Abstract
The dissection of RNA editing mechanisms in Physarum mitochondria has been hindered by the absence of a soluble in vitro system. Based on our studies in isolated mitochondria, insertion of non-encoded nucleotides into Physarum mitochondrial RNAs is closely linked to transcription. Here we have fractionated mitochondrial lysates, enriching for run-on RNA synthesis, and find that editing activity co-fractionates with pre-formed transcription elongation complexes. The establishment of this soluble transcription-editing system allows access to the components of the editing machinery and permits manipulation of transcription and editing substrates. Thus, the availability of this system provides, for the first time, a means of investigating roles for cis-acting elements, trans-acting factors and nucleotide requirements for the insertion of non-encoded nucleotides into Physarum mitochondrial RNAs. This methodology should also be broadly applicable to the study of RNA processing and editing mechanisms in a wide range of mitochondrial systems.
INTRODUCTION
Gene expression can be affected by RNA editing through insertion, deletion or substitution of individual residues. Sequence alterations due to editing have been described in organisms across a broad phylogenetic spectrum, including mammals, plants, viruses, snails, Drosophila, kinetoplastid and amoebid protozoa, chytriomycete fungi, and slime moulds (1) and are particularly prevalent in mitochondria (2). In the acellular slime mould Physarum polycephalum, RNA editing creates open reading frames in mitochondrial mRNAs, and is responsible for the formation of conserved primary, secondary and presumably tertiary structures of mitochondrial tRNAs and rRNAs (3–6). These alterations involve the addition of non-encoded C, U, A and G residues as either single or dinucleotide insertions, as well as infrequent C to U changes. The means by which these site-specific changes are specified is still unknown.
To investigate the mechanism(s) of Physarum editing, we previously developed an isolated mitochondrial system capable of carrying out accurate and efficient insertional RNA editing (7). Using this system we have demonstrated that nascent RNAs are editing substrates (7), that editing occurs very close to the site of RNA synthesis in a 5′→3′ direction (8,9), and that transcription and editing are physically and/or mechanistically coupled in some way (9). These results have led us to propose that insertional editing in Physarum polycephalum is a co-transcriptional process (10).
Although use of isolated mitochondria has allowed us to investigate many aspects of Physarum editing (11), this system has a number of limitations for mechanistic studies. Primary drawbacks include the inaccessibility of the transcription and editing machineries to most manipulations and the presence of substantial nucleotide pools. We therefore decided to develop a more purified in vitro system in which to study editing. Based on the close association between transcription and editing in Physarum mitochondria, we reasoned that it might be possible to purify the editing machinery further by following endogenous transcription activity, which is easily assayed. Here we describe the establishment of a soluble system that should be readily adaptable to other mitochondrial editing systems. These partially purified transcription elongation complexes carry out both run-on RNA synthesis and insertional editing in an efficient manner. This transcription/editing system eliminates many of the problems intrinsic to the use of isolated mitochondria and therefore represents a significant advance in our efforts to dissect Physarum editing mechanisms.
MATERIALS AND METHODS
All experimental procedures were carried out as described by Visomirski-Robic and Gott (7), except as noted.
Physarum cultures
Physarum strain M3C was maintained as microplasmodia in semi-defined medium plus hemin (SDMH) (12). Cultures were grown in 250 ml baffled flasks at 150 r.p.m. on a gyratory shaker at 26°C. For large scale cultures, 2 l baffled flasks containing 300 ml SDMH were innoculated and shaken in a gyratory shaker at 210 r.p.m. for 48 h at room temperature. Typically, 2–3 l cultures at mid-log phase were used for preparation of transcription elongation complexes.
Plasmids and in vitro transcription of control RNAs
PCR-derived cDNA and mitochondrial DNA clones were generated as previously described (4). The PCR products were cloned into pBSM13+ (Stratagene) for in vitro transcription with T7 RNA polymerase and isolation of single-stranded DNA (ssDNA) used in S1 nuclease protection experiments. All inserts were sequenced in their entirety. Control RNAs were produced using the Ambion Maxiscript in vitro transcription kit and linearized templates. The control RNAs used in Figure 4A were derived from the following regions of the α-ATPase sequence: nt 877–1631 (edited) and nt 848–1577 (unedited).
Figure 4.
