Abstract
Background:
Cancer cells exhibit selective metabolic reprogramming to promote proliferation, invasiveness, and metastasis. Sphingolipids such as sphingosine and sphinganine have been reported to modulate cell death processes in cancer cells. However, the potential of extracellular sphinganine and its mimetic compounds as inducers of cancer cell death has not been thoroughly investigated.
Methods:
We obtained extracellular conditioned medium from HCT-116 cells treated with the previously reported anticancer composition, goat urine DMSO fraction (GUDF). The extracellular metabolites were purified using a novel and in-house developed vertical tube gel electrophoresis (VTGE) technique and identified through LC-HRMS. Extracellular metabolites such as sphinganine, sphingosine, C16 sphinganine, and phytosphingosine were screened for their inhibitory role against intracellular kinases using molecular docking. Molecular dynamics (MD) simulations were performed to study the inhibitory potential of a novel designed modified mimetic sphinganine (MMS) (Pubchem CID: 162625115) upon c-Src kinase. Furthermore, inhibitory potential and ADME profile of MMS was compared with luteolin, a known c-Src kinase inhibitor.
Results:
Data showed accumulation of sphinganine and other sphingolipids such as C16 sphinganine, phytosphingosine, and ceramide (d18:1/14:0) in the extracellular compartment of GUDF-treated HCT-116 cells. Molecular docking projected c-Src kinase as an inhibitory target of sphinganine. MD simulations projected MMS with strong (-7.1 kcal/mol) and specific (MET341, ASP404) binding to the inhibitory pocket of c-Src kinase. The projected MMS showed comparable inhibitory role and acceptable ADME profile over known inhibitors.
Conclusion:
In summary, our findings highlight the significance of extracellular sphinganine and other sphingolipids, including C16 sphinganine, phytosphingosine, and ceramide (d18:1/14:0), in the context of drug-induced cell death in HCT-116 cancer cells. Furthermore, we demonstrated the importance of extracellular sphinganine and its modified mimetic sphinganine (MMS) as a potential inhibitor of c-Src kinase. These findings suggest that MMS holds promise for future applications in targeted and combinatorial anticancer therapy.
Key Words: Sphingolipids, colon cancer, cell death, tyrosine kinase, inhibitors, MD simulations
Introduction
Colorectal cancer is one of the leading cancer types at 4th position in terms of incidence and 3rd in terms of mortality [1]. Cancer cells are known for uncontrolled cell proliferation which is due to the virtue of aberrant signal transduction [2, 3]. Deregulated metabolic adaptations and altered growth signaling pathways are considered crucial hallmarks of various types of tumors including colon cancer.
Different types of oncogenes and their associated cellular signaling pathways contribute to the alteration of cellular and energy metabolism which is one of the important attributes of cancer cells [3, 4]. Tumor display reprogrammed metabolic activities that promote cancer progression and drug resistance. Metabolic heterogeneity is a critical factor in the success and failure of anticancer therapies [2- 5].
Many human diseases, including cancer, result from an alteration in lipid metabolic enzymes and their pathways including sphingolipids [6, 7]. The changes in the fate and composition of sphingolipids of cancer cells in the microenvironment influence oncogenic functions, signaling, and drug resistance [8-10].
The metabolomics study on intracellular and extracellular compartments of cancer cells treated by drugs is pivotal to understanding the metabolic adaptations in cancer cells leading to drug resistance [11-16]. At the same time, data on extracellular metabolite profiling of sphingolipids in colon cancer cells treated by drugs is highly limited specifically in the context of modulation of the c-Src kinase [17, 18].
Limited data hinted at the involvement of intracellular and extracellular sphinganine in the apoptotic cell death of cancer cells [19-25]. Due to a lack of suitable methodologies, evidence of the abundance of sphinganine and other sphingolipid bases in colon cancer cells treated by anticancer drugs is limited. Among several prominent intracellular signal transducer kinases, the c-Src kinase is a crucial tyrosine kinase that controls the growth and proliferation of various types of cancer including colon cancer cells [26-29]. On the contrary, c-Src kinase contributes to the evasion of apoptosis and in turn drug resistance [30-33]. However, the molecular link between sphingolipid bases such as sphinganine and sphingosine with the modulation of the c-Src kinase is highly lacking and specifically in the context of colon cancer cells and apoptotic cell death. In recent, the small molecular inhibitors of c-Src kinase and other intracellular kinases are conceived as a new class of anticancer drugs to attenuate proliferation and induce cell death in cancer cells including colon cancer [34-38]
Additionally, the mimetic class of anticancer drugs as inhibitors of intracellular signal transducers such as c-Src kinase appears to be a safe and feasible avenue in cancer therapies [39-42]. However, the extrapolation of understanding sphingolipid metabolites in the context of the development of inhibitors of c-Src kinase as a modified mimetic of sphinganine is highly scarce.
In our study, we utilized a novel and in-house VTGE methodology to assess the levels of extracellular sphingolipids, including sphinganine, in HCT-116 cancer cells treated with anticancer compositions. Additionally, we employed molecular docking and MD simulations to evaluate the relevance of sphinganine and its modified mimetic form (MMS) as inhibitors of c-Src kinase.
Materials and Methods
Materials
Cell culture reagents were purchased from Invitrogen India Pvt. Ltd, Bangalore, India. and Himedia Laboratories Pvt. Ltd, Mumbai, India. HCT-116 colon cancer cells were procured from the National Centre of Cell Science (NCCS) Pune, India. DMSO, pBR322 DNA, agarose, acrylamide, and other chemicals were of analytical grade and obtained from Himedia Laboratories Pvt. Ltd. Mumbai, India and Merck India Pvt. Ltd, Mumbai, India.
Preparation of extracellular conditioned medium
HCT-116 colon cancer cells were cultured and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) with high glucose supplemented with 10% heat-inactivated FBS/penicillin (100 units/ml)/streptomycin (100 µg/ml) at 37°C in a humidified 5% CO2 incubator. Further, 60-70% of confluent HCT-116 cells were harvested and plated onto a six-well plate at a seeding density of 1.5 X 105 cells per well in a complete DMEM medium. After 16-18 h overnight growth, HCT-116 cells were treated with goat urine DMSO fraction (GUDF) (50 µg/ml) final concentration. In brief, GUDF was prepared in DMSO solvent by using standardized steps including vortexing, centrifugation, and filtration by 0.45-micron syringe filter membrane to ensure that GUDF is sterile and compatible with cell culture. This was adopted from a previously published procedure by our lab [43-46]. Next, treatment by GUDF and DMSO control was allowed for 72 h. At the end of incubation, HCT-116 cells were harvested with the help of a routine procedure and evaluated for loss of proliferation and viability due to GUDF by using a Trypan blue dye exclusion assay. We observed by phase contrast microscopy to assess alterations in the morphology of HCT-116 cells due to GUDF. To substantiate the elaborated effects of GUDF upon HCT-116 cells including cell death has been previously published by our lab. However, the importance of this paper is an extension to the previous observations by exploring metabolites in the extracellular conditioned medium of HCT-116 cells treated by GUDF. Therefore, the extracellular conditioned medium of GUDF and DMSO-treated HCT-116 cells were collected in separate sterile tubes and processed by centrifugation to ensure that viable and dead cells are not left in the tube so that only the extracellular nature of metabolites will be analyzed [43- 46].
Extracellular metabolite profiling
The above collected extracellular conditioned medium of HCT-116 cells treated by DMSO and GUDF (750 µl) was mixed with 4X loading buffer (0.5 M Tris, pH 6.8, and Glycerol). Next, the conditioned medium along with the loading buffer was loaded on the vertical tube gel electrophoresis (VTGE) purification system (Figure S1) with a matrix of 15% acrylamide gel (acrylamide: bisacrylamide, 30:1). The fractionated extracellular metabolites were collected in the running buffer (96 mM glycine of pH at 8.3). The detailed procedure was adopted from previously published in-house VTGE-assisted purification of metabolites [44, 43, 45, 46]. Furthermore, LC-HRMS analyses of VTGE-purified extracellular metabolite elutes were performed by Agilent TOF/Q-TOF Mass Spectrometer station Dual AJS ESI ion source. During LC separation, RPC18 Hypersil GOLD C18 100 x 2.1 mm-3 µm and mobile phase of 100% Water (0.1% FA in water) and 100% Acetonitrile (90% ACN +10% H2O+ 0.1% FA) were used in the proportion of 95% and 5% [39]. Mass spectrometry was performed in a positive mode and analyzed as per the procedure adopted from previously reported methodology [43, 46].
