-targeting CRISPR-Cas nucleases (8,9,11,12) supports this model. Similarly, any phage protein whose function requires it to be localized within the phage nucleus must be specifically translocated from the cytoplasm into the phage nucleus after translation. Finally, replicated phage genomic DNA must be translocated through the nuclear shell for packaging into pro-capsids that are docked onto the exterior of the nuclear shell (4,5,13,14).
The requirement for translocation of mRNAs, proteins and genomic DNA through the phage nuclear shell implies the existence of additional shell components embedded within or associated with the ChmA lattice that mediate these activities. In prior work, we used proximity labeling and localization analysis in the chimallivirus PhiPA3 to identify proteins that physically associate with ChmA and localize to the nuclear shell (15). One of these proteins, termed ChmB, interacts directly with ChmA both in vitro and in vivo, and associates with the virion portal protein in vitro. ChmB’s network of protein–protein interactions and its distinctive 3D structure suggest that it may form pores in the phage nuclear shell that enable the docking of pro-capsids for genome packaging. Further data showing that the expression of dominant-negative ChmB mutants compromises early steps in phage nucleus growth and maturation further suggests that ChmB may participate in mRNA and/or protein translocation through the nuclear shell (15).
Here, we show that another conserved phage nuclear shell-associated protein, which we term ChmC, adopts a nucleic acid-binding fold, binds RNA in vitro, and forms RNA-protein condensates through a conserved asparagine-rich C-terminal region. In phage-infected cells, ChmC specifically binds phage mRNAs. Targeting ChmC using dCas13-based translational knockdown reveals that the protein plays critical roles in the translocation of mRNAs from the phage nucleus to the cytoplasm, production of phage proteins, and production of new virions. Together, these data suggest that ChmC acts as a chaperone for phage-encoded mRNAs, likely aiding their translocation through the nuclear shell to promote translation and infection progression.
Materials and methods
Bacterial strains, growth conditions and phage preparations
For PhiPA3 phage, P. aeruginosa K2733 (PA01 efflux pump knockout; (ΔMexAB-OprMΔMexCD-OprJΔMexEF-OprNΔMexXY-OprM)) was used as the host. For Goslar phage, E. coli MC1000 (derived from E.coli MG1655) was used as the host. Both bacterial strains were cultured in Luria-Bertani (LB) media or LB top agar (0.35% agar) at 30°C (P. aeruginosa) or 37°C (E. coli). To amplify phages, 100 μl of liquid culture at OD600= 0.6 was mixed with 20 μl of high-titer phage lysate and incubated at room temperature for 20 minutes, then the mixture was added to 5 ml of warm LB top agar, poured onto LB plates, and incubated overnight. The next day, 5 ml of Phage Buffer (10 mM Tris–HCl pH 7.5, 10 mM MgSO4, 68 mM NaCl, and 1 mM CaCl2) was added to each plate and incubated at room temperature for 5 h. The phage buffers were then collected and lysates were centrifuged at 15 000 rpm for 10 minutes. Supernatants were stored at 4°C with 0.01% chloroform.
Plasmid constructions and transformation
Genes of interest were PCR-amplified with 25 bp homology arms from high-titer phage lysates and ligated into respective plasmid backbones using NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs). Recombinant plasmids were transformed into E. coli DH5α and plated on LB agar containing appropriate antibiotics (25 μg/ml gentamicin sulfate, 100 μg/ml ampicillin, 100 μg/ml spectinomycin, or 100 μg/ml chloramphenicol). After plasmids were confirmed by DNA sequencing, chemically competent organisms of interest were transformed and selected on LB plates with relevant antibiotics. Selected colonies were grown in LB media with the antibiotics and stored in 25% glycerol at –80°C.
Fluorescence microscopy of single cell infections
1% agarose pads were prepared on concavity slides with desired inducing reagents of arabinose or IPTG. For imaging P. aeruginosa, pad mixes also contained FM4-64 (1 μg/ml) to stain cell membranes and DAPI (1 μg/ml) to stain DNA. Strains of interest were resuspended from overnight incubated LB plates into 25% LB to an OD600= 0.3. 5 μl of each resuspension was spotted on a concave slide and incubated in a humidor for 2 h in 37°C (E. coli stains) or 30°C (P. aeruginosa strains). For phage infections, 10 μl of phages (1010 pfu/ml) were added to cells (resulting in a multiplicity of infection (MOI) of ∼7) and incubated until the desired time point. For imaging E. coli, dyes were added by spotting 7 μl of the mix containing (2 μg/ml DAPI, 4 μg/ml FM4-64, 25% LB). The slides were sealed with a coverslip and fluorescent microscopy was performed using a DeltaVision Spectris Deconvolution Microscope (Applied Precision). Regions of interests were imaged using at least 8 Z-axis stacks from the middle focal plane in 0.15 μm increments. Final images were created by DeltaVision SoftWoRx Image Analysis Program and its deconvolution algorithm and analyzed by Fiji (16).
Protein structure prediction
To model the structure of PhiPA3 or Goslar ChmC tetramers or PhiPA3 nvRNAP, we used AlphaFold multimer (17,18) using ColabFold (19) installed locally on a Linux workstation with NVIDIA RTX 3090 GPU (https://github.com/YoshitakaMo/localcolabfold).
Protein purification and characterization
For protein expression, E. coli strain Rosetta 2 (DE3) pLysS (EMD Millipore) cells were transformed with plasmids and grown overnight in LB plus appropriate antibiotics. The next day, cultures (1 l 2× YT media plus antibiotics in 2 l shaker flasks) were started and grown at 37°C until they reached an OD600 of 0.7, then induced with 0.25 mM IPTG and moved to 20°C for 16 h. The cells were collected by centrifugation and resuspended in a buffer containing 25 mM Tris–HCl pH 7.5, 10% glycerol, 1 mM NaN3, 300 mM NaCl, 5 mM imidazole and 5 mM β-mercaptoethanol. The proteins were purified using Ni2+ affinity chromatography (Ni-NTA agarose, Qiagen) and then passed over an anion-exchange column (Hitrap Q HP, Cytiva). Eluted fractions were concentrated and passed over a size exclusion column (Superdex 200, Cytiva) in GF buffer (Buffer A and 300 mM NaCl and 1 mM dithiothreitol). Fractions corresponding to the peak of interest were concentrated using ultrafiltration (Amicon Ultra, EMD Millipore) to reach a concentration of 10 mg/ml and stored at 4°C.
For analysis of molecular weight in solution using size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS), 100 μl of purified protein at a concentration of 5 mg/ml was injected onto a size exclusion column (Superdex 200 Increase 10/300 GL, Cytiva) in GF buffer, then light scattering and refractive index profiles were collected using miniDAWN TREOS and Optilab T-rEX detectors (Wyatt Technology). SEC-MALS data were analyzed using ASTRA software version 8.
DNA and RNA binding assays
For measurement of DNA and RNA binding affinity by fluorescence polarization, 30 nM of a 22-base DNA (sequence ATTGTACCACTATTCCGAACAA) or RNA (sequence AUUGUACCACUAUUCCGAACAA) was mixed with the indicated concentration of purified PhiPA3 ChmC in FP buffer (20 mM HEPES pH 7.5, 75 mM KCl, 2 mM DTT, 5% glycerol, 0.02% NP40 substitute, 0.15 mg/ml BSA), and incubated for 10 minutes at room temperature. For RNA binding, reactions were supplemented with 1.25 mM RNaseOUT (ThermoFisher Scientific #10777019) Fluorescence polarization was measured with a Tecan Infinite M1000 PRO fluorescence reader, and data was analyzed by GraphPad Prism using a cooperative binding model.
Knockdown of the phage proteins with dCas13d
31 nucleotide-long RNA guides that target the ribosome binding side or translational start of the gene of interest were designed and cloned into entry vectors with Ruminococcus flavefaciens Cas13d with mutations R239A, H244A, R858A, H863A (dCas13d). dCas13d is expressed under a tetR/tetA promoter, and guide RNAs are expressed under a J23119 promoter (20,21). Plasmid selection was performed with chloramphenicol while appropriate anhydrotetracycline (aTc) concentration allows the expression of dCas13d. Plasmids were transformed to host MC1000 by electroporation and selected with 100 μg/ml chloramphenicol. For knockdown of ChmC in Goslar infections, E. coli strains were induced with 50 nM aTc and incubated at 37°C for 2 h before infection.
Western blot
Samples were collected for western blot simultaneously with sample collection for TMT-tag mass spectrometry. Briefly, overnight liquid cultures of E. coli MC1000 strains were diluted to OD600= 0.1 in fresh media with appropriate antibiotics and grown to OD600= 0.6. 10 ml of pad mix (1% agarose, 25% LB, 50 nM aTc and 30 μg/ml chloramphenicol) was poured into a 6 cm petri dish. When the mixture solidified, 200 μl of each E. coli strain at OD600= 0.1 was spread on a pad. Petri dishes were placed in a 37°C humidor and incubated for 2 h. Cells were infected with 100 μl of Goslar phage (titer 109 PFU/ml). At 0, 30, 60, and 90 minutes post infection cells were resuspended with 1 ml of 25% LB media and collected into 1.7 ml tubes, then pelleted by centrifugation at 4000 rpm at 4°C, then washed with 25% LB. Pellets were aliquoted into five tubes for the final wash, then washed pellets were stored at –80°C.
For cell lysis, samples were thawed on ice for 5 min and gently mixed with 500 μl of lysis buffer (10% Glycerol, 25 mM Tris pH 7.5, 150 mM NaCl supplemented with 4mg/ml lysozyme, 20 μg/ml DNase I, 2× Complete Protease Inhibitor, 0.4 mM PMSF). Suspensions were incubated on ice for 1 hour, then sonicated for 30 s (Branson Sonifier) with duty cycle 40, output level 4 on ice. Suspensions were centrifuged at 15 000 rpm for 30 min at 4°C. 40 μl of supernatant for each sample was mixed with 2× Laemmli buffer and boiled for 5 min at 95°C. 10 μl of each sample was loaded onto a 4–20% Mini-PROTEAN TGX Precast Protein Gel (Bio-Rad) and run at 180 V for 45 min. Proteins were transferred to PVDF membranes using a Trans-Blot Turbo RTA Mini 0.2 μm PVDF Transfer Kit (Bio-Rad) according to the manufacturer's instructions, using a Trans-Blot Turbo Transfer System (Bio-Rad) at Turbo setting. Membranes were blocked (5% Non-Fat Dry Milk in TBST) for an hour in room temperature on a shaker. The solution was replaced with blocking buffer with appropriate dilutions of primary antibody (1:500 Rabbit anti-ChmC (Genscript, custom-generated) or 1:2000 Rabbit anti-OmpA, (Joe Pogliano, custom-generated)) and incubated overnight on a shaker at 4°C. The next day, membranes were washed 3 times with TBS-T for 15 min and incubated with solution containing secondary antibody (1:10000 HRP Goat anti-rabbit IgG, Thermo Fisher Scientific #65-6120) in blocking buffer at room temperature for 1 h. The membrane was washed 3 times for 15 min each. For signal detection from HRP, Amersham ECL Select Western Blotting Detection Reagent (Cytiva) was used according to manufacturer's instructions. Membranes were imaged with a ChemiDoc Imaging System (Bio-Rad) in Protein Blot - Chemiluminescence setting. Anti-OmpA membranes were imaged with 15 s exposure, while anti-ChmC membranes were imaged with 300 s exposure.