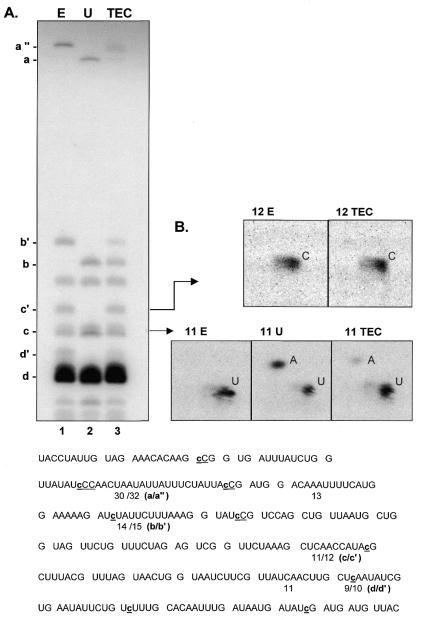
RNA editing in TEC preparations. (A) [α-32P]GTP-labeled RNAs were gel purified after S1 nuclease protection with an α-ATPase-specific probe, digested with ribonuclease T1 and the resulting oligonucleotides separated on a denaturing 20% polyacrylamide gel. Oligonucleotide fragments containing editing sites are indicated to the left. For oligonucleotides overlapping sites of nucleotide insertion, each apostrophe designates the presence of an added nucleotide. (B) Nearest neighbor analysis of RNase T1 fragments. RNase T1 oligonucleotides c (11 nt) and c′ (12 nt) were eluted from the gel shown in (A) and digested to mononucleotides, and the resulting 3′ NMPs were separated via two-dimensional thin-layer chromatography as described in Materials and Methods. Bottom, sequence of the RNase T1 oligonucleotides present in the S1 nuclease protected region. The size of each [α-32P]GTP-labeled fragment visible on the gel is indicated, with the fragments containing editing sites designated with letters corresponding to the unedited (a, b, c, d) and edited (a″, b′, c′, d′) sequence. Nucleotide insertion sites are shown in lower case letters within the sequence. Note that the unedited control RNA contains two 11 nt RNase T1 fragments, while the edited control RNA has an 11mer and a 12mer.
Protein gels
Equal aliquots (21 µl) of Sepharose 4B column fractions were mixed with 7 µl 4× loading dye (4× = 0.25 M Tris pH 6.8, 8% SDS, 20% β-mercaptoethanol, 40% glycerol, BPB) and heated to 95°C for 3 min prior to separation on a 5% stacking:8% resolving polyacrylamide:bis (29:1), SDS gel in Tris–glycine buffer (13). Bands were visualized by staining with Coomassie Brilliant Blue R250:methanol:H2O:glacial acetic acid (0.25:45:45:10, w/v/v/v) followed by destaining in methanol: H2O:glacial acetic acid (45:45:10, v/v/v).
Isolation of mitochondrial transcription elongation complexes
Mitochondria isolation was performed as previously described by Visomirski-Robic and Gott (7). Unless otherwise stated, all steps were carried out at 4°C. Typically, mitochondrial pellets (∼40–80 mg of mitochondrial protein) were resuspended in 1.5–2 ml of 2× TEDMG buffer (2× = 20 mM Tris–HCl pH 8.0, 1 mM EDTA, 2 mM DTT, 10 mM MgCl2, 20% glycerol). After addition of the protease inhibitors PMSF and leupeptin, mitochondria were lysed by the addition of NP-40 in the presence of KCl. Mitochondrial lysates were adjusted to a final concentration of 10–20 mg/ml mitochondrial protein in 10 mM Tris–HCl (pH 8.0 at room temperature), 0.5 mM EDTA, 1 mM DTT, 5 mM MgCl2, 10% glycerol, 0.5 mM PMSF, 1 µg/ml leupeptin, 1% NP-40, 250 mM KCl in a total volume of 3–4 ml. The mitochondrial lysate was then spun at 39 000 r.p.m. for 60 min (130 000 g) in a TLS 55 rotor using a Beckman table-top ultracentrifuge. The cleared lysate was loaded onto a 45 ml (0.9 × 70 cm) Sepharose 4B column previously equilibrated with 1× TEDMG buffer and 1 ml fractions were collected and assayed for transcription activity as described below. Total protein present in each fraction was determined using the BioRad DC protein assay as directed by the manufacturer. Active fractions were pooled and dialyzed against PEDMG buffer (20 mM potassium phosphate pH 7.8, 0.1 mM EDTA, 1 mM DTT, 0.5 mM MgCl2, 50% glycerol), aliquoted and frozen at –80°C (14). Aliquots prepared in this way are stable for several months without significant loss in activity.
Run-on transcription assay
Synthesis of run-on transcripts by column fractions was assayed in 20 mM Tris–HCl pH 8.0, 10 mM MgCl2, 100 µg/ml BSA, 2 mM DTT, 500 µM each unlabled nucleotide, 5 µM labeled nucleotide (2.5 µCi/reaction), using 4 µl of column fraction in each 20 µl reaction. Reactions were incubated at 30°C for 30 min, terminated by spotting on 2 cm2 DE81 filter papers, and the filters washed 4× for 5 min in 500 ml of 0.3 M ammonium formate pH 7.8, 10 mM sodium pyrophosphate, briefly rinsing with water between each wash (15). After the fourth wash, the filter papers were rinsed once with 10 ml of 95% ethanol, air-dried and counted using a liquid scintillation counter.