Molecular Docking
We identified selective sphingolipids such as sphinganine, hexadecasphinganine, and phytosphingosine in the extracellular compartment of HCT-116 cells treated by GUDF over DMSO control. To explore the biological relevance of sphingolipids including sphinganine in the intracellular signaling pathways, we started to explore the potential targets of sphinganine and other sphingolipids using molecular docking.
Potential intracellular sphingolipids including sphinganine (PubChem CID: 91486), sphingosine (PubChem CID: 5280335), phytosphingosine (PubChem CID: 5281120), hexadecasphinganine (PubChem CID: 656816), modified mimetic sphinganine (MMS) (PubChem CID: 162625115) and known c-Src kinase inhibitors such as luteolin (PubChem CID: 5280445 and PP2 (PubChem CID: 4878) were retrieved from the PubChem database and downloaded in SDF format. Before performing molecular docking, all ligands were energy minimized to obtain stable conformation using Maestro software with the steepest descent method and MMFF94s force field. In this study, potential protein targets including c-Src kinase (2SRC), MAPK phosphatase Pyst1 (1MKP), PKB kinase domain (1GZN), ERK1 (6GES), and BRAF-MEK1-14-3-3 (6Q0K) and others enzymes were downloaded from the PDB database. The preparation of proteins and molecular docking process by AutoDock Vina software was adopted from the standardized protocol [47, 48]. AutoDock Vina comes with the feature of calculating grid maps automatically. Confirmation of the binding position of the potential inhibitory sphingolipids and known inhibitors into the cavity of the c-Src kinase and the calculation of binding parameters such as bond distance was done by Discovery Studio Visualizer3 (DSV3) [49].
Molecular Dynamics (MD) Simulations
The 20ns MD simulations for the complexes of sphinganine (PubChem CID: 91486), and luteolin (PubChem CID: 5280445), a known inhibitor with c-Src kinase (PDB ID: 2SRC), were performed with the help of Desmond software to confirm the binding stability and strength of the complex. Desmond has inbuilt functions to add pressure, volume system, temperature, and many functionalities to accomplish protein-ligand binding. The first complex between sphinganine and c-Src kinase was immersed in 9071 and the second was 7350 TIP3P explicit water molecules respectively within a cubic box of 10 Å spacing using periodic boundary conditions. MD simulation study was carried out with a run of 20ns at a temperature of 300K in considering certain parameters such as integrator as MD. The conformational changes upon binding of sphinganine with c-Src kinase were recorded in 1000 trajectory frames generated during the 20ns MD simulation. Root Mean Square Deviation (RMSD) was calculated to reveal the binding stability of sphinganine and luteolin with c-Src complexes. The steepest descent method was used to minimize the complex system energetically with the OPLS-2005 force field. The 20ns time scale MD simulations for each complex were performed at constant NPT (N=Number of Atoms, P=Pressure, and T=Temperature) ensemble. Throughout the equilibrations, systems were coupled with the Martyna-Tobias-Klein barostat method for controlling pressure at 1 atm, and the temperature was regulated by using velocity rescaling Nose Hoover chain thermostat method at 300 K. The M-SHAKE algorithm was used to constrain the bond length of hydrogen atoms. Rest steps were adopted as per previously published procedure [43- 48].
ADMET profiling
Sphinganine, sphingosine, MMS, and known c-Src kinase inhibitors such as luteolin and PP2 were tested for drug-induced liver injury, cell toxicity, carcinogenicity, maximal recommended therapeutic dose (MRTD), and druglikeness using vNN-ADMET and SWISS-ADMET web servers [50, 51].
Statistical Analysis
Data are presented as the mean ± SD of at least three independent experiments. Differences are considered statistically significant at P < 0.05, using a Student’s t-test.
Results
Extracellular sphingolipid profiling
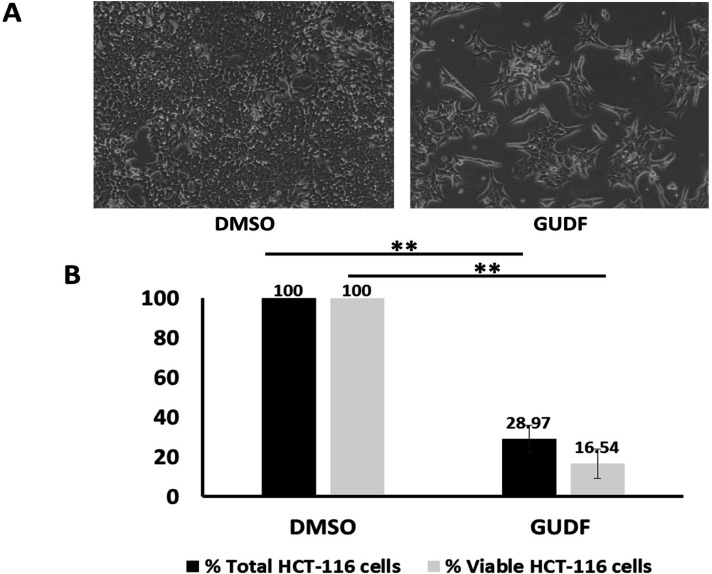
Hence, we selected HCT-116 colon cancer cells as a model and treated them with goat urine DMSO fraction (GUDF) anticancer compositions enriched with tripeptides. This paper is incremental evidence from previous data that we have shown the proliferative arrest and apoptotic cell death in colon cancer cells by GUDF. In line with our previous findings, GUDF showed a significant reduction of up to 28.97% of total HCT-116 cells normalized over DMSO control. At the same time, GUDF induced loss of HCT-116 cell viability up to 16.54% compared to DMSO control (Figure 1B). Microscopy photographs of GUDF-treated HCT-116 cells indicated a very clear reduction in total cells and cell death of HCT-116 cells in comparison with DMSO control (Figure 1A). Besides these observations, we have previously reported effects of GUDF leading to the induction of apoptotic cell death in HCT-116 cells [43- 46]. Hence, we selected GUDF-treated HCT-116 cells as one of the potential sources of profiling of extracellular metabolites during cell death in the case of HCT-116 cells. However, an argument is possible regarding the scope of other cancer cell types and other forms of drugs.
Figure 1.
