Abstract
RsrI [N6-adenine] DNA methyltransferase (M·RsrI), which recognizes GAATTC and is a member of a restriction–modification system in Rhodobacter sphaeroides, was purified to >95% homogeneity using a simplified procedure involving two ion exchange chromatographic steps. Electrophoretic gel retardation assays with purified M·RsrI were performed on unmethylated, hemimethylated, dimethylated or non-specific target DNA duplexes (25 bp) in the presence of sinefungin, a potent inhibitory analog of AdoMet. M·RsrI binding was affected by the methylation status of the DNA substrate and was enhanced by the presence of the cofactor analog. M·RsrI bound DNA substrates in the presence of sinefungin with decreasing affinities: hemimethylated > unmethylated > dimethylated >> non-specific DNA. Gel retardation studies with DNA substrates containing an abasic site substituted for the target adenine DNA provided evidence consistent with M·RsrI extruding the target base from the duplex. Consistent with such base flipping, an ∼1.7-fold fluorescence intensity increase was observed upon stoichiometric addition of M·RsrI to hemimethylated DNA containing the fluorescent analog 2-aminopurine in place of the target adenine. Pre-steady-state kinetic and isotope- partitioning experiments revealed that the enzyme displays burst kinetics, confirmed the catalytic competence of the M·RsrI–AdoMet complex and eliminated the possibility of an ordered mechanism where DNA is required to bind first. The equilibrium dissociation constants for AdoMet, AdoHcy and sinefungin were determined using an intrinsic tryptophan fluorescence-quenching assay.
INTRODUCTION
DNA methyltransferases (MTases) catalyze the transfer of a methyl group from AdoMet to specific bases in or near particular sequences of duplex DNA (reviewed in 1). DNA methylation plays diverse roles in the biology of eukaryotes and prokaryotes. In eukaryotes, DNA methylation is involved in gene regulation, development, oncogenesis and the organization of chromatin structure (reviewed in 2). In prokaryotes, MTases participate in DNA mismatch repair (3) and chromosome replication (4). In addition, MTases, along with partner restriction endonucleases, comprise restriction–modification (R-M) systems. R-M systems serve an ‘immune’ function that protects bacteria from invasion by exogenous DNA, e.g. by bacteriophage infection. Typically, MTases of R-M systems methylate the host DNA to render it refractory to cleavage by the partner endonuclease. In addition to their biological roles, MTases are convenient models for the study of sequence-specific enzyme–DNA interactions.
Two groups of DNA MTases are defined by the atom of the target base that they modify. The 5-methylcytosine (5mC) MTases catalyze the transfer of a methyl group from AdoMet to the C5 carbon of cytosine to form 5mC. Members of the second group, the amino MTases, methylate the exocyclic nitrogens at the C4 carbon of cytosine or C6 of adenine to form N4-methylcytosine (N4mC) or N6-methyladenine (N6mA), respectively. The amino MTases are subdivided into three classes, α, β and γ, based on the arrangement of nine variably conserved amino acid sequence motifs and by structural analysis (5,6). Comparisons of the crystal structures of the C5 MTases, [M·HhaI (7) and M·HaeIII (8)] and amino MTases of the α [DpnM (9)], β [M·RsrI (10) and M·PvuII (11)] and γ [M·TaqI (12)] classes reveal that these enzymes all have a bilobal, two-domain structure. Proteolysis of M·EcoRI (13) also supports this observation. The motifs of amino MTases involved in cofactor binding (I–III and X) and catalysis (IV–VIII) are located within one domain that is linked to the DNA target recognition domain (TRD). Furthermore, despite the organizational differences at the primary amino acid sequence level, all the crystallized DNA MTases share a common tertiary core structure, termed the methylase fold (11). This consensus fold comprises a seven strand central β-sheet (composed of five parallel β-strands followed by an anti-parallel β-hairpin) with three α-helices on each side of the β-sheet.
The amino MTases transfer the methyl group directly to the nitrogen of the amino group without formation of a covalent intermediate (11,14). Methylation of the C5 carbon of cytosine involves extrusion of the target base from DNA and the formation of a covalent 5,6-dihydrocytosine intermediate (7,15). Amino MTases likely also use base extrusion or base flipping to allow the target base to reach into the active site.
M·RsrI (β class), like its well-characterized isoenzyme M·EcoRI (γ class), recognizes the duplex DNA sequence GAATTC and catalyzes the transfer of a methyl group from AdoMet to the exocyclic amino group of the central adenine to form GAmATTC and AdoHcy (16). Direct, optimized sequence alignments show that the RsrI and EcoRI endonucleases share 50% amino acid identity with one another, whereas the partner MTases, M·RsrI and M·EcoRI, share only 16% identity (16). This lack of amino acid similarity presents an interesting opportunity to study DNA binding and catalysis by enzymes from two different classes that act on the same target DNA sequence. We have partially characterized M·RsrI (16,17).
We report here a streamlined purification procedure for M·RsrI, electrophoretic gel retardation studies to define its binding to DNA, fluorescence studies to determine its cofactor binding constants and base flipping properties and burst and isotope-partitioning experiments to define some of its kinetic properties. We show that M·RsrI, like M·EcoRI, does not use an ordered BiBi kinetic mechanism where DNA binds first and also likely uses base flipping as part of its catalytic mechanism. The enzyme displays pre-steady-state burst kinetics, suggesting that an early event, presumably methyl transfer to DNA, is faster than later steps leading to product release. M·RsrI and M·EcoRI have similar (µM) cofactor dissociation constants. M·RsrI has a 2- to 3-fold higher affinity for hemimethylated DNA than unmethylated DNA, unlike M·EcoRI, which displays no binding preference for unmethylated versus hemimethylated DNA. The dissociation constants for DNA for both enzymes are in the low nanomolar range. Electrophoretic gel retardation assays with analog-containing DNA suggest that M·RsrI, unlike M·EcoRI, may use minor groove contacts for recognition. Despite these differences their dissociation constants with 2-aminopurine (2A)-containing DNA are in the low nanomolar range. We relate these biochemical results to the structure of the enzyme (10).
MATERIALS AND METHODS
Enzymes, radioisotopes and chemicals
Sinefungin was a gift from Margaret Neidenthal (Eli Lilly Laboratories). HPLC-purified oligodeoxyribonucleotide (ODN) 25mers were synthesized at Midland Certified Reagent Co. or purchased unpurified from Gibco BRL or the W.M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign. The duplex 14mer CBM (18), formed from the HPLC-purified ODN d(GGCGGAATTCGCGG) and its complement d(CCGCGAmATTCCGCC), where mA is N6-methyladenine, was purchased from the Bioresource Center of the University of California at San Francisco. HiTrap SP or HiTrap Q columns and the FPLC system were from Amersham Pharmacia Biotech. PhastSystem 20% gels, buffer strips, silver staining reagents and the PhastSystem were also from Amersham Pharmacia Biotech. λ DNA and restriction, modification and PCR enzymes were purchased from either Gibco BRL, Promega, New England Biolabs or Stratagene. S-adenosylmethionine (AdoMet) and S-adenosylhomocysteine (AdoHcy) were purchased from Sigma and purified (18). Slide-A-Lyzer cassettes were from Pierce Chemical Co. [γ-32P]ATP and [methyl-14C]AdoMet were from Amersham and Dupont NEN, respectively.
Buffers
Cell opening buffer (COB): 20 mM potassium phosphate, pH 6.4, 10 mM EDTA, 10% glycerol, 20 mM mercaptoethanol (β-ME), 10 mM KCl and 500 µM phenylmethylsulfonyl fluoride (PMSF). One milliliter of Sigma protease inhibitor cocktail P8340 (lacking EDTA) was added per 40 ml of COB. The final concentrations of the inhibitors were 2.5 µM 2-aminoethylbenzenesulfonyl fluoride, 37 µM pepstatin A, 34 µM trans-epoxysuccinyl-l-leucylamido(4-guanidino)butane (E-64), 98 µM bestatin, 54 µM leupeptin and 2 µM aprotinin.