Transcription elongation complex (TEC) transcription and RNA isolation
Transcription reactions using dialyzed TEC preparations were carried out in 20 mM Tris–HCl pH 8.0, 10 mM MgCl2, 100 µg/ml BSA, 2 mM DTT, 500 µM unlabeled nucleotides for 40 min at 30°C; labeled nucleotide and protein concentrations varied between experiments and are described in the appropriate sections. Labeling reactions in Figures 1 and 4 were chased for 10 min with 500 µM of the limiting nucleotide, GTP. In the experiment shown in Figure 1, equal volumes of each fraction were used in transcription assays with the exception of lane 5 (mito S130). Because the mito S130 fraction contained significantly less protein (700 µg/ml), this reaction was scaled up 10-fold so that even small amounts of RNA synthesis would be detected. Transcription reactions were terminated by the addition of EDTA and SDS to a final concentration of 20 mM and 0.1%, respectively. Samples were extracted twice with an equal volume of phenol–CIA [phenol:chloroform:isoamyl alcohol (25:24:1, v/v/v)], and nucleic acids were precipitated with 1/10 volume 4 M ammonium acetate and 2.5 volumes ethanol. Samples (except those in Figs 1 and 3C) were treated with 10–20 U of RNase free-DNase I (Boehringer Mannheim) in a final concentration of 10 mM Tris–HCl pH 7.5, 6.25 µM DTT, 12.5 µM magnesium acetate and 0.25 U/µl RNasin (Boehringer Mannheim) in a total volume of 20–40 µl. The reactions were terminated by addition of SDS (0.6% final), acetic acid (to 12 mM), and EDTA (to 30 mM), extracted as described above and precipitated.
Figure 1.
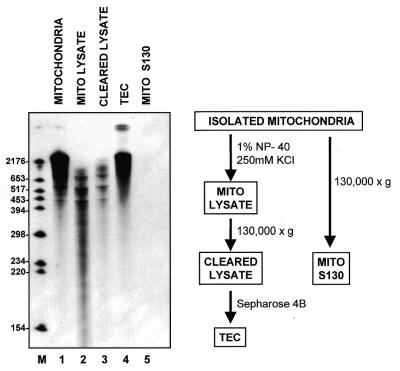
RNA synthesis in mitochondrial fractions. Run-on transcription in isolated mitochondria (lane 1), lysed mitochondria (lane 2), cleared mitochondrial lysate (lane 3), pooled TECs (lane 4) and the cleared supernatant from unlysed mitochondria (lane 5) was assayed as described in Materials and Methods. Note that the mito S130 sample (lane 5) represents the amount of RNA synthesized in a transcription reaction that had been scaled up ten-fold relative to the samples in lanes 1–4, supporting the conclusion that the clearing step is sufficient to pellet all unlysed mitochondria.
Figure 3.
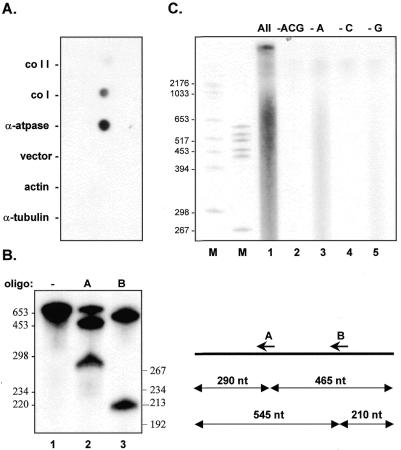
Characterization of TEC. (A) Dot blot hybridization analysis of RNAs synthesized by TEC. DNAs from the nuclear genes actin and tubulin, the mitochondrial genes coI, coII and α-ATPase, and the cloning vector were immobilized and hybridized to labeled transcripts synthesized by TEC as described in Materials and Methods. (B) RNase H digestion of S1 nuclease protected α-ATPase mRNA synthesized in the absence of a cold nucleotide chase. Lane 1, no oligonucleotide, no RNase H; lane 2, oligonucleotide A in the presence of RNase H; lane 3, oligonucleotide B in the presence of RNase H. Sizes of the expected cleavage products are shown in the diagram to the right. (C) Nucleotide requirements for transcription by TEC. Run-on RNA synthesis in the presence of all four ribonucleotides (lane 1); 20 µM [α-32P]UTP only (lane 2); 20 µM [α-32P]UTP, 500 µM CTP and GTP (lane 3); 20 µM [α-32P]UTP and 500 µM ATP and GTP (lane 4); or 20 µM [α-32P]UTP and 500 µM CTP and ATP (lane 5).
Dot blot hybridization
Denatured plasmid DNA was immobilized on a nylon membrane (GeneScreen, NEN Research Products) as described previously (7). The membrane was blocked through preincubation with hybridization buffer (1% BSA, 0.5 M sodium phosphate pH 7.2, 15% formamide, 1 mM EDTA, 7% SDS) for 4.5 h at 37°C. Hybridization of [α-32P]GTP-labeled TEC RNA (5 µM GTP, 160 µg/ml protein in a 60 µl reaction) was performed using the same hybridization buffer at 37°C for 17 h. The membrane was washed twice at room temperature with 5× SSC, 0.1% SDS at room temperature, then once with 2× SSC, 0.1% SDS at room temperature prior to autoradiography.
ATP quantitation
The concentration of ATP present in two different TEC preparations was determined using the ATP bioluminescent assay kit (FL-AA, Sigma) as directed by the manufacturer using the supplied standards. This coupled firefly luciferase assay is linear in the range of 2 × 10–12–8 × 10–5 M ATP.
