Abstract
Mammalian mitochondrial DNA end-binding activity is nearly indistinguishable from that of nuclear Ku. This observation led to the hypothesis that mitochondrial DNA end-binding activity is in part dependent upon Ku80 gene expression. To test this hypothesis, we assayed for Ku activity in mitochondrial extracts prepared from the xrs-5 hamster cell line that lacks Ku80 mRNA expression. Mitochondrial protein extracts prepared from this cell line lacked the DNA end-binding activity found in similar extracts prepared from wild-type cells. Azacytidine-reverted xrs-5 cells that acquired nuclear DNA end-binding activity also acquired mitochondrial DNA end-binding activity. Western blot analysis of human mitochondrial protein extracts using a monoclonal antibody specific for an N-terminal epitope of Ku80 identified a protein with an apparent molecular weight of 68 kDa. This mitochondrial protein was not detected by a monoclonal antibody specific for an epitope at the C-terminal end of Ku80. Consistently, while both the N- and C-terminal Ku80 monoclonal antibodies supershifted the nuclear DNA end-binding complex on an electrophoretic mobility shift assay, only the N-terminal monoclonal antibody supershifted the mitochondrial DNA end-binding complex. To confirm that the 68 kDa Ku protein was not a consequence of nuclear protein contamination of mitochondrial preparations, highly purified intact nuclei and mitochondria were treated with proteinase K which traverses the pores of intact nuclei but gains limited access into intact mitochondria. Ku80 in purified intact nuclei was sensitive to treatment with this protease, while the 68 kDa Ku protein characteristic of purified intact mitochondria was resistant. Further, immunocytochemical analysis revealed the co-localization of the N-terminal specific Ku80 monoclonal antibody with a mitochondrial-targeted green fluorescence protein. Mitochondrial localization of the C-terminal Ku80 monoclonal antibody was not observed. These data are consistent with the hypothesis that a C-terminally truncated form of Ku80 is localized in mammalian mitochondria where it functions in a DNA end-binding activity.
INTRODUCTION
The Ku protein was originally identified as an autoantigen in patients with scleroderma-polymyositis overlap syndrome (1). The protein was purified using antisera from these patients and shown to be a heterodimer of two subunits of 70 and 80 kDa proteins referred to as Ku70 and Ku80, respectively (2). As a heterodimer, Ku binds with high affinity to DNA ends, single-strand nicks and breaks, and hairpin loops independent of sequence (3). The characteristic DNA end-binding (DEB) activity of Ku is detected by an electrophoretic mobility shift assay (EMSA) in which Ku binding to a linear radioactive DNA probe is detected in the presence of a vast excess of non-radioactive circular DNA (4). A role for the DEB activity of Ku in mammalian cells was established when Taciolli et al. (5) identified Ku80 as the product of the XRCC5 gene. This gene was previously shown to be deficient in several hamster cell lines that were sensitive to ionizing radiation and chemical agents that induce DNA double strand breaks (6). In the same year, Taciolli et al. (5) and Rathmell and Chu (4,7) implicated the DEB activity of Ku in V(D)J recombination and non-homologous DNA end-joining (NHEJ), the most common pathway for double-strand break repair (DSBR) in mammalian cells (8).
Ku is an abundant protein, with an estimated 400 000 Ku molecules per cell (9). The relative abundance of this protein is consistent with reports that Ku is involved in multiple cellular processes. As the DEB component of the DNA-dependent protein kinase (DNA-PK) (10,11), Ku plays an essential role in NHEJ and V(D)J recombination (8). HDF1, the yeast Ku70 homolog, is also required for proper maintenance of telomere length in that organism (12–14). A similar role for Ku70 and Ku80 in the maintenance of telomeres in mammalian cells has been established. Ku70 physically interacts with telomeres in mammalian cells (15), and both Ku70 and Ku80 deficient cells exhibit excessive telomere end-to-end fusions (15,16). Further, Ku70 and Ku80 null mice have been developed and used to study the role of Ku in vivo (17–20). In addition to ionizing radiation sensitivity and an inability to support V(D)J recombination, these mice also exhibited stunted growth and premature senescence that may or may not be related to their role in DSBR (21). Taken together, these data suggest that Ku is involved in multiple cellular processes that stabilize DNA.
More recently, a DEB activity was detected in highly purified mitochondrial protein extracts prepared from hamster, rat and human cell lines (22). Western blot and EMSA super-shift experiments revealed that the human mitochondrial DEB activity contained a 68 kDa protein that was immunologically related to Ku80 (22). These experiments did not, however, shed light on whether this mitochondrial DEB protein was encoded by the Ku80 gene or was instead encoded by a novel gene with sequence similarity to Ku80. Therefore, the goal of this study was to test the hypothesis that Ku80 is localized in mammalian mitochondria and participates in a DEB activity.
We report here that Ku80 is required for mammalian mitochondrial DEB activity, and that in human and monkey cells the mitochondrial Ku80 protein lacks a C-terminal epitope. This conclusion was based on the following findings. (i) The hamster cell line xrs-5 that lacks detectable Ku80 mRNA expression also lacks mitochondrial DEB activity. (ii) Reversion of wild-type nuclear DEB activity in xrs-5 cells by treatment with 5-azacytidine was always associated with reversion of wild-type mitochondrial DEB activity. (iii) A monoclonal antibody (Mab) specific for an N-terminal epitope on the human Ku80 protein recognized both nuclear and mitochondrial proteins, whereas a Mab specific for a C-terminal epitope on Ku80 identified the nuclear, but not the mitochondrial, form. Furthermore, while the N-terminal-specific Mab supershifted both nuclear and mitochondrial DEB complexes, the C-terminal Mab only supershifted the nuclear DEB complex. (iv) The truncated Ku80 in intact human mitochondria was resistant to proteinase K treatment whereas Ku80 in intact nuclei was not. (v) Immunocytochemical analysis revealed co-localization of a mitochondrial-targeted green fluorescent protein (GFP) with the truncated Ku80. These data provide strong support for the conclusion that Ku80 is localized in mammalian mitochondria and functions in a DEB activity. The presence of a Ku80-dependent DEB activity in mitochondrial extracts suggests that this protein plays a role in mitochondrial DNA dynamics.