GUDF Induces Proliferative Arrest and Cell Death in HCT-116 Colon Cancer Cells. (A) HCT-116 colon cancer cells were treated by DMSO and GUDF (50 µg/ml) for 72 hr. Routine microscopy was performed at 100X to observe the cell number and cellular morphology. (B) HCT-116 colon cancer cells were treated by DMSO and GUDF (50 µg/ml) for 72 hr. Percentage total cell and viable HCT-116 colon cancer cells estimated by Trypan blue dye exclusion assay. Trypan blue dye exclusion assay was performed to estimate viable and dead cells. Data are represented as mean ± SD. Each experiment was conducted independently three times. The bar graph without an asterisk denotes that there is no significant difference compared to the DMSO control. * Significantly different from DMSO control at the P-value < 0.05. ** Significantly different from DMSO control at P-value < 0.01. *** Significantly different from DMSO control at P-value < 0.001
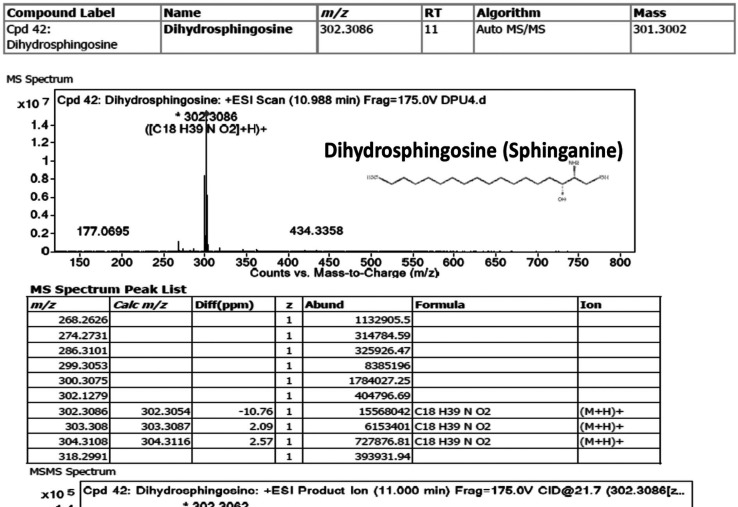
Since objectives of this study are to study the biological relevance of sphingolipids in the extracellular components of HCT-116 cells and then extrapolation of specific sphingolipids such as sphinganine as a mimetic class of inhibitors of intracellular kinases such as c-Src enzymes. In this direction, we employed a novel and in-house developed VTGE-assisted approach to purify metabolite components of the extracellular conditioned medium of HCT-116 cells treated by both GUDF and DMSO. We found a significant abundance of specific forms of sphingolipid bases such as sphinganine (dihydrosphingosine) (RT-11, mass-301.3002, chemical formula-C18 H39 N O2), C16 sphinganine (hexadecasphinganine) (RT-9.65, mass-273.2669, chemical formula-C16 H35 N O2), phytosphingosine (RT-9.995, mass-317.292, chemical formula-C18 H39 N O3) and ceramide (d18:1/14:0) (RT-19.819, mass-509.4777, chemical formula-C32 H63 N O3) in case of GUDF treated HCT-116 cells (Table 1). At the same time, the levels of sphinganine, C16 sphinganine, phytosphingosine, and ceramide (d18:1/14:0) are found to be undetectable in the extracellular conditioned medium of DMSO-treated HCT-116 cells. LC-HRMS based MS/MS showed specific positive ESI product ions of these sphingolipids such as sphinganine (m/z 302.3086, 406.3483, 675.4956), C16 sphinganine (m/z 274.2745, 688.4899, 918.8047), phytosphingosine (m/z 150.1122, 256.2632) and Ceramide (d18:1/14:0) (m/z 256.2626, 512.4994, 905.6919) (Figure 2, Figure S2, S3, S4, respectively). The abundance characteristics and matching score of these sphingolipids in the extracellular compartment of GUDF-treated HCT-116 cells are substantial in comparison to DMSO control.
Table 1.
List of Sphingolipids Detected in the Extracellular Conditioned Medium of HCT-116 Cells Treated by GUDF and DMSO. Here, extracellular conditioned medium of HCT-116 was first purified by VTGE tool and later analyzed by LC-HRMS in positive ESI mode
| Name of Sphingolipids | RT | Mass | Abundance | Chemical formula | DB Diff (ppm) |
|---|---|---|---|---|---|
| Extracellular conditioned medium of HCT-116 cells treated by GUDF | |||||
| Dihydrosphingosine (Sphinganine) | 11 | 301.3002 | 15568042 | C18 H39 N O2 | -6.87 |
| C16 Sphinganine | 9.65 | 273.2669 | 8575711 | C16 H35 N O2 | -0.14 |
| Phytosphingosine | 9.995 | 317.292 | Detectable | C18 H39 N O3 | 0.35 |
| Ceramide (d18:1/14:0) | 19.819 | 509.4777 | Detectable | C32 H63 N O3 | 4.96 |
| Extracellular conditioned medium of HCT-116 cells treated by DMSO | |||||
| C16 Sphinganine | Undetectable | ||||
| Phytosphingosine | Undetectable | ||||
| Dihydrosphingosine | Undetectable | ||||
| C14 Ceramide (d18:1/14:0) | Undetectable | ||||
Figure 2.
Sphinganine is Secreted and Accumulated in HCT-116 Colon Cancer Cells Treated by GUDF Positive Mode ESI MS and MS/MS fragment ion spectra of sphinganine in GUDF treated HCT-116 cancer cells. Sphinganine is known as a sphingolipid base
Molecular docking
In our observations, we significantly found an abundance of sphingolipids such as sphinganine in the extracellular compartment of HCT-116 cells displaying drug-induced cell death. Hence, we asked questions in terms of intracellular and extracellular adaptations that could contribute to the export of sphinganine, but sphingosine was not detected. However, literature suggested the opposite effects of sphinganine over sphingosine and the role of sphinganine in cancer cell death.
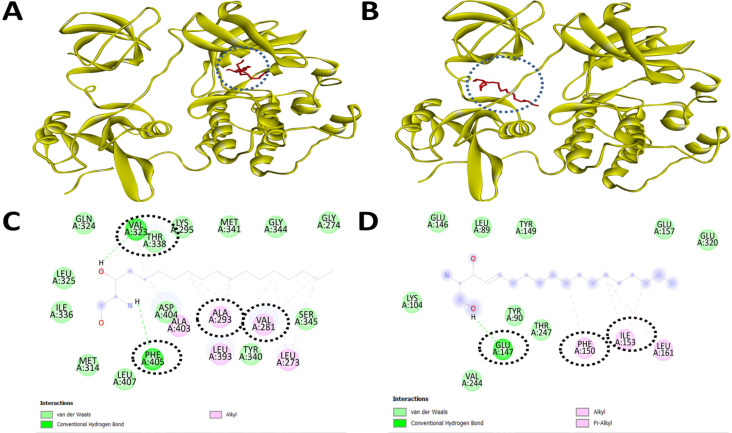
Therefore, we started the initial screening of potential intracellular kinases including c-Src kinase which can be a potential target for inhibition of activation by these forms of sphingolipids. Based on the molecular docking data, we found interesting observations that sphingolipids such as sphinganine and sphingosine bind to c-Src kinase appreciably over other selected intracellular signal transducer proteins (Table 2). Detailed analysis of binding pockets of sphinganine and sphingosine with c-Src kinase revealed contrasting amino acid residues such as inhibitory pocket residues (ALA293, LYS295, VAL323, THR338, TYR340, MET341, ASP404, PHE405) and activator site residues (TYR90, GLU147, PHE150, ILE151, LEU161) (Figure 3). Such molecular data on the differential binding of sphinganine and sphingosine with c-Src kinase prompted the exploration of detailed comparison with known inhibitors of c-Src kinase in terms of binding affinity and interacting amino acid residues. Visualization of the inhibitory binding pocket of sphinganine (ALA293, LYS295, VAL323, THR338, TYR340, MET341, ASP404, PHE405) showed a similar binding pocket compared to a known c-Src kinase inhibitor luteolin and PP2 (Table 3, and Figure 4).
Table 2.
Molecular Docking based Screening of Potential Inhibitory Protein Targets of Sphingolipids Detected in the Extracellular Compartment of Cancer Cells Treated by Anticancer Drug. Molecular docking is performed by AutoDock Vina
| Sr. No | Target Proteins (PDB ID) | Sphinglipids (PubChem ID) | Binding energy (-kcal/mol) | RMSD value l.b. |
RMSD value u.b |
|---|---|---|---|---|---|
| 1 | c-Src kinase (2SRC) | Sphingosine (PubChem ID: 528035) | -5.7 | 0 | 0 |
| 2 | c-Src kinase (2SRC) | Sphinganine (PubChem ID: 91486) | -5.6 | 0 | 0 |
| 3 | c-Src kinase (2SRC) | Phytospingosine (PubChem ID: 122121) | -5.5 | 0 | 0 |
| 4 | c-Src kinase (2SRC) | Hexadecasphinganine (PubChem ID: 656816) | -5.3 | 0 | 0 |
| 5 | MAPK phosphatase Pyst1 (1MKP) | Sphingosine (PubChem ID: 528035) | -3.9 | 0 | 0 |
| 6 | MAPK phosphatase Pyst1 (1MKP) | Sphinganine (PubChem ID: 91486) | -3.9 | 0 | 0 |
| 7 | MAPK phosphatase Pyst1 (1MKP) | Phytospingosine (PubChem ID: 122121) | -3.4 | 0 | 0 |
| 8 | MAPK phosphatase Pyst1 (1MKP) | Hexadecasphinganine (PubChem ID: 656816) | -3.2 | 0 | 0 |
| 9 | PKB kinase domain (1GZN) | Sphingosine (PubChem ID: 528035) | 0 | 0 | 0 |
| 10 | PKB kinase domain (1GZN) | Sphinganine (PubChem ID: 91486) | 0 | 0 | 0 |
| 11 | PKB kinase domain (1GZN) | Phytospingosine (PubChem ID: 122121) | 0 | 0 | 0 |
| 12 | PKB kinase domain (1GZN) | Hexadecasphinganine (PubChem ID: 656816) | 0 | 0 | 0 |
| 13 | ERK1 (6GES) | Sphingosine (PubChem ID: 528035) | 0 | 0 | 0 |
| 14 | ERK1 (6GES) | Sphinganine (PubChem ID: 91486) | 0 | 0 | 0 |
| 15 | ERK1 (6GES) | Phytospingosine (PubChem ID: 122121) | 0 | 0 | 0 |
| 16 | ERK1 (6GES) | Hexadecasphinganine (PubChem ID: 656816) | 0 | 0 | 0 |
| 17 | BRAF-MEK1-14-3-3 (6Q0K) | Sphingosine (PubChem ID: 528035) | 0 | 0 | 0 |
| 18 | BRAF-MEK1-14-3-3 (6Q0K) | Sphinganine (PubChem ID: 91486) | 0 | 0 | 0 |
| 19 | BRAF-MEK1-14-3-3 (6Q0K) | Phytospingosine (PubChem ID: 122121) | 0 | 0 | 0 |
| 20 | BRAF-MEK1-14-3-3 (6Q0K) | Hexadecasphinganine (PubChem ID: 656816) | 0 | 0 | 0 |
Figure 3.