S1 nuclease digestion
S1 nuclease protection experiments were performed as described by Visomirski-Robic and Gott (7), except that 6 µg of ssDNA was used to protect RNA during the first round of S1 digestion, 4 µg of ssDNA was used in the second round and 2 µg of ssDNA was used in the third round (when necessary). Digestions were performed at 26°C for 1.5 h using S1 mapping buffer (0.75 M NaCl, 0.05 M sodium acetate, pH 4.5, 4.5 mM ZnSO4) containing 200–300 U S1 nuclease (Boehringer Mannheim). The ssDNA probes used in Figure 3B (nt 877–1631, edited α-ATPase) and Figure 4A (nt 1035–1305, edited α-ATPase) were prepared as described by Visomirski-Robic and Gott (7).
RNase H digestion
To ascertain the extent of RNA synthesis in TEC preparations, labeled RNA was synthesized in the absence of a cold nucleotide chase in 20 mM Tris–HCl pH 8.0, 10 mM MgCl2, 100 µg/ml BSA, 2 mM DTT, 500 µM each unlabled nucleotide, 20 µM [α-32P]ATP (150 µCi/reaction), 60 µg mitochondrial protein in a 90 µl reaction. A 755 nt region of the α-ATPase mRNA was isolated via two rounds of S1 protection and purified on a 4% denaturing polyacrylamide gel containing 7 M urea/1× TBE. The band was eluted in 10 mM Tris–HCl pH 7.5, 250 mM sodium acetate, 1 mM EDTA, 0.25% SDS for 14 h at room temperature, extracted with phenol, then CIA, and ethanol precipitated. After DNase I treatment and gel purification as described above, 1300 c.p.m. of S1 protected RNA was mixed with 2 µl of 10 µM oligonucleotide A or B in a total volume of 13.5 µl, heated to 95°C for 2 min, spun briefly, then put on ice. RNase H digestions were carried out for 45 min at 37°C in 20 mM Tris–HCl pH 7.5, 100 mM KCl, 10 mM MgCl2, 0.1 mM DTT, 5% sucrose using 2 U of RNase H (Gibco BRL) in a final volume of 25 µl. Reactions were stopped by the addition of EDTA (16 mM final) and SDS (to 0.08%), followed by phenol and CIA extractions, and ethanol precipitation.
RNase T1 digestion
To determine whether RNAs synthesized by TEC preparations are edited, RNA was labeled in 20 mM Tris–HCl pH 8.0, 10 mM MgCl2, 100 µg/ml BSA, 2 mM DTT, 500 µM each unlabeled nucleotide, 20 µM [α-32P]GTP, 90 µg/ml protein for 40 min at 30°C, then chased for 10 min with 500 µM GTP. A 270 nt region of the α-ATPase mRNA was isolated via three rounds of S1 protection and purified on a 4% denaturing polyacrylamide gel. Edited and unedited control transcripts were isolated in parallel using two rounds of S1 nuclease protection with the same ssDNA probe under conditions described by Visomirski-Robic and Gott (7). The gel-purified S1-protected RNA fragments were resuspended in 5 µl of dH2O, incubated at 95°C for 2 min, then put directly on ice. After the addition of 1 µl of 1 mg/ml tRNA (Sigma) to each sample, RNAs were digested for 45 min at 37°C with 1 µl of 100 U/µl RNase T1 (Boehringer Mannheim). Reactions were stopped by the addition of an equal volume of gel dye (7 M urea, 1× TBE, xylene cyanol, BPB). Samples were heated for 2 min at 85°C and RNase T1 oligonucleotides were separated on 20% polyacrylamide, 7 M urea, 50 mM Tris, 50 mM boric acid, 1 mM EDTA pH 8.3 (TBE) gels prior to autoradiography.
Nearest neighbor analysis
RNase T1 fragments isolated from the denaturing polyacrylamide gels in Figure 4A were resuspended in 4 µl dH2O and digested by the addition of 1 µl of a ribonuclease mixture containing 150 U/ml RNase T2 (Sigma), 10 U/µl RNase T1 (Boehringer Mannheim), 0.1 mg/ml RNase A (Sigma) in 15 mM ammonium acetate pH 4.5. Reactions were incubated at 37°C for 30 min, then 65°C for 5 min. Samples were spotted onto Macherey-Nagel chromatography sheets (cat. no. 106016 Polygram CEL400 UV254) with unlabeled 3′ NMP standards and subjected to chromatography in two dimensions (16). First dimension: isobutyric acid, concentrated NH4OH, 0.1 M EDTA, H2O (66:1:1:33, v/v/v/v); second dimension: 0.1 M sodium phosphate pH 6.8, ammonium sulfate, n-propanol (100:60:2, v/w/v).