Mass spectrometry of phage infections
For P. aeruginosa, overnight bacterial cultures in LB media were diluted to OD600= 0.1 in fresh media, then further grown to OD600= 0.5. Regrown cultures were diluted 1:10 into 50 ml total volume in 250 ml flasks and grown in LB supplemented with 0.2 mM CaCl2. Cells were infected with phage PhiPA3 at a multiplicity of infection (MOI) of 3 when they reached OD600= 0.3, then collected at the indicated time points. Cultures were pelleted by centrifugation at 4000 rpm at 4°C. Cell pellets were washed with 25% LB to remove any free phage. After the last wash, pellets were snap-frozen in liquid nitrogen and stored at –80°C.
For E. coli, overnight cultures were diluted to OD600= 0.1 in fresh media, then further grown to OD600= 0.6. 10 ml of pad mix (1% agarose, 25% LB, 50 nM aTc and 30 ug/ml chloramphenicol) was poured into a 6 cm petri dish for each infection. When cultures reached desired OD600, each strain was prepared as 200 μl of OD600= 0.1 and spread on the prepared pads. Petri dishes were placed in a 37°C humidor and incubated for 2 h, then 100 μl of Goslar (titer 109 PFU/ml) was spread on the pad and incubated at 37°C for the indicated times. At the time of collection, 1 ml of 25% LB media was added and cells were carefully resuspended. Cells were pelleted by centrifugation at 4000 rpm at 4°C, washed with 25% LB, then snap-frozen and stored at –80°C for mass spectrometry.
Plaque assays for phage infectivity
For plaque assays, 500 μl of saturated overnight bacterial cultures were mixed with 4.5 ml of 0.35% LB top agar. This mixture was poured on an LB plate that contains 100 μg/ml chloramphenicol and 50 nM aTc. After 30 min of incubation at room temperature for the mixture to dry, 3 μl of 10-fold serial dilutions of Goslar lysates (109 pfu/ml) were spotted on each plate. Plates were incubated at 37°C for 16 h, then plaques were counted.
GFP pulldowns
GFP pulldowns were performed with GFP-Trap Magnetic Agarose beads (Proteintech). P. aeruginosa strains carrying pHERD30T plasmids expressing GFP-tagged proteins of interest were grown in LB with 25 μg/ml gentamicin sulfate. Saturated overnight cultures were diluted to OD600= 0.1 in fresh media, then grown at 30°C until they reached OD600= 0.5. Cultures were diluted 1:10 in 50 ml LB supplemented with arabinose, gentamicin sulfate, and calcium chloride. Once the cells reached at OD600= 0.3, they were infected with PhiPA3 at MOI 3. The cultures were collected at 45 min post-infection, centrifuged, and the resulting cell pellets were stored at –80°C.
To perform the GFP pulldown, frozen cell pellets were thawed and incubated for 1 h with 500 μl lysis buffer (10% glycerol, 25 mM Tris–HCl pH 7.5, 150 mM NaCl, 4 mg/ml lysozyme, 20 μg/ml DNase I, 2× cOmplete Protease Inhibitor, 0.4 mM PMSF). The cell suspensions were then sonicated (10 rounds x 20 pulses/round, Duty Cycle 40, Output 4), and the resulting lysed cells were centrifuged (30 min at 15000 rpm at 4ºC). Beads were washed 3 times with a wash buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA) and added to the supernatant of the cell lysate. The mixture was rotated end-to-end for 1 hour at 4°C. The beads incubated in the cell lysate were washed 3 times with 1 ml wash buffer. After the last wash, beads were stored at –80°C for later analysis by SDS-PAGE and mass spectrometry.
eCLIP-Seq
Overnight bacterial cultures in LB media were diluted to OD600= 0.1 and grown to OD600= 0.5 at 30°C. The cultures were diluted 1:10 into 50 ml total volume in 250 ml flasks and grown in LB supplemented with 0.2 mM CaCl2 and 0.1% Arabinose. Cells were infected with phage PhiPA3 at a multiplicity of infection (MOI) of 3 when they reached OD600= 0.3. After 45 min of infection, cultures were collected and centrifuged with 4000 rpm at 4°C for 8 min. Cells were washed with PBS 2 times and resuspended in 10ml fresh PBS. The samples were spread on 10 cm petri-dish to coat the surface and UV crosslinking was performed with 400 mJ/cm2 at 254 nm. Samples were collected from the petri dish and pelleted at 4C at 4000 rpm for 10 min and snap-frozen in liquid nitrogen to be stored at -80°C.
Each sample was treated with immunoprecipitation protocol with GFP-Trap beads. After the RNA-bound proteins were collected, cells were treated with FastAP (ThermoFisher) and T4 PNK (NEB), then barcoded RNA adapters were ligated to the 3′ end (T4 RNA Ligase, NEB). Samples were separated by SDS-PAGE and transferred to nitrocellulose membranes. The regions corresponding to the approximately expected size of sfGFP-alone and ChmC-sfGFP were excised, and the membrane was suspended in a buffer with proteinase K (NEB). RNA isolation was performed with phenol/chloroform extraction and purified on spin columns (Zymo Research). Reverse-transcription was performed with AffinityScript (Agilent). cDNAs were treated with ExoSAP-IT (Affymetrix) to remove the excess oligonucleotides. Second DNA adapters (containing 5 [N5] or 10 [N10] random bases at the 5′-end) were ligated to the 5′-end of the cDNA (T4 RNA Ligase, NEB). The DNA was amplified by PCR and purified with PippinPrep system (Sage Science) and sequenced with Illumina HiSeq 4000. Libraries were analyzed for fragment size distribution on a D1000 Screentape (Agilent). Reads were processed and mapped to the PhiPA3 genome. Normalization of the eCLIP data was performed using the input samples prior to pulldown.
Reads were processed according to the protocol as previously described (22). Briefly, sequenced reads from both IP and corresponding size-matched input (SMInput) were trimmed of adapters and mapped to repeat elements first to remove non-uniquely mapped reads, then to the genome composed of both P. aeruginosa PA01 (NCBI RefSeq #GCF_000006765.1) and PhiPA3 (NCBI RefSeq #GCF_001502095.1). PCR collapsing was then performed, and CLIPper (23) was used to call peak clusters on each set of these uniquely mapped, deduplicated (usable) reads. Annotations from P. aeruginosa PA01 and PhiPA3 were used to construct a custom CLIPper index. Reads within these clusters were normalized against the SMinput sample using scripts (available at https://github.com/yeolab/eclip), and peaks found to be enriched above a log2(fold change) of 3 and –log10(Fisher Exact or chi-square P-value) threshold of 3 were deemed significant and merged using IDR (24) to produce a set of reproducible peaks from replicates. Metagene plots were generated from usable reads, using a strategy laid forth by (25). Briefly, read densities from SMInput samples were subtracted from corresponding IP signals across the set of annotated start codons of expressed phage genes, using SMInput data as a proxy for gene expression. eCLIP-Seq data have been deposited at the NCBI GEO repository (https://www.ncbi.nlm.nih.gov/geo/) with dataset identifier GSE243675.
mRNA FISH
We designed mRNA FISH probes to three Goslar genes: HOV27_gp022 (NCBI protein record YP_009820707.1; reverse complement of bases 16 719–15 826 of NCBI nucleotide record NC_048170.1), HOV27_gp053 (NCBI protein record YP_009820738.1; reverse complement of bases 58 494–55 117 of NCBI nucleotide record NC_048170.1), and HOV27_gp217 (NCBI protein record YP_009820902.1; bases 199 310–200 200 of NCBI nucleotide record NC_048170.1). Probe oligonucleotides were designed according to (26) and contained from 5′ to 3′: (1) a 20-nt forward region for PCR amplification; (2) a 20-nt adapter sequence complementary to an adapter oligonucleotide; (3) a 40-nt target sequence complementary to one of the genes listed above; (4) two additional repeats of the 20-nt adapter sequence; (5) a 20-nt reverse region for PCR amplification. We used three adapter oligonucleotides that contained from 5′ to 3′: (i) a 20-nt adapter sequence complementary to the probe oligonucleotide; (ii) a 40-nt readout sequence complementary to a readout oligonucleotide. We used three readout oligonucleotides that contained a 40-nt readout sequence complementary to the adapter oligonucleotide, with both 5′ and 3′ fluorescent labels (Cy3, Cy5, or Alexa750). All oligonucleotide sequences are listed in Supplementary Table S4).
E. coli strains were grown on LB agar plates overnight, then suspended in fresh LB media at an OD600 of 0.1 in the morning. Once the cultures were grown to mid-log phase (OD600= 0.6), samples were diluted to OD600= 0.1 and 200 μl was spread onto an agarose pad (1% agarose, 25% LB, 50 nM aTc and 30 μg/ml chloramphenicol) in a 6 cm petri dish for infection. Petri dishes were incubated at 37°C for 2 h, then 100 μl of Goslar phage (109 PFU/ml) was spread on the pad and incubated for 70 min at 37°C. To collect the cells, 1 ml of 25% LB media was added and cells were carefully resuspended from the surface of the pads. Cells were pelleted by centrifugation at 4000 rpm at 4°C. The pellet was resuspended in 500 μl 25% LB media and buffered paraformaldehyde (100 μl 24% paraformaldehyde and 20 μl 1 M NaPO4 pH 7.4) was added. The tubes were mixed by inversion and allowed to fix for 10 min at room temperature. After the incubation, fixed cells were pelleted by centrifugation at 4000 rpm at room temperature and washed three times with PBS. The final pellet was resuspended in 200 μl PBS, then 50 μl was gently pipetted onto a poly-l-lysine coated cover slip. After 10 min, the solution was gently pipetted out from the side of the cover slip and 100 μl of freshly made 4% PFA in PBS was pipetted onto the cover slip to fix the cells to the cover slips. The cover slips were incubated at room temperature for 10 min, then the fixing solution was carefully pipetted out from the side. The slides were stored in 80% ethanol at 4°C.