Sphinganine Displays Strong binding to the Inhibitory Site of the c-Src Kinase, while Sphingosine Showed Binding to the Active Site of the c-Src Kinase. Molecular docking and inhibitory interaction by sphinganine and sphingosine upon c-Src kinase with the help of Autodock Vina. (A). A ribbon structure with a full 3D view between sphinganine and c-Src kinase. (B). A ribbon structure with a full 3D view between sphingosine and c-Src kinase. (C). Discovery Studio Visualizer assisted 2-D image of docked molecular structure between sphinganine and c-Src kinase. (D). Discovery Studio Visualizer assisted 2-D image of docked molecular structure between sphingosine and c-Src kinase
Table 3.
Molecular Docking and Visualization of Complexes of Sphinganine, Sphingosine, Modified Mimetic Sphinganine (MMS), PP2 (A known c-Src kinase inhibitor) and Luteolin (A known c-Src kinase inhibitor) with c-Src Kinase (PDB ID: 2SRC). Visualization of complexed were done by Discovery Studio Visualizer 3 (DSV3)
| Name of Protein and its ligand | Maximum binding affinity (-kcal/mol) | Binding Residues | No. of Bonds (H-Bonds, van der waals, Alkyl) | Bond Distances (Å) |
|---|---|---|---|---|
| Sphinganine | -5.7 | ALA293, LYS295, VAL323, THR338, TYR340, MET341, ASP404, PHE405 | 8 | 3.16, 2.96, 3.06, 2.86, 2.57, 3.25 2.98. 2.53 |
| Sphingosine | -5.6 | TYR90, GLU147, PHE150, ILE151, LEU161 | 5 | 3.65, 2.65 3.62, 3.65 3.43 |
| modified mimetic sphinganine (MMS) (PubChem CID: 162625115) | 7.1 | PHE278, VAL281, MET341, LEU407, ALA408, TYR416 | 6 | 3.75, 3.62 2.89, 3.92 3.56, 2.86 |
| PP2 (A known c-Src kinase inhibitor) PubChem CID: 4878 | -8.6 | VAL281, VAL323, THR338, GLU339, MET341, LEU393, ALA403, ASP404 | 8 | 4.32, 3.23, 2.16, 2.54, 3.35, 4.12 2.97. 2.45 |
| Luteolin (A known c-Src kinase inhibitor) Luteolin (PubChem CID: 5280445) | -8.4 | VAL273, VAL281 ALA293, LYS295 MET341, ASP404 |
6 | 2.92, 3.85 3.12, 2.67 2.72, 3.12 |
Figure 4.
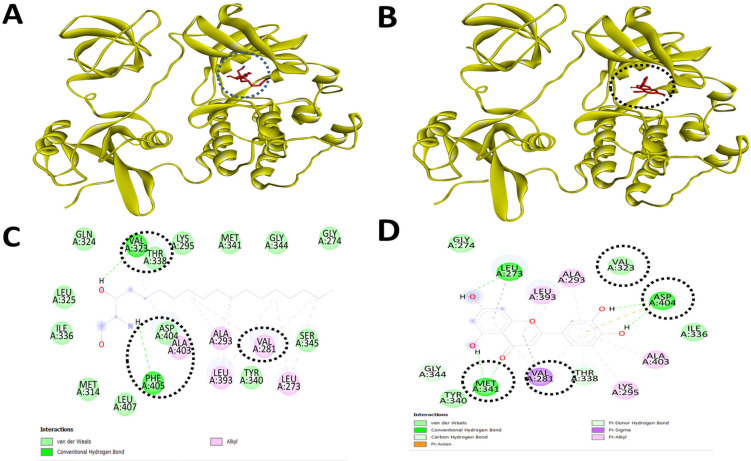
Sphinganine and Luteolin, a Known Inhibitor of c-Src Kinase Show Similar Binding to the Inhibitory Site of the c-Src Kinase. Molecular docking and inhibitory interaction by sphinganine and luteolin upon c-Src kinase with the help of Autodock Vina. (A). A ribbon structure with a full 3D view between sphinganine and c-Src kinase. (B). A ribbon structure with a full 3D view between luteolin and c-Src kinase. (C). Discovery Studio Visualizer assisted 2-D image of docked molecular structure between sphinganine and c-Src kinase. (D). Discovery Studio Visualizer assisted 2-D image of docked molecular structure between luteolin and c-Src kinase
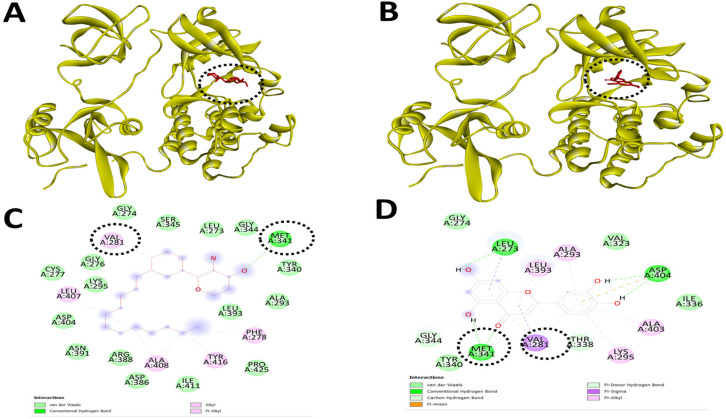
With these interesting observations on sphinganine and existing views that sphinganine is a potential inducer of apoptotic cell death, we extended the approach to design modified mimetic sphinganine (MMS) as a potential inhibitor of the c-Src kinase. One potential MMS compound is selected based on the binding affinity (-7.1 kcal/mol) and specific inhibitory binding pocket. We deposited this MMS structure as PubChem CID: 162625115 and [PubChem SID: 461454363]. Molecular docking and visualization of the inhibitory complex showed a specific and strong binding upon c-Src kinase as an inhibitor with notable residues such as PHE278, VAL281, MET341, LEU407, ALA408, and TYR416. Among these inhibitory domains amino acid resides, MET341 and others including VAL273, VAL28, ALA293, LYS295, MET341, and ASP404 were similar in the case of known c-Src kinase inhibitors such as luteolin and PP2 (Table 3 and Figure 5).
Figure 5.