SP low salt buffer: 20 mM potassium phosphate, pH 6.4, 10% glycerol, 20 mM β-ME, 1 mM EDTA, 50 mM KCl and 0.4 mM PMSF.
SP high salt buffer: 20 mM potassium phosphate, pH 6.4, 10% glycerol, 20 mM β-ME, 1 mM EDTA, 2 M KCl and 0.4 mM PMSF.
Dialysis and Q low salt buffer: 20 mM Tris–HCl, pH 8.0, 10% glycerol, 20 mM β-ME, 1 mM EDTA, 50 mM KCl and 0.4 mM PMSF.
Q high salt buffer: 20 mM Tris–HCl, pH 8.0, 10% glycerol, 20 mM β-ME, 1 mM EDTA, 2 M KCl and 0.4 mM PMSF.
Enzyme dilution buffer: 20 mM Tris–HCl, pH 7.5 or 8.0, 10% glycerol, 20 mM β-ME, 1 mM EDTA, 50 mM KCl and 0.2 mg/ml acetylated BSA (Promega). Buffers were filtered through Millipore HA-45 filters.
Cloning of pTrc99A::rsrIM
The plasmid pTrc99A::rsrIM was generated by ligation of a NcoI and XbaI double-digested PCR product of the rsrIM gene into NcoI and XbaI double-digested pTrc99A (Pharmacia) vector. The wild-type gene was placed behind the isopropyl-β-d-thiogalactoside (IPTG)-inducible trc promoter of pTrc99A. The PCR primers 5′Rsrm [d(CCTGCCATGGCAAACCGATCTCA)] and 3′Mrsr [d(GTCTAGATTATGAAGCAACATCTCCC)] were used to amplify the rsrIM gene (∼1 kb) from source plasmid pTZ18U-rsrIRM (17). The restriction site of NcoI or XbaI in each primer is underlined. The 100 µl PCR reaction contained 300 nM of the forward and reverse primer, 200 µM dNTPs, 1× PCR buffer supplied by the manufacturer, 3.5 U Expand High Fidelity Polymerase (Boehringer-Mannheim) and 0.75 µg DNA template. A four-step PCR thermocycling protocol was used: (i) 97°C for 5 min; (ii) 10 cycles of 97°C for 1 min, 42°C for 1 min and 72°C for 1 min; (iii) 15 cycles of 97°C for 1 min, 42°C for 1 min and 72°C for 1 min, with a 20 s extension for each cycle; (iv) 72°C for 7 min (for completion of extension of PCR products). The ligation reaction was transformed by electroporation into Escherichia coli DH1 (F– supE44 hsdk17 recA1 endA1 gyrA96 thi-1 relA1). Plasmids of several clones were isolated by alkaline lysis (19). The sequence of the desired plasmid was confirmed.
Production of wild-type RsrI MTase and preparation of cell extract by sonication
An overnight culture of DH1 harboring pTrc99A::rsrIM was added to 4 l of LB broth supplemented with 200 µg/ml ampicillin (Ap) and subsequently grown at 37°C to an optical density of 0.4 at 600 nm, as determined by a Hewlett Packard 8451 diode array spectrometer, and induced by the addition of 1 mM IPTG. Cells were grown after induction for 4 h at 37°C and harvested by centrifugation for 20 min at 4000 g at 4°C. Eight grams of wet cell paste was then resuspended in COB (2.5 ml/g wet cell paste) and sonicated using a Heat Systems Ultrasonic Processor W-385 macrotip at 25% amplitude four times for 30 s with intervening 2 min incubations on ice to prevent overheating. The lysate was centrifuged immediately for 40 min at 14 000 g at 4°C. The clarified extract (∼20 ml) was collected and quickly frozen on dry ice and stored at –80°C. Frozen extracts were suitable for enzyme purification for at least 2 months.
Purification of wild-type M·RsrI
A Pharmacia Biotech FPLC system was used for all chromatography steps. Clarified cell extract was passed over two tandem 5 ml Pharmacia HiTrap SP columns pre-equilibrated in SP low salt buffer. The columns were then washed with 15 column volumes of SP low salt buffer (150 ml). A gradient of 150 ml of 0–40% SP high salt buffer eluted M·RsrI at 400–600 mM KCl. Fractions containing M·RsrI activity were pooled (∼15 ml) and dialyzed using Slide-A-Lyzer dialysis cassettes (Pierce Chemical Co.) in 4 l of dialysis buffer for at least 1 h at 4°C. The dialysate was applied to two tandem 5 ml HiTrap Q columns pre-equilibrated in Q low salt buffer. The columns were washed with 15 column volumes of the same buffer. M·RsrI eluted at 100–200 mM KCl during elution with a gradient of 150 ml of 0–40% Q high salt buffer. Fractions containing M·RsrI were pooled and sterile glycerol was added to a final concentration of 50% (v/v). Aliquots of the enzyme were stored at –20°C and were stable for at least 6 months. Protein concentrations were determined by the Bradford assay (20) or, for purified fractions, from the calculated extinction coefficient of 48 700 M–1cm–1 at 280 nm (17). To assess the purity at each stage, Coomassie Blue stained SDS–PAGE gels were analyzed by densitometric analysis using the public domain NIH Image program v.1.61.
MTase assays
Serial dilutions (2.5-fold) of clarified lysate or pooled M·RsrI HiTrap SP or Q column fractions were prepared in enzyme dilution buffer using a sterile 96-well plate. An aliquot of 1 or 2 µl of each dilution was added to a solution containing 50 mM Tris–HCl, pH 7.5, 5 mM EDTA, 5 mM β-ME, 100 µM AdoMet and 1 µg unmethylated bacteriophage λ DNA (20 µl final volume) and incubated for 1 h at 37°C. The reactions were stopped by incubation at 65°C for 15 min. Samples were cooled on ice for 5 min, with subsequent addition of Gibco React 3 buffer (to 1×) and 40 U EcoRI endonuclease to a final reaction volume of 50 µl. The reaction mixtures were incubated at 37°C for 30 min, inactivated by incubation at 65°C for 15 min and analyzed by agarose gel electrophoresis. One unit of M·RsrI activity is defined as the maximal dilution of enzyme that will completely protect 1 µg λ DNA from digestion by R·EcoRI.
Fluorescence measurements
A Perkin-Elmer LS 50 luminescence spectrophotometer was used for fluorescence measurements. For cofactor affinity determinations an excitation wavelength of 295 nm was used and emission spectra from 320–370 nm were collected. The change in fluorescence intensity at the maximum emission of 337 nm was monitored. The excitation and emission slit was 7.5 nm and the scan speed was 100 nm/min for 500 µl volumes of 280 nM M·RsrI, 100 mM Tris–HCl, pH 7.5, 10 mM EDTA, 10 mM dithiothreitol (DTT) and 10 mM NaCl. M·RsrI was equilibrated in the assay buffer at 22°C until the fluorescence intensity remained constant. Increasing concentrations of from 2 to 40 µM each ligand were added at 3–4 min intervals. Data from two to four determinations were averaged. Excitation was performed at 295 nm to avoid cofactor fluorescence quenching due to the inner filter effect (21), which was negligible (≤0.5% at the highest cofactor concentrations). The data were analyzed using the Stern–Volmer (22) or modified Stern–Volmer (23) plots. Data were evaluated by least squares linear regression analysis using Kaleidagraph 3.0 (Synergy Software).
Steady-state fluorescence measurements with DNA containing 2A were performed essentially as described (24) except that a slit width of 7.5 nm for excitation and emission and a 0.5 ml cuvette were used. The solutions contained 100 mM Tris–HCl, pH 7.5 with M·RsrI or pH 8.0 with M·EcoRI, 10 mM EDTA, 10 mM DTT, 10 mM NaCl, 10 µM sinefungin and 0.2 mg/ml BSA, 450 nM enzyme and 400 nM 2A duplex.