Before hybridization, cover slips were incubated 5% SDS in PBS + RNAse inhibitor for 10 min at room temperature. The solution was switched to Pre-Hybe buffer (20 ml 100% formamide (Ambion #AM9342), 5 ml 20× SSC (Ambion #AM9763), 25ml DI water, 50 μl Tween-20 (Sigma #P9416), 10 μl RNAse inhibitor (New England Biolabs #M0314)) and incubated for 10 min at room temperature. Next, the cover slips were incubated in Hybe buffer (500 μl 100% formamide, 100 μl 20× SSC, 200 μl 50% dextran sulfate (Sigma #D8906)) before the hybridization with probe oligos. Oligos were resuspended in Tris-EDTA buffer and added to Hybe buffer (80 μl Hybe buffer, 2 μl of 1:5 diluted probes, 18 μl DI water, 2 μl RNAse inhibitor). Each cover slip was placed on parafilm with 100 μl of probes containing Hybe buffer mixture in a 6 cm petri dish. Additional parafilm was placed on top to prevent water evaporation and petri dishes were incubated at 47°C for 20 h. Next day, the Hybe buffer mixture was switched with 2× SSC (supplemented with RNAse inhibitor) and placed at 4°C until hybridization with adapter oligos.
Before hybridization with adapters, each cover slip was washed with fresh 2× SSC (supplemented with RNAse inhibitor). Adapter solution was prepared (1 μl of each adapter (P1A1, P1A2, P1A3; see Supplementary Table S4), 1 ml 35% formamide in 2× SSC, 0.1% Tween-20) and 2 ml placed on a clean parafilm for each cover slip. The cover slips were incubated facing the parafilm with adapter solution for 30 min at room temperature. Each cover slip was washed with Wash buffer (30% formamide in 2× SSC, 0.1% Tween-20) for 15 min, followed by an additional three washes with 2× SSC (supplemented with RNAse inhibitor). Readout buffer was prepared with readout oligos (1 μl for each readout oligo, 2 ml 35% Hybe Buffer, 1 μl RNAse inhibitor) and 2 ml was pipetted onto a new parafilm for each cover slip. Each cover slip was incubated with readout oligos for 30 min in room temperature in the dark. Each cover slip was washed with Wash buffer and washed 3 times with 2× SSC supplemented with RNAse inhibitor. Each cover slip was stored in the dark at 4°C until imaging.
The microscope setup and the image acquisitions were performed as previously described (26) with a custom-built microscope using a 60x Nikon objective lens. Images were collected at 405, 560, 647 and 750 nm emission wavelengths, and raw images were exported using Napari software (27). Images were analyzed with Fiji ImageJ and statistical analysis and graphing were performed with Graphpad Prism.
Protein identification by mass spectrometry
Frozen cell pellets were thawed and resuspended in 100 μl water. 10 μl of resuspended cells were mixed with 200 μl of 6M guanidine-HCl, vortexed and subjected to 3 cycles of 100°C for 5 min followed by cooling to room temperature. Boiled cell lysates were mixed with 1.8 ml of pure methanol and incubated at –20°C for 20 min. The mixture was centrifuged at 14000 rpm for 10 min at 4°C. All liquid was removed and pellet was resuspended in 200 μl of 8 M urea in 0.2 M ammonium bicarbonate and incubated at 37°C for 1 hour with constant agitation. 4 μl of 500 mM TCEP (Tris(2-carboxyethyl) phosphine) and 20 μl 400 mM chloro-acetamide were added to the samples.
Protein concentration was measured by BCA assay and 600 μl of 200 mM ammonium bicarbonate was added to bring the urea concentration to 2 M. 1 μg of sequencing-grade trypsin was added for each 100 μg of protein in the sample and incubated at 42°C for overnight. The next day, 50 μl of 50% formic acid was added (final pH = 2), then samples were desalted with C18 solid phase extraction (Waters Sep-Pak C18 12 cc Vac Cartridge # WAT036915) according to the manufacturer's protocol. Samples were resuspended in 1 ml phosphate-buffered saline and peptide concentration of each sample was measured with BCA (bicinchoninic acid assay).
Trypsin-digested peptides were analyzed by ultra-high pressure liquid chromatography (UPLC) coupled with tandem mass spectroscopy (LC-MS/MS) using nano-spray ionization. Nanospray ionization was performed with Orbitrap fusion Lumos hybrid mass spectrometer (Thermo) interfaced with nano-scale reverse-phase UPLC (Thermo Dionex UltiMate 3000 RSLC nano System) using a 25 cm, 75-micron ID glass capillary packed with 1.7-μm C18 (130) BEH beads (Waters corporation). Peptides transferred from C18 column into the mass spectrometer by a linear gradient (5–80% buffer B) using buffers A (98% H2O, 2% acetonitrile, 0.1% formic acid) and B (100% acetonitrile, 0.1% formic acid) at a flow rate of 375 μl/min over 3 h. Mass spectrometer parameters were; MS1 survey scan using the orbitrap detector (mass range (m/z): 400–1500 (using quadrupole isolation), 120000 resolution setting, spray voltage of 2200 V, Ion transfer tube temperature of 275°C, AGC target of 400 000, and maximum injection time of 50 ms) which was followed by a data dependent scans (top speed for most intense ions, with charge state set to only include + 2–5 ions, and 5 s exclusion time, while selecting ions with minimal intensities of 50 000 at in which the collision event was carried out in the high energy collision cell (HCD Collision Energy of 30%), and the fragment masses were analyzed in the ion trap mass analyzer (With ion trap scan rate of turbo, first mass m/z was 100, AGC Target 5000 and maximum injection time of 35 ms). Protein identification and quantitation were carried out using Peaks Studio X (Bioinformatics Solutions Inc.). Mass spectrometry data have been deposited at the PRIDE repository (https://www.ebi.ac.uk/pride/) with dataset identifier PXD045260.
LFQ mass spectrometry
For label-free quantitation (LFQ) mass spectrometry analysis of non-targeting versus ChmC knockdown, E. coli MC1000 cultures containing appropriate CRISPRi-ART plasmids (non-targeting or ChmC guide #2) were diluted to OD600= 0.1 in fresh LB media and spread on to pad mix (1% agarose, 25% LB, 50 nM aTc and 30 ug/ml chloramphenicol). After 2 h of incubation at 37°C, cells were infected on agarose pads with Goslar phages at a multiplicity of infection (MOI) of 3. Infected cells were incubated at 37°C for 90 min, then collected from agarose pads by scraping into 1.7 ml microcentrifuge tubes. Cells were centrifuged at 4000 RPM at 4°C, and washed 3 times with 25% LB. Cell pellets were stored at –80°C.
Tryptic mass spectrometry was performed on each sample as above. For label free quantitation, peptides identified in each sample were divided into host (MC1000/MG1655) and Goslar phage peptides. From each data set, shared E. coli OmpA peptides in all samples were selected, and the sum of these peptides’ peak areas in each sample was used for normalization between all eight samples (four non-targeting, four ChmC knockdown). Normalized peak areas of Goslar proteins from each data set were calculated with the calculated OmpA normalization coefficient for each sample (Supplementary Table S6). Log2(fold change) and –log2(P) values were calculated for all proteins that were detected in at least two replicates for each sample (non-targeting versus ChmC knockdown). Putative virion structural proteins were annotated by using either NCBI annotations or through detectable sequence or predicted-structure homology to virion structural proteins of related bacteriophages. LFQ mass spectrometry data have been deposited at the PRIDE repository (https://www.ebi.ac.uk/pride/) with dataset identifier PXD045260.
Condensate analysis
Macromolecular condensation assays were conducted in vitro using phase separation buffer (20 mM HEPES pH 7.4, 50 mM NaCl) at a temperature of 25°C, a protein concentration of 30 μM, and an RNA concentration of 2 μM (for samples including RNA). To prepare samples, unlabeled PhiPA3 ChmC was pre-mixed with Cy5-labeled ChmC (linked to an engineered N-terminal cysteine using maleimide linkage) at a ratio of 1:10 and diluted to 60 μM in phase separation buffer. For samples without RNA co-incubation, an equal volume of phase separation buffer was added to each protein sample to reach the final working concentration of 30 μM. For samples with RNA co-incubation, an equal volume of RNA at 2× final concentration (4 μM for 40 base RNA; 166 nM for 2.3 kb RNA) in phase separation buffer was added and mixed gently. Samples were mixed in protein LoBind tubes (Eppendorf) and then immediately transferred into a 96-well non-binding plate (Greiner Bio-one). Samples were imaged immediately after transfer into the 96-well plate using a CQ1 confocal quantitative image microscope (Yokogawa) with a 20x-PH objective. The fluorescent signal was captured under a laser at 640 nm. For live imaging, the entire field was automatically captured every 10 min. For quantitation, condensates were identified by particle analysis in ImageJ (16). After thresholding, individual particles were counted and their areas measured. For each sample showing particles, the coefficient of variation was calculated as the standard deviation of particle area divided by the mean particle area.
Results
Identification of the abundant and early-expressed jumbo phage protein ChmC
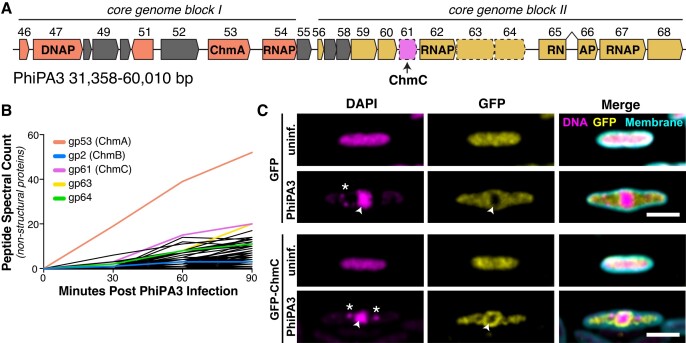
We previously used proximity ligation to identify proteins in the nucleus-forming jumbo phage PhiPA3 that physically associate with the major nuclear shell protein ChmA or with a phage nucleus-localized protein, UvsX (gp175) (15). Through this analysis, we identified a minor nuclear shell component (ChmB) and several uncharacterized proteins with no known function (15). One of these proteins was PhiPA3 gp61, which is located in a highly conserved cluster of genes that includes chmA (gp53), several subunits of the phage-encoded ‘non-virion RNA polymerase’ (nvRNAP: gp62, gp65-66, and gp67), and two additional phage nucleus-associated proteins, gp63 and gp64 (Figure 1A). PhiPA3 gp61 is of particular interest as its homolog is the second highest-expressed non-structural protein in Pseudomonas chlororaphis cells infected with the related jumbo phage 201Phi2-1 (gp123) (4). To test whether PhiPA3 gp61 is also highly expressed, we infected P. aeruginosa cells with PhiPA3 and performed mass spectrometry proteomics to identify the timing and expression of phage proteins. We confirmed that both ChmA and gp61 are highly abundant in PhiPA3 infections (Figure 1B, Supplementary Tables S1 and S2). Based on its conservation, abundance, and association with the phage nuclear shell, we term this protein Chimallin C (ChmC).