MMS and Luteolin, a Known Inhibitor of c-Src Kinase Show Similar Binding to the Inhibitory Site of the c-Src Kinase. Molecular docking and inhibitory interaction by MMS and luteolin upon c-Src kinase with the help of Autodock Vina. (A). A ribbon structure with a full 3D view between MMS and c-Src kinase. (B). A ribbon structure with a full 3D view between luteolin and c-Src kinase. (C). Discovery Studio Visualizer assisted 2-D image of docked molecular structure between MMS and c-Src kinase. (D). Discovery Studio Visualizer assisted 2-D image of docked molecular structure between luteolin and c-Src kinase
MD simulations
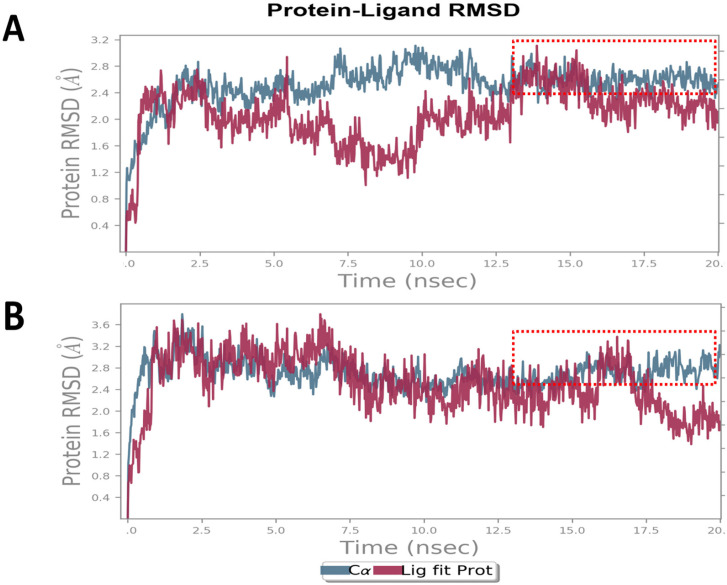
Furthermore, to evaluate the stability of the MMS and c-Src kinase inhibitory complex, we performed MD simulations and compared them with luteolin, a known c-Src kinase inhibitor. RMSD plot of MMS (Figure 6A) showed a deviation in the range of 1.2 to 3.0 Å that is well acceptable to denote the stability of the complex. Noticeably, the RMSD of c-Src kinase was found to be stable and linear at the end of simulations between 15 to 20ns. The RMSD plot of c-Src kinase in a complex in the case of MMS showed a highly comparable RMSD plot of c-Src in a complex with luteolin (Figure 6B) with a deviation in the range of 1.5 to 3.5 Å.
Figure 6.
MMS and Luteolin, a Known Inhibitor of c-Src Kinase Show Similar and Stable Inhibitory Complexes. MD simulations for 20ns derived Root Mean Square Deviation (RMSD) plot of c-Src kinase in complex with (A) MMS and (B) luteolin
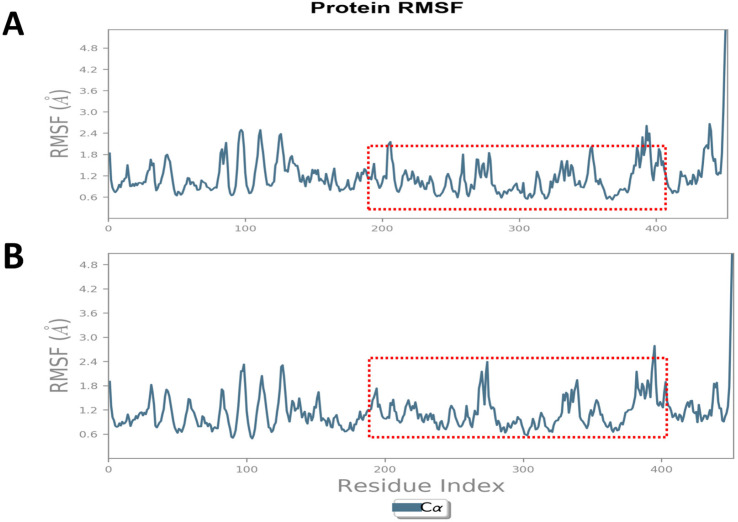
We analyzed the RMSF plot of c-Src in complex with MMS (Figure 7A) and luteolin (Figure 7B) and data indicated the least fluctuations (0.5 to 1.8 Å) in the regulatory domain (300 to 390 a.a) of the c-Src kinase that also serves as the inhibitory binding pocket of these inhibitors. MMS showed similar or even lesser fluctuations of amino acid residues accountable for potential regulatory and inhibitory roles in c-Src kinase.
Figure 7.
MMS and Luteolin, a Known Inhibitor of c-Src Kinase Display the Least Fluctuation of Ligand-Protein Complexes. MD simulations for 20ns derived Root Mean Square Deviation (RMSD) plot of c-Src kinase in complex with (A) MMS and (B) luteolin
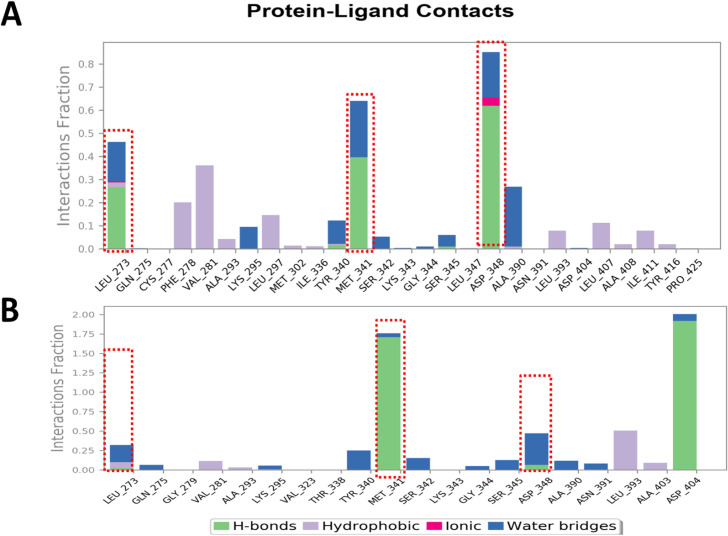
We evaluated the c-Src kinase-MMS (Figure 8A) and c-Src kinase-luteolin (Figure 8B) contact map to estimate the important amino acid residues for their role in the stability of complexes. Data appreciably indicated that key regulatory amino acid residues including MET341 contributed to the stable protein-ligand contact and the contact map of MMS almost matched with the luteolin that is known to bind upon the inhibitory domain of the c-Src kinase.
Figure 8.
Protein-Ligand Contact Map of MMS and Luteolin, a Known Inhibitor of c-Src Kinase Shows Key Residues of the Regulatory Domain of the c-Src Kinase. MD simulations for 20 ns derived Ligand-Protein contact map of c-Raf kinase in complex with (A) MMS and (B) Luteolin
ADMET profiling
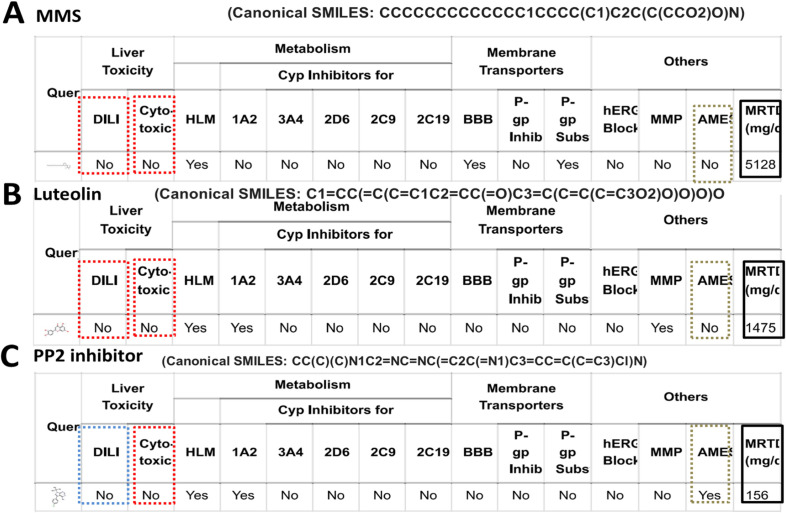
Given the opposite effects of sphinganine over sphingosine and the intent to know the toxicity, druglikeness, and leadlikeness of MMS and other inhibitors of c-Src kinase, we performed vNN-ADMET profiling. ADMET data clearly and surprisingly indicate that sphinganine is projected for cytotoxicity and distinct over sphingosine (Figure S5). At the same time, MMS appears to be safer in terms of lack of cytotoxicity, negative AMES, and no drug-induced liver injury. Interestingly, the ADMET profile of MMS compared with known inhibitors of c-Src kinase suggested better attributes in terms of AMES test and MRTD values where MMS showed more than ten times acceptable MRTD values (5128 mg/day) over PP2 (156 mg/day) and luteolin (1475 mg/day) (Figure 9, 10). We also analyzed the ADME profile by SWISSADME server and attributes such as druglikeness and leadlikness were found to be within well-acceptable parameters (Figure S6).
