ABSTRACT
The integration of HPV DNA into human chromosomes plays a pivotal role in the onset of papillomavirus-related cancers. HPV DNA integration often occurs by linearizing the viral DNA in the E1/E2 region, resulting in the loss of a critical viral early polyadenylation signal (PAS), which is essential for the polyadenylation of the E6E7 bicistronic transcripts and for the expression of the viral E6 and E7 oncogenes. Here, we provide compelling evidence that, despite the presence of numerous integrated viral DNA copies, virus-host fusion transcripts originate from only a single integrated HPV DNA in HPV16 and HPV18 cervical cancers and cervical cancer-derived cell lines. The host genomic elements neighboring the integrated HPV DNA are critical for the efficient expression of the viral oncogenes that leads to clonal cell expansion. The fusion RNAs that are produced use a host RNA polyadenylation signal downstream of the integration site, and almost all involve splicing to host sequences. In cell culture, siRNAs specifically targeting the host portion of the virus-host fusion transcripts effectively silenced viral E6 and E7 expression. This, in turn, inhibited cell growth and promoted cell senescence in HPV16+ CaSki and HPV18+ HeLa cells. Showing that HPV E6 and E7 expression from a single integration site is instrumental in clonal cell expansion sheds new light on the mechanisms of HPV-induced carcinogenesis and could be used for the development of precision medicine tailored to combat HPV-related malignancies.
IMPORTANCE
Persistent oncogenic HPV infections lead to viral DNA integration into the human genome and the development of cervical, anogenital, and oropharyngeal cancers. The expression of the viral E6 and E7 oncogenes plays a key role in cell transformation and tumorigenesis. However, how E6 and E7 could be expressed from the integrated viral DNA which often lacks a viral polyadenylation signal in the cancer cells remains unknown. By analyzing the integrated HPV DNA sites and expressed HPV RNAs in cervical cancer tissues and cell lines, we show that HPV oncogenes are expressed from only one of multiple chromosomal HPV DNA integrated copies. A host polyadenylation signal downstream of the integrated viral DNA is used for polyadenylation and stabilization of the virus-host chimeric RNAs, making the oncogenic transcripts targetable by siRNAs. This observation provides further understanding of the tumorigenic mechanism of HPV integration and suggests possible therapeutic strategies for the development of precision medicine for HPV cancers.
KEYWORDS: human papillomavirus, integration, oncogene expression, carcinogenesis, clonal expansion
INTRODUCTION
Human papillomavirus (HPV) is a small non-enveloped DNA virus with a compact circular genome. It replicates and assembles exclusively in the nucleus, and its life cycle is intricately tied to the differentiation of keratinocytes. While most HPV infections typically result in benign warts, high-risk genotypes, like HPV16 and HPV18, are responsible for cervical cancers (1, 2) and an increasing number of head and neck and oropharyngeal cancers (3).
High-risk HPVs (HR-HPV) express their oncogenes, E6 and E7, as a bicistronic RNA from a viral early promoter. This RNA undergoes alternative splicing and uses a viral early polyadenylation signal (PAS) located downstream of the viral E2 open reading frame (ORF) (4, 5). The HR-HPV E6 ORF has an intron and E6 protein can be expressed only from a bicistronic E6E7 RNA that retains the E6 intron. Splicing of the E6 intron creates E6*I RNA, a spliced isoform of the E6E7 bicistronic RNA that is abundant in cervical cancer tissues and their derived cell lines. This spliced E6*I RNA is responsible for the expression of viral E7 protein (6–8).
The key event for carcinogenesis in persistent HR-HPV infections is the integration of HPV DNA into the host genome (9–12) which leads to the elevated expression of the viral E6 and E7 oncogenes, disrupting cell cycle control by inducing degradation of cellular tumor suppressor p53 and pRb proteins (13, 14). Integrated HPV DNA that is linearized in the E1/E2 ORF regions alleviates E2 repression of E6 and E7 expression (15–17). However, disrupting the E1/E2 regions results in the loss of a critical viral early polyadenylation signal (PAS), which is needed for the polyadenylation of the E6E7 bicistronic transcripts and for expression of the viral oncogenes (4). To be expressed from integrated HPV DNA, the E6E7 transcripts need to have a host PAS downstream of the integrated viral DNA. Several studies have reported that host sequences are co-expressed from the HPV integration sites in HPV-positive cervical cancer cell lines and tissues and that a host PAS is detectable in the E6E7 transcripts in CaSki cells and an HPV16 cervical carcinoma (18–23). Moreover, an early report showed that the E6E7 transcripts are expressed from a single integration site in CaSki cells and 13 HPV16/18 cancers, whereas other silent HPV integration sites in CaSki cells are presumably suppressed by host DNA methylation (24).