Kinetics and isotope-partitioning experiments
For burst assays stock M·RsrI (57 µM) was diluted to 15 µM in 100 mM Tris–HCl, pH 7.5, 10 mM DTT, 10 mM EDTA, 10 mM NaCl, 0.2 mg/ml BSA and 42 µM [methyl-14C]AdoMet to saturate the enzyme with cofactor. An aliquot (4 µl) of this pre-mix was then diluted 50-fold into 196 µl of 100 mM Tris–HCl, pH 7.5, 10 mM DTT, 10 mM EDTA, 10 mM NaCl, 0.2 mg/ml BSA, 42 µM [methyl-14C]AdoMet and 3.6 µM 14mer duplex CBM (18). The final concentration of M·RsrI was 0.3 µM and that of DNA 3.6 µM. The isotope-partitioning assays were performed in the same manner as the burst assays except that the pre-mix (4 µl) was added to 196 µl of solution containing 42 µM unlabeled AdoMet. As a control, 4 µl of the pre-mix was added to 184 µl of 100 mM Tris–HCl, pH 7.5, 5 mM DTT, 5 mM EDTA, 5 mM NaCl, 0.2 mg/ml BSA and 42 µM unlabeled AdoMet to dilute the specific activity of the cofactor before adding the DNA. After mixing, 12 µl of 60 µM CBM DNA in TE, pH 8.0, was added to a final concentration of 3.6 µM in 200 µl. Reaction aliquots (40 µl) were spotted at 5, 10, 15, 20 and 30 s on DE-81 filter paper. Filters were processed to quantify the methyl groups transferred to the DNA (25). All experiments were performed in triplicate at 22°C. Data were evaluated as described in Fluorescence measurements.
Purification and annealing of oligomers
The duplex EcoRI site-containing 25mer sequence d(GGCAGACACGGANTTCCACAGACGG) was used, where N was either deoxyadenosine, N6-methyldeoxyadenosine (mA), 2A or a stable 1′,2′-dideoxyribose abasic site (ab). Another 25mer duplex containing the reverse site sequence [d(GGCAGACAGCTTAAGCCACAGACGG)] was used as a non-specific DNA control. These lyophilized, HPLC-purified single-strand oligomers were resuspended in NaCl/Tris/EDTA buffer (10 mM Tris–HCl, 1 mM EDTA and 100 mM NaCl, pH 8.0). The duplex concentrations were determined spectrophotometrically using the nearest neighbor method (26). The oligonucleotides containing methylated bases were purified to >95% homogeneity using reverse phase HPLC with an Altex reverse phase C18 column on a Beckman 110A HPLC system at room temperature at a flow rate of 1 ml/min. A gradient from 5% acetonitrile in 0.1 M TEAA, pH 7.0, to 40% acetonitrile in 0.1 M TEAA, pH 7.0, over 30 min was generated and each oligomer eluted at ∼21% acetonitrile. The oligomers were lyophilized and resuspended in NaCl/Tris/EDTA buffer, pH 8.0, and stored at –20°C. The DNA duplexes are listed in Table 2. A 1:1 molar ratio of each oligonucleotide in a 50 µl reaction was added to a final concentration of 10 µM. The mixture was heated to 90°C for 5 min, allowed to cool to 4°C over 120 min in a Perkin Elmer 480 Thermocycler and stored at –20°C.
Table 2. Ligand dissociation constants.
| Ligand | KD (µM) | Fluorescence quenching (%) |
|---|---|---|
| AdoMet | 6.1 (0.1)a | 54.4 (1.8) |
| Sinefungin | 4.2 (0.1) | 46.5 (0.4) |
| AdoHcy | 9.6 (0.1) | 49.5 (1.0) |
aThe values in parentheses represent ± 1 SD from the mean of three experiments.
Labeling of oligomers
Reactions (30 µl) containing 10 U T4 polynucleotide kinase (Gibco BRL), 2.5 µM [γ-32P]ATP (6000 Ci/mmol) and 1 µM annealed duplex in 1× kinase buffer supplied by the manufacturer were incubated for 30 min at 37°C. The reaction was stopped with 0.5 µl of 0.5 M EDTA (final concentration 8 mM) and applied to a Micro Bio-Spin Bio-Gel P-6 column (equilibrated in 150 mM NaCl, 17.5 mM sodium citrate, pH 7.0) to remove unreacted ATP. Polyacrylamide gel electrophoresis under non-denaturing conditions (15%) in 0.5× TBE (44.5 mM Tris–borate, pH 8.2, 10 mM EDTA) confirmed the presence of ≥96% duplex. Each duplex was subsequently digested with 12 U R·EcoRI at 22°C for 1 h to test for methylation of the EcoRI site and analyzed by non-denaturing electrophoresis (data not shown). Only the unmethylated duplexes were susceptible to digestion.
Electrophoretic gel retardation assays
DNA binding reactions (20 µl) contained 100 mM Tris–HCl, pH 7.5, 10 mM EDTA, 10 mM DTT, 19 mM NaCl, with or without 200 µM sinefungin, 0.2 mg/ml BSA, 5% glycerol, 5 nM labeled DNA duplex and varying concentrations of M·RsrI. M·RsrI (57 µM) was freshly diluted into dilution buffer (100 mM Tris–HCl, pH 7.5, 10 mM EDTA, 10 mM DTT, 10 mM NaCl, 0.2 mg/ml BSA and 10% glycerol). NaCl/Tris/EDTA buffer, pH 8.0, was used to dilute the 1 µM DNA stock to 50 nM. Two microliters of dilution buffer or a dilution of protein was added to 16 µl of binding buffer and the mixture incubated for 5 min at 22°C before adding 2 µl of 50 nM labeled substrate DNA to the samples. Samples were incubated for 20 min at 22°C, loaded onto a running 10% polyacrylamide gel equilibrated in 0.5× TBE and subjected to 100–125 V for 90–120 min at 4°C. Gels were dried and exposed to a phosphorimager screen for analysis. Quantitation was performed with a Molecular Dynamics PhosphorImager 425S using Image Quant software v.3.3. Data from three experiments were averaged and fitted to equation 1 (27):
P = Pmax/(1 + {KDapp/[E]}) 1
where Pmax is the maximum DNA bound and [E] is the protein concentration. Data were analyzed by non-linear least squares regression analysis as described above.
Effect of sinefungin on DNA binding
Binding reactions (20 µl) contained 100 mM Tris–HCl, pH 7.5, 10 mM EDTA, 10 mM DTT, 19 mM NaCl, 0.2 mg/ml BSA, 5% glycerol, 5 nM top strand hemimethylated DNA duplex (mTB), 15 nM M·RsrI and varying concentrations of sinefungin. M·RsrI (57 µM) was freshly diluted into dilution buffer. The 1 µM DNA stock was diluted to 50 nM with NaCl/Tris/EDTA buffer, pH 8.0. Two microliters of a dilution of sinefungin in binding buffer were added to 16 µl of binding buffer containing M·RsrI and the mixture incubated for 5 min at 22°C before adding 2 µl of DNA to the samples. The final concentrations of DNA and enzyme were 5 and 15 nM, respectively. Samples and data were processed as described for the gel retardation assays.
RESULTS
Purification
M·RsrI was overproduced in E.coli and purified using a two-column FPLC purification procedure that supplants an earlier seven-step protocol (17). The enzyme was 95% homogeneous as judged by SDS–PAGE analysis (data not shown) and was ∼94% active as determined by the pre-steady-state burst assay described below.