MATERIALS AND METHODS
Cell culture
The Chinese hamster ovary-derived, Ku80-deficient cell line xrs-5, African green monkey kidney cell line CV1 and the human fibrosarcoma-derived cell line HT1080 were obtained from the American Tissue Culture Collection. The Ku80-deficient cell line xrs-6 and the xrs-6-HamKu86 cell lines were obtained from the European Collection of Cell Culture. xrs-6-HamKu86 cells have been transfected with a hamsterKu80 transgene, resulting in wild-type levels of nuclear DEB activity (23). Chinese hamster ovary-derived V79 cells were kindly supplied by Gilbert Chu (Stanford University). xrs-5 cells were cultured in minimal essential medium alpha (Gibco). V79, HT1080 and CV1 cell lines were cultured in Dulbecco’s modified Eagle’s medium (Gibco). Tissue culture media was supplemented with 9% fetal bovine serum, penicillin and streptomycin.
5-azacytidine treatment of xrs-5 cells
Reversion of wild-type nuclear DEB activity in xrs-5 cells was achieved by culturing the cells in the presence of 5-azacytidine (Sigma) essentially as described elsewhere (24). Approximately 106 cells were treated with freshly prepared 5-azacytidine (4 µM) in growth media for 24 h. Fresh drug and media were added to the cells and incubation continued for an additional 24 h period. Following the 48-h incubation, cells were trypsinized and re-plated at a cloning density of 102 cells per plate. Independent clones were expanded and analyzed for nuclear and mitochondrial DEB activity.
Isolation of subcellular organelles
Nuclei. Nuclei were isolated from three confluent 150 mm tissue culture dishes of human HT1080 cells. After washing in ice-cold phosphate buffered saline (PBS), cells were scraped from the dishes using a rubber policeman and treated with 200 strokes of a Dounce homogenizer in buffer containing 10 mM KCl, 10 mM MgCl2, 10 mM Tris pH 7.4, and 10 mM DTT in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF). Nuclei were collected by centrifugation at 2600 g for 5 min, then washed and re-centrifuged twice more.
Mitochondria. Mitochondria were isolated from 20 confluent 150 × 20 mm tissue culture dishes of human HT1080 cells. Cells were washed in PBS and resuspended in a buffer containing 0.3 M sucrose, 1 mM EGTA, 5 mM MOPS, 5 mM KH2PO4, 0.1% bovine serum albumin (BSA), pH 7.4. Cells were homogenized with 200 strokes in a Dounce homogenizer. Unlysed cells and nuclei were sedimented at 2600 g, and a crude mitochondrial fraction collected from the supernatant. Mitochondria were further purified on a 30% Percoll gradient in 0.3 M sucrose by centrifugation for 30 min at 24 000 g. Isolation of the mitochondrial fraction was monitored by measuring cytochrome c oxidase activity (25).
Preparation of nuclear and mitochondrial protein extracts
Purified intact nuclei or mitochondria were lysed in buffer consisting of 10 mM KCl, 10 mM MgCl2, 10 mM Tris pH 7.4, 10 mM DTT and 350 mM NaCl. Buffer was supplemented with pepstatin (0.7 µg/ml), aprotinin (1 µg/ml), leupeptin (0.2 µg/ml) and PMSF (1 mM). Preparations were incubated for 1 h on ice. Membrane and protein fractions were separated by centrifugation at 70 000 r.p.m. for 30 min in a Beckman Ultra-TL100 centrifuge. Supernatant was removed and dialyzed against 25 mM Tris, pH 7.4, 1 mM EDTA, 1 mM PMSF, 1 mM DTT and 10% glycerol.
Electrophoretic mobility shift assay (EMSA)
A 94 bp DNA fragment (f94) was liberated from the plasmid pCR2.1 (Invitrogen) using the restriction endonucleases HindIII and XbaI. The f94 was then end-labeled with [32P]dATP in the presence of non-radioactive dCTP, dGTP and dTTP by the Klenow fragment of Escherichia coli DNA polymerase. In a 10 µl total volume, 0.2 ng probe f94 was incubated with 0.5 µg protein extract in the presence of 200 ng circular φX174 DNA or 200 ng linear (HincII digested φX174) DNA for 30 min at 14°C in a reaction buffer containing 12 mM HEPES, 5 mM MgCl2, 4 mM Tris pH 7.9, 100 mM KCl, 0.6 mM EDTA, 0.6 mM DTT and 12% glycerol. Following incubation, 2 µl of 5× gel-loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol, 30% glycerol) was added to each sample before electrophoreses on a 4% polyacrylamide gel in 1× TBE (90 mM Tris–borate, 2 mM EDTA) running buffer at 4°C and 20 V/cm. Radioactivity was detected using a phoshorimager (Molecular Dynamics, Foster City, CA).
Supershift assay
Supershift analysis was performed using two Mabs specific for human Ku80. The first Mab recognizes an epitope contained within the N-terminal amino acids 8 to 221 of Ku80 (clone S10B1; 26). The second Mab is specific for a C-terminal epitope that maps between amino acids 610 and 705 of Ku80 (clone 111; 26,27). Both Mabs were purchased from Lab Vision Corporation (Fremont, CA). Standard EMSAs were performed on human HT1080 nuclear and mitochondrial protein extracts as described above. Following the 30 min incubation, 10 µl of Mab S10B1 or 4 µl of Mab 111 was added to the reaction and allowed to incubate at 14°C for an additional 1 h before electrophoreses. Binding of Ku80 Mabs to nuclear and mitochondrial DEB proteins was visualized using a phosphorimager.