However, even after more than three decades of HPV research, the mechanism(s) of E6E7 expression in tumor cells remains elusive. The reported studies did not directly address the role(s) of the host sequences downstream of the integration site in the expression of viral E6 and E7 oncogenes. Nor is it clear whether HR-HPV-related cancers can be caused purely by episomal HPV DNAs as suggested based on measurements of the E2 and E6 DNA ratio by quantitative PCR (25–27). Other reports, which used a 3′ rapid amplification of cDNA ends (RACE)-based assay to amplify papillomavirus oncogene transcripts (APOT) (22, 28), suggest that episomal DNA may be not involved because there is a positive correlation of the frequency of the HPV DNA integration and progression of cervical lesions. We used genome-wide sequencing to identify HPV virus-host chimeric junctions to better understand HPV integration in cervical cancer tissues and cancer-derived cell lines. We compared chimeric junction reads (CJRs) from paired DNA-seq and RNA-seq to investigate the expression and post-transcriptional processing of virus-host RNA transcripts and the corresponding integrated HPV DNA in both cervical cancer tissues and three well-established cervical cancer cell lines. We found that only one of the integrated HPV DNA copies produces stable transcripts irrespective of the total number of the integrated HPV DNA copies present in the host genome.
RESULTS
HPV virus-host chimeric junctions and E6E7 expression measured by genome-wide DNA-seq and RNA-seq analyses of cervical cancer tissues
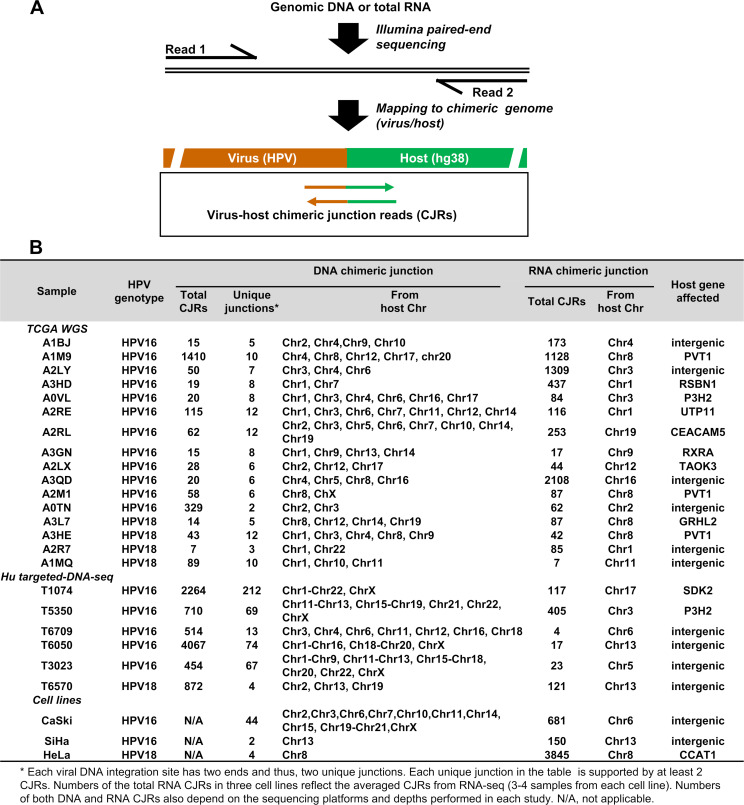
Seventy-four cervical cancer tissues (67 from TCGA WGS [the cancer genome atlas database with whole genome sequencing] and 7 from targeted DNA-seq) (29, 30) were analyzed using paired DNA-seq and RNA-seq data. Having both virus-host DNA and RNA CJRs (Fig. 1A) makes it possible to identify viral RNAs expressed from HPV DNAs integrated at specific sites. Of these 74 samples, 55 (74%) were either HPV16-positive (43 tissues) or HPV18-positive (12 tissues) and 19 (26%) were negative for both HPV16 and HPV18 in our assays. We did not look for other high-risk HPV types in the HPV16- and HPV18-negative tissues. Among the 55 HPV16- and HPV18-positive tissues, 33 (60%) had HPV16 (23 tissues) or HPV18 (10 tissues) integration sites that were identified by both DNA and RNA CJRs. Of these junction sites, 15 showed microhomology of the virus and host sequences (Table S1). For the remaining 22 (40%) samples, most of which had low-throughput sequencing data, we failed to obtain DNA or RNA CJRs or both. Twenty-two out of 33 (17 HPV16 and 5 HPV18) samples (~67%) had HPV DNA integrated in multiple sites on different chromosomes, and the remaining 11 (6 HPV16 and 5 HPV18) (~33%) samples exhibited only one integration site. All the RNA CJRs that were detected in the samples came only from a single chromosomal HPV DNA integration site regardless of whether the DNA analysis detected a single or multiple chromosomal HPV DNA integration sites in the sample (Fig. 1B; Tables S1 and S2). In all 33 tissues, the integrated HPV DNAs from which we found the RNA CJRs arose by linearizing the viral DNA in the E1 or E2 region.