DNA binding
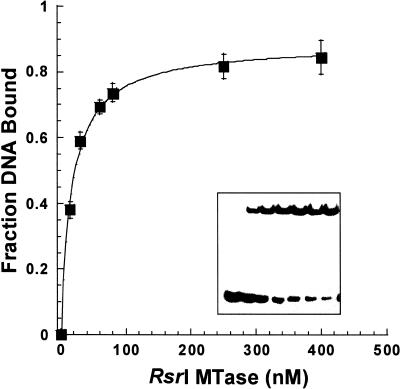
We assessed the ability of the enzyme to discriminate between canonical DNA in different states of methylation by an electrophoretic gel retardation assay. Such assays are widely used to evaluate protein–nucleic acid interactions (28) and have been applied to other MTases, e.g. T4 Dam (27), M·EcoRV (29) and M·EcoRI (30). The assays used nanomolar concentrations of purified M·RsrI and 0.5 or 5 nM 25mer duplex DNA substrates containing unmethylated, hemimethylated, dimethylated, abasic, 2A-substituted or non-cognate sites (Table 1). Twenty-five base pair duplexes were essential for the formation of electrophoretically stable complexes; shorter (14–16 bp) duplex-containing complexes dissociated during electrophoresis (data not shown). Some assays were conducted in the presence of sinefungin, a potent competitive inhibitor of DNA–MTase interactions (18). A representative gel-shift image from an experiment using 5 nM unmethylated DNA and increasing concentrations of M·RsrI in the presence of sinefungin is shown in Figure 1. A single shifted band is formed. Figure 1 shows a binding curve derived from the averaging of densitometric analysis data from three gel retardation assays. The KDapp value (17.3 ± 1.5 nM) is defined as the M·RsrI concentration at half-maximal binding. The maximal binding (Pmax) for most DNA substrates was 80–90%; specific complex formation was observed with the fully methylated substrate but with lower saturation in binding (40–50%), as observed with M·EcoRV (29). Failure to achieve complete binding of the DNA is commonly observed using non-equilibrium methods, e.g. gel retardation and filter binding assays (31), and in gel retardation assays is probably due to partial dissociation of the complex prior to or during electrophoresis. Although not a true equilibrium method, gel retardation provides a good approximation of relative dissociation constants since reproducible, stable complex formation was observed for all the canonical DNA substrates.
Table 1. Apparent M·RsrI–DNA dissociation constants.
| No. | DNA substratea | Duplex sequenceb | [DNA] | KDapp (± 1 SD)c with sinefungin | KDapp (± 1 SD) without sinefungin | KDapp ratiod |
|---|---|---|---|---|---|---|
| 1 | Unmethylated | 5′-GAATTC-3′ | 5 nM | 17 (1.5) | 311 (50.3) | 18 |
| TB | 3′-CTTAAG-5′ | [23 (0.5)]e | [ND]f | |||
| 2 | Top strand methylated | 5′-GAmATTC-3′ | 5 nM | 6 (1.6) | 17 (3.4) | 3 |
| mTB | 3′-CTTAAG-5′ | [3 (0.4)] | [ND] | |||
| 3 | Bottom strand methylated | 5′-GAATTC-3′ | 5 nM | 10 (4.0) | 56 (17.3) | 6 |
| TmB | 3′-CTTmAAG-5′ | [3 (0.4)] | [ND] | |||
| 0.5 nM | 6 (2.2) | ND | ND | |||
| 4 | Dimethylated | 5′-GAmATTC-3′ | 5 nM | 156 (22.6) | 117 (16.6) | 1 |
| mTmB | 3′-CTTmAAG-5′ | [ND] | [ND] | |||
| 5 | No site | 5′-CTTAAG-3′ | 5 nM | >>800g | >>800 | ND |
| NS | 3′-GAATTC-5′ | [>>800]g | ||||
| 6 | Abasic (ab) top strand/bottom strand methylated | 5′-GA(ab)TTC-3′ | 0.5 nM | 2 (0.8) | ND | ND |
| abTmB | 3′-CTTmAAG-5′ | |||||
| 7 | Abasic (ab) top strand/bottom strand unmethylated | 5′-GA(ab)TTC-3′ | 0.5 nM | 5 (1.0) | ND | ND |
| abTB | 3′-CTTAAG-5′ | |||||
| 8 | 2-Aminopurine (2A) top strand/unmethylated bottom strand | 5′-GA(2A)TTC-3′ | 5 nM | [47 (0.2)] | [ND] | ND |
| 2ATB | 3′-CTTAAG-5′ | |||||
| 9 | 2A top strand/bottom strand methylated | 5′-GA(2A)TTC-3′ | 5 nM | [57 (3.2)] | [ND] | ND |
| 2ATmB | 3′-CTTmAAG-5′ |
aT, top strand; B, bottom strand.
bOnly the hexanucleotide target sequence is shown. Target adenines are in bold. All ODNs were 25 bp long and have the same sequences flanking the canonical EcoRI site. See Materials and Methods for the sequences.
cValues in parentheses represent ± 1 SD from the mean of values from two or three individual experiments
dWith/without sinefungin.
eBracketed values are KDapp values derived from electrophoretic gel retardation assays containing 20 mM potassium glutamate instead of 19 mM NaCl in the reaction mixture.
fND, not determined.
gBinding not observed at concentrations <800 nM M·RsrI.
Figure 1.
Binding isotherm of M·RsrI with unmethylated DNA and sinefungin. The DNA (5 nM) was sequence TB (Table 1). The curve was generated by fitting the average of three gel retardation assays as described in Materials and Methods. Error bars represent ± 1 SD. (Inset) Representative gel retardation image with [M·RsrI] increasing from 0 to 400 nM.
The apparent dissociation constants for one non-canonical and several canonical substrates are presented in Table 1. M·RsrI bound 2- to 3-fold better to hemimethylated duplexes (mTB and TmB, where T indicates the top strand, B the bottom strand and m an N6-methylated target base) than to the unmethylated duplex (TB) when sinefungin was present. M·RsrI had 9-fold greater affinity for the unmethylated duplex (TB) than the dimethylated duplex (mTmB) in the presence of the analog. In the absence of sinefungin, M·RsrI had 3- to 6-fold lower affinity for the hemimethylated substrates and 18-fold lower affinity for the unmethylated duplex when compared to dissociation constants obtained in the presence of sinefungin. M·RsrI showed no inhibitor-induced preference for the dimethylated substrate. The enzyme failed to bind to the control duplex lacking a site under all conditions tested, with an apparent dissociation constant >>0.8 µM. In summary, M·RsrI binds DNA substrates in the presence of sinefungin with decreasing affinities: hemimethylated > unmethylated > dimethylated > non-specific DNA. It binds DNA substrates in the absence of sinefungin with decreasing affinities: hemimethylated > dimethylated > unmethylated > non-specific DNA. Thus, both bound cofactor, as mimicked by its surrogate sinefungin, and the methylation state of the canonical DNA site influenced DNA binding.
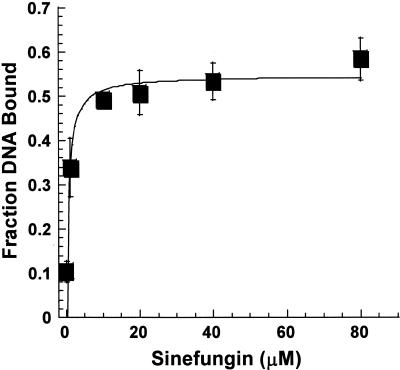
We determined the effect of increasing amounts of sinefungin on ternary complex formation with hemimethylated DNA to determine the concentration of sinefungin needed to maximize binding (29). M·RsrI (15 nM) and 5 nM top strand hemimethylated duplex (mTB) were incubated with increasing amounts of sinefungin. This enzyme:DNA ratio, which shifts only ∼0.1 of the DNA, allowed us to determine the effects of sinefungin with high sensitivity. Saturation of DNA binding occurred at concentrations of sinefungin >10 µM. The binding isotherm presented in Figure 2 shows that the 200 µM sinefungin used in other experiments (Table 1) saturates the enzyme since a KDapp value of 0.8 ± 0.4 µM was obtained.