Western blot analysis
Western blot analysis was performed using the Ku80-specific Mabs S10B1 and 111. Nuclear or mitochondrial protein extract was loaded on a 10% polyacrylamide gel and electrophoresed at 15 V/cm. The gel was then electrophoretically transferred to nitrocellulose membrane (BioRad). The Ku80 Mabs were both diluted 1:500 in 5% BSA in tris-buffered saline (TBS) and incubated with the membrane blot for 1 h at room temperature. After washing the membrane three times in 0.1% BSA in TBS, the membrane was incubated with a 1:5000 dilution of alkaline phosphatase-conjugated goat anti-mouse IgG (Sigma) in 5% BSA in TBS for an additional 1 h. Membranes were washed once more before adding the alkaline phosphatase substrate 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (Sigma).
Proteinase K assay
Intact nuclei and percoll-purified mitochondrial fractions were prepared from HT1080 cells as described above. The intact organelles were then resuspended in 200 µl of their respective homogenization buffers. Two 30 µl aliquots were removed from each and set on ice. To one aliquot, an additional 30 µl of proteinase K (20 µg/µl) in 0.6 M mannitol and 10 mM Tris, pH 7.4 was added. To the remaining aliquot, 30 µl of control buffer alone was added. Samples were incubated for 30 min at 0°C before adding 12 µl 5× SDS gel-loading buffer (250 mM Tris pH 6.8, 500 mM DTT, 10% SDS, 0.5% bromophenol blue, 50% glycerol) and boiled for 10 min. Western blot analysis was performed on the samples using Ku80 polyclonal antisera (Serotec, Raleigh, NC) as described above.
Immunocytochemistry
CV1 cells were grown to confluence on glass cover-slips, then transiently transfected using DEAE dextran (28) with the pEGFP-N1-derived plasmid carrying the mitochondrial targeting sequence of cytochrome c oxidase in frame with the GFP (29). Following transfection, cells were fixed in 3.7% formaldehyde in PBS and permeablized by incubation in 0.1% Triton X-100 in PBS as described elsewhere (30). Cells were washed in PBS before incubating for 1 h at 37°C with primary antibody (Ku80 Mabs S10B1 or 111) at a dilution of 1:500 in PBS in 200 µl total volume. Cells were washed three times in PBS before adding the secondary antibody for 1 h at 37°C (goat anti-mouse IgG conjugated to rhodamine; Pierce) diluted 1:500 in PBS. Cells were then washed three times in PBS before visualizing fluorescence.
Cells were viewed using a BioRad MRC-1024 confocal microscope. A 60× water immersion objective was used. Samples were excited at 488 nm and emissions due to rhodamine and fluorescein collected sequentially. Images were analyzed using the program Adobe Photoshop 4.0.1.
RESULTS
Ku80 mRNA expression is required for mitochondrial DEB activity
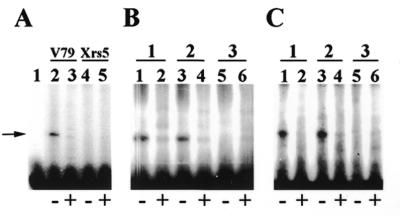
To test the hypothesis that Ku80 was required for mammalian mitochondrial DEB activity, nuclear and mitochondrial protein extracts were prepared from the hamster cell line xrs-5. The xrs-5 cell line lacks detectable Ku80 mRNA expression and is deficient in nuclear DEB activity (6,23). Nuclear protein extracts were prepared from this cell line and the lack of nuclear DEB activity on an EMSA was confirmed (data not shown). As Figure 1A indicates, mitochondrial protein extracts prepared from V79 cells have DEB activity (lane 2), whereas similar extracts prepared from xrs-5 cells do not (lane 4). The gel-shifted DEB complex (indicated by the arrow) seen in Figure 1 is specific for DNA ends, since inclusion of non-radioactive linear DNA (indicated by the + in Fig. 1A, lanes 3 and 5) competes this activity away. Four independent mitochondrial protein extracts were prepared from the xrs-5 cell line and all were tested for DEB activity. In no case was mitochondrial DEB activity detected (data not shown). This finding suggests that Ku80 mRNA expression is essential for mitochondrial DEB activity.
Figure 1.
Ku80 mRNA expression is required for mitochondrial DEB activity. (A) EMSAs performed on wild-type V79 (lanes 2 and 3) and Ku80-deficient xrs-5 (lanes 4 and 5) mitochondrial protein extracts. Lane 1, probe f94 alone with no extract. Minus (–) and plus (+) signs refer to conditions without and with linear competitor DNA, respectively. The arrow points to migration of DEB complex. (B) DEB activity in nuclear extracts prepared from three 5-azacytidine-treated clones of xrs-5. (C) DEB activity in mitochondrial extracts prepared from three 5-azacytidine-treated clones of xrs-5.
It has been shown that treatment of xrs-5 cells with the demethylating agent 5-azacytidine leads to activation of a transcriptionally silent wild-type Ku80 gene, and hence the generation of clones that express Ku80 mRNA (24). We therefore treated xrs-5 cells with 5-azacytidine and screened several clones for nuclear DEB activity. Using this strategy, three out of eight clones tested had nuclear DEB activity, a frequency similar to that observed by Meechan et al. (24). Nuclear DEB activities in two of the reverted xrs-5 cell lines (clones 1 and 2) and in one of the xrs-5 cell lines that did not revert (clone 3) are depicted in Figure 1B. Mitochondrial protein extracts prepared from xrs-5 clones 1, 2 and 3 are depicted in Figure 1C. As Figure 1C indicates, clones 1 and 2 that had nuclear DEB activity also had mitochondrial DEB activity, while clone 3 that lacked nuclear DEB activity also lacked mitochondrial DEB activity. In all cases DEB activity was competed away by the inclusion of unlabeled linear DNA in the reaction (indicated by the + in Fig. 1). To independently confirm the results presented above, mitochondrial protein extracts were prepared from the Ku80-deficient xrs-6 cell line, and from xrs-6-HamKu86 cells, which express a hamster Ku80 transgene (both cell lines were obtained from the European Collection of Cell Culture, see Materials and Methods). As expected, a mitochondrial protein extract prepared from the xrs-6 cell line lacked DEB activity. In contrast, a mitochondrial protein extract prepared from the xrs-6-HamKu86 cell line possessed DEB activity (data not shown). These data confirm that expression of the Ku80 gene is required for mitochondrial DEB activity.