Fig 1.
DNA-seq and RNA-seq analysis of integrated HPV DNA and its expression in cervical cancer tissues and cell lines. Diagram (A) shows the virus-host CJRs determined from one of the two ends of a virus DNA integration site. The sequencing reads were fully mapped to the chimeric virus (HPV16 or HPV18)-host hg38 genomes. CJRs are the reads from an integration junction with minimal ≥20 nts from each side of the HPV genome and the host genome. (B) The mapped HPV integration sites on different chromosomes and viral RNA transcription from a single HPV DNA integration site in the cervical cancer tissues and cell lines. Chr1-Chr22, from chromosome 1 to chromosome 22.
An integration event that breaks the circular HPV genome in the E1/E2 region would prevent viral early RNA transcripts from using the viral early polyadenylation signal (PAS) downstream of the E2 ORF for polyadenylation. Thus, the integrated viral DNA would not be expected to express the viral E6E7 RNA or, by extension, the E6 and E7 oncoproteins (4, 5). How can the integrated HPV DNA in these 33 tissues, including the 22 with HPV DNA integrated in multiple sites on different chromosomes, express viral E6 and E7 in the absence of a viral early PAS? We asked whether the integrated HPV DNA used the viral early promoter P97 (HPV16) or P55/102 (HPV18) and whether the E6E7 RNAs used a nearby host PAS. The distribution of RNA reads was analyzed and visualized on the chimeric virus-host genomes using the Integrative Genomic Viewer (IGV) program, which showed that all the E6E7 RNA used the viral early promoter from the integrated HPV DNA and 32 of the 33 used a host PAS downstream of the integration site. In seven of these tissues, E6E7 RNAs also used the normal viral PAS (Table S1 and Table S2). We believe that these seven tissue samples (five HPV16 and two HPV18; six from the samples in which we detected HPV DNA from multiple integration sites and one in which we detected HPV DNA from a single integration site) probably included a cancer border region with pre-cancer lesions which could contain replicating episomal HPV DNA. We also found one tissue, A3NI, in which the E6E7 RNA from the integrated HPV18 DNA used the viral PAS (Table S1). This tissue had DNA CJRs from five different chromosomes, but we were unable to detect RNA CJRs (Fig. S1A). However, in the A3NI tissue, there was an HPV16 integration-induced amplification of a 33.2 kb region of Chr9 which included both the MAG and CD22 genes (Fig. S1A; Table S3 for details of HPV16 integration junctions).
Characterization of the expressed viral RNA from a single chromosomal HPV DNA integration site in TCGA cervical cancer tissues
We mapped all the DNA-seq reads and RNA-seq reads to the chimeric HPV-host genomes and correlated the chimeric virus-host DNA CJRs with RNA CJRs (including both integration junctions and splice junctions). In the 11 cervical cancer tissues which had only one chromosomal HPV DNA integration site (Table S1), we showed (Fig. S1C, D, F and G) how the viral E6E7 RNAs were transcribed and processed. In the tissue A1M8 (which had a single chromosomal HPV16 integration site), there was a single truncated HPV16 DNA (~3.2 kb) integrated in a rearranged region covering the SORBS1, ALDH18A1, and ENTPD1 genes on Chr10 (Fig. S1B and E), with one end of the linearized HPV16 DNA, at nt 6,801 in L1, joined to Chr10 nt 95,509,128 and the other viral DNA end, at nt 2,136 in E1, joined to Chr10 nt 95,758,768, which led to a ~250 kb deletion in the rearranged region of Chr10. The integrated viral DNA expressed viral E6E7 RNA using the viral early promoter P97 and a host 3ʹ splice site at Chr10 nt 95,756,720 to reach a host PAS at 95,753,225, ~5.5 kb downstream of the integration site (Fig. S1E). Similarly, in A1MI (which had a single chromosomal HPV18 integration site), the truncated HPV18 DNA (~4.9 kb), which was integrated in a LINC-PINT gene on Chr7 (also Fig. S1B and H), with one end of the linearized HPV16 DNA, at nt 4,891 in L2, joined to Chr7 nt 130,952,063 and the other viral DNA end, at nt 2,922 in E2, joined to Chr7 nt 130,934,029, which led to a ~18 kb deletion in the LINC-PINT gene. The integrated viral DNA expressed the viral E6E7 RNA using the viral early promoter P55/102 and a host 3ʹ splice site at Chr7 nt 130,939,239 to reach a host PAS at Chr7 nt 130,940,881, ~6.9 kb downstream of the integration site (Fig. S1H). In both tissues, three virus-host chimeric transcripts were found which arose by alternative RNA splicing (Fig. S1E and H).