Figure 2.
Stimulation of DNA binding by sinefungin. M·RsrI (15 nM) was incubated with increasing amounts of sinefungin. The data from three experiments were analyzed as described in Materials and Methods.
Base flipping
We tested the affinity of M·RsrI for substrates containing an abasic site (abTB and abTmB) in place of the target adenine to investigate whether M·RsrI might extrude or flip its target base from the helix to accomplish methylation. Base flipping expels the target base from the interior of the DNA helix into the active site of the enzyme and has been directly observed in the crystal structures of two C5-cytosine MTases, HhaI (7) and HaeIII (8). Fluorescence-based assays (24,32), gel retardation assays with DNA substrates containing abasic sites or mismatches (33) and permanganate susceptibility assays (34) suggest that base flipping occurs in amino MTase enzymes. The structure of M·TaqI bound to DNA, which has recently been solved, reveals how the γ class of adenine MTases achieve base flipping and specificity (E.Weinhold, personal communication). Gel retardation assays performed with M·HhaI showed a 10-fold greater affinity for DNA substrates containing an abasic site with respect to the normal G:C base pair at the site of methylation (35). We used a lower concentration of DNA for assays with abasic substrates because the apparent KD was <5 nM. M·RsrI bound an abasic unmethylated (abTB) and hemimethylated substrate (abTmB) 1.2- to 3.0-fold more tightly, respectively, than the normal hemimethylated duplex (TmB). The methyl group on the adenine opposite the abasic site enhanced affinity; substrate containing it was bound 2.5-fold better than the abasic unmethylated substrate, in keeping with the preference for regular hemimethylated DNA to unmethylated DNA noted earlier. The increased affinity for substrates containing an abasic site suggests that the enzyme employs base extrusion.
Electrophoretic gel retardation assays with substrates containing 2A were also conducted to test their suitability as ligands for M·RsrI and for use in fluorescence-based studies of base flipping. This fluorescent adenine analog fluoresces with an emission maximum at 370 nm when excited at 310 nm. An increase in fluorescence has been observed with 2A-substituted DNA upon binding of M·EcoRI (24) and M·TaqI (32) and this change has been attributed to eversion of the fluorophore from the DNA into the active site of the enzyme. Binding assays with 2A-containing duplex DNA substrates (2ATB and 2ATmB), as well as other duplex substrates under slightly different conditions (substituting 20 mM potassium glutamate for NaCl), gave approximately the same affinity values. M·RsrI bound ∼19-fold less well to the 2A-substituted hemimethylated duplex (2ATmB) than to the bottom strand hemimethylated equivalent (TmB). However, M·RsrI bound only 2-fold less well with 2A-substituted unmethylated duplex (2ATB) when compared to the unmethylated substrate (TB).
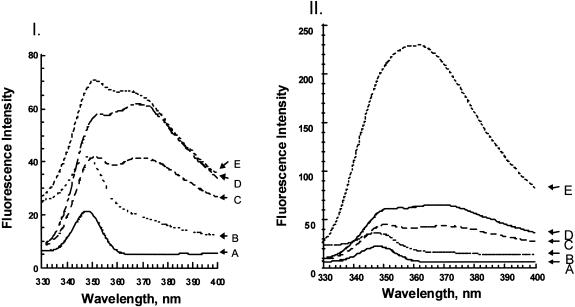
Since M·RsrI formed a complex with the 2A-substituted hemimethylated duplex (2ATmB) in the gel retardation assay, we also applied a fluorescence-based assay to assess base flipping. As a positive control, fluorescence data were collected using M·EcoRI with this substrate. Addition of 450 nM M·EcoRI or M·RsrI to 400 nM 2A-containing hemimethylated DNA was compared. Representative fluorescence spectra are shown in Figure 3. In the absence of enzyme, the addition of the complementary hemimethylated strand to the 2A-containing top strand results in an ∼1.5-fold decrease in fluorescence intensity and an emission maximum shift from 368 to 371 nm with respect to the 2A-containing top strand by itself (Fig. 3, curves C and D in I and II). The control with M·EcoRI added showed an ∼6-fold increase in fluorescence intensity and an 8 nm blue shift to 362 nm (Fig. 3, curves C–E in II). M·RsrI addition produces a bilobal peak with λmax at 351 and 364 nm, similar to the spectra for the 2A duplex (Fig. 3, curves C–E in I). A similar bilobal spectrum has been observed with M·TaqI (32). M·RsrI shows an ∼1.7-fold increase in fluorescence intensity and a 7 nm blue shift to λmax = 364 nm (Fig. 3, curves C–E in I). The differences in intensity enhancement between M·EcoRI and M·RsrI under similar buffer conditions and protein concentrations likely indicates differences in the active site environments of the expelled 2A probe.
Figure 3.
Steady-state fluorescence emission spectra of M·RsrI (I) and M·EcoRI (II) with 2ATmB DNA. (A) Buffer only, Raman peak at 348 nm; (B) enzyme only, ∼450 nM M·RsrI (λmax = 348 nm) or M·EcoRI (λmax = 348 nm); (C) 400 nM 2A-containing DNA duplex (λmax = ∼371 nm for both sets of spectra); (D) 400 nM single-strand 2A-containing DNA (λmax = 368 nm for both sets of spectra); (E) addition of ∼450 nM purified M·RsrI (λmax = 351nm and 364 nm) or M·EcoRI (λmax = 362 nm) to duplex 2-A-containing DNA.
AdoMet, AdoHcy and sinefungin binding
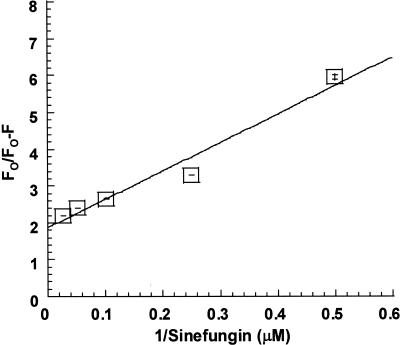
M·RsrI contains six tryptophans, at positions 75, 84, 88, 140, 161 and 208. All of these, except for the one at 208, lie in the predicted catalytic region (containing motifs IV–VIII) (5) opposite the AdoMet binding region (containing motifs I, II, III and X). The intrinsic fluorescent-properties of M·RsrI were exploited in a fluorescence-quenching assay to determine the dissociation constants for AdoMet, AdoHcy and sinefungin. The Stern–Volmer plot (22) and Lehrer’s modified Stern–Volmer plot (23) were used to analyze the quenching data (Fig. 4). The Lehrer plot for sinefungin binding in Figure 4 mimics those with AdoMet and AdoHcy, which are not shown. Stern–Volmer plots for AdoMet, AdoHcy and sinefungin exhibited a negative deviation from linearity, a result expected when only a fraction of the tryptophans are accessible to quenching by ligand binding (data not shown). Ligand binding may selectively affect the quantum yields of fluorophores in proximity to the binding site while leaving others unperturbed (36). Lehrer’s plot was therefore used to analyze these quenching processes (23). The data in Figure 4 and those generated with AdoMet and AdoHcy (data not shown) give a linear plot. The linearity of the Lehrer plot suggests that cofactor binding is the dominant fluorescence-quenching phenomenon over the range of concentrations tested (2–40 µM). AdoHcy, AdoMet and sinefungin interact as ligands with M·RsrI and can be considered ‘specific’ quenchers (37). Fluorescence quenching due to collisional or alternate binding processes might be manifested in the Lehrer plot as deviations from linearity, especially at low 1/[Q] values (38). The KD values and percentage of quenchable M·RsrI fluorescence derived from the Lehrer plots are reported in Table 2. The maximal emission peak for all the enzyme–substrate complexes was 337 nm, suggesting that the accessible tryptophans are in a hydrophobic environment.