Western blot analysis of mitochondrial Ku80
The human mitochondrial protein that cross-reacted with polyclonal Ku80 antisera migrated on a 10% polyacrylamide gel with an apparent molecular weight of 68 kDa (22). Several groups have described the presence of a C-terminally truncated Ku80 protein in human cells with a similar apparent molecular weight (31–33). Therefore, experiments were performed to determine whether this human mitochondrial protein was a C-terminal truncated form of Ku80. Nuclear and mitochondrial protein extracts were prepared from HT1080 cells and western blot analysis performed using Mabs S10B1 and 111 that recognize N-terminal and C-terminal epitopes of Ku80, respectively. As Figure 2 indicates, Mab S10B1 specifically recognized an 86 kDa nuclear protein, and a 68 kDa mitochondrial protein. In contrast, Mab 111 recognized the 86 kDa nuclear protein, but failed to detect any mitochondrial proteins (Fig. 2). Additional western blot experiments were performed using three independently prepared human HT1080 mitochondrial protein extracts. In no case did Mab 111 recognize any mitochondrial proteins (data not shown). These results indicate that the mitochondrial 68 kDa protein lacks the epitope recognized by Mab 111 that maps between amino acids 610 and 705 of the Ku80 protein.
Figure 2.
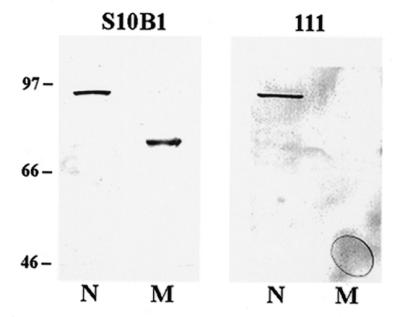
Mammalian mitochondrial Ku80 lacks a C-terminal epitope. Ten micrograms of nuclear (N) and mitochondrial (M) protein extracts prepared from human HT1080 cultured fibroblasts were loaded on a 10% PAGE. Western blot analysis was performed using the Ku80 Mabs designated S10B1 (specific for an N-terminal epitope of Ku80) and 111 (specific for a C-terminal epitope of Ku80). The positions of marker proteins and their molecular weights (in kDa) are indicated.
Mitochondrial membrane protects variant Ku80 from proteinase K digestion
Proteinase K is a broad-spectrum serine protease with a relatively small mass that allows it to traverse the nuclear pore. Thus, addition of proteinase K to a preparation of intact nuclei will lead to proteolysis of nuclear proteins. In contrast, mitochondria contain no such pores and, hence, proteins residing inside intact mitochondria are resistant to degradation by proteinase K. This characteristic of proteinase K was utilized by Willer et al. (34) to distinguish between two forms of the yeast CDC9-encoded DNA ligase; one form targeted to the nucleus and the other to the mitochondria. We similarly used this approach to further test the hypothesis that an alternate form of Ku80 is present in intact mitochondria. To conduct this analysis, intact nuclear and percoll-purified intact mitochondrial fractions were prepared (Materials and Methods), then treated with or without proteinase K for 30 min at 0°C. After treatment, SDS-containing polyacrylamide gel-loading buffer was added to the organelles, which were then boiled for 10 min. Samples were subjected to western blot analysis using polyclonal Ku80 antisera. As Figure 3A indicates, treatment of intact nuclei with proteinase K (+) resulted in the complete loss of immunoreactive protein. In contrast, substantial amounts of Ku80 immunoreactive protein remained following treatment of mitochondria with proteinase K. The inability of the mitochondria to provide complete protection from proteinase K treatment is presumably due to the fact that mitochondria become ‘leaky’ during percoll purification (35). However, as Figure 3A indicates, mitochondrial Ku80 was far more resistant to proteinase K treatment than nuclear Ku80. The results of this experiment were confirmed by preparing a crude intact mitochondrial fraction that was not percoll-purified, but instead pretreated with 20 µg/µl proteinase K for 30 min on ice to remove nuclear protein, then pelted at 15 000 r.p.m. to wash out the protease. The washed mitochondria were resuspended in homogenization buffer or lysis buffer (Materials and Methods) and treated with or without 20 µg/µl proteinase K for 30 min at 0°C before conducting western blot analysis. As seen in Figure 3B, crude intact mitochondria were completely resistant to treatment with proteinase K (compare lanes 1 and 2). When the mitochondrial membrane was disrupted by the addition of 350 mM NaCl (lysis buffer), the inherent sensitivity of this protein to proteinase K was demonstrated (compare lanes 3 and 4).
Figure 3.