RESULTS AND DISCUSSION
Isolation of pre-formed transcription elongation complexes
The overall goal of this work was to develop a soluble system in which to study Physarum RNA editing. Because our previous studies demonstrated that editing is closely associated with transcription (8,9) and we have been unable to demonstrate editing of synthetic RNA transcripts in vitro, we have focused our recent efforts on developing a coupled transcription/editing system. Previous studies (7) have demonstrated that high molecular weight run-on transcripts are synthesized in isolated mitochondria under our labeling conditions (see Fig. 1, lane 1). However, RNAs synthesized in mitochondrial lysates (lane 2) and soluble (S130) extracts (lane 3) are significantly shorter than those synthesized by mitochondria under the same conditions. This is likely to be due, at least in part, to the presence of nucleolytic activities, since we have found that radiolabeled RNA transcripts are rapidly degraded in S130 extracts (data not shown). Our strategy, therefore, was to enrich for transcription (and editing) activities while removing nucleases and other possible contaminants, such as transcription inhibitors, that would interfere with the synthesis of high molecular weight RNAs.
In the initial studies described here, we have purposefully utilized relatively crude preparations of TEC to increase the chances of retaining editing activity. These TECs consist of the endogenous ∼60 kb Physarum mitochondrial genome (17) and its associated proteins (including RNA polymerases) and nascent RNAs in the process of being synthesized from a variety of genes (7). We have taken advantage of the size of these complexes, using a gel filtration strategy somewhat similar to that used to enrich for TECs from yeast mitochondria (14). Mitochondrial S130 extracts are fractionated using Sepharose 4B gel filtration chromatography and assayed for transcription in the absence of added template. TECs and other large complexes are present in the excluded volume, while smaller proteins and free nucleotides elute in much later fractions.
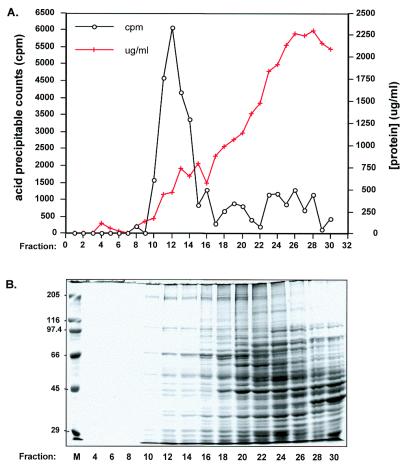
A typical gel filtration profile is shown in Figure 2A. Because we are interested in isolating pre-formed TECs, each fraction is assayed for incorporation of labeled RNA precursors into acid precipitable material in the absence of added DNA template. As expected, a peak of run-on transcription is found in the fractions corresponding to the excluded volume of the Sepharose 4B column. In the early stages of this study, a second peak of transcription activity was observed in much later fractions (fractions 24–30). A similar phenomenon was noted by Levens et al. (14) during preparation of yeast mitochondrial TECs. They attributed the appearance of this second peak to the DNase activity present in the mitochondrial lysates, with cleavage of the endogenous DNA template allowing DNA-bound RNA polymerases to enter the column. Consistent with this interpretation, we have found that the amount of transcription activity present in the second peak is a function of the length of time spent preparing the Physarum mitochondrial S130 extracts (data not shown). As the processing time decreased, this peak decreased to the levels shown in Figure 2A, with a concomitant increase in activity in the fractions excluded from the column. To confirm that we were assaying run-on transcription rather than reinitiation, we also examined the sensitivity of this RNA synthesis to heparin (18). Whereas transcription of poly dA/dT templates by partially purified preparations of the Physarum mitochondrial RNA polymerase is completely eliminated by the addition of 5 µg/ml heparin, transcription by TECs is unaffected by levels of heparin up to 100 µg/ml (E.Byrne, unpublished data). These results indicate that the vast majority of the RNA synthesis that we observe is due to elongation of pre-formed TEC.
Figure 2.
Fractionation of cleared mitochondrial lysates using Sepharose 4B. (A) Circles, transcription activity in the absence of added template assayed as described in Materials and Methods. Crosses, protein concentration of each fraction. (B) SDS–polyacrylamide gel electrophoresis of even numbered Sepharose 4B fractions, with bands visualized by staining with Coomassie blue.
Substantial purification is achieved by this fractionation procedure. Although the pooled TECs contain <5% of the total mitochondrial protein (Table 1), nearly all of the run-on transcription activity is found in these fractions (Table 1 and Fig. 2A). Indeed, the total number of units of enzymatic activity appears to increase during purification, most likely as a result of the elimination of nucleases and/or transcription inhibitors, as described below. While the specific activity does not change significantly between isolated mitochondria and the S130 extracts, there is an increase in the specific activity of pooled TECs of ∼35-fold relative to the starting material. To examine the overall distribution of proteins after chromatography, alternate gel-filtration column fractions were electrophoresed on an SDS–polyacrylamide gel. A typical example is shown in Figure 2B. Generally, fractions 10–14 contain the majority of the transcription activity and are pooled for further use. Although these fractions still contain multiple polypeptides, it is clear that only a small subset of the mitochondrial proteins are present in any abundance. Because fractions 10–14 are found in the excluded volume, most of the smaller proteins present in these fractions are likely to be associated with high molecular weight complexes, particularly given that the vast majority of small proteins are found in the later fractions, as expected. Therefore, although it is clear that these isolated TEC preparations are still a complex mixture of proteins, significant purification was achieved by this chromatography step.