Figure 4.
Lehrer plot of quenching of M·RsrI fluorescence by sinefungin. Error bars represent ± 1 SD.
Kinetic and isotope-partitioning analyses
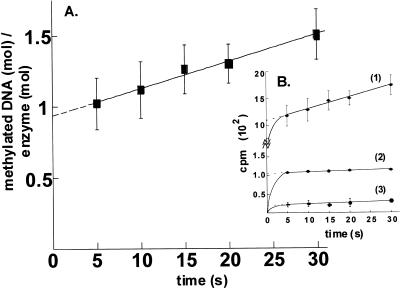
We conducted an active site titration to test for a burst when M·RsrI was incubated with saturating amounts of AdoMet (42 µM) and DNA (3.6 µM). Under these conditions, the burst magnitude and the amount of active enzyme can be determined by a linear extrapolation of the steady-state rate curve. M·RsrI displays burst kinetics with a 14mer, canonical site-containing hemimethylated substrate, forming 0.94 ± 0.03 mol product/mol M·RsrI with a kcat of 1.9 ± 0.1 × 10–2 s–1 (Fig. 5A). These findings indicate that the enzyme is at least 94% active and that a step after DNA methylation per se is rate limiting. Similar burst kinetics have been observed for other amino MTases, e.g. M·PvuII (39) and M·EcoRI (25).
Figure 5.
Kinetic analysis of M·RsrI. (A) Burst. M·RsrI was saturated with high specific activity AdoMet and diluted 50-fold into a solution containing the same specific activity AdoMet at the same concentration and DNA. (B) Isotope partition. Curve 1, re-plot of curve in (A); curve 2, enzyme was saturated with high specific activity AdoMet as in (A) but diluted into a solution containing the same concentration of unlabeled AdoMet and excess DNA; curve 3, control. Enzyme saturated with high specific activity AdoMet was diluted 50-fold into unlabeled AdoMet of the same concentration before addition of DNA. Error bars represent ± 1 SD.
We also conducted an isotope-partitioning analysis of M·RsrI to examine the competency of the M·RsrI–AdoMet complex. The isotope-partitioning experiment is similar to that for the burst determination, except that a pre-saturated binary complex of M·RsrI with [14C]AdoMet was diluted 50-fold into a mixture containing excess DNA and unlabeled AdoMet to determine what fraction of the bound labeled AdoMet would react before exchanging with unlabeled cofactor. The formation of radiolabeled product reflects the propensity of the enzyme-bound labeled AdoMet to undergo catalysis without dissociation (40). As a control, the specific activity of the labeled AdoMet was diluted before adding the DNA. Since the steady-state portion of the isotope-partitioning and control experiments were determined using labeled AdoMet with a 50-fold reduction in specific activity, unlike the burst experiment, the slopes of the lines are not identical. The results are reported as radioactive product (c.p.m.) formed as a function of time in Figure 5B. A comparison of the intercept of curve 2 (∼110 c.p.m.) with curve 3 (∼20 c.p.m.) and curve 1 (∼1100 c.p.m.) of Figure 5B clearly demonstrates that M·RsrI retains a fraction (10%), but not all, of its bound, high specific-activity labeled AdoMet before the first turnover. The detection of a burst in the isotope- partitioning assay despite the presence of ∼50 fold excess unlabeled AdoMet (Fig. 5B, line 2) provides evidence that the pre-bound AdoMet is catalytically competent and excludes an ordered BiBi mechanism whereby DNA must bind first. If DNA were required to bind to M·RsrI before AdoMet for catalysis, all of the pre-bound, catalytically non-productive AdoMet would dissociate, resulting in a curve similar to the control (Fig. 5B, line 3). Further steady-state kinetic analysis will be required to distinguish between an ordered (AdoMet first) or random kinetic mechanism for M·RsrI. Furthermore, the turnover numbers for the isotope-partitioning (kcat = 1.3 ± 0.2 × 10–2 s–1) and control (kcat = 1.4 ± 0.4 × 10–2 s–1) experiments calculated using the 50-fold diluted specific activity of AdoMet agree well with the turnover number for the burst experiment (1.9 ± 0.1 × 10–2 s–1).
DISCUSSION
DNA binding in vitro
We determined that the affinity of M·RsrI for DNA is a function of the state of methylation of the EcoRI site and of bound cofactor (Table 1). Like the N6mA MTases T4 Dam (27) and M·EcoRV (29), and the C5 MTases HhaI (41) and M·MspI (42), M·RsrI bound hemimethylated DNA substrates best. This result is unlike M·EcoRI (30) and the EcoK type I MTase (43), both of which failed to discriminate between hemimethylated and unmethylated DNA. M·RsrI, like M·EcoRV and M·MspI, also binds weakly to the dimethylated substrate and did not bind well to non-canonical DNA. M·RsrI is also sensitive to methylation of the target sequence in a DNA binding assay in vivo (44). O’Gara et al. (41) observed, in the structure of M·HhaI–DNA–AdoHcy, that enhanced affinity for hemimethylated DNA may be mediated by a single amino acid, Glu239, whose side chain is in van der Waal’s contact with the 5-mC in the strand complementary to that containing the target methylatable cytosine. A similar situation may pertain with M·RsrI and hemimethylated DNA.
Sinefungin causes M·RsrI to bind DNA with 3- to 5-fold greater affinity to hemimethylated duplexes and with 18-fold greater affinity to unmethylated DNA. Interestingly, methylated DNA is bound no better in the presence of this AdoMet analog. These results suggest that bound cofactor affects site-specific binding by M·RsrI. Increased binding in the presence of AdoMet, AdoHcy or sinefungin is a general property of DNA MTases. For instance, M·EcoRI binds unmethylated DNA ∼100-fold more tightly in the presence of sinefungin (18). T4 Dam (27) binds both unmethylated and hemimethylated substrates ∼2.5- to 3-fold better in the presence of AdoMet.
Decreased binding to 2A-containing substrates suggests that M·RsrI may contact the minor groove during DNA recognition or catalysis. The amino group in the minor groove might also affect the ability of the enzyme to distort DNA upon binding and indirect effects due to alterations in the local structure of the DNA due to incorporation of the analog cannot be excluded (45). The N6mA MTase T4 Dam bound 6-fold less well to a 2A-substituted unmethylated 24mer duplex than to the normal unmethylated substrate (27). Conversely, M·EcoRI showed a 2.1-fold increase in affinity for 2A-containing 14mer substrates (46). The structures of the ternary complexes of M·HaeIII and M·HhaI mediate recognition via major groove contacts and extrusion of the base through the minor groove of DNA (7,8). Footprinting studies with M·EcoRV (47), M·SssI and M·HhaI (48) suggest that a majority of interactions of these enzymes with DNA occur with bases and phosphates in the major groove. However, studies with base analog-containing substrates with M·EcoRI (49) and M·EcoRV (50) showed that interactions likely also occur in the minor groove.
Base flipping
Electrophoretic gel retardation assays with DNA substrates containing an abasic site suggested that M·RsrI, like other DNA MTases, flips its target base from the DNA helix. M·RsrI bound an abasic hemimethylated substrate 3-fold more tightly than the adenine-containing hemimethylated control. This degree of binding enhancement is similar to that observed with M·EcoRI and M·EcoRV, which display 4- and 6-fold greater affinities for an abasic hemimethylated substrate, respectively (46,51). Enhanced affinity for DNA substrates containing abasic substitutions at the target base pair presumably reflects the reduced energy needed to expel the deoxyribose, in contrast to the Watson–Crick hydrogen-bonded and stacked nucleotide residue, from the helix upon enzyme binding (35).