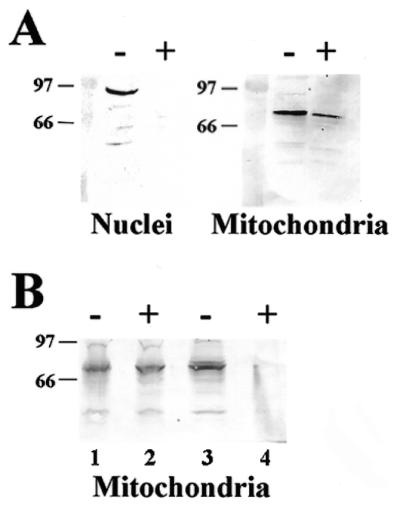
Ku80 present in intact mitochondria is resistant to proteinase K, while that present in the nucleus is not. (A) Western blot analysis of intact nuclei and intact percoll-purified mitochondria incubated in the absence (–) or presence (+) of 20 µg/µl proteinase K. The positions of marker proteins and their molecular weights are indicated (in kDa). (B) Western blot analysis of intact crude mitochondria (lanes 1 and 2) and lysed crude mitochondria (lanes 3 and 4) pre-incubated in the absence (–) or presence (+) of 20 µg/µl proteinase K.
Mitochondrial Ku80 is part of the EMSA DEB complex
Mab-mediated supershift analysis was performed to determine if the human mitochondrial truncated form of Ku80 detected by western blot analysis (Fig. 2) was physically associated with the mitochondrial DEB complex. Nuclear and mitochondrial protein extracts were prepared from HT1080 cells and standard EMSAs performed in the presence or absence of Mabs S10B1 and 111 (Materials and Methods). As Figure 4, lanes 1 and 5 indicate, nuclear (N) and mitochondrial (M) extracts possessed DEB activity (the respective mobilities of the DEB complexes are indicated by the large and small arrowheads). Addition of Mab S10B1 supershifted the EMSA DEB complex present in both the nuclear and mitochondrial extracts (Fig. 4, lanes 2 and 6, respectively). In contrast, Mab 111 caused a supershift of the nuclear DEB complex (Fig. 4, lane 3) but did not alter the mobility of the mitochondrial DEB complex (Fig. 4, lane 7). As in Figure 1, inclusion of unlabeled linear DNA competed away the DEB complex in both nuclear (Fig. 4, lane 4) and mitochondrial (Fig. 4, lane 8) extracts. These data demonstrate that the truncated form of Ku80 detected by western blot analysis in human mitochondrial protein extracts is involved in DEB activity.
Figure 4.
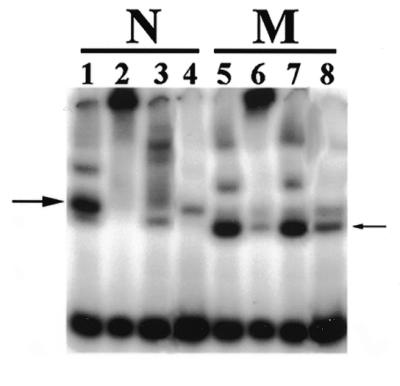
A C-terminal truncated form of Ku80 is associated with mitochondrial DEB activity. A standard EMSA was performed (Materials and Methods) using HT1080 nuclear (N) or mitochondrial (M) protein extracts. Two micrograms of Ku80 Mab S10B1 (N-terminal antibody) was added to the EMSA in lanes 2 and 6. Ku80 Mab 111 (0.8 µg) (C-terminal antibody) was added to the EMSA in lanes 3 and 7. Linear competitor DNA was substituted for circular DNA in lanes 4 and 8. The large arrowhead indicates the mobility of nuclear DEB complex. The small arrowhead indicates the mobility of mitochondrial DEB complex.
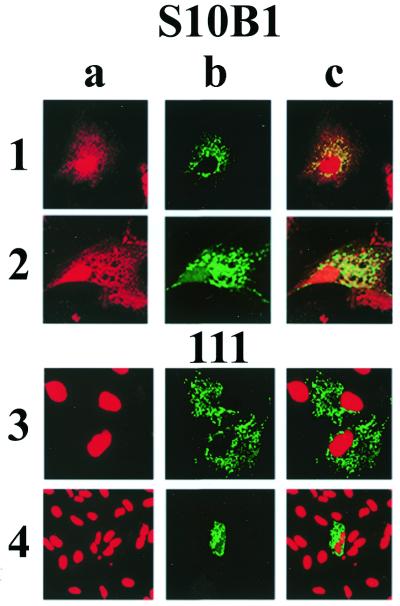
Ku80 Mab co-localizes with mitochondrial-targeted GFP
The data presented thus far provide support for the hypothesis that a C-terminally truncated form of Ku80 is localized in mammalian mitochondria where it catalyzes a DEB activity. We wished to provide independent confirmation that the presence of the 68 kDa form of Ku80 in mitochondrial protein extracts was not due to contamination by non-mitochondrial subcellular fractions. Immunocytochemical analysis of CV1 cells using the S10B1 and 111 Ku80 Mabs was thus performed. CV1 cells were transiently transfected with a pEGFP-N1 vector that carries the mitochondrial targeting sequence of cytochrome c oxidase in frame with the gene for the GFP (29). Since transfection efficiency was <100%, not all cells expressed the GFP. Cells were then fixed and permeablized before incubating with the primary Ku80 Mabs followed by incubation with secondary goat anti-mouse antibody conjugated to rhodamine (Materials and Methods). Representative examples of this analysis using the Ku80 N-terminal S10B1 Mab are shown in rows 1 and 2 of Figure 5. The S10B1 Mab recognized protein present in both nuclear and cytoplasmic compartments (Fig. 5, panels 1a and 2a). Fluorescence due to the presence of mitochondrial GFP outlines the boundaries of the mitochondria within these cells (Fig. 5, panels 1b and 2b). Panels 1c and 2c of Figure 5 display the merged images from panels (1a + 1b) and (2a + 2b), respectively, and demonstrate that the cytoplasmic S10B1-mediated rhodamine fluorescence co-localized with that of the GFP, indicating that it is mitochondrial. Thus the Ku80 protein recognized by this antibody is found in both the nucleus as well as in the mitochondria. An identical analysis was conducted using the Ku80 Mab 111 that recognized nuclear, but not mitochondrial, Ku80 on both western and EMSA supershift analysis. The results from this analysis are presented in Figure 5, rows 3 and 4. Panels 3a and 4a indicate that the Mab 111 recognizes a nuclear protein. Panels 3b and 4b again display the intracellular distribution of the mitochondrial GFP protein in these cells. Finally, panels 3c and 4c of Figure 5 display the merged images from panels (3a + 3b) and (4a + 4b), respectively. This analysis was consistent with earlier experiments indicating that the Ku80 C-terminal-specific 111 Mab recognizes Ku80 in the nucleus, but not in the mitochondrial compartment (Fig. 5, panels 3a and 4a). Taken together the results presented in Figure 5 indicate that the truncated 68 kDa form of Ku80 exists within the mitochondrial compartment of these cells.