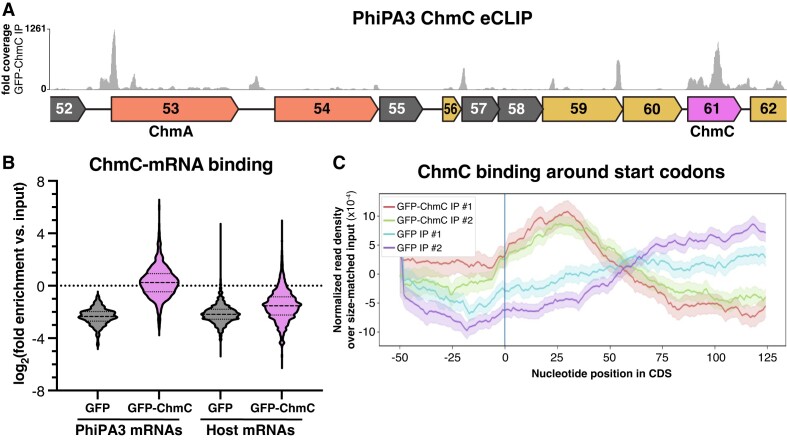
Figure 1.
PhiPA3 ChmC is associated with the phage nucleus. (A) Map of the PhiPA3 genome spanning jumbo phage conserved blocks I and II (3). Genes in conserved block I are colored salmon, and genes in conserved block II are colored goldenrod. Genes conserved with the closely related jumbo phage PhiKZ but not all jumbo phages are colored gray. gp47 is a putative DNA polymerase (DNAP), and gp53 is the major nuclear shell protein Chimallin (ChmA). Gp54, gp64, gp65-66 (interrupted by a self-splicing intron), and gp67 are subunits of the phage-encoded non-virion RNA polymerase (RNAP). Dotted outlines indicate three proteins (gp61/ChmC, gp63, and gp64) shown to be associated with the phage nuclear shell (15). (B) Mass spectrometry proteomics analysis of PhiPA3-infected P. aeruginosa, showing spectral counts of non-structural phage proteins. gp53 (ChmA), gp2 (ChmB), gp61 (ChmC), gp63 and gp64 are shown in colors and labeled. These data are from a single replicate from each time point. See Supplementary Tables S1 and S2 for full mass spectrometry results. (C) Localization of sfGFP (top) or sfGFP-fused PhiPA3 ChmC (bottom) in PhiPA3-infected P. aeruginosa cells at 75 min post infection. Magenta: DAPI nucleic acid dye; cyan: FM4-64 membrane dye; yellow: GFP. Arrowheads indicate phage nuclei, and asterisks indicate phage bouquets. Scale bar = 2 μm.
Confirming our earlier microscopic observations (15), we find that ectopically-expressed PhiPA3 ChmC fused to GFP localizes both to the cytoplasm and the phage nuclear shell in late-stage PhiPA3 infections of P. aeruginosa (Figure 1C). Fluorescent microscopy is unable to determine whether nuclear shell-associated ChmC is localized within the nucleus or just outside the ChmA shell, but our prior identification of ChmC through proximity-labeling with the nuclear-localized protein UvsX suggests that the protein is at least partially localized within the phage nucleus (15). We next purified GFP-tagged ChmC from P. aeruginosa cells infected with PhiPA3 and used mass spectrometry to identify associated proteins (Supplementary Figure S1, Supplementary Table S3). In this analysis, we identified the putative phage nucleus pore protein ChmB (gp2) and one subunit of each phage-encoded RNA polymerase: gp62 is part of the non-virion RNA polymerase, and gp77 is part of the virion RNA polymerase. We also observed association with 19 ribosomal and ribosome-associated proteins plus the host RNA polymerase α, β, and β′ subunits. The enrichment of ribosomal proteins and RNA polymerase subunits in this experiment suggests that ChmC may be a non-specific RNA binding protein. The observed association of ChmC with the putative pore-forming protein ChmB, moreover, suggests that these two proteins may functionally cooperate at the nuclear shell.
ChmC adopts a nucleic acid-binding fold and binds RNA
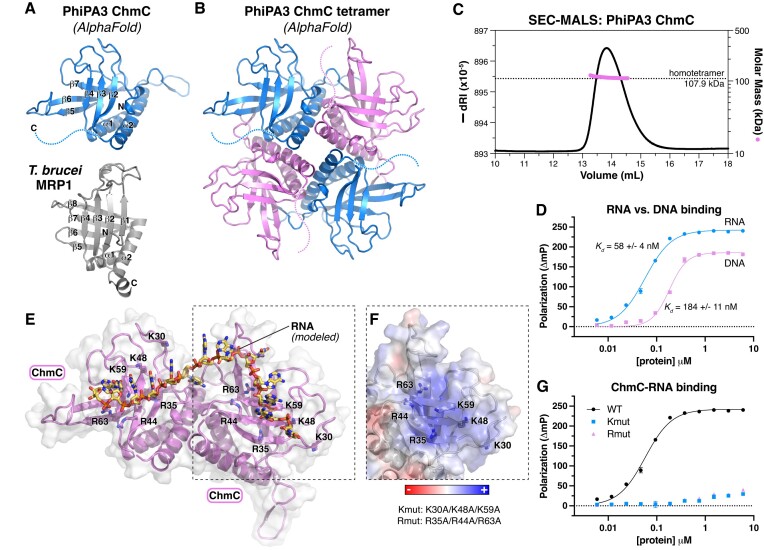
ChmC is conserved across jumbo phages but shows no identifiable sequence similarity to known proteins. To gain insight into ChmC’s structure and potential function, we used AlphaFold2 (17) to predict its 3D structure with high confidence (Figure 2A, B, Supplementary Figure S2A, B). Analysis of the resulting model using the DALI protein structure comparison server revealed a predicted Whirly domain fold (also termed a PUR domain) common to multiple families of single-stranded RNA and DNA binding proteins (28–30). The Whirly fold typically comprises a tandem repeat of β-β-β-β-α secondary structure elements, and is exemplified by the Trypanosoma brucei MRP1 protein (Figure 2A) (29). The predicted structure of PhiPA3 ChmC shows a tandem repeat of β-β-β-α elements, with an overall 3D structure highly reminiscent of the Whirly domain (Cα r.m.s.d. of 4.7 Å for ChmC versus T. brucei MRP1 over 120 residues) (Figure 2A).
Figure 2.
ChmC adopts an RNA-binding Whirly fold (A) AlphaFold predicted structure of PhiPA3 ChmC (blue) at top oriented equivalently to the structure of T. brucei MRP1 (PDB ID 2GJE; gray) (29) at bottom, with secondary structure elements labeled. (B) AlphaFold predicted structure of a PhiPA3 ChmC homotetramer, with alternating subunits colored blue and pink. See Supplementary Figure S2A-B for details of AlphaFold predictions. (C) Size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) analysis of purified PhiPA3 ChmC. (D) Single-stranded RNA (blue circles) and DNA (pink squares) binding of PhiPA3 ChmC as measured by fluorescence polarization. Data points are shown as average ± standard deviation of triplicate technical replicates, and curves are fit with a cooperative binding model (Hill coefficient for RNA binding 1.6 ± 0.1; Hill coefficient for DNA binding 2.4 ± 0.3). (E) Structural model of PhiPA3 ChmC binding single-stranded RNA, generated by overlaying two adjacent subunits of the ChmC AlphaFold 2 model shown in panel (B) with structure of a T. brucei MRP1:MRP2 heterotetramer bound to RNA (PDB ID 2GJE; see Supplementary Figure S2C) (29). Lysine and arginine residues mutated to generate the Kmut (K30A/K48A/K59A) and Rmut (R35A/R44A/R63A) are shown as sticks and labeled. (F) Surface charge representation of one ChmC subunit, oriented equivalently to panel (E) (dotted line). See Supplementary Figure S2D. (G) Single-stranded RNA binding of PhiPA3 ChmC wild-type (black circles, reproduced from panel (D)), Kmut (blue squares), and Rmut (pink triangles), as measured by fluorescence polarization. Data points are shown as average ± standard deviation of triplicate technical replicates. See Supplementary Figure S2E for DNA binding.
Whirly domain proteins typically form higher-order complexes including homo- and heterotetramers, exemplified by the 2:2 heterotetramers formed by T. brucei MRP1 and MRP2 (29). We used AlphaFold 2 to predict the structure of PhiPA3 ChmC oligomers, and obtained a confident prediction of a ChmC homotetramer that is strikingly similar to the T. brucei MRP1:MRP2 heterotetramer structure (Figure 2B, Supplementary Figure S2A, C). We expressed and purified full-length PhiPA3 ChmC in E. coli and analyzed its oligomeric state by size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS). Supporting our structure prediction, we found that ChmC forms a stable homotetramer in solution (Figure 2C).
Given the known roles of Whirly domain proteins in nucleic acid binding, we tested the ability of purified PhiPA3 ChmC to bind single-stranded (ss) DNA or RNA in vitro. Using a fluorescence polarization assay, we found that ChmC binds both ssDNA and ssRNA, but that the protein binds ssRNA with a higher affinity (Kd = 58 ± 4 nM) than it binds ssDNA (Kd = 184 ± 11 nM) (Figure 2D; see Supplementary Figure S2E for binding to single-stranded versus double-stranded DNA). We modeled a ChmC-RNA complex by overlaying the PhiPA3 ChmC tetramer model onto the structure of T. brucei MRP1:MRP2 bound to RNA (29) (Figure 2E). Based on this model, we designed two multi-site mutants to disrupt nucleic acid binding, termed Kmut (K30A/K48A/K59A) and Rmut (R35A/R44A/R63A) (Figure 2E, F, Supplementary Figure S3A, B), and found that both mutants completely disrupt RNA and ssDNA binding in vitro (Figure 2G, Supplementary Figure S3C). Based on these data, we conclude that ChmC adopts a homotetramer of Whirly domain folds and binds RNA.
ChmC forms condensates with RNA
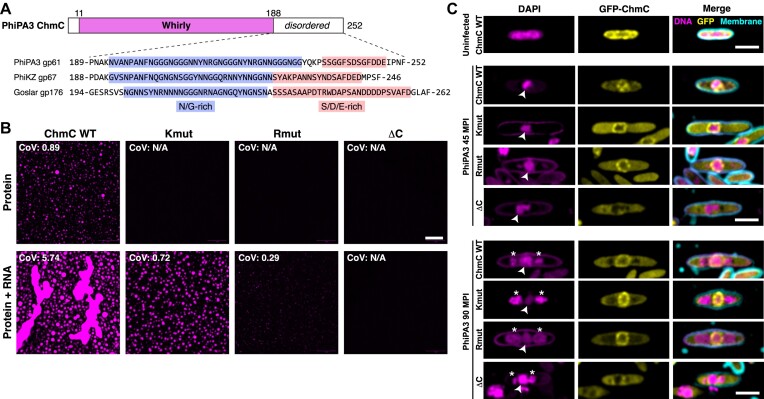
AlphaFold2 structure predictions of ChmC homologs from different phages consistently showed low confidence scores (pLDDT) for the C-terminal 50–70 residues of the protein, indicating that this region is likely disordered in solution. Across several ChmC orthologs, this region contains a ∼40-residue subregion enriched in asparagine and glycine residues (N/G-rich), followed by a ∼15-residue subregion enriched in serine, aspartate, and glutamate residues (Figure 3A). N/G-rich regions have been shown to promote macromolecular condensate formation, likely through dipole-dipole interactions (31,32). Disordered regions rich in serine, aspartate and glutamate, meanwhile, have been termed ‘electronegative clusters’ (ENCs) which can stabilize RNA binding proteins in solution and suppress nonspecific RNA binding (33). Indeed, both the PSPredictor (34) and catGRANULE (35) algorithms strongly predicted that ChmC forms condensates, and that this propensity is driven by the protein's C-terminal disordered region (Supplementary Figure S4A). Together with our finding that ChmC binds RNA in vitro, these predictions suggested that ChmC might form macromolecular condensates with RNA through multivalent RNA binding combined with low-affinity interactions through its C-terminal disordered region, similar to many other RNA binding proteins in both prokaryotes (36,37) and eukaryotes (38–40).