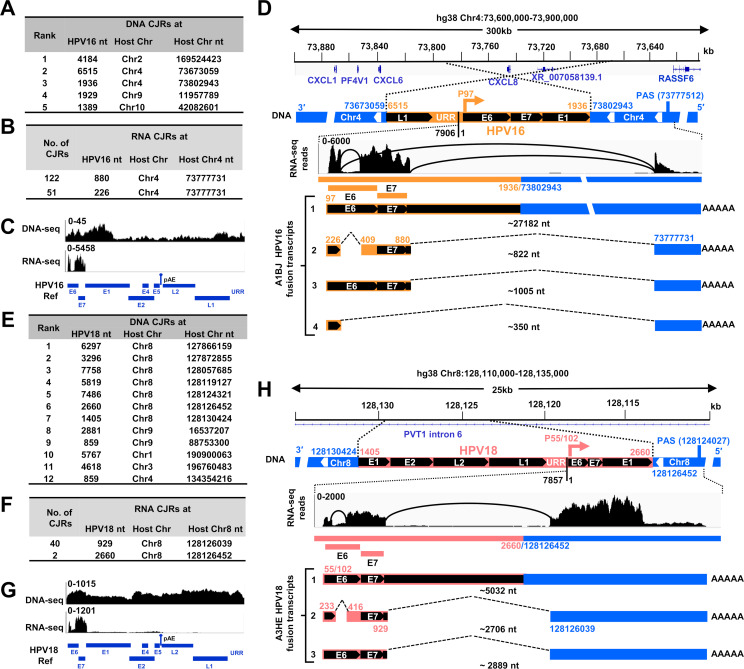
Four TCGA tissues A1BJ (HPV16), A2RE (HPV16), AIM9 (HPV16), and A3HE (HPV18) had multiple HPV DNA copies integrated on different chromosomes, but only one DNA copy expressed E6E7 RNA (Fig. 2; Fig. S1I through N). The tissue A1BJ had HPV16 DNA integrated in four different chromosomes (Fig. 2A). The combined viral DNA reads from four different chromosomes covered the full-length HPV16 genome, but only the viral DNA integrated in Chr4 expressed E6E7 RNA (Fig. 2B and C), suggesting that integration involves HPV genomes that are linearized in different places on the viral genome. The truncated HPV16 DNA (3,327 bp) was integrated in a rearranged region covering CXCL8 and XR_007058139.1 genes on Chr4, with one end of the linearized HPV16 DNA, at nt 6,515 in L1, joined to Chr4 nt 73,673,059 and the other viral DNA end, at nt 1,936 in E1, joined to Chr4 nt 73,802,943. This integration led to a ~130 kb deletion in the rearranged host region and a 4,579 bp deletion from the viral genome. Mapping the total RNA-seq reads to the region of the host genome for this integrated HPV16 DNA and analyzing the splice junctions (by Sashimi plots) showed that the integrated viral DNA expressed a bicistronic E6E7 RNA using the viral P97 early promoter and a host 3ʹ splice site at Chr4 nt 73,777,731 to reach a host PAS at Chr4 nt 73,777,512, ~25 kb downstream of the viral integration site (Fig. 2D). Four HPV16-host fusion RNA isoforms, which arose by alternative RNA splicing, were detected in the A1BJ tissue. The abundant transcript-2 will produce viral E7, the rare transcript-3 E6, transcript-4 an E6*-host fusion, and transcript-1 is an unspliced pre-mRNA (Fig. 2D).
Fig 2.