Figure 5.
Immunocytochemical analysis reveals mitochondrial localization of Ku80. CV1 cells were transiently transfected with a mitochondrial-targeted GFP, then fixed and permeablized on glass cover-slips. Fixed cells were incubated for 1 h at 37°C with Ku80 Mabs S10B1 (rows 1 and 2) or 111 (rows 3 and 4) at a 1:50 dilution. Cells were then incubated for an additional 1 h with anti-mouse IgG conjugated to rhodamine. Column a represents the rhodamine fluorescence (anti-Ku80 antibody). Column b represents the GFP fluorescence (mitochondrial GFP). Column c represents merged images from (1a +1b), (2a + 2b), (3a + 3b) and (4a + 4b).
DISCUSSION
Ku80 is required for DEB activity in mammalian mitochondrial protein extracts. This conclusion was based on the results of several independent lines of evidence. The hamster cell line xrs-5 lacks Ku80 mRNA expression and consequently is deficient in both nuclear and mitochondrial DEB activity. Reversion of wild-type nuclear DEB activity with 5-azacytidine was always associated with reversion of wild-type mitochondrial DEB activity in these cells. Similarly, mitochondrial DEB activity was detected in the xrs-6-HamKu86 cell line that expressed the hamster gene encoding Ku80. Mabs specific for Ku80 revealed that human mitochondria contain a C-terminally truncated Ku80 protein that is associated with DNA ends during an EMSA. We demonstrated that this truncated Ku80 protein was present both in human mitochondrial protein extracts as well as in intact mitochondria. Immunocytochemical analysis of fixed CV1 cells also identified the presence of a C-terminally truncated Ku80 protein in the mitochondria. Taken together, these data provide clear evidence that Ku80 is involved in mammalian mitochondrial DEB activity.
Our initial studies of mitochondrial DEB activity utilized the cell line XR-V15B that possesses an in-frame deletion in the gene encoding Ku80 resulting in the loss of amino acids 371 to 417 of the encoded protein (36). Despite this deletion, Ku80 mRNA is expressed at wild-type levels in these cells (36). Although this deletion disrupts nuclear DEB activity, the mechanism of this disruption is unknown. Originally, the deletion found in XR-V15B Ku80 was thought to span a domain critical for Ku80 heterodimerization with Ku70 (37). However, a more detailed mapping of Ku80 functional domains revealed that amino acids 449 to 477 of Ku80 were required for heterodimerization with Ku70 (38). This domain lies C-terminal to the deletion found in the XR-V15B Ku80 protein. Our previous work confirmed that the XR-V15B cell line lacked DEB activity in the nucleus, but found that mitochondrial DEB activity was wild-type. Although this finding suggested that Ku80 was not involved in mitochondrial DEB activity, we were unable to rule out the possibility that the expressed Ku80 mRNA in this cell line encoded a Ku80 protein that had activity in the mitochondria. Because we were unable to rule out this possibility, we obtained the xrs-5 cell line that lacks detectable Ku80 mRNA expression. The xrs-5 cell line provided a more appropriate tool to ascertain the role of Ku80 in mammalian mitochondrial DEB activity. As indicated, results from the present study do confirm that Ku80 is required for mitochondrial DEB activity.
Several different groups have described a variant form of Ku80 in human cells with a similar apparent molecular weight to that described here (31–33). A variant human Ku80 protein that possessed a C-terminal truncation was originally characterized by Paillard and Strauss (31). Han et al. (32) detected both full-length Ku80 and a C-terminally truncated form of Ku80 in nuclear extracts prepared from the human promyelocytic leukemic cell line HL60. This group concluded that the truncated protein was most likely generated by a post-translational modification of Ku80. Muller et al. (33) detected a similar C-terminally truncated Ku80 protein in whole cell extracts prepared from human B lymphocytes, and also concluded that it was a post-translational product of Ku80.
Our studies on the human C-terminally truncated Ku80 indicate that it resides in the mitochondria, and suggests that the processing of the C-terminus occurs prior to disruption of the cells during purification. The CV1 cells used for immunocytochemical analysis (Fig. 4) were rapidly fixed in formaldehyde prior to incubating with Ku80-specific antibodies. Presumably this fixing procedure would not allow time for proteolytic processing to occur. The full-length protein was detected in the nuclei of these cells, but only the C-terminally truncated form in the mitochondria. Similarly, isolated intact mitochondria contained only the C-terminally truncated Ku80 (Fig. 5). During the preparation of our nuclear and mitochondrial extracts, a broad spectrum of protease inhibitors was included, and the entire procedure carried out below 4°C. Our data indicates that Ku80 is localized in mammalian mitochondria, and that a specific biological event is apparently responsible for generating this truncated form of the protein either prior to or upon entry into that organelle. The role, if any, of this C-terminal truncation in protein function in the mitochondria or targeting to the mitochondria remains unclear.