Table 1. Purification of transcription elongation complexes from Physarum mitochondria.
| Total protein (mg) | Units (U) | Specific activity (U/mg) | Fold | Yield (%) | |
|---|---|---|---|---|---|
| Isolated mito | 50 | 750 | 15 | 1.6 | 147 |
| Mito lysate | 54.3 | 508.4 | 9.4 | 1 | 100 |
| Lysate S130 | 29.3 | 463.1 | 15.8 | 1.7 | 91 |
| Pooled TEC | 1.7 | 885 | 520.6 | 55.4 | 174 |
One unit activity is defined as 1 µmol of UMP incorporated at 30°C in 30 min under standard assay conditions.
To determine whether this fractionation succeeded in removing the contaminating activities that limited the synthesis of large transcripts in S130 extracts, RNAs synthesized by TECs were analyzed on a denaturing polyacrylamide gel (Fig. 1, lane 4). Although similar amounts of DNA were present at each purification step (data not shown), the ability to synthesize high molecular weight RNAs is restored only after gel-filtration chromatography. Importantly, the synthesis of these RNAs is not the result of contamination by intact mitochondria, since a control experiment (Fig. 1, lane 5) indicates that the centrifugation step is sufficient to remove any unlysed mitochondria from the S130 extracts used for chromatography. Thus, this simple gel filtration strategy provides sufficient purification to separate TECs from other mitochondrial components that might interfere with RNA synthesis.
Characterization of transcription elongation complexes and their RNA products
Enriched transcription complexes transcribe multiple mitochondrial genes (Fig. 3A and data not shown). To ascertain the level of RNA synthesis from individual genes, labeled RNAs made by TEC were hybridized to immobilized DNAs derived from five different Physarum genes (Fig. 3A). As expected, there was no detectable hybridization of labeled RNAs to the Physarum nuclear genes actin and α-tubulin, indicating the lack of nuclear contamination in our mitochondrial TEC preparations. Somewhat surprisingly, however, the relative abundance of run-on transcripts differs from what was observed previously for isolated mitochondria (7). For instance, in intact mitochondria the cytochrome c oxidase subunit 1 (coI) mRNA is expressed at significantly higher levels than the α-ATPase mRNA, whereas the opposite is true for TEC preparations. Because equivalent differences in labeled transcript abundance are also observed in S1 nuclease protection experiments (data not shown), we believe that these results reflect real differences in the distribution of RNA polymerases between the two in vitro systems. This may be due to subtle differences in transcription complex stability or losses in specific protein components, since TECs are subjected to higher salt concentrations during purification, or due to minor changes in culture conditions required for large scale growth of Physarum microplasmodia (see Materials and Methods).
To determine the extent of RNA synthesis in our TEC preparations, we have utilized oligonucleotide-directed RNase H digestion. Since transcripts from multiple genes are labeled during transcription by TECs, a previously developed hybrid protection protocol (7,8) was used to isolate RNAs derived from a 755 nt region of the highly expressed α-ATPase mRNA. Uniformly labeled RNAs were annealed to antisense ssDNA and digested with S1 nuclease. The protected RNA was gel purified (Fig. 3B, lane 1) and digested with RNase H in the presence of oligonucleotide A (lane 2) or oligonucleotide B (lane 3). Incubation in the presence of either oligonucleotide resulted in cleavage of the RNA at the predicted position, resulting in fragments of 290 and 465 nt with oligonucleotide A and bands of 545 and 210 nt with oligonucleotide B. Based on the fact that both fragments in lane 2 are labeled, we can set the lower limit for the extent of RNA synthesis in these preparations as being >465 nt (i.e., the distance between the binding site for oligonucleotide A and the 3′-end of the protected fragment), with RNA synthesis likely to proceed well beyond 465 nt given the level of labeling of the 5′ (290 nt) fragment. Consistent with this interpretation, when S1-protected fragments from different regions of the α-ATPase mRNA are digested with RNase T1 and the resulting oligonucleotides separated in two dimensions, all expected spots are visible, indicating that the entire length of the protected fragment is labeled. These results are in contrast to what is observed in isolated mitochondria, where RNA synthesis is limited to roughly 250–300 nt under our standard mitochondrial labeling conditions (9). This difference is most likely due to changes in the physical state of the template and/or protein composition between the two in vitro systems.