Studies with the fluorescent probe 2A suggested that the fluorescence-based assay for examining the base flipping phenomenon may not be equally suitable for all enzymes. The decreased affinity of M·RsrI for 2A-containing substrates may reduce the ability of the enzyme to extrude the analog. The 7 nm blue shift and 1.7-fold increase in fluorescence intensity upon M·RsrI binding to the 2A substrate suggest a conformational change in the DNA, possibly indicative of base extrusion. The differences between M·EcoRI and M·RsrI (Fig. 5) may indicate differences in the environments of the expelled fluorescent probe. The 7 nm blue shift upon M·RsrI addition suggests that the environment surrounding 2A has a hydrophobic character since the quantum yield of 2A ribonucleotides diminishes drastically, with a concomitant blue shift in the emission spectrum, when placed in increasingly hydrophobic solvents (52). The crystal structure of M·RsrI contains several aromatic residues (Y68, W84, W88, F105, F249 and F250) in proximity (≈10 Å) to residues of motif IV (65–68) (10). Thus, the weak fluorescence enhancement may be due to quenching of the flipped fluorescent 2A base by hydrophobic residues in the active site.
Cofactor binding
Changes in the intrinsic fluorescence emission spectra of proteins upon ligand binding have long been used to estimate the equilibrium binding constants of the complexes. The KD values for M·RsrI were 4.2, 6.1 and 9.6 µM with sinefungin, AdoMet and AdoHcy, respectively, comparable to those obtained for other type II MTases. The KD values reported for E.coli Dam MTase are 2.0, 6.5 and 7.0 µM for sinefungin, AdoMet and AdoHcy, respectively (53). Similarly, dissociation constants determined for M·TaqI are 0.34, 2.0 and 2.4 µM (54). Similar fluorescence-quenching studies with M·EcoRI and AdoMet and sinefungin reported KD values of 10 and 15 µM, respectively (21). The KD value (4.2 µM) obtained for sinefungin is similar to the value obtained for the Kapp of sinefungin derived from gel retardation assays (0.8 ± 0.4 µM). That 45–55% of the accessible intrinsic fluorescence of M·RsrI is quenched for all three ligands suggests a similar binding mechanism for each. Structural comparisons of binary complexes of M·TaqI with AdoMet, AdoHcy or sinefungin show differences in conformation between the substrate, product and inhibitor molecules, respectively, although the polypeptide chain conformation remains essentially unchanged (54). Different cofactor configurations may account for the quantitative differences in both the dissociation constants and quenching observed for M·RsrI. Two tryptophans, W84 and W88, are near the catalytic DPPY motif and the motifs involved in AdoMet binding (10). These two tryptophans most likely contribute to the observed decrease in M·RsrI fluorescence upon cofactor or inhibitor binding.
Kinetic studies
Pre-steady-state and steady-state kinetic analysis of the N6mA MTase M·EcoRI has shown that the enzyme follows an ordered BiBi mechanism where AdoMet binds before DNA (25), requiring bound AdoMet for target site recognition. Another N6mA MTase, M·EcaI, follows a random, rapid equilibrium BiBi mechanism (55), also requiring bound AdoMet for site-specific DNA recognition. Isotope-partitioning analysis of M·RsrI excludes the possibility of an ordered BiBi mechanism whereby DNA binds first. In contrast, the 5mC MTases M·MspI (56), M·HhaI (15,57) and Dnmt1 bind DNA before AdoMet (58). In 5mC MTases the mechanism (15) involves a covalent DNA intermediate, a 5,6-dihydrocytosine–cysteine adduct that may dictate that DNA is bound before AdoMet. The amino MTase reaction occurs by direct methyl group transfer from pre-bound AdoMet by nucleophilic attack of an activated amino group of either N4-cytosine or N6-adenine (11,59) and its mechanism of methyl transfer may account for the different order of substrate addition.
Comparisons with M·EcoRI
The isocatalytic enzymes M·EcoRI and M·RsrI share similar pre-steady-state kinetic properties; both display burst kinetics and bind AdoMet before binding DNA. Fluorescence- quenching studies and gel retardation assays with M·EcoRI and M·RsrI reveal similar dissociation constants for cofactors (µM) and DNA binding (nM). However, M·RsrI lacked the several-fold increase in steady-state fluorescence displayed by M·EcoRI with a 2A-containing DNA substrate, suggesting differences in the environment of enzyme-bound 2A. The decreased binding affinity of M·RsrI for the 2A-containing duplex, in contrast to M·EcoRI, indicates differences in the recognition properties of the two enzymes, in spite of their similar catalytic and cofactor binding properties. Interestingly, M·EcoRI, unlike M·RsrI, lacks apparent discrimination between hemimethylated and unmethylated DNA. As with some other DNA modification enzymes, M·EcoRI bends DNA and ‘flips’ the target base (at a rate equal to kmethyation) for methylation (60,61). We have not determined whether M·RsrI can bend DNA, but gel retardation assays using a duplex containing an abasic site suggest the protein ‘flips’ the base. Both enzymes likely have a similar structural architecture and organization of conserved AdoMet binding and catalytic motifs shared with other DNA and AdoMet-dependent MTases (5).
Conclusion
A biochemical characterization of M·RsrI assessed the DNA recognition, cofactor binding and catalytic properties of the enzyme. M·RsrI discrimination in DNA binding was based on the methylation status and sequence of the DNA and the presence of cofactor. The apparent binding constants for DNA binding and for AdoMet, AdoHcy and sinefungin binding were comparable to those obtained with other type II C5 and amino MTases. Binding assays with canonical substrates containing an abasic site or a fluorescent base at the target adenine base suggested that M·RsrI extrudes the target adenine into the active site of the enzyme. M·RsrI does not use an ordered kinetic mechanism in which DNA binds before AdoMet. The enzyme shows burst kinetics, suggesting that product release is rate limiting.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Ms Ha Huynh for her assistance in the purification of wild-type enzyme and Mr Chad Thomas for aiding with preparation of the manuscript. The authors also thank Dr Mair Churchill for critical reading of the manuscript and Elmar Weinhold for sharing data prior to publication. This work was supported by NIH grant GM25621 to R.I.G., NSF grant MCB 9603567 to N.O.R. and a grant from the University of Illinois Research Board.
REFERENCES
- 1.Cheng X. and Blumenthal,R.M. (1999) S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions. World Scientific, Singapore, Singapore.