RNA transcription, splicing, and polyadenylation from a single HPV DNA integration site in the HPV16 cervical cancer tissue A1BJ and HPV18 cervical cancer tissue A3HE. (A and E) Distributions of the DNA CJRs mapped to the chimeric HPV16- or HPV18-host chromosomes in the frequency order of the determined unique junction reads in the tissues A1BJ (A) and A3HE (E). (B and F) The mapped RNA CJRs (≥2 CJRs) from RNA-seq of the tissues A1BJ (B) and A3HE (F) to the chimeric virus-host genome. (C and G) Mapping and visualization of HPV-specific reads from DNA-seq and RNA-seq to the HPV16-hg38 genome (C) and to the HPV18-hg38 genome (G) by IGV. (D) The mapped HPV16 DNA integration site in the intergenic region between CXCL6 and RASSF6 genes on Chr4 in the tissue A1BJ. Expression of viral E6E7 RNA from the integrated DNA is shown as a chimeric virus-host RNA, with RNA-seq reads-coverage (0–6,000 scale) and Sashimi plots for splice directions illustrated by IGV (middle). Numbers above or below the diagrams indicate the virus (yellow) or host (blue) genome nucleotide (nt) positions, the viral early promoter P97 (arrows), the host polyadenylation signal (PAS), and the RNA splice sites. Four isoforms of HPV16-host fusion RNA identified were transcribed from the viral early promoter and polyadenylated by using a host PAS at Chr4 nt 73,777,512. This integration of HPV16 DNA led to ~4.5 kb loss of the viral genome region from nt 1,937 to nt 6,514. URR, upstream regulatory region; 7906/1, viral genome tail-to-head junction. The number below each isoform RNA indicates the estimated length of the RNA without a pA tail. (H) Diagram showing the mapped HPV18 DNA in the intron 6 region of PVT1 (GENCODE V44) on Chr8 in the tissue A3HE and the expression of the virus-host fusion RNA from the viral early promoter P55/102. A host PAS at Chr8 nt 128,124,027 downstream of the HPV18 integration site was used for polyadenylation of the virus-host fusion RNA and production of three RNA isoforms via alternative RNA splicing. See other details in (D).
Similarly, the tissues A2RE and A1M9 expressed E6E7 from only one of multiple integrated viral DNA copies on different chromosomes. In A2RE, a truncated HPV16 DNA (~2.7 kb) was integrated in a rearranged region covering UTP11, POU3F1, MIR3659HG, LINC02786, and XR_001737985.1 genes on Chr1 (Fig. S1I through K). Integration led to a ~125 kb deletion of the rearranged region, with one end of the linearized HPV16 DNA at nt 5,181 in L1 joined to Chr1 nt 38,147,258 and the other viral DNA end at nt 1,663 in E1 joined to Chr1 nt 38,021,662. The integrated viral DNA expressed viral E6E7 RNA from the viral early promoter P97 and RNA splicing involved two host 3ʹ splice sites. The RNA used a UTP11 PAS at Chr1 nt 38,024,800, ~3.1 kb downstream of the viral integration site (Fig. S1K). In A1M9 (Fig. S1L through N), the truncated HPV16 DNA (~6.6 kb) was integrated in an intron of the PVT1 gene on Chr8. The integration led to a 23 bp deletion in the intron, with one end of the linearized HPV16 DNA at nt 4,386 in L2 joined to Chr8 nt 127,981,141 and the other viral DNA end at nt 3,116 in E2 joined to Chr8 nt 127,981,163. The integrated viral DNA expressed viral E6E7 RNA from the viral early promoter P97 and either two or four host 3ʹ splice sites to reach a host PAS at Chr8 nt 127,999,309, ~18 kb downstream of the viral integration site (Fig. S1N). Alternative RNA splicing produced four and six isoforms of the E6E7 RNA, respectively, in A2RE and A1M9 tissues (Fig. S1K and N).
In the tissue A3HE, integrated HPV18 DNA was found in five different chromosomes. The combined viral DNA reads covered the entire HPV18 genome, but only one integration site, on Chr8, expressed detectable E6E7 RNA (Fig. 2E through H). The viral DNA had a 1,255 bp duplication in the E1 region and was integrated in the PVT1 gene, with one end of the HPV18 DNA at E1 nt 1,405 joined to Chr8 nt 128,130,424 and, on the other end, E1 nt 2,660 was joined to Chr8 nt 128,126,452. The integrated HPV18 DNA expressed viral E6E7 RNA using the early viral promoter P55/P102 and a host 3ʹ splice site at Chr8 nt 128,126,039 to reach a host PAS at Chr8 nt 128,124,027, ~2.4 kb downstream of the viral integration site (Fig. 2H). Three E6E7 transcripts were detected; transcript-2 was used for the translation of the viral E7 protein (Fig. 2H).