Figure 9.
ADMET Profile of MMS, Luteolin and PPI (an inhibitor of c-Src kinase) Predicts Favorable Attributes on DILI, Cytotoxicity, Carcinogenicity, and MRTD Values. ADMET profile is generated using v-NN/ADMET server. (A) MMS and (B) Luteolin, (C) PPI
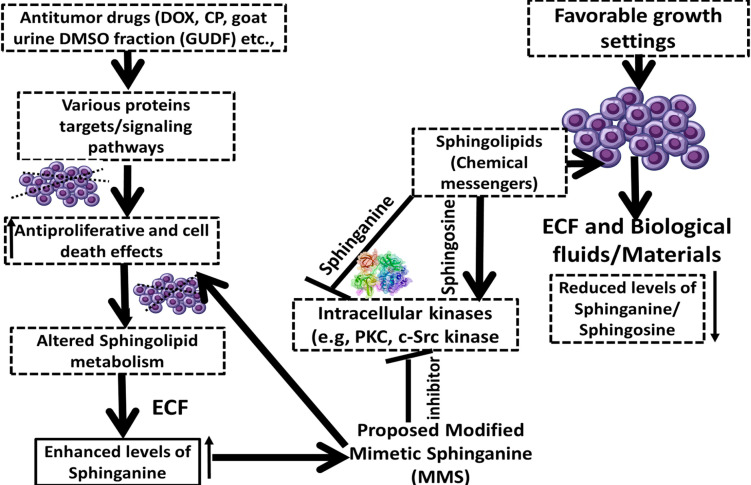
Figure 10.
A Proposed Model on the Role of Extracellular Secretion of Sphinganine in Drug-Induced Cell Death in Colon Cancer Cells. A proposition on the potential of modified mimetic sphinganine (MMS) as a potential inhibitor of the c-Src kinase
Discussion
Implications of sphingolipids such as sphinganine and sphingosine are highlighted in the context of proliferation, apoptotic cell death, and drug resistance in cancer cells [3- 5, 7, 8- 10]. In essence, possibilities of sphinganine and sphingosine as chemical messengers that can modulate the action of intracellular transducer kinases are projected [7-12, 14-16]. At the same time, data is highly limited to linking the extracellular secretion of sphingolipids such as sphinganine an inhibitor of intracellular kinases such as c-Src in the case of colon cancer cells treated by anticancer compositions.
Sphingolipids are known as the complex lipid that comprises various forms including organic aliphatic amino alcohol sphingosine and sphinganine [5-10]. These sphingolipids contribute to various cellular regulation of growth, proliferation, and cell death processes in cancer cells by making integration with key intracellular transducers such as c-Src kinase and PKC [7-12, 14-16].
There are views that levels of these sphingolipids such as sphingosine and sphinganine are reduced in the extracellular fluids such as plasma, saliva, and urine [10- 18]. At the same time, data is lacking in the context of the abundance of sphingosine and sphinganine during drug-induced cell death in cancer cells such as colon cancer.
A view is pertinent that the availability of sphingoid bases such as sphingosine and sphinganine could be linked with the complex dietary sphingolipids that can be converted into sphingosine and sphinganine in the colon tissue environment [5-10]. At the same time, the de novo process of synthesis of sphinganine and sphingosine could be limited in the human body. Therefore, the availability of sphinganine and sphingosine in the extracellular fluid could be a potential factor that can modulate the signaling pathways connected with cell death. The attributes of sphinganine in the extracellular fluids of cancer cells are seen as an indicator of induced cancer cell death by anticancer drugs [20-25].
The relevance of sphinganine and other forms of sphingolipids such as other sphingolipids such as C16 sphinganine, phytosphingosine, and ceramide (d18:1/14:0) is suggested in the context of pro-apoptotic signaling molecules during drug-induced stress and cell death in the colon and other cancer cell types [20-25, 29]. In another way, the possibility of the involvement of sphingosine and sphinganine at the intracellular levels by acting as modulations of intracellular kinases is compelling. However, data is limited on the molecular interactions of sphingosine and sphinganine with c-Src kinase which is known to contribute to the inhibition of apoptosis in cancer cells.
A pertinent question may be raised regarding the relevance of GUDF as a source of anticancer drug compositions. In this paper, we have attempted to link the relevance of extracellular secretion of sphingolipid bases such as sphinganine in the context of drug-induced cancer cell death. Since we have earlier shown the detailed procedure for the extraction and purification of GUDF enriched with tripeptides and further evaluated the cell death effects of GUDF upon HCT-116 colon cancer cells [43- 45]. Previously, we have standardized in-house VTGE-assisted methodology for the profiling of intracellular and extracellular metabolites of cancer cells treated by drugs such as GUDF. In the present paper, therefore, we have extended the observations of GUDF-induced cell death in HCT-116 cancer cells with the potential metabolic adaptations in the form of the secretion of sphingolipid bases such as sphinganine. This paper warrants further investigations to employ different cancer cells and anticancer drugs that may involve the extracellular secretion of sphingolipid bases such as sphinganine during cell death. Based on the above observations and premises, we explore the molecular interactions of sphingolipid bases such as sphinganine with intracellular kinases including c-Src kinase, and continue with the design of potential inhibitors of the c-Src kinase.
Crystallography and molecular data on c-Src kinase also suggest that the regulatory domain of c-Src kinase ranges from 270-410 a.a residue. Additionally, several reported inhibitors of c-Src kinase including luteolin and PPI are known to establish strong binding upon key residues such as MET-341 and ASP404 [26- 30]. Importantly, MMS displayed specific nature of interactive bonds such as H-bonds and binding affinity that almost resembled known inhibitors of c-Src kinase such as luteolin and PPI [31-33].
The role of sphingosine and its phosphorylated form sphingosine 1-phosphate is documented to drive the proliferation, survival, and drug resistance of colon cancer cells [19- 25]. These forms of sphingolipids such as sphingosine and its phosphorylated form sphingosine 1-phosphate are suggested to exert as an activator of various oncoproteins including PKC and COX enzymes leading in favor of proliferation and survival of colon and other types of cancer cells [5-10]. However, the role of sphinganine and sphinganine 1-phosphate is proposed as anti-growth and proliferation and at the same time inducer of cell death in cancer cells including colon cancer cells [5-10]. Sphinganine and its analog are indicated to display autophagic and apoptotic cell death in colon cancer cells [6- 8].
The role of c-Src kinase inhibitors such as luteolin, PP2, Ruxolitinib, ZINC3214460, and ZINC1380384 is suggested to suppress proliferation and induce cell death in various types of cancer cells [19- 25]. However, a link between the secretion of sphingolipids such as sphinganine and the inhibition of c-Src kinase is entirely lacking. Furthermore, mimetic of sphinganine that can inhibit c-Src kinase and in turn lead to cancer cell death is not explored.
The analysis of the inhibitory binding pocket of c-Src kinase in complex with potential inhibitors indicated prominent amino acid residues such as THR340, MET341, and ASP404 as a major contributor to the protein-ligand stability [26- 33]. In literature, the relevance of inhibitors of the c-Src kinase is emphasized as anticancer drugs such as Ruxolitinib, PP2, and luteolin that showed specific inhibitory amino acid residues such as THR338 and MET341 [30-33]. Due to the crucial role of c-Src kinase in pro-cancer signaling pathways, therapeutic targeting of c-Src is strongly considered [31-33].
The involvement of c-Src kinase in the blockade of apoptosis is reported in tumors and hence, considered a promising target so that induction of apoptosis and autophagic cell death in cancer cells could be achieved [30-33]. The role of phospholipids such as sphingosine has been documented as an inducer of transducer intracellular kinases such as PKC and in turn a favorable messenger of the growth and proliferation of cancer cells [26- 33]. The levels of phospholipids such as Sphinganine are suggested to be enhanced during various forms of cancer cell death [25- 30]. In a comparative study, sphinganine is shown as a potent inducer of cell cycle arrest and cell death in breast cancer cells over sphingosine [34- 38].