- 2.Jackson-Grusby L. and Jaenisch,R. (1996) Semin. Cancer Biol., 7, 261–268. [DOI] [PubMed] [Google Scholar]
- 3.Palmer B.R. and Marinus,M.G. (1994) Gene, 143, 1–12. [DOI] [PubMed] [Google Scholar]
- 4.Reisenauer A., Kahng,L.S., McCollum,S. and Shapiro,L. (1999) J. Bacteriol., 181, 5135–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malone T., Blumenthal,R.M. and Cheng,X. (1995) J. Mol. Biol., 253, 618–632. [DOI] [PubMed] [Google Scholar]
- 6.Wilson G.G. (1992) Methods Enzymol., 216, 259–279. [DOI] [PubMed] [Google Scholar]
- 7.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 8.Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) Cell, 82, 143–153. [DOI] [PubMed] [Google Scholar]
- 9.Tran P.H., Korszun,Z.R., Cerritelli,S., Springhorn,S.S. and Lacks,S.A. (1998) Structure, 6, 1563–1575. [DOI] [PubMed] [Google Scholar]
- 10.Scavetta R.D., Thomas,C.B., Walsh,M., Szegedi,S., Joachimiak,A., Gumport,R.I. and Churchill,M.E.A. (2000) Nucleic Acids Res., 28, 3950–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong W., O’Gara,M., Blumenthal,R.M. and Cheng,X. (1997) Nucleic Acids Res., 25, 2702–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labahn J., Granzin,J., Schluckebier,G., Robinson,D.P., Jack,W.E., Schildkraut,I. and Saenger,W. (1994) Proc. Natl Acad. Sci. USA, 91, 10957–10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reich N.O., Maegley,K.A., Shoemaker,D.D. and Everett,E. (1991) Biochemistry, 30, 2940–2946. [DOI] [PubMed] [Google Scholar]
- 14.Ho D.K., Wu,J.C., Santi,D.V. and Floss,H.G. (1991) Arch. Biochem. Biophys., 284, 264–269. [DOI] [PubMed] [Google Scholar]
- 15.Wu J.C. and Santi,D.V. (1987) J. Biol. Chem., 262, 4778–4786. [PubMed] [Google Scholar]
- 16.Kaszubska W., Aiken,C., O’Connor,C.D. and Gumport,R.I. (1989) Nucleic Acids Res., 17, 10403–10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaszubska W., Webb,H.K. and Gumport,R.I. (1992) Gene, 118, 5–11. [DOI] [PubMed] [Google Scholar]
- 18.Reich N.O. and Mashhoon,N. (1990) J. Biol. Chem., 265, 8966–8970. [PubMed] [Google Scholar]
- 19.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Bradford M.M. (1976) Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 21.Maegley K.A., Gonzalez,L.,Jr, Smith,D.W. and Reich,N.O. (1992) J. Biol. Chem., 267, 18527–18532. [PubMed] [Google Scholar]
- 22.Stern O. and Volmer,M. (1919) Phys. Z., 20, 183–188. [Google Scholar]
- 23.Lehrer S.S. (1971) Biochemistry, 10, 3254–3263. [DOI] [PubMed] [Google Scholar]
- 24.Allan B.W. and Reich,N.O. (1996) Biochemistry, 35, 14757–14762. [DOI] [PubMed] [Google Scholar]
- 25.Reich N.O. and Mashhoon,N. (1991) Biochemistry, 30, 2933–2939. [DOI] [PubMed] [Google Scholar]
- 26.Gray D.M., Hung,S.H. and Johnson,K.H. (1995) Methods Enzymol., 246, 19–34. [DOI] [PubMed] [Google Scholar]
- 27.Malygin E.G., Petrov,N.A., Gorbunov,Y.A., Kossykh,V.G. and Hattman,S. (1997) Nucleic Acids Res., 25, 4393–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carey J. (1991) Methods Enzymol., 208, 103–117. [DOI] [PubMed] [Google Scholar]
- 29.Szczelkun M.D. and Connolly,B.A. (1995) Biochemistry, 34, 10724–10733. [DOI] [PubMed] [Google Scholar]
- 30.Reich N.O., Olsen,C., Osti,F. and Murphy,J. (1992) J. Biol. Chem., 267, 15802–15807. [PubMed] [Google Scholar]
- 31.Klig L.S., Carey,J. and Yanofsky,C. (1988) J. Mol. Biol., 202, 769–777. [DOI] [PubMed] [Google Scholar]
- 32.Holz B., Klimasauskas,S., Serva,S. and Weinhold,E. (1998) Nucleic Acids Res., 26, 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mernagh D.R., Taylor,I.A. and Kneale,G.G. (1998) Biochem. J., 336, 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serva S., Weinhold,E., Roberts,R.J. and Klimasauskas,S. (1998) Nucleic Acids Res., 26, 3473–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Gara M., Horton,J.R., Roberts,R.J. and Cheng,X. (1998) Nature Struct. Biol., 5, 872–877. [DOI] [PubMed] [Google Scholar]
- 36.Samworth C.M., Degli Esposti,M. and Lenaz,G. (1988) Eur. J. Biochem., 171, 81–86. [DOI] [PubMed] [Google Scholar]
- 37.Lange R., Anzenbacher,P., Muller,S., Maurin,L. and Balny,C. (1994) Eur. J. Biochem., 226, 963–970. [DOI] [PubMed] [Google Scholar]
- 38.Eftink M.R. and Ghiron,C.A. (1981) Anal. Biochem., 114, 199–227. [DOI] [PubMed] [Google Scholar]
- 39.Adams G.M. and Blumenthal,R.M. (1997) Biochemistry, 36, 8284–8292. [DOI] [PubMed] [Google Scholar]
- 40.Rose I.A. (1980) Adv. Enzymol. Relat. Areas Mol. Biol., 64, 47–59. [Google Scholar]
- 41.O’Gara M., Roberts,R.J. and Cheng,X. (1996) J. Mol. Biol., 263, 597–606. [DOI] [PubMed] [Google Scholar]
- 42.Dubey A.K. and Roberts,R.J. (1992) Nucleic Acids Res., 20, 3167–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powell L.M., Dryden,D.T. and Murray,N.E. (1998) J. Mol. Biol., 283, 963–976. [DOI] [PubMed] [Google Scholar]
- 44.Szegedi S.S. and Gumport,R.I. (2000) Nucleic Acids Res., 28, 3972–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aiken C.R. and Gumport,R.I. (1991) Methods Enzymol., 208, 433–457. [DOI] [PubMed] [Google Scholar]
- 46.Allan B.W., Beechem,J.M., Lindstrom,W.M. and Reich,N.O. (1998) J. Biol. Chem., 273, 2368–2373. [DOI] [PubMed] [Google Scholar]
- 47.Szczelkun M.D., Jones,H. and Connolly,B.A. (1995) Biochemistry, 34, 10734–10743. [DOI] [PubMed] [Google Scholar]
- 48.Renbaum P. and Razin,A. (1995) J. Mol. Biol., 248, 19–26. [DOI] [PubMed] [Google Scholar]
- 49.Brennan C.A., Van Cleve,M.D. and Gumport,R.I. (1986) J. Biol. Chem., 261, 7279–7286. [PubMed] [Google Scholar]
- 50.Newman P.C., Nwosu,V.U., Williams,D.M., Cosstick,R., Seela,F. and Connolly,B.A. (1990) Biochemistry, 29, 9891–9901. [DOI] [PubMed] [Google Scholar]
- 51.Cal S. and Connolly,B.A. (1997) J. Biol. Chem., 272, 490–496. [DOI] [PubMed] [Google Scholar]
- 52.Ward D.C. and Reich,E. (1969) J. Biol. Chem., 244, 1228–1237. [PubMed] [Google Scholar]
- 53.Bergerat A. and Guschlbauer,W. (1990) Nucleic Acids Res., 18, 4369–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schluckebier G., Kozak,M., Bleimling,N., Weinhold,E. and Saenger,W. (1997) J. Mol. Biol., 265, 56–67. [DOI] [PubMed] [Google Scholar]
- 55.Szilak L., Der,A., Deak,F. and Venetianer,P. (1993) Eur. J. Biochem., 218, 727–733. [DOI] [PubMed] [Google Scholar]
- 56.Bhattacharya S.K. and Dubey,A.K. (1999) J. Biol. Chem., 274, 14743–14749. [DOI] [PubMed] [Google Scholar]
- 57.Lindstrom W.M. Jr, Flynn,J. and Reich,N.O. (2000) J. Biol. Chem., 275, 4912–4919. [DOI] [PubMed] [Google Scholar]
- 58.Flynn J., Glickman,J.F. and Reich,N.O. (1996) Biochemistry, 35, 7308–7315. [DOI] [PubMed] [Google Scholar]
- 59.Pogolotti A.L. Jr, Ono,A., Subramaniam,R. and Santi,D.V. (1988) J. Biol. Chem., 263, 7461–7464. [PubMed] [Google Scholar]
- 60.Garcia R.A., Bustamante,C.J. and Reich,N.O. (1996) Proc. Natl Acad. Sci. USA, 93, 7618–7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allan B.W., Garcia,R., Maegley,K., Mort,J., Wong,D., Lindstrom,W., Beechem,J.M. and Reich,N.O. (1999) J. Biol. Chem., 274, 19269–19275. [DOI] [PubMed] [Google Scholar]