One of the drawbacks to using isolated mitochondria to study editing is the presence of significant endogenous nucleotide pools, particularly ATP and GTP. In contrast, transcription in isolated TEC is largely dependent upon the addition of each of the four exogenously supplied nucleotides (Fig. 3C and data not shown). Since some RNA synthesis is observed when ATP is omitted from TEC transcription reactions (Fig. 3C, lane 3), we analyzed the ATP content of two different TEC preparations to obtain an estimate of the remaining nucleotide pools. Using a quantitative bioluminescence assay (see Materials and Methods), we found that the concentration of ATP in these samples was in the range of 15–18 nM, roughly three orders of magnitude less than the concentration of the limiting nucleotide in our standard transcription reactions. Based on the extremely low levels of transcription observed when CTP (lane 4), GTP (lane 5) or UTP (data not shown) is omitted, we infer that the concentration of the other three nucleotides is considerably below 15 nM. Thus, this method of isolating TEC results in a substantial reduction in the percentage of total nucleotide in our transcription reactions that is contributed by endogenous nucleotide pools. This has allowed us to rigorously examine the effects of altered nucleotide concentrations on nucleotide insertion in Physarum (in preparation) and enhances our ability to incorporate modified nucleotides into run-on transcripts (unpublished data).
Editing activity co-purifies with transcription elongation complexes
In light of the differences between isolated mitochondria and TEC preparations, it was important to ascertain whether the editing machinery is retained during purification. To determine whether RNAs synthesized by TECs are edited, we have examined the extent of editing at ∼50% of the 54 single C insertion sites within the highly expressed α-ATPase gene using a previously developed hybrid protection assay (8). In this assay, labeled RNAs are isolated via S1 nuclease protection, digested with RNase T1, and the resulting oligonucleotides are run on a denaturing 20% polyacrylamide gel alongside similarly treated uniformly-labeled control transcripts having either edited or unedited sequence. As can be seen in the experiment shown in Figure 4A, RNAs synthesized by TEC yield RNase T1 fragments comigrating with those from the edited control sample (a″, b′, c′, d′). Importantly, nearest neighbor analyses of each of these bands, as well as RNase T1 fragments from all other regions tested, yielded the same labeling patterns as the edited control fragments (see below and data not shown). A substantial fraction (>50%) of the coI mRNA is also edited at the GU dinucleotide insertion site (data not shown), indicating that TEC preparations retain significant editing activity and that this activity is capable of adding non-encoded nucleotides at both single and dinucleotide insertion sites.
Although editing is reasonably efficient in this system, the extent of editing varies from site to site within all mRNA regions examined. This is illustrated by the five C insertion sites resolved in Figure 4A. The largest RNase T1 fragment in this region of the α-ATPase mRNA contains two editing sites, with a 32 nt fragment found in the edited control (oligonucleotide a″) and a 30mer present in the unedited control (oligonucleotide a) (Fig. 4A, lanes 1 and 2, respectively). Roughly 70% of the RNA synthesized by TEC contains either one or two added nucleotides within this RNase T1 fragment (lane 3). Even higher levels of editing are observed for oligonucleotide c′ (∼95%), as discussed below. In contrast, other sites, such as those found within fragments b/b′ and d/d′ are less extensively edited. However, as was also observed in isolated mitochondria (9), we do not see any correlation between the extent of editing and the location of an insertion site along the message; i.e., there is no 5′→3′ (or 3′→5′) polarity. For instance, site c/c′, which is almost completely edited, is preceded by site b/b′ and followed by site d/d′, both of which are edited to a lower extent. Therefore, it is highly unlikely that the differences in editing efficiency between sites are due to the loss of the editing activity as the transcription/editing machinery progresses down the template. Interestingly, the extent of editing at a given site appears to be context dependent, and we have recently found that editing efficiency can be manipulated in a predictable manner by altering reaction conditions (in preparation).
Editing is accurate in this soluble transcription/editing system
Insertion of non-encoded nucleotides is also accurate in these TEC preparations. To determine the site of addition and the identity of the added nucleotides in transcripts synthesized in this system, each RNase T1 fragment was gel-purified and subjected to nearest neighbor analysis. In the example shown in Figure 4B, the 12 nt fragments (c′) from lanes 1 (edited control, 12E) and 3 (transcription complexes, 12TEC) were isolated from the gel shown in Figure 4A, digested to 3′ NMPs, and the nucleotides were separated using two-dimensional thin layer chromatography. Based on the sequence of this RNase T1 fragment (CUCAACCAUAcpG, where c is the site of C insertion and p represents the labeled phosphate within the fragment), we would expect the transfer of labeled phosphate to a C residue if this site is accurately edited in [α-32P]GTP-labeled RNA. As can be seen in Figure 4B, both samples contained only labeled 3′ CMP, indicating that the expected nucleotide was added at the correct site in TECs. RNA fingerprinting experiments involving other regions of the α-ATPase mRNA and secondary analyses of additional RNase T1 fragments are entirely consistent with these findings (data not shown).
The extent of editing at the insertion site within fragment c can also be estimated based on the labeling pattern of the isolated 11 nt fragments from GTP-labeled RNA (Fig. 4B). Since there are two 11mers in unedited RNA (CUCAACCAUApG and UUAUCAACUUpG), secondary digests of the 11mers from unedited control RNA (11U) yield equal amounts of labeled A and U, whereas only labeled U is seen in the edited control sample (11E), which contains only the latter 11 nt fragment. Our finding that 95% of the label in the 11mer from the TEC sample (11TEC) is found as 3′ UMP indicates that RNA made by isolated TEC is almost completely edited at the editing site within fragment c. Thus, it appears that the editing machinery is associated with most, if not all, of the pre-formed transcription complexes in these preparations.