There is precedence in mammalian and yeast cells for alternate or identical forms of the same DNA repair proteins to localize in both nuclear and mitochondrial compartments. This precedence is logical as the two subcellular compartments contain DNA and would therefore be expected to possess similar processes that maintain the DNA. For example, both the mammalian DNA ligase III and the yeast CDC9 genes contain at least two sites where transcription is initiated. Transcription initiation from one site generates an mRNA that encodes a nuclear-targeted ligase, while initiation from a second site generates an mRNA that encodes a mitochondrial-targeted ligase (29,34). Alternate splicing of the RNA encoding 8-oxoguanine DNA glycosylase generates at least two mRNA species, one encoding a nuclear form, the other encoding a mitochondrial form (39). Post-translational modification is similarly known to regulate subcellular localization of the base-excision DNA repair protein N-glycosylase (40). These proteins play similar roles in both the nucleus and mitochondria.
Targeting of cytoplasmic protein to subcellular locations is a complex phenomenon that is not entirely understood. Most known mitochondrial targeting sequences are located on the N-terminus of the protein and consist of a cluster of positively charged amino acids that form an α-helical structure (41). There is also evidence, however, for C- to N-terminal protein transport into the mitochondria initiated by C-terminal targeting sequences (42,43). Folsch et al. (42) demonstrated that the mitochondrial import machinery showed no preference for marker protein fused with N- or C-terminal targeting sequence. Further, the C-terminal mitochondrial targeting sequence of the DNA helicase Hmi1p is cleaved off upon entry into the mitochondrial matrix (43). Targeting mechanisms other than N- or C-terminal targeting sequences have been identified. For example, mitochondrial heme lyases possess a mitochondrial targeting sequence in the third quarter of the protein (44). Phosphorylation of the cytochrome P4502B1 at position S128 redirects this protein from the endoplasmic reticulum to the mitochondria (45). These examples demonstrate that multiple distinct mechanisms of targeting to the mitochondria are found in biological systems. The human mitochondrial Ku80 lacks the C-terminal amino acids of the full-length protein, consistent with the cleavage of a C-terminal mitochondrial-targeting sequence. However, sequence inspection of human and hamster Ku80 revealed no obvious mitochondrial targeting sequence at either terminus. Targeting of Ku80 to the mitochondria may rely on novel mechanisms. Our laboratory is currently investigating the mechanism of Ku80 targeting to the mitochondria.
There are a number of possible functions for a Ku80-mediated DEB activity in mammalian mitochondria. This activity could function to bind to and target damaged mitochondrial DNA for degradation. This hypothesis is reasonable given the high copy number of mitochondrial DNA, and the observation that significantly reduced levels of mitochondrial DNA has no effect on cytochrome c oxidase expression in mitochondria (46). Alternatively, the mitochondrial DEB activity could participate in a DNA repair pathway, as is the case in the nucleus. Mammalian mitochondria possess a DNA end-joining activity (47) that is catalyzed by a mitochondrial form of DNA ligase III (29). The DNA end-joining activity seen in mitochondrial extracts is strikingly similar to DNA end-joining activity present in nuclear extracts (48). It is thus conceivable that the mitochondrial DEB activity plays an accessory role in ligase III-mediated DNA end-joining in mitochondria in vivo.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Uma Lakshmipathy for advice concerning immunofluorescence experiments and editorial comments. This work was supported in part by grants from the NIH (R29 CA61906), the American Cancer Society (DHP-171) and the American Heart Association (9951198Z). G.C. is supported by a fellowship from the American Heart Association.
REFERENCES
- 1.Mimori T., Akizuki,M., Yamagata,H., Inada,S., Yoshida,S. and Homma,M. (1981) J. Clin. Invest., 68, 611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mimori T., Hardin,J.A. and Steitz,J.A. (1986) J. Biol. Chem., 261, 2274–2278. [PubMed] [Google Scholar]
- 3.Dynan W.S. and Yoo,S. (1998). Nucleic Acids Res., 26, 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rathmell W.K. and Chu,G. (1994) Mol. Cell Biol., 14, 4741–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taccioli G.E., Gottlieb,T.M., Blunt,T., Priestley,A., Demengeot,J., Mizuta,R., Lehmann,A.R., Alt,F.W., Jackson,S.P. and Jeggo,P.A. (1994) Science, 265, 1442–1445. [DOI] [PubMed] [Google Scholar]
- 6.Jeggo P.A. and Kemp,L.M. (1983) Mutat. Res., 112, 313–327. [DOI] [PubMed] [Google Scholar]
- 7.Rathmell W.K. and Chu,G. (1994) Proc. Natl Acad. Sci. USA, 91, 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeggo P.A. (1998) Adv. Genet., 38, 185–219. [DOI] [PubMed] [Google Scholar]
- 9.Mimori T. and Hardin,J.A. (1986) J. Biol. Chem., 261, 10375–10379. [PubMed] [Google Scholar]