Figure 3.
ChmC forms phase-separated condensates with RNA (A) Top: Domain structure of PhiPA3 ChmC, with Whirly domain colored magenta and regions predicted to be disordered in white. Bottom: Sequence of the C-terminal predicted disordered domain in ChmC from PhiPA3 (gp61), PhiKZ (gp67), and Goslar (gp176). Asparagine/glycine (N/G) rich regions are highlighted in blue, and serine/aspartate/glutamate (S/D/E) rich regions are highlighted in salmon. See Supplementary Figure S4A for catGRANULE analysis of all three proteins, and Supplementary Figure S3D-E for analysis of ChmC ΔC binding DNA and RNA. (B) Fluorescence microscopy imaging of PhiPA3 ChmC (wild type, Kmut, Rmut, or ΔC; 10% Cy5-labeled) at 30 μM protein concentration, either alone (top row) or with 83 nM of a 2.3 kb RNA (5.8 μg/ml; bottom row). All images were taken 30 min after final dilution and mixing with RNA. For all conditions that showed condensate formation, the coefficient of variation (CoV) was calculated as the standard deviation of particle area divided by the mean particle area (WT protein alone n = 423; WT + 40 base RNA n = 222; Kmut + 40 base RNA n = 247; Rmut + 40 base RNA n = 5; WT + 2.3 kb RNA n = 552; Kmut + 2.3 kb RNA n = 368; Rmut + 2.3 kb RNA n = 147). Scale bar = 30 μm. See Supplementary Figure S4B for DIC imaging. (C) Localization of GFP-tagged PhiPA3 ChmC (wild type, Kmut, Rmut, or ΔC in PhiPA3-infected P. aeruginosa cells at 45 and 90 min post infection (MPI). Yellow: GFP; magenta: DAPI nucleic acid; cyan: FM4-64 membrane dye. Arrowheads indicate phage nuclei, and asterisks indicate phage bouquets. Scale bar = 2 μm.
To directly test the propensity for PhiPA3 ChmC to form condensates, we engineered a cysteine residue at the protein's N-terminus to enable fluorescent labeling using a maleimide-linked Cy5 dye. In a low-salt buffer (50 mM NaCl), we observed that purified ChmC forms uniform small droplets, and that the addition of a 2.3 kb RNA (encoding the major capsid protein gp136) stimulates formation of large droplets that showed hallmarks of liquid-liquid phase separation, including dynamic growth and fusion of droplets (Figure 3B, Supplementary Figure S4B). Disrupting ChmC’s ability to bind RNA using the Kmut or Rmut multisite mutations dramatically reduced, but did not eliminate formation of condensates in the presence of RNA (Figure 3B). Since both the Kmut and Rmut proteins retain some positively-charged residues on the RNA binding surface, it is likely that these proteins retain some ability to bind RNA, albeit with low affinity beyond the detection limit of fluorescence polarization. Removal of the C-terminal disordered region (residues 204–251 removed; ChmC-ΔC), meanwhile, completely eliminated formation of condensates in both the absence and presence of RNA (Figure 3B). Importantly, the ChmC-ΔC protein formed homotetramers (Supplementary Figure S3A, B) and retained the ability to bind RNA in vitro (Supplementary Figure S3D, E).
We next examined localization of ChmC mutants in PhiPA3-infected P. aeruginosa cells. The RNA binding mutant proteins (Kmut and Rmut) showed localization to the nuclear shell in infected P. aeruginosa cells, equivalent to wild-type protein. A limitation of this assay is that the mutant proteins are expressed alongside wild-type phage-encoded ChmC, and may form hetero-oligomers with wild-type ChmC in infected cells. Despite this limitation, both the Kmut and Rmut showed a localization pattern distinct from wild-type ChmC, retaining some detectable nuclear shell localization but showing more uniform cytoplasmic distribution than wild-type protein (Figure 3C). Meanwhile, removal of the ChmC C-terminus completely disrupted nuclear shell binding activity, suggesting that the protein's ability to form condensates (with or without RNA) is important for its localization to the nuclear shell.
ChmC associates with viral mRNAs in infected cells
To test whether ChmC associates with RNA, particularly phage mRNAs, in infected cells, we performed eCLIP-seq (enhanced UV crosslinking and immunoprecipitation, followed by deep sequencing) in PhiPA3-infected P. aeruginosa cells expressing GFP-tagged ChmC. We obtained ∼1.6M uniquely-mapping sequence reads for GFP-tagged PhiPA3 ChmC across two independent replicates (1161658 + 423427 reads), and 1.5M reads for control GFP samples (1070428 + 423357 reads). We mapped all reads to the host (P. aeruginosa PA01) and phage genomes (Figure 4A, Supplementary Figure S5A), then calculated the enrichment for each gene for immunoprecipitated samples compared to matched input samples. All samples were collected 45 min after phage infection, after the host genome is fully degraded and the phage genome has begun replicating (4,5,13).
Figure 4.
ChmC binds phage mRNAs. (A) Fold sequence coverage from eCLIP analysis of GFP-ChmC, in the region of the PhiPA3 genome encoding ChmA and ChmC. See Supplementary Figure S5 for eCLIP validation. (B) Log2(fold enrichment versus input for all PhiPA3 and host (P. aeruginosa) genes detected in eCLIP analysis of GFP (gray) and GFP-ChmC (pink). (C) Metagene analysis showing enrichment of GFP-ChmC binding near start codons of PhiPA3 genes (replicate #1 red, replicate #2 green).
Chimalliviruses encode two multi-subunit RNA polymerases: a ‘virion RNA polymerase’ (vRNAP) that is packaged in the virion and is responsible for transcription immediately after infection, and a ‘non-virion RNA polymerase’ (nvRNAP) that is responsible for transcription in the middle and late stages of infection (41–43). A prior RNA-seq analysis of the jumbo phage PhiKZ defined the operon structure of this phage and found that while vRNAP-transcribed early genes have a defined promoter sequence, genes transcribed by the nvRNAP do not show a reproducible promoter sequence (44). This analysis also identified a number of noncoding antisense RNAs, which were proposed to regulate translation of phage genes (44). Since a similar RNA-seq analysis has not been performed for PhiPA3, our analysis of ChmC eCLIP data is limited to annotated protein- and tRNA-encoding genes. We first analyzed the relative enrichment of phage mRNAs versus host mRNAs in the ChmC eCLIP immunoprecipitates versus input RNA. We found that phage mRNAs were slightly enriched (median log2(fold enrichment) of 0.186) compared to input samples, while host-encoded mRNAs were significantly depleted (median log2(fold enrichment) of –1.54) compared to matched input samples (Figure 4B). Overall, these data suggest that ChmC preferentially associates with phage mRNAs over host mRNAs, but that within phage mRNAs, ChmC shows little to no specificity. This preference may arise from ChmC’s localization within the phage nucleus, and/or from an inherent specificity for particular sequences or structures in phage mRNAs.
We next visually inspected ChmC eCLIP sequence coverage on the PhiPA3 genome. While mRNAs for some highly expressed genes (judging from the abundance of sequence reads in eCLIP input samples) were bound by ChmC across the entire open reading frame (e.g. the chmC gene itself; Figure 4A), we noticed that the majority of binding occurred in defined peaks near the start codons of genes. While the operon structure of PhiPA3 is not annotated, many peaks occurred near the start codons of genes that appear to be within polycistronic mRNAs. That is, ChmC binding occurs not only near the 5′ end of an mRNA (e.g. chmA; Figure 4A), but also likely occurs near internal start codons in mRNAs that encode multiple genes (e.g. gp57, gp59 and gp60; Figure 4A). We performed a metagene analysis and found that across annotated PhiPA3 genes, ChmC shows enriched binding in a ∼50-bp region centered 25–30 bp downstream of the start codon (Figure 4C). We performed motif analysis to identify any defined sequences bound by ChmC, but could not unambiguously identify a preferred binding motif. Finally, we also observed high sequence coverage in short regions that do not correlate with annotated genes, including peaks between the chmA (gp53) and gp54 genes (Figure 4A) and between gp203 and gp204 (Supplementary Figure S5A). These data suggest that PhiPA3 encodes small regulatory RNAs similarly to the related jumbo phage PhiKZ (45), and that ChmC binds many of these RNAs in addition to binding protein-coding mRNAs.
ChmC knockdown halts phage nucleus development and virion assembly
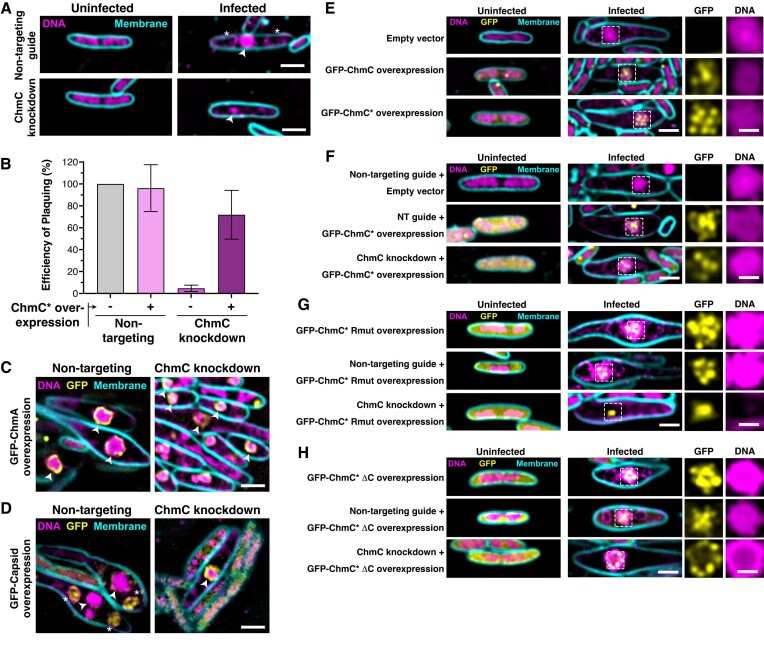
The jumbo phage nucleus prevents Cas9-based targeting of the phage genome due to its inaccessibility to most host-encoded proteins, including CRISPR-Cas nucleases (8,9). Because phage mRNAs are transported into the host cytoplasm for translation, however, jumbo phages are susceptible to RNA-targeting CRISPR-Cas nucleases (8,9). In related work, we used catalytically-dead Ruminococcus flavefaciens Cas13d (dCas13d) and a guide RNA overlapping the translation start site of an mRNA to efficiently inhibit translation of proteins encoded by the E. coli nucleus-forming phage Goslar; we term this method CRISPRi-ART (CRISPR interference by Antisense RNA Targeting) (20,21). To determine the biological roles of ChmC, we designed guide RNAs that target the Goslar chmC gene (gp176) and found that when expressed alongside dCas13d, these guide RNAs block ChmC translation in infected cells as judged by western blotting (Supplementary Figure S6A, B). Infection of E. coli MC1000 cells expressing dCas13d and chmC-targeting guide RNAs significantly reduced phage titer, with the most effective guide (guide 3) reducing the efficiency of plaquing to ∼4% of the efficiency observed in cells encoding a non-targeting guide RNA (Supplementary Figure S6B, C). By microscopy, we observed that ChmC translational knockdown resulted in a strong reduction in phage bouquet formation in infected cells, indicative of a failure to assemble new virions (Supplementary Figure S6D). At 110 mpi, phage nuclei were also smaller and contained less DNA compared to control cells upon ChmC translational knockdown (Figure 5A, Supplementary Figure S6E–G). We could rescue these phenotypes by overexpressing a recoded ChmC (referred to as ChmC*) resistant to dCas13d-mediated knockdown (Figure 5B, Supplementary Figure S7).