The major goal of this work was to establish a soluble transcription/editing system that would allow us to dissect editing mechanisms further. Our data indicate that, unlike mitochondrial lysates, crude TEC preparations contain little or no nucleolytic activity and synthesize high molecular weight RNAs. Importantly, the transcripts produced in this system are accurately edited, with high levels of editing observed at multiple sites. This more purified editing system offers a number of significant advantages over isolated mitochondria for the study of Physarum editing mechanisms. First, it permits access to nascent RNAs and the mitochondrial DNA template, potentially crucial targets of the editing machinery. Secondly, the lack of a mitochondrial membrane allows us to test the effects of inhibitors, antibodies, and various enzymes on the editing reaction. Thirdly, the virtual absence of endogenous ribonucleotide pools allows us to manipulate relative substrate concentrations and facilitates the use of modified nucleotides in transcription/editing reactions. Fourthly, these preparations provide us with a starting point for further purification of the components of the transcription/editing machinery.
The development of this in vitro system will undoubtedly have a substantial impact on the study of RNA editing in Physarum. In addition, given the coupling observed between transcription and other RNA processing events such as splicing and polyadenylation (19,20), this general strategy should be widely applicable to other mitochondrial RNA processing and editing systems. These include organisms in which in vitro RNA editing cannot be obtained reproducibly, those that carry out editing at very low efficiencies, and in vitro systems that are limited by processivity.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Timothy Nilsen, Dr David Setzer and the members of our laboratory for advice and helpful comments on the manuscript, Drs Linda Visomirski-Robic and Donal Luse for helpful discussions, and Dr Yi-Tao Yu for his suggestions regarding 2-D TLC. This work was supported by grants to J.M.G. from NSF (MCB-9630672) and NIH (GM54663).
REFERENCES
- 1.Gott J.M. and Emeson,R.B. (2000) Annu. Rev. Genet., 34, 499–531. [DOI] [PubMed] [Google Scholar]
- 2.Smith H.C., Gott,J.M. and Hanson,M.R. (1997) RNA, 3, 1105–1123. [PMC free article] [PubMed] [Google Scholar]
- 3.Mahendran R., Spottswood,M.R. and Miller,D.L. (1991) Nature, 349, 434–438. [DOI] [PubMed] [Google Scholar]
- 4.Gott J.M., Visomirski,L.M. and Hunter,J.L. (1993) J. Biol. Chem., 268, 25483–25486. [PubMed] [Google Scholar]
- 5.Mahendran R., Spottswood,M.S., Ghate,A., Ling,M.L., Jeng,K. and Miller,D.L. (1994) EMBO J., 13, 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antes T., Costandy,H., Mahendran,R., Spottswood,M. and Miller,D. (1998) Mol. Cell Biol., 18, 7521–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visomirski-Robic L.M. and Gott,J.M. (1995) RNA, 1, 681–691. [PMC free article] [PubMed] [Google Scholar]
- 8.Visomirski-Robic L.M. and Gott,J.M. (1997) Proc. Natl Acad. Sci. USA, 94, 4324–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visomirski-Robic L.M. and Gott,J.M. (1997) RNA, 3, 821–837. [PMC free article] [PubMed] [Google Scholar]
- 10.Gott J.M. (2000) In Bass,B. (ed.), RNA Editing: Frontiers in Molecular Biology. Oxford University Press, Oxford, UK, in press.
- 11.Gott J.M. and Visomirski-Robic,L.M. (1998) In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 395–411.
- 12.Daniel J.W. and Baldwin,H.H. (1964) In Prescott,D.M. (ed.), Methods in Cell Physiology, Vol. 1. Academic Press, New York, NY, pp. 9–41.
- 13.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Levens D., Morimoto,R. and Rabinowitz,M. (1981) J. Biol. Chem., 256, 1466–1473. [PubMed] [Google Scholar]
- 15.Rowen L. and Kornberg,A. (1978) J. Biol. Chem., 253, 758–764. [PubMed] [Google Scholar]
- 16.Keith G. (1995) Biochimie, 77, 142–144. [DOI] [PubMed] [Google Scholar]
- 17.Jones E.P., Manhendran,R., Spottswood,M.R., Yang,Y.-C. and Miller,D.L. (1990) Curr. Genet., 17, 331–337. [DOI] [PubMed] [Google Scholar]
- 18.Walter G., Zillig,W., Palm,P. and Fuchs,E. (1967) Eur. J. Biochem., 3, 194–210. [DOI] [PubMed] [Google Scholar]
- 19.Cramer P., Caceres,J.F., Cazalla,D., Kadener,S., Muro,A.F., Baralle,F.E. and Kornblihtt,A.R. (1999) Mol. Cell, 4, 251–258. [DOI] [PubMed] [Google Scholar]
- 20.Minvielle-Sebastia L. and Keller,W. (1999) Curr. Opin. Cell Biol., 11, 352–357. [DOI] [PubMed] [Google Scholar]