However, the idea to design the structure of MMS is derived from the data derived from the molecular interactions of sphinganine within the inhibitory domain of the c-Src kinase. But the derived MMS structure needed to be evaluated for its ADMET profile including cytotoxicity, carcinogenicity, druglikeness, and pharmacokinetics. Data projected that MMS has highly comparable and even better in terms of some parameters such as MRTD value over known inhibitors such as luteolin and PPI. Another point needs to be highlighted that sphinganine is predicted for cytotoxicity by ADMET profiling. Remarkably, MMS derived from sphinganine is predicted to show the absence of cytotoxicity, drug-induced liver injury, and carcinogenetic. A notable observation from a comparison of the ADMET profile of sphingosine and sphinganine showed a striking difference in terms of cytotoxicity in the case of sphinganine over sphinganine. This observation can explain partially the reasons behind the extracellular secretion of sphinganine by cancer cells as one of the possible metabolic adaptations to avoid the accumulation of cytotoxic sphinganine and a potential inhibitor of c-Src kinase during drug-mediated cell death and stress.
In conclusion, extracellular secretion of sphinganine and other sphingolipids such as C16 sphinganine, phytosphingosine, and ceramide (d18:1/14:0) is reported for HCT-116 cancer cells under GUDG-induced stress and cell death. Furthermore, sphinganine and sphingosine display opposite effects in proliferation and cell death attributes of cancer cells possibly by modulating key enzymes such as c-Src kinase. This study helped in the proposition of MMS as a potent inhibitor of the c-Src kinase that is not reported. Molecular docking and MD simulations helped to link the biological relevance of extracellular sphinganine as an inhibitor of c-Src kinase and that finally clued for the development of MMS as a potential inhibitor. We propose that MMS could be explored as a good c-Src kinase inhibitor and a promising anticancer agent.
Author Contribution Statement
Rasika Nandangiri: Methodology, Data collection, Data Interpretation and analysis, Manuscript draft preparation; Seethamma T N: Methodology, Data collection; Ajay Kumar Raj: Methodology, Data collection, Data Interpretation and analysis; Kiran Bharat Lokhande: Methodology (MD Simulations), Data collection (MD Simulations), Data Interpretation and analysis (MD Simulations), Manuscript Draft Preparation; Kratika Khunteta: Methodology, Data Collection, Data interpretation and analysis, Manuscript draft preparation Ameya Hebale: Methodology, Data collection, Data Interpretation and analysis, Manuscript draft preparation; Vaidehi Patel: Methodology, Data Collection, Data interpretation and analysis, Manuscript draft preparation Haet Kothari: Methodology, Data collection, Data Interpretation and analysis, Manuscript draft preparation; Sachin C. Sarode: Manuscript Draft Preparation, Editin; Nilesh Kumar Sharma: Conceptualization/Idea, Data Interpretation and analysis Manuscript Draft Preparation, Editing of Manuscript.
Acknowledgements
Funding statement
This research work was funded by Intra-mural seed grant (DPU/01/12/2020) by Dr. D. Y. Patil Vidyapeeth, Pune, India. The authors also acknowledge facilities and support from DST-FIST sponsored Research Lab, Central Research Facility (DPU, Pune) and SAIF, IIT, Mumbai, India.
If it was approved by any scientific Body/ if it is part of an approved student thesis
This paper was a part of approved student thesis.
Any conflict of interest
The authors declare no conflict of interests.
How the ethical issue was handled (name the ethical committee that approved the research)
This research work does not involve human subjects and hence not applicable.
Supplementary materials
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 4.Elia I, Haigis MC. Metabolites and the tumour microenvironment: From cellular mechanisms to systemic metabolism. Nat Metab. 2021;3(1):21–32. doi: 10.1038/s42255-020-00317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawamori T, Osta W, Johnson KR, Pettus BJ, Bielawski J, Tanaka T, et al. Sphingosine kinase 1 is up-regulated in colon carcinogenesis. Faseb J. 2006;20(2):386–8. doi: 10.1096/fj.05-4331fje. [DOI] [PubMed] [Google Scholar]
- 6.Coward J, Ambrosini G, Musi E, Truman JP, Haimovitz-Friedman A, Allegood JC, et al. Safingol (l-threo-sphinganine) induces autophagy in solid tumor cells through inhibition of pkc and the pi3-kinase pathway. Autophagy. 2009;5(2):184–93. doi: 10.4161/auto.5.2.7361. [DOI] [PubMed] [Google Scholar]
- 7.Yin J, Miyazaki K, Shaner RL, Merrill AH JR, Kannagi R. Altered sphingolipid metabolism induced by tumor hypoxia - new vistas in glycolipid tumor markers. FEBS Lett. 2010;584(9):1872–8. doi: 10.1016/j.febslet.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosa R, Marciano R, Malapelle U, Formisano L, Nappi L, D’Amato C, et al. Sphingosine kinase 1 overexpression contributes to cetuximab resistance in human colorectal cancer models. Clin Cancer Res. 2013;19(1):138–47. doi: 10.1158/1078-0432.CCR-12-1050. [DOI] [PubMed] [Google Scholar]
- 9.Furuya H, Shimizu Y, Tamashiro PM, Iino K, Bielawski J, Chan OTM, et al. Sphingosine kinase 1 expression enhances colon tumor growth. J Transl Med. 2017;15(1):120 . doi: 10.1186/s12967-017-1220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-Larrauri A, Presa N, Dominguez-Herrera A, Ouro A, Trueba M, Gomez-Muñoz A. Role of bioactive sphingolipids in physiology and pathology. Essays Biochem. 2020;64(3):579–89. doi: 10.1042/EBC20190091. [DOI] [PubMed] [Google Scholar]
- 11.Scherer M, Leuthäuser-Jaschinski K, Ecker J, Schmitz G, Liebisch G. A rapid and quantitative lc-ms/ms method to profile sphingolipids. J Lipid Res. 2010;51(7):2001–11. doi: 10.1194/jlr.D005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q, Gao P, Wang X, Duan Y. The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Sci Rep. 2014;4:6802. doi: 10.1038/srep06802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faedo RR, da Silva G, da Silva RM, Ushida TR, da Silva RR, Lacchini R, et al. Sphingolipids signature in plasma and tissue as diagnostic and prognostic tools in oral squamous cell carcinoma. Biochim Biophys Acta Mol Cell Biol Lipids. 2022;1867(1):159057. doi: 10.1016/j.bbalip.2021.159057. [DOI] [PubMed] [Google Scholar]
- 14.Farshidfar F, Kopciuk KA, Hilsden R, McGregor SE, Mazurak VC, Buie WD, et al. A quantitative multimodal metabolomic assay for colorectal cancer. BMC Cancer. 2018;18(1):26. doi: 10.1186/s12885-017-3923-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Hu JH, Liu CC, Liu M, Liu Z, Sun LX. Lc-ms based cell metabolic profiling of tumor cells: A new predictive method for research on the mechanism of action of anticancer candidates. RSC Adv. 2018;8(30):16645–56. doi: 10.1039/c8ra00242h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhadwal P, Dahiya D, Shinde D, Vaiphei K, Math RGH, Randhawa V, et al. Lc-hrms based approach to identify novel sphingolipid biomarkers in breast cancer patients. Sci Rep. 2020;10(1):4668 . doi: 10.1038/s41598-020-61283-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tobias F, Hummon AB. Lipidomic comparison of 2d and 3d colon cancer cell culture models. J Mass Spectrom. 2022;57(8):e4880. doi: 10.1002/jms.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez-Rivera G, Valdés A, Cifuentes A. Metabolomic characterization of the antiproliferative activity of bioactive compounds from fruit by-products against colon cancer cells. Methods Mol Biol. 2023;2571:45–55. doi: 10.1007/978-1-0716-2699-3_5. [DOI] [PubMed] [Google Scholar]