- 10.Dvir A., Peterson,S.R., Knuth,M.W., Lu,H. and Dynan,W.S. (1992) Proc. Natl Acad. Sci. USA, 89, 11920–11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb T.M. and Jackson,S.P. (1993) Cell, 72, 131–142. [DOI] [PubMed] [Google Scholar]
- 12.Porter S.E., Greenwell,P.W., Ritchie,K.B. and Petes,T.D. (1996) Nucleic Acids Res., 24, 582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gravel S., Larrivee,M., Labrecque,P. and Wellinger,R.J. (1998) Science, 280, 741–744. [DOI] [PubMed] [Google Scholar]
- 14.Nugent C.I., Bosco,G., Ross,L.O., Evans,S.K., Salinger,A.P., Moore,J.K., Haber,J.E. and Lundblad,V. (1998) Curr. Biol., 8, 831–834. [DOI] [PubMed] [Google Scholar]
- 15.Hsu H.L., Gilley,D., Blackburn,E.H. and Chen,D.J. (1999) Proc. Natl Acad. Sci. USA, 96, 12454–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey S.M., Meyne,J.M., Chen,D.J., Kurimasa,A., Li,G.C., Lehnert,B.E. and Goodwin,E.H. (1999) Proc. Natl Acad. Sci. USA, 96, 14899–14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu C., Bogue,M.A., Lim,D.S., Hasty,P. and Roth,D.B. (1996) Cell, 86, 379–389. [DOI] [PubMed] [Google Scholar]
- 18.Gu Y., Seidl,K.J., Rathbun,G.A., Zhu,C., Manis,J.P., van der Stoep,N., Davidson,L., Cheng,H.L., Sekiguchi,J.M. and Frank,K. (1997) Immunity, 7, 653–665. [DOI] [PubMed] [Google Scholar]
- 19.Gu Y., Jin,S., Gao,Y., Weaver,D.T. and Alt,F.W. (1997) Proc. Natl Acad. Sci. USA, 94, 8076–8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogel H., Lim,D.-S., Karsenty,G., Finegold,M. and Hasty,P. (1999) Proc. Natl Acad. Sci. USA, 96, 10770–10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Featherstone C. and Jackson,S.P. (1999) Mutat. Res., 434, 3–15. [DOI] [PubMed] [Google Scholar]
- 22.Coffey G., Lakshmipathy,U. and Campell,C. (1999) Nucleic Acids Res., 27, 3348–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singleton B.K., Priestley,A., Steingrimsdottir,H., Gell,D., Blunt,T., Jackson,S.P., Lehmann,A.R. and Jeggo,P.A. (1997) Mol. Cell Biol., 17, 1264–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meechan P.J., Haraf,D.J., Diamond,A.M. and Grdina,D.J. (1994) Radiat. Res., 140, 437–440. [PubMed] [Google Scholar]
- 25.Storrie B. and Madden,E.A. (1990) Methods Enzymol., 182, 203–225. [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Chou,C.H., Blankson,J., Satoh,M., Knuth,M.W., Eisenberg,R.A., Pisetsky,D.S. and Reeves,W.H. (1993) Mol. Biol. Rep., 18, 15–28. [DOI] [PubMed] [Google Scholar]
- 27.Reeves W.H. (1985) J. Exp. Med., 161, 18–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopata M., Cleveland,D.W. and Sollner-Webb,B. (1984) Nucleic Acids Res., 12, 5707–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakshmipathy U. and Campbell,C. (1999) Mol. Cell Biol., 19, 3869–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brini M., Marsault,R., Bastianutto,C., Pozzan,T. and Rizzuto,R. (1994) Cell Calcium, 16, 259–268. [DOI] [PubMed] [Google Scholar]
- 31.Paillard S. and Strauss,F. (1993) Proteins: Struct. Funct. Gen., 15, 330–337. [DOI] [PubMed] [Google Scholar]
- 32.Han Z. Johnston,C., Reeves,W.H., Carter,T., Wyche,J.H. and Hendrickson,E.A. (1996) J. Biol. Chem., 271, 14098–14104. [DOI] [PubMed] [Google Scholar]
- 33.Muller C., Dusseau,C., Calsou,P. and Salles,B. (1998) Oncogene, 16, 1553–1560. [DOI] [PubMed] [Google Scholar]
- 34.Willer M., Rainey,M., Pullen,T. and Stirling,C.J. (1999) Curr. Biol., 9, 1085–1094. [DOI] [PubMed] [Google Scholar]
- 35.Mickelson J.R., Greaser,M.L. and Marsh,B.B. (1980) Anal. Biochem., 109, 255–260. [DOI] [PubMed] [Google Scholar]
- 36.Errami A., Smider,V., Rathmell,W.K., He,D.M., Hendrickson,E.A., Zdzienicka,M.Z. and Chu,G. (1996) Mol. Cell Biol., 16, 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cary R.B., Chen,F., Shen,Z. and Chen,D.J. (1998) Nucleic Acids Res., 26, 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osipovich O., Durum,S.K. and Muegge,K. (1997) J. Biol. Chem., 272, 27259–27265. [DOI] [PubMed] [Google Scholar]
- 39.Nishioka K., Ohtsubo,T., Oda,H., Fujiwara,T., Kang,D., Sugimachi,K. and Nakebeppu,Y. (1999) Mol. Biol. Cell, 10, 1637–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You H.J., Swanson,R.L., Harrington,C., Corbett,A.H., Jinks-Robertson,S., Senturker,S., Wallace,S.S., Boiteux,S., Dizdaroglu,M. and Doetsch,P.W. (1999) Biochemistry, 38, 11298–11306. [DOI] [PubMed] [Google Scholar]
- 41.Roise D. (1997) J. Bioenerg. Biomembr., 29, 19–27. [DOI] [PubMed] [Google Scholar]
- 42.Folsch H., Gaume,B., Brunner,M., Neupert,W. and Stuart,R.A. (1998) EMBO J., 17, 6508–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee C.M., Sedmun,J., Neupert,W. and Stuart,R.A. (1999) J. Biol. Chem., 274, 20937–20942. [DOI] [PubMed] [Google Scholar]
- 44.Diekert K., Kispal,G., Guiard,B. and Lill,R. (1999) Proc. Natl Acad. Sci. USA, 96, 11752–11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anandatheerthavarada H.K., Biswas,G., Mullick,J., Sepuri,N.B.V., Otvos,L., Pain,D. and Avadhani,N.G. (1999) EMBO J., 18, 5494–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barazzoni R., Short,K.R. and Nair,K.S. (2000) J. Biol. Chem., 275, 3343–3347. [DOI] [PubMed] [Google Scholar]
- 47.Lakshmipathy U. and Campbell,C. (1999) Nucleic Acids Res., 27, 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramsden D.A. and Gellert,M. (1998) EMBO J., 17, 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]