Characterization of the expressed viral RNAs from a single integrated viral DNA copy in Chinese cervical cancer tissues
We also correlated the chimeric virus-host CJRs from paired DNA-seq and RNA-seq data from seven Chinese cervical cancer tissues (Table S2). All seven tissues had multiple HPV DNA copies integrated in multiple chromosomes. We found RNA CJRs in six of the seven tissues. In each of the six samples, the viral RNAs were produced by only one of the integrated viral DNAs (Table S2). The results from these six tissues are summarized in Fig. 1B; Table S2 and Fig. S1O through X.
Tissue sample T1074 had HPV16 DNA integrated on all 23 chromosomes (Fig. S1O through Q); however, only one copy (3,964 bp) that was integrated in the last intron of SDK2 gene on Chr17 produced detectable E6E7 bicistronic RNA (Fig. 1B; Fig. S1O and P). This integration led to a 109 bp deletion in the last intron of SDK2 (Fig. S1Q), with one end of the HPV16 DNA, in L1 at nt 5,595, joined to Chr17 nt 73,344,270. The other end, in E1 at nt 1,652, was joined to Chr17 nt 73,344,162. The integrated HPV16 DNA expressed viral E6E7 RNA using the viral early promoter P97 and a host 3ʹ splice site at Chr17 nt 73,338,940 to reach the host SDK2 PAS at Chr17 nt 73,334,404, ~9.8 kb downstream of the viral DNA integration site, resulting in the production of four detectable isoforms of E6E7 RNA. Transcript-3 expressed E7 (Fig. S1Q). This viral DNA also led to the expression of SDK2-viral RNA fusion. RNA splicing from SDK2 exon 44 to a viral 3ʹ splice site at nt 5,639 in the integrated HPV16 DNA and polyadenylation at the viral late PAS at nt 7,321 produced a ~8 kb transcript-1 which probably encodes a SDK2-L1 fusion protein (Fig. S1Q).
In tissues T3023, T5350, T6050, and T6709 (Fig. S1R through U), in which HPV16 DNA was integrated in multiple chromosomal sites, only one integrated viral DNA produced detectable E6E7 RNA. In T3023, T5350, and T6709, the expressed viral DNA was integrated in an intergenic or intronic region. In T6050, the expressed viral DNA was integrated in the 3′ UTR region of KLF12. All E6E7 transcripts from these integrated viral DNAs used the viral early promoter P97 and a host PAS downstream of the integration site to express and polyadenylate the E6E7 RNA (Fig. S1R through U). Alternative RNA splicing led to the production of the several detectable isoforms of E6E7 RNA, with abundant E6*I RNA used to produce E7 (6, 31).
In the tissue T6570, HPV18 DNA was integrated in three different chromosomes, but only the viral DNA on Chr13 was found to express stable viral E6E7 RNA (Fig. S1V through X). The integration of the truncated HPV18 DNA led to the duplication of the viral DNA in an intergenic region between KLF12 and KLF5 and the deletion of the LINC00392 gene on Chr13. However, the duplicated copies of the HPV genome (HPV18a and HPV18b, each 7,681 bp), which had a 176 bp deletion in the E2 region, were separated by a rearranged ~20 kb remnant of a ~307 kb region of the host genome, of which ~287 kb were deleted. In both copies of the integrated viral DNA, one end of the HPV18 DNA, in E2 at nt 3,631, was joined to Chr13 nt 73,350,892, and the other end, in E2 at nt 3,455, was joined to Chr13 nt 73,658,495 (Fig. S1X, top). E6E7 RNA was expressed only from HPV18a, using the early viral promoter P55/102 (32) and was polyadenylated using a viral PAS at nt 4,235 from the HPV18b site (Fig. S1X, middle). The E6E7 RNA from HPV18a was produced using a host 3ʹ splice site in the rearranged host DNA remanent; this sequence is not present downstream of HPV18b. Although HPV18b had the same viral early promoter, it did not give rise to any stable/detectable transcripts, presumably because there was no accessible downstream PAS. The E6E7 RNA transcribed from the integrated HPV18a was alternatively spliced, resulting in the production of three detectable virus-host chimeric transcripts (Fig. S1X, bottom). Transcript-2 was the major E6*I isoform used for the translation of the viral E7 protein (6, 8).