Figure 5.
ChmC knockdown impairs phage nucleus development and infection progression (A) Microscopy of E. coli MC1000 cells, either uninfected (left) or infected with Goslar (110 min post infection (MPI); right).(B) Efficiency of plaquing of Goslar phage infecting E. coli MC1000 cells expressing dCas13d and either a non-targeting guide RNA or a guide RNA targeting the 5′ end of the ChmC (gp176) gene (guide 3; see Supplementary Figure S6 for analysis of three guide RNAs). Data are presented as mean +/- standard error of the mean of four biological replicates, calculated as a percentage of Goslar phage plaque forming units with no dCas13d/guide expression. See Supplementary Figure S7 for example plaque images and microscopy of knockdown/rescue cells. (C) Microscopy of E. coli MC1000 expressing GFP-tagged ChmA plus dCas13d and either a non-targeting guide RNA (left) or ChmC-targeting guide 3 (ChmC knockdown; right) (110 MPI). DNA is stained with DAPI and shown in magenta; membranes are stained with FM4-64 and shown in cyan; GFP is shown in yellow. Arrowheads indicate the phage nucleus. Scale bar = 2 μm. See Supplementary Figure S8A for further images. (D) Microscopy of E. coli MC1000 expressing GFP-tagged capsid protein plus dCas13d and either a non-targeting guide RNA (left) or ChmC-targeting guide 3 (ChmC knockdown; right) (110 MPI). DNA is stained with DAPI and shown in magenta; membranes are stained with FM4-64 and shown in cyan; GFP is shown in yellow. Arrowheads indicate the phage nucleus, and asterisks indicate phage bouquets. See Supplementary Figure S8B for further images. (E) Microscopy of E. coli MC1000 cells expressing no protein (Empty vector, top row) or GFP-tagged ChmC (gp176, wild-type sequence in middle row and recoded sequence in bottom row), infected with Goslar (110 MPI). Yellow: GFP; magenta: DAPI (nucleic acid); cyan: FM4-64 (membrane). Scale bar = 2 μm for main panels, 1 μm for individual GFP and DNA channels representing zoomed views of the boxed regions. (F) Microscopy of E. coli MC1000 cells expressing dCas13d plus a non-targeting guide RNA (top and middle rows) or a ChmC-targeting guide (bottom row), expressing either no protein (Empty vector, top row) or GFP-tagged recoded ChmC (middle and bottom rows), and infected with Goslar (110 MPI). Yellow: GFP; magenta: DAPI nucleic acid; cyan: FM4-64 membrane dye. Scale bar = 2 μm for main panels, 1 μm for individual GFP and DNA channels representing zoomed views of the boxed regions. (G) As panel (E), except cells in the middle and bottom row are expressing ChmC Rmut (see Supplementary Figure S9 for Rmut design and analysis by Goslar plaquing assay) (H) As panel (E), except cells in the middle and bottom row are expressing ChmC ΔC (see Supplementary Figure S9 for ΔC design and analysis by Goslar plaquing assay).
We next imaged Goslar-infected E. coli cells expressing GFP-tagged ChmA (gp246) or the major capsid protein (gp41) at 110 min post-infection. Infected cells expressing dCas13d and a non-targeting guide RNA showed characteristic expansion of the cell diameter around the developing phage nucleus, which was labeled by GFP-tagged ChmA (Figure 5C, Supplementary Figure S8A). Infected cells expressing dCas13d and a chmC-targeting guide RNA showed little to no expansion of the cell diameter, and phage nuclei were markedly smaller than in control cells (Figure 5C, Supplementary Figure S8A-B). ChmC knockdown also caused infected cells to fail to form phage bouquets, and the major capsid protein was visibly attached to the nuclear shell, typical of an earlier developmental stage when capsids localize to the nuclear shell for genomic DNA packaging (Figure 5D, Supplementary Figure S8C) (4,13). Together, these data suggest that knockdown of ChmC significantly slows or halts Goslar infections at an early stage, preventing full maturation of the phage nucleus and production of viable phage progeny.
We next examined the localization of ectopically-expressed GFP-ChmC*, both in the context of an unperturbed Goslar infection (Figure 5E) and with phage-encoded ChmC knocked down by dCas13d (Figure 5F). In both cases, we observed that GFP-ChmC* localizes to the phage nucleus. In contrast to PhiPA3 ChmC, which localizes across the nuclear shell, Goslar ChmC* forms discrete puncta both within the nucleus and along the nuclear shell. These puncta could represent hubs of transcription within the nucleus, hubs for translocation of phage mRNAs through the nuclear shell, or both. We generated mutants of Goslar ChmC* analogous to the PhiPA3 ChmC-Rmut and ΔC mutants (Supplementary Figure S9A-B). In infected cells, both mutants localized as puncta within the phage nucleus in cells expressing a non-targeting guide RNA (Figure 5G, H). In cells expressing a chmC-targeting guide RNA, however, ChmC*-Rmut localized as a single punctum that lacked any DNA (Figure 5G). We interpret this as indicative of a failure in nuclear shell assembly around the initially-injected phage genome, resulting in an aborted nuclear shell containing ChmC but with no detectable DNA content. Consistent with this failure to assemble the phage nucleus, ChmC*-Rmut failed to rescue Goslar phage titer when ChmC was knocked down (Supplementary Figure S9C-D). In contrast, ChmC*-ΔC supported growth of the phage nucleus when ChmC was knocked down, and localized as puncta on the perimeter of the phage nucleus (Figure 5H). Despite the observation that it supports phage nucleus development, however, ChmC*-ΔC failed to rescue Goslar phage titer when ChmC was knocked down (Supplementary Figure S9C, E). Overall, our data on ChmC*-Rmut suggest that ChmC’s RNA binding activity is required for early development of the phage nucleus, and our data on ChmC*-ΔC suggests that the protein's ability to form RNA-protein condensates is important for a later, but no less important, step in the phage life cycle.
ChmC knockdown causes mRNA localization defects and a global reduction of phage protein levels
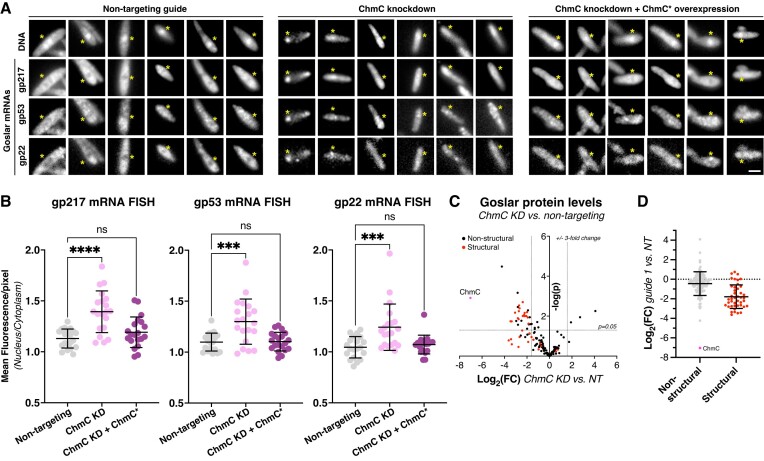
Our data on ChmC mRNA binding and localization are consistent with roles in stabilizing phage mRNAs and/or promoting their translocation through the phage nuclear shell for translation. To directly test for a role in mRNA nuclear-cytoplasmic translocation, we performed mRNA fluorescence in situ hybridization (mRNA FISH) on three Goslar genes encoding putative virion structural proteins: gp22, gp53 and gp217. We designed mRNA FISH probes for each gene (Supplementary Table S4) and imaged fixed E. coli cells 90 min after infection with Goslar phage. We tested mRNA localization in three conditions: (i) with dCas13d and a non-targeting guide RNA; (ii) with dCas13d and a ChmC-targeting guide RNA (ChmC knockdown) and (iii) ChmC knockdown plus ChmC* overexpression. With the non-targeting guide RNA, all three mRNAs were notably depleted in the area occupied by the phage nucleus, suggesting that phage mRNAs are actively translocated out of the nucleus (Figure 6A, B). Upon ChmC knockdown, mRNAs accumulated in the nucleus (Figure 6A, B), and upon ChmC* overexpression mRNAs were again depleted in the nucleus (Figure 6A, B). These data directly implicate ChmC in mRNA translocation from the phage nucleus to the cytoplasm of the infected cell.
Figure 6.
ChmC knockdown causes a global reduction in phage protein levels. (A) Fluorescence microscopy of Goslar-infected E. coli cells expressing dCas13d and a non-targeting guide RNA (Non-targeting; left panels), a chmC-targeting guide RNA (ChmC knockdown; center panels), or a chmC-targeting guide RNA plus a separate recoded chmC gene (ChmC* overexpression; right panels). DNA is stained with DAPI, and Goslar genes encoding gp217, gp53, and gp22 are visualized by mRNA FISH (see Materials and Methods). The position of the phage nucleus, located based on DAPI fluorescence, is indicated with a yellow asterisk in each panel. Each column represents a single cell; six cells are shown per condition. Scale bar = 2 μm. (B) Quantification of mRNA FISH, represented as the relative fluorescence intensity per pixel within the phage nucleus versus the cytoplasm, for mRNAs encoding gp217 (left), gp53 (center), and gp22 (right). At least 20 cells per condition were measured. Individual data points are shown, and bars represent average plus/minus standard deviation. ns: not significant; ***P ≤ 0.001; ****P ≤ 0.0001 (Dunnett's multiple comparisons test). See Supplementary Figure S10A for the same data graphed as fraction of overall fluorescence per cell within the phage nucleus. (C) Waterfall plot showing changes in expression level (log2(fold change)) in ChmC knockdown versus non-targeting conditions, from label-free quantitation mass spectrometry (see Supplementary Table S5 for data). ChmC is shown as a pink dot and labeled. Putative virion structural proteins are shown in red; all other proteins are shown in black. Dotted lines indicate a ±3-fold change (x axis) and a P-value ≤0.05. (D) Log2(fold change) for non-structural proteins (gray; ChmC colored pink and labeled) and putative virion structural proteins (red) when comparing ChmC knockdown to non-targeting conditions. Individual data points are shown, and bars represent average plus/minus standard deviation. Average log2(fold change) for non-structural proteins = –0.44, and for structural proteins = –1.78. See Supplementary Figure S10B for mass spectrometry analysis of Goslar protein expression versus time of infection.