- 19.Merrill AH Jr, Sereni AM, Stevens VL, Hannun YA, Bell RM, Kinkade JM Jr. Inhibition of phorbol ester-dependent differentiation of human promyelocytic leukemic (hl-60) cells by sphinganine and other long-chain bases. J Biol Chem. 1986;261(27):12610–5. [PubMed] [Google Scholar]
- 20.Hannun YA, Bell RM. Regulation of protein kinase c by sphingosine and lysosphingolipids. Clin Chim Acta. 1989;185(3):333–45. doi: 10.1016/0009-8981(89)90224-6. [DOI] [PubMed] [Google Scholar]
- 21.Ahn EH, Schroeder JJ. Sphingoid bases and ceramide induce apoptosis in ht-29 and hct-116 human colon cancer cells. Exp Biol Med (Maywood) 2002;227(5):345–53. doi: 10.1177/153537020222700507. [DOI] [PubMed] [Google Scholar]
- 22.Ahn EH, Chang CC, Schroeder JJ. Evaluation of sphinganine and sphingosine as human breast cancer chemotherapeutic and chemopreventive agents. Exp Biol Med (Maywood) 2006;231(10):1664–72. doi: 10.1177/153537020623101012. [DOI] [PubMed] [Google Scholar]
- 23.Bu S, Kapanadze B, Hsu T, Trojanowska M. Opposite effects of dihydrosphingosine 1-phosphate and sphingosine 1-phosphate on transforming growth factor-beta/smad signaling are mediated through the pten/ppm1a-dependent pathway. J Biol Chem. 2008;283(28):19593–602. doi: 10.1074/jbc.M802417200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Maurer BJ, Liu YY, Wang E, Allegood JC, Kelly S, et al. N-(4-hydroxyphenyl)retinamide increases dihydroceramide and synergizes with dimethylsphingosine to enhance cancer cell killing. Mol Cancer Ther. 2008;7(9):2967–76. doi: 10.1158/1535-7163.MCT-08-0549. [DOI] [PubMed] [Google Scholar]
- 25.Yamane M. Palmitoyl-ceramide accumulation with necrotic cell death in a549 cells, followed by a steep increase in sphinganine content. Biochim Open. 2015;1:11–27. doi: 10.1016/j.biopen.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3(5):629–38. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 27.Alvarado JJ, Betts L, Moroco JA, Smithgall TE, Yeh JI. Crystal structure of the src family kinase hck sh3-sh2 linker regulatory region supports an sh3-dominant activation mechanism. J Biol Chem. 2010;285(46):35455–61. doi: 10.1074/jbc.M110.145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao R, Mi N, Zhang H. 3d-qsar study of c-src kinase inhibitors based on docking. J Mol Model. 2010;16(2):361–75. doi: 10.1007/s00894-009-0530-1. [DOI] [PubMed] [Google Scholar]
- 29.Morad SA, Cabot MC. Ceramide-orchestrated signalling in cancer cells. Nat Rev Cancer. 2013;13(1):51–65. doi: 10.1038/nrc3398. [DOI] [PubMed] [Google Scholar]
- 30.Byun S, Lee KW, Jung SK, Lee EJ, Hwang MK, Lim SH, et al. Luteolin inhibits protein kinase c(epsilon) and c-src activities and uvb-induced skin cancer. Cancer Res. 2010;70(6):2415–23. doi: 10.1158/0008-5472.CAN-09-4093. [DOI] [PubMed] [Google Scholar]
- 31.Duan Y, Chen L, Chen Y, Fan XG. C-src binds to the cancer drug ruxolitinib with an active conformation. PLoS One. 2014;9(9):e106225. doi: 10.1371/journal.pone.0106225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jha V, Macchia M, Tuccinardi T, Poli G. Three-dimensional interactions analysis of the anticancer target c-src kinase with its inhibitors. Cancers (Basel) 2020;12(8):2327. doi: 10.3390/cancers12082327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Zhang TJ, Tu S, Zhang ZH, Meng FH. Identification of novel src inhibitors: Pharmacophore-based virtual screening, molecular docking and molecular dynamics simulations. Molecules. 2020;25:18. doi: 10.3390/molecules25184094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee M, Kim JY, Koh WS. Apoptotic effect of pp2 a src tyrosine kinase inhibitor, in murine b cell leukemia. J Cell Biochem. 2004;93(3):629–38. doi: 10.1002/jcb.20215. [DOI] [PubMed] [Google Scholar]
- 35.Ling LU, Lin H, Tan KB, Chiu GN. The role of protein kinase c in the synergistic interaction of safingol and irinotecan in colon cancer cells. Int J Oncol. 2009;35(6):1463–71. doi: 10.3892/ijo_00000465. [DOI] [PubMed] [Google Scholar]
- 36.Wu Z, Chang PC, Yang JC, Chu CY, Wang LY, Chen NT, et al. Autophagy blockade sensitizes prostate cancer cells towards src family kinase inhibitors. Genes Cancer. 2010;1(1):40–9. doi: 10.1177/1947601909358324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Navarra M, Celano M, Maiuolo J, Schenone S, Botta M, Angelucci A, et al. Antiproliferative and pro-apoptotic effects afforded by novel src-kinase inhibitors in human neuroblastoma cells. BMC Cancer. 2010;10:602 . doi: 10.1186/1471-2407-10-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez J, Hesling C, Prudent J, Popgeorgiev N, Gadet R, Mikaelian I, et al. Src tyrosine kinase inhibits apoptosis through the erk1/2- dependent degradation of the death accelerator bik. Cell Death Differ. 2012;19(9):1459–69. doi: 10.1038/cdd.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller KD, Pniewski K, Perry CE, Papp SB, Shaffer JD, Velasco-Silva JN, et al. Targeting acss2 with a transition-state mimetic inhibits triple-negative breast cancer growth. Cancer Res. 2021;81(5):1252–64. doi: 10.1158/0008-5472.CAN-20-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dahl E, Villwock S, Habenberger P, Choidas A, Rose M, Klebl BM. White paper: Mimetics of class 2 tumor suppressor proteins as novel drug candidates for personalized cancer therapy. Cancers (Basel) 2022;14:18. doi: 10.3390/cancers14184386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melo G, Silva CAB, Hague A, Parkinson EK, Rivero ERC. Anticancer effects of putative and validated bh3-mimetic drugs in head and neck squamous cell carcinomas: An overview of current knowledge. Oral Oncol. 2022;132:105979. doi: 10.1016/j.oraloncology.2022.105979. [DOI] [PubMed] [Google Scholar]
- 42.Ehrler JHM, Brunst S, Tjaden A, Kilu W, Heering J, Hernandez-Olmos V, et al. Compilation and evaluation of a fatty acid mimetics screening library. Biochem Pharmacol. 2022;204:115191. doi: 10.1016/j.bcp.2022.115191. [DOI] [PubMed] [Google Scholar]
- 43.Kumar A, Kothari J, Lokhande K, Seethamma T, Swamy KV, Sharma Ph D. Fmasc N. Novel antiproliferative tripeptides inhibit ap-1 transcriptional complex. Int J Pept Res Ther. 2021;27:1–20. [Google Scholar]
- 44.Sharma nk, ajay kumar, asawari waghmode., inventors. Design of vertical tube electrophoretic system and method to fractionate small molecular weight compounds using polyacrylamide gel matrix. Date of publication: 01/03/2019. (patent application number no: Ina 201921000760). Publication typeina, the patent official journal no- 19/2018, page no-9035. Published. 2019
- 45.Kumar A, Patel S, Bhatkar D, Sarode SC, Sharma NK. A novel method to detect intracellular metabolite alterations in mcf-7 cells by doxorubicin induced cell death. Metabolomics. 2021;17(1):3. doi: 10.1007/s11306-020-01755-2. [DOI] [PubMed] [Google Scholar]
- 46.Raj AK, Lokhande KB, Prasad TK, Nandangiri R, Choudhary S, Pal JK, et al. Intracellular ellagic acid derived from goat urine dmso fraction (gudf) predicted as an inhibitor of c-raf kinase. Curr Mol Med. 2024;24(2):264–79. doi: 10.2174/1566524023666230113141032. [DOI] [PubMed] [Google Scholar]
- 47.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998:19. [Google Scholar]
- 48.Trott O, Olson AJ. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DSV3. Discovery studio visualizer v3.0. Accelrys software inc.; 2010. [Google Scholar]
- 50.Schyman P, Liu R, Desai V, Wallqvist A. Vnn web server for admet predictions. Front Pharmacol. 2017;8:889. doi: 10.3389/fphar.2017.00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daina A, Michielin O, Zoete V. Swissadme: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.