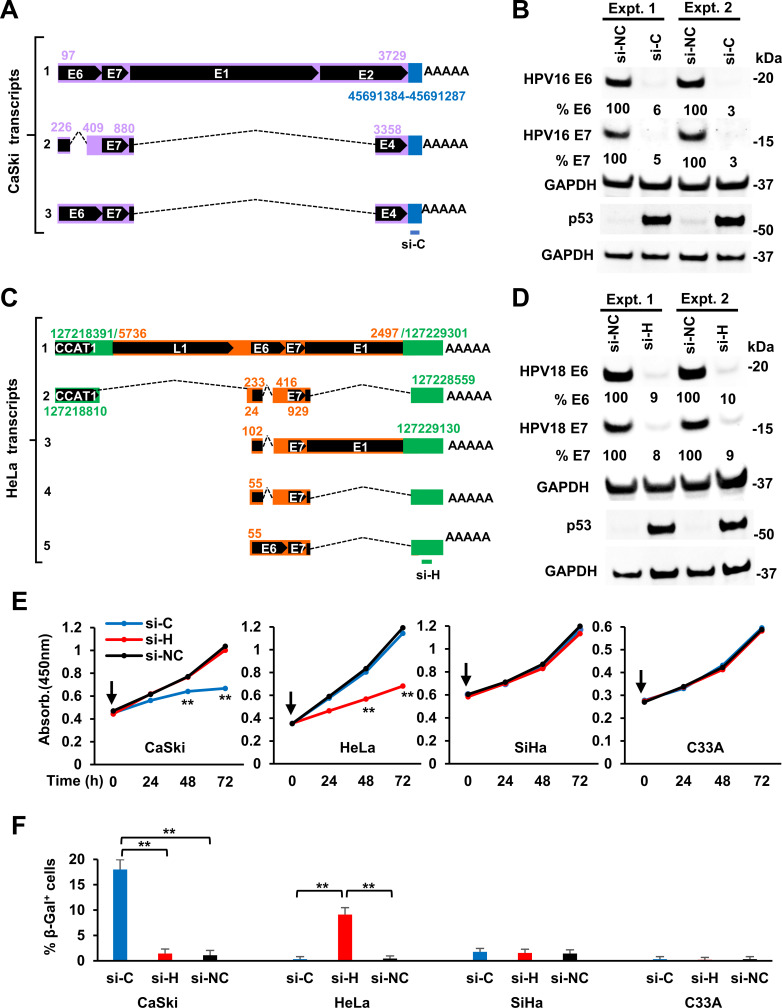
In CaSki cells, E6E7 RNA is primarily expressed from a single copy of integrated HPV16 DNA on chromosome 6
To ask whether the expression of HPV oncogenes from a single viral DNA integration site is responsible for clonal growth of the cells in cervical cancer (33, 34), we characterized the transcriptionally active HPV16 DNA integrated in the cervical cancer-derived cell lines, CaSki and SiHa and HPV18 DNA in HeLa cells where HPV DNA integrations have been extensively studied (18, 19, 21, 35–37).
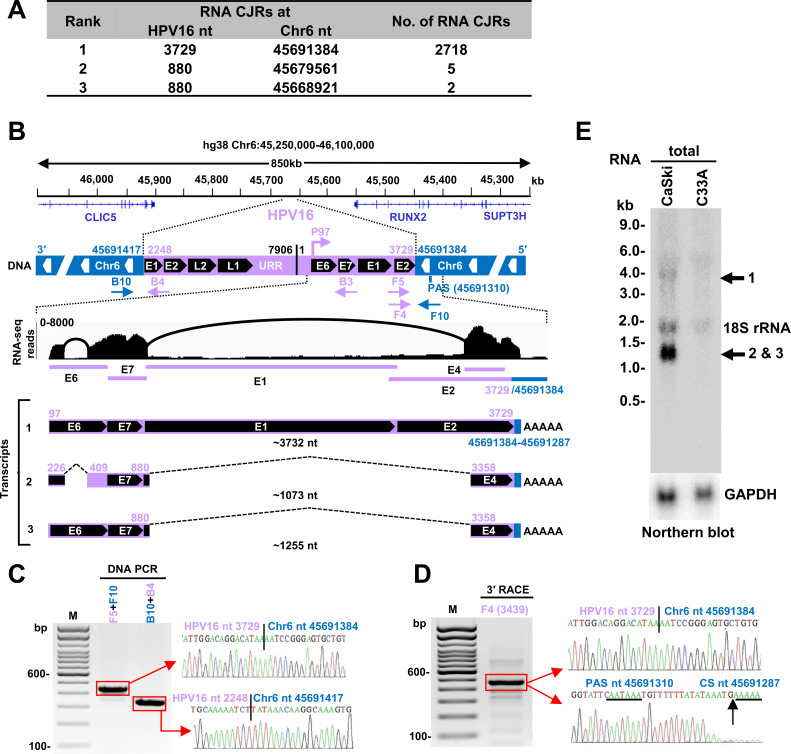
CaSki cells have been reported to contain ~600 copies of HPV16 DNA per cell (19, 21, 36, 38). However, whole-genome sequencing (WGS) (39) identified 44 different HPV16 integration sites on 12 different chromosomes (Chr2, 3, 6, 7, 10, 11, 14, 15, 19, 20, 21, and X) (Fig. 1B). The differences in the number of HPV DNA copies reported in different studies may result from instability and evolution of CaSki cell lines used in different laboratories, qPCR assays, sequencing platforms, and coverage depths (30, 39, 40). We found that two of the host-virus RNA junctions (Fig. 3A) were in an intergenic region between the CLIC5 and RUNX2 genes (Fig. 3B) on Chr6, as reported (30, 39). There is a repeat region on Chr6 from nt 45,691,285 to 45,691,466 that appears to be unstable, perhaps as a result of HPV infection (41). We found an integrated HPV16 DNA with 1,482 bp duplication in the E1 to E2 regions that has one end, nt 2,248 in E1, joined to Chr6 nt 45,691,417. The other end of HPV16, nt 3,729 in E2, was joined to Chr6 nt 45,691,384, leading to a 34 bp deletion of host DNA (Fig. 3B). We further confirmed both junctions by DNA PCR and Sanger sequencing (Fig. 3C).