Finally, we tested the effects of ChmC knockdown on global phage protein levels 110 min post-infection using mass spectrometry with label-free quantitation. Overall, we measured the levels of 171 of 247 annotated Goslar proteins in non-targeting versus ChmC knockdown conditions. Of these, 127 proteins showed a reduction in protein levels after ChmC knockdown, and 43 proteins were reduced more than three-fold (Figure 6C, Supplementary Table S5). By contrast, 44 proteins showed increased levels upon ChmC knockdown, with only four proteins showing a more than three-fold increase. Virion structural proteins, which tend to be expressed late in infections (Supplementary Figure S10B, Supplementary Table S6), were more strongly reduced on average than non-structural proteins (Figure 6D).
Discussion
The nucleus-like compartment assembled by chimalliviruses introduces major challenges to the phage life cycle, principally the need to translocate mRNAs out of the phage nucleus and to translocate specific phage proteins into the nucleus. Here, we identify an abundant and early-expressed protein, ChmC, that is conserved across chimalliviruses and is encoded in a conserved block of genes alongside several subunits of the phage's non-virion RNA polymerase. ChmC adopts a nucleic acid binding fold, binds phage mRNAs, and forms condensates with RNA in vitro. PhiPA3 ChmC localizes to the phage nuclear shell, while Goslar ChmC forms puncta that localize both within the phage nucleus and along the nuclear shell. Targeted knockdown of Goslar ChmC results in reduced phage nucleus size and a failure to form phage bouquets in infected cells, mRNA localization defects, a global reduction of phage protein levels, and a dramatic reduction in efficiency of plaquing. Together, these data show that ChmC plays crucial roles in the life cycle of chimalliviruses, likely by promoting mRNA translocation through the nuclear shell.
Our structure predictions and biochemical characterization indicate that ChmC adopts a Whirly domain fold. This fold was first identified in single-stranded DNA/RNA binding proteins in plants where they are involved in transcriptional responses to stress (30,46). In bacteria, Whirly-related proteins are primarily involved in binding and compacting the nucleoid through their non-specific DNA binding activity (47–50). ChmC preferentially binds RNA over DNA in vitro, and shows enriched binding near the start codons of many phage mRNAs. While we do not detect any sequence motifs specifically recognized by ChmC, the protein may nonetheless specifically recognize particular mRNA sequences or structures that determine this binding pattern. Overall, these findings demonstrate that while ChmC is structurally related to other Whirly domain proteins, it has adopted distinct RNA regulatory roles in chimalliviruses.
Many eukaryotic viruses encode RNA binding proteins with diverse roles in the viral life cycle. Many such proteins also form RNA-protein condensates like ChmC (51). Coronaviruses like SARS, MERS, and SARS-CoV-2 encode an RNA-binding nucleocapsid (N) protein that promotes viral RNA production and suppresses host responses through its ability to form RNA-protein condensates, in addition to packaging viral genomic RNA into virions (52). Reminiscent of ChmC’s likely role in mRNA translocation through the nuclear shell, Influenza NEP (nuclear export protein) and HIV Rev are both RNA binding proteins that promote export of viral RNAs produced in the host-cell nucleus into the cytoplasm (53).
Our data show that ChmC is crucial for the proper progression of infection in chimalliviruses, likely by aiding the translocation of phage mRNAs through the nuclear shell to promote translation. In E. coli cells infected with phage Goslar, ChmC forms puncta both within the phage nucleus and along the nuclear shell itself. These puncta are not simply RNA-protein condensates, since shell-associated puncta are also formed by ChmC-ΔC, which cannot form condensates with RNA. We hypothesize that these ChmC puncta may represent hubs for the translocation of mRNAs through the nuclear shell. These hubs are likely built around shell-penetrating pores, potentially the same ChmB pores that mediate capsid docking and genome packaging in late-stage infections (15). The location of chmC within a conserved block of genes encoding nvRNAP subunits further suggests a direct functional link between mRNA transcription and translocation through the nuclear shell, mediated by ChmC. Further work will be required to establish whether ChmB or other proteins are required for this process.
The phage nucleus of chimalliviruses is a fascinating example of convergent evolution, representing a functional analog of the eukaryotic nucleus complete with physical segregation of the genome from the cytoplasm and specific mechanisms for mRNA export and protein import. Our work reveals the first identified RNA binding protein necessary for replication of a DNA-genome bacteriophage, with potential roles in mRNA production, export, and translation to support the unique life cycle of nucleus-forming jumbo phages.
Supplementary Material
Acknowledgements
The authors thank Elizabeth Villa and members of the Corbett and Pogliano labs for helpful discussions.
Contributor Information
Eray Enustun, Department of Molecular Biology, University of California San Diego, La Jolla, CA 92093, USA.
Emily G Armbruster, Department of Molecular Biology, University of California San Diego, La Jolla, CA 92093, USA.
Jina Lee, Department of Molecular Biology, University of California San Diego, La Jolla, CA 92093, USA.
Sitao Zhang, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA.
Brian A Yee, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA.
Kseniya Malukhina, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA.
Yajie Gu, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA.
Amar Deep, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA.
Jack T Naritomi, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA.
Qishan Liang, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA.
Stefan Aigner, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA.
Benjamin A Adler, California Institute for Quantitative Biosciences (QB3), University of California, Berkeley, CA 94720, USA; Innovative Genomics Institute, University of California, Berkeley, CA 94720, USA.
Brady F Cress, Innovative Genomics Institute, University of California, Berkeley, CA 94720, USA.
Jennifer A Doudna, California Institute for Quantitative Biosciences (QB3), University of California, Berkeley, CA 94720, USA; Innovative Genomics Institute, University of California, Berkeley, CA 94720, USA; Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA; Department of Chemistry, University of California, Berkeley, CA 94720, USA; Howard Hughes Medical Institute, University of California, Berkeley, CA 94720, USA; Environmental Genomics and Systems Biology Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA; MBIB Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Vorrapon Chaikeeratisak, Department of Biochemistry, Faculty of Science, Chulalongkorn University, Bangkok 10330, Thailand.
Don W Cleveland, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA; Moores Cancer Center, University of California at San Diego, La Jolla, CA, USA.
Majid Ghassemian, Biomolecular and Proteomics Mass Spectrometry Facility, University of California San Diego, La Jolla, CA 92093, USA.
Bogdan Bintu, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA.
Gene W Yeo, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA; Moores Cancer Center, University of California at San Diego, La Jolla, CA, USA.
Joe Pogliano, Department of Molecular Biology, University of California San Diego, La Jolla, CA 92093, USA.
Kevin D Corbett, Department of Molecular Biology, University of California San Diego, La Jolla, CA 92093, USA; Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA; Moores Cancer Center, University of California at San Diego, La Jolla, CA, USA.
Data availability
Mass spectrometry data is available at the PRIDE repository (https://www.ebi.ac.uk/pride/) with dataset identifier PXD045260, DOI: 10.6019/PXD045260. eCLIP-Seq data is available at the GEO repository (https://www.ncbi.nlm.nih.gov/geo/), with dataset identifier GSE243675.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institutes of Health [R01 GM129245 to J.P., R35 GM144121 to K.D.C., R01 HG004659 to G.W.Y., S10 OD021724 for shared mass spectrometry resources]; Howard Hughes Medical Institute Emerging Pathogens Initiative (to J.P. and K.D.C., in part); J.A.D. is an Investigator of the Howard Hughes Medical Institute; Q.L. was supported by an individual predoctoral fellowship from the American Heart Association; B.A.A. was supported by m-CAFEs Microbial Community Analysis & Functional Evaluation in Soils (m-CAFEs@lbl.gov), a Science Focus Area led by Lawrence Berkeley National Laboratory based upon work supported by the US Department of Energy, Office of Science, Office of Biological & Environmental Research [DE-AC02-05CH11231]; B.F.C. was supported by U.S. Department of Energy, Office of Science, through the Genomic Science Program, Office of Biological and Environmental Research, under the Secure Biosystems Design Initiative project Intrinsic Control for Genome and Transcriptome Editing in Communities (InCoGenTEC). Funding for open access charge: Howard Hughes Medical Institute [Emerging Pathogens Initiative to K.D.C., J.P.].
Conflict of interest statement. J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, Scribe Therapeutics, Intellia Therapeutics, and Mammoth Biosciences. J.A.D. is a scientific advisory board member of Vertex, Caribou Biosciences, Intellia Therapeutics, Scribe Therapeutics, Mammoth Biosciences, Algen Biotechnologies, Felix Biosciences, The Column Group and Inari. J.A.D. is Chief Science Advisor to Sixth Street, a Director at Johnson & Johnson, Altos and Tempus, and has research projects sponsored by Apple Tree Partners and Roche. G.W.Y. is an SAB member of Jumpcode Genomics and a co-founder, member of the Board of Directors, on the SAB, equity holder, and paid consultant for Locanabio and Eclipse BioInnovations. G.W.Y. is a distinguished visiting professor at the National University of Singapore. G.W.Y.’s interests have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. J.P. has an equity interest in Linnaeus Bioscience Incorporated and receives income. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict-of-interest policies. The Regents of the University of California have filed a provisional patent application for CRISPR technologies on which B.A.A., B.F.C., E.J.A., J.L., J.A.P. are J.A.D. are inventors.
References
- 1. Hampton H.G., Watson B.N.J., Fineran P.C.. The arms race between bacteria and their phage foes. Nature. 2020; 577:327–336. [DOI] [PubMed] [Google Scholar]
- 2. Stanley S.Y., Maxwell K.L.. Phage-encoded anti-CRISPR defenses. Annu. Rev. Genet. 2018; 52:445–464. [DOI] [PubMed] [Google Scholar]
- 3. Prichard A., Lee J., Laughlin T.G., Lee A., Thomas K.P., Sy A., Spencer T., Asavavimol A., Cafferata A., Cameron M.et al.. Identifying the core genome of the nucleus-forming bacteriophage family and characterization of Erwinia phage RAY. Cell Rep. 2023; 42:112432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chaikeeratisak V., Nguyen K., Khanna K., Brilot A.F., Erb M.L., Coker J.K.C., Vavilina A., Newton G.L., Buschauer R., Pogliano K.et al.. Assembly of a nucleus-like structure during viral replication in bacteria. Science. 2017; 355:194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaikeeratisak V., Nguyen K., Egan M.E., Erb M.L., Vavilina A., Pogliano J.. The phage nucleus and tubulin spindle are conserved among large Pseudomonas phages. Cell Rep. 2017; 20:1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knipe D.M., Prichard A., Sharma S., Pogliano J.. Replication compartments of eukaryotic and bacterial DNA viruses: common themes between different domains of host cells. Annu. Rev. Virol. 2022; 9:307–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laughlin T.G., Deep A., Prichard A.M., Seitz C., Gu Y., Enustun E., Suslov S., Khanna K., Birkholz E.A., Armbruster E.et al.. Architecture and self-assembly of the jumbo bacteriophage nuclear shell. Nature. 2022; 608:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mendoza S.D., Nieweglowska E.S., Govindarajan S., Leon L.M., Berry J.D., Tiwari A., Chaikeeratisak V., Pogliano J., Agard D.A., Bondy-Denomy J.. A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nature. 2020; 577:244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Malone L.M., Warring S.L., Jackson S.A., Warnecke C., Gardner P.P., Gumy L.F., Fineran P.C.. A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat. Microbiol. 2020; 5:48–55. [DOI] [PubMed] [Google Scholar]
- 10. Nieweglowska E.S., Brilot A.F., Méndez-Moran M., Kokontis C., Baek M., Li J., Cheng Y., Baker D., Bondy-Denomy J., Agard D.A.. The ϕPA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice. Nat. Commun. 2023; 14:927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guan J., Oromí-Bosch A., Mendoza S.D., Karambelkar S., Berry J.D., Bondy-Denomy J.. Bacteriophage genome engineering with CRISPR-Cas13a. Nat. Microbiol. 2022; 7:1956–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mayo-Muñoz D., Smith L.M., Garcia-Doval C., Malone L.M., Harding K.R., Jackson S.A., Hampton H.G., Fagerlund R.D., Gumy L.F., Fineran P.C.. Type III CRISPR-Cas provides resistance against nucleus-forming jumbo phages via abortive infection. Mol. Cell. 2022; 82:4471–4486. [DOI] [PubMed] [Google Scholar]
- 13. Vorrapon C., Kanika K., Nguyen Katrina T., Egan MacKennon E., Eray E., Emily A., Jina L., Kit P., Elizabeth V., Joe P.. Subcellular organization of viral particles during maturation of nucleus-forming jumbo phage. Sci. Adv. 2022; 8:eabj9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Birkholz E.A., Laughlin T.G., Armbruster E., Suslov S., Lee J., Wittmann J., Corbett K.D., Villa E., Pogliano J.. A cytoskeletal vortex drives phage nucleus rotation during jumbo phage replication in E. coli. Cell Rep. 2022; 40:111179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Enustun E., Deep A., Gu Y., Nguyen K.T., Chaikeeratisak V., Armbruster E., Ghassemian M., Villa E., Pogliano J., Corbett K.D.. Identification of the bacteriophage nucleus protein interaction network. Nat. Struct. Mol. Biol. 2023; 30:1653–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B.et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A.et al.. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Evans R., O’Neill M., Pritzel A., Antropova N., Senior A., Green T., Žídek A., Bates R., Blackwell S., Yim J.et al.. Protein complex prediction with AlphaFold-Multimer. 2022; bioRxiv doi:04 October 2021, preprint: not peer reviewed 10.1101/2021.10.04.463034. [DOI]
- 19. Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M.. ColabFold: making protein folding accessible to all. Nat. Methods. 2022; 19:679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adler B.A., Al-Shimary M.J., Patel J.R., Armbruster E.G., Colognori D., Charles E.J., Miller K.V., Lahiri A., Trinidad M.T., Boger R.et al.. Genome-wide charachterization of diverse bacteriophages enabled by RNA-binding CRISPRi. 2023; bioRxiv doi:18 September 2023, preprint: not peer reviewed 10.1101/2023.09.18.558157. [DOI]
- 21. Armbruster E.G., Lee J., Hutchings J., VanderWal A.R., Enustun E., Adler B.A., Aindow A., Deep A., Rodriguez Z.K., Morgan C.J.et al.. Sequential membrane- and protein-bound organelles compartmentalize genomes during phage infection. 2023; bioRxiv doi:21 September 2023, preprint: not peer reviewed 10.1101/2023.09.20.558163. [DOI]
- 22. Blue S.M., Yee B.A., Pratt G.A., Mueller J.R., Park S.S., Shishkin A.A., Starner A.C., Van Nostrand E.L., Yeo G.W.. Transcriptome-wide identification of RNA-binding protein binding sites using seCLIP-seq. Nat. Protoc. 2022; 17:1223–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lovci M.T., Ghanem D., Marr H., Arnold J., Gee S., Parra M., Liang T.Y., Stark T.J., Gehman L.T., Hoon S.et al.. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol. 2013; 20:1434–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Q., Brown J.B., Huang H., Bickel P.J.. Measuring reproducibility of high-throughput experiments. Aoas. 2011; 5:1752–1779. [Google Scholar]
- 25. Yee B.A., Pratt G.A., Graveley B.R., Van Nostrand E.L., Yeo G.W.. RBP-Maps enables robust generation of splicing regulatory maps. RNA. 2019; 25:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Su J.-H., Zheng P., Kinrot S.S., Bintu B., Zhuang X.. Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell. 2020; 182:1641–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ahlers J., Althviz Moré D., Amsalem O., Anderson A., Bokota G., Boone P., Bragantini J., Buckley G., Burt A., Bussonnier M.et al.. napari: a multi-dimensional image viewer for Python Zenodo. 2023;
- 28. Graebsch A., Roche S., Niessing D. X-ray structure of Pur-α reveals a Whirly-like fold and an unusual nucleic-acid binding surface. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:18521–18526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schumacher M.A., Karamooz E., Zíková A., Trantírek L., Lukes J.. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006; 126:701–711. [DOI] [PubMed] [Google Scholar]
- 30. Desveaux D., Allard J., Brisson N., Sygusch J.. A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat. Struct. Biol. 2002; 9:512–517. [DOI] [PubMed] [Google Scholar]
- 31. Mitrea D.M., Cika J.A., Guy C.S., Ban D., Banerjee P.R., Stanley C.B., Nourse A., Deniz A.A., Kriwacki R.W.. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife. 2016; 5:e13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pak C.W., Kosno M., Holehouse A.S., Padrick S.B., Mittal A., Ali R., Yunus A.A., Liu D.R., Pappu R.V., Rosen M.K.. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell. 2016; 63:72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zaharias S., Zhang Z., Davis K., Fargason T., Cashman D., Yu T., Zhang J.. Intrinsically disordered electronegative clusters improve stability and binding specificity of RNA-binding proteins. J. Biol. Chem. 2021; 297:100945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chu X., Sun T., Li Q., Xu Y., Zhang Z., Lai L., Pei J.. 2022) Prediction of liquid–liquid phase separating proteins using machine learning. BMC Bioinf. 23:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bolognesi B., Lorenzo Gotor N., Dhar R., Cirillo D., Baldrighi M., Tartaglia G.G., Lehner B.. A concentration-dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep. 2016; 16:222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Azaldegui C.A., Vecchiarelli A.G., Biteen J.S.. The emergence of phase separation as an organizing principle in bacteria. Biophys. J. 2021; 120:1123–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nandana V., Schrader J.M.. Roles of liquid–liquid phase separation in bacterial RNA metabolism. Curr. Opin. Microbiol. 2021; 61:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin Y., Protter D.S.W., Rosen M.K., Parker R.. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 2015; 60:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmidt H.B., Görlich D. Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem. Sci. 2016; 41:46–61. [DOI] [PubMed] [Google Scholar]
- 40. Chong P.A., Vernon R.M., Forman-Kay J.D.. RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 2018; 430:4650–4665. [DOI] [PubMed] [Google Scholar]
- 41. deYMartín Garrido N., Orekhova M., Lai Wan Loong Y.T.E., Litvinova A., Ramlaul K., Artamonova T., Melnikov A.S., Serdobintsev P., Aylett C.H.S., Yakunina M.. Structure of the bacteriophage PhiKZ non-virion RNA polymerase. Nucleic Acids Res. 2021; 49:7732–7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yakunina M., Artamonova T., Borukhov S., Makarova K.S., Severinov K., Minakhin L.. A non-canonical multisubunit RNA polymerase encoded by a giant bacteriophage. Nucleic Acids Res. 2015; 43:10411–10420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Iyer L., Anantharaman V., Krishnan A., Burroughs A.M., Aravind L.. Jumbo phages: a comparative genomic overview of core functions and adaptions for biological conflicts. Viruses. 2021; 13:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ceyssens P.-J., Minakhin L., Van den Bossche A., Yakunina M., Klimuk E., Blasdel B., De Smet J., Noben J.-P., Bläsi U., Severinov K.et al.. Development of giant bacteriophage ϕKZ is independent of the host transcription apparatus. J. Virol. 2014; 88:10501–10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wicke L., Ponath F., Coppens L., Gerovac M., Lavigne R., Vogel J.. Introducing differential RNA-seq mapping to track the early infection phase for Pseudomonas phage φKZ. RNA Biol. 2021; 18:1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krupinska K., Desel C., Frank S., Hensel G.. WHIRLIES are multifunctional DNA-binding proteins with impact on plant development and stress resistance. Front. Plant Sci. 2022; 13:880423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luijsterburg M.S., Noom M.C., Wuite G.J.L., Dame R.T.. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 2006; 156:262–272. [DOI] [PubMed] [Google Scholar]
- 48. Dillon S.C., Dorman C.J.. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010; 8:185–195. [DOI] [PubMed] [Google Scholar]
- 49. Hołówka J., Zakrzewska-Czerwińska J.. Nucleoid associated proteins: the small organizers that help to cope with stress. Front. Microbiol. 2020; 11:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Prikryl J., Watkins K.P., Friso G., van Wijk K.J., Barkan A.. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 2008; 36:5152–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brocca S., Grandori R., Longhi S., Uversky V.. Liquid–liquid phase separation by intrinsically disordered protein regions of viruses: roles in viral life cycle and control of virus–host interactions. Int. J. Mol. Sci. 2020; 21:9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ye Q., Lu S., Corbett K.D.. Structural basis for SARS-CoV-2 nucleocapsid protein recognition by single-domain antibodies. Front. Immunol. 2021; 12:719037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gales J.P., Kubina J., Geldreich A., Dimitrova M.. Strength in diversity: nuclear export of viral RNAs. Viruses. 2020; 12:1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data is available at the PRIDE repository (https://www.ebi.ac.uk/pride/) with dataset identifier PXD045260, DOI: 10.6019/PXD045260. eCLIP-Seq data is available at the GEO repository (https://www.ncbi.nlm.nih.gov/geo/), with dataset identifier GSE243675.