Fig 3.
Transcription and RNA splicing from a single HPV16 DNA integration site identified in CaSki cells. (A) Virus-host RNA junctions with ≥2 CJRs that mapped to Chr6. (B) The integrated HPV16 DNA on Chr6 in CaSki cells and its expression. The integrated HPV 16 DNA between Chr6 nt 45,691,417 and nt 45,691,384 has a redundant E1 and E2 region on each end with the indicated upstream regulatory region (URR) and tail-to-head junction (nt 7,906/1). The fusion RNA is expressed from the viral early promoter P97 and polyadenylated by using a host PAS on Chr6 nt 45,691,310. Arrows below the genome region are the primers used to validate the expression of the virus-host fusion RNA. RNA-seq reads-coverage (0–8,000 scale) in the integrated virus-host genome region and Sashimi plots for splice junctions were visualized by IGV (middle). Three RNA isoforms, including the pre-mRNA (transcript-1) and the spliced mRNA transcript-2 and -3, are shown (bottom). The number in nt below each isoform RNA indicates the estimated length of the RNA without a pA tail. (C) Validation of the virus-host DNA junctions at the virus integration site using the primer sets F5 + F10 and B10 + B4. The PCR products in the red rectangles were gel purified and sequenced, with the sequencing chromatograms on the right. (D) The cleavage and polyadenylation sites in the virus-host fusion transcripts were mapped by 3′ RACE using an HPV16 primer F4. The sequencing chromatograms on the right show the virus-host junction, PAS (underlined) site, and cleavage site (CS, vertical arrow). (E) Northern blot analysis of total CaSki RNA (10 µg) using an antisense oligo probe B3 (B) derived from HPV16 nt 855–836. Total RNA from HPV-negative C33A cells served as a negative control. GAPDH served as an RNA loading control. Major isoforms of the virus-host transcripts detected are shown on the right.
Mapping the total RNA-seq reads showed the integrated HPV16 DNA expressed viral E6E7 RNA using the viral early promoter P97 and a host PAS at Chr6 nt 45,691,310, which is 74 nt downstream of the integration junction (Fig. 3B) to produce three isoforms of spliced E6E7 RNA. We used 3ʹ RACE to confirm the RNA cleavage site at Chr6 nt 45,691,287, 17 nt downstream of the mapped host PAS, as the RNA polyadenylation of all three isoforms of the E6E7 transcripts in CaSki cells (Fig. 3D). These three transcripts were verified by RT-PCR (data not shown) and by Northern blot analyses (Fig. 3E) with transcript-2 (E6*I) being the most abundant, as has been reported (6, 42).
E6E7 RNA in SiHa cells is expressed from chromosome 13 using a host 3′ splice site ~56 kb downstream of the integrated viral DNA
SiHa cells have been reported to have one or two copies of integrated HPV16 DNA per cell (18, 19, 36). Two CJRs of one integrated HPV16 DNA copy were found by WGS (39) in an intergenic region of Chr13 between the KLF5 and LINC00392 genes. Both junctions were verified by PCR and sequencing (Fig. S2A). The truncated HPV16 DNA (7,655 bp) which had a 252 bp deletion in E2 ORF has one end in E2, at nt 3,385, linked to Chr13 nt 73,214,733 and the other end links E2 at nt 3,133 to Chr13 nt 73,513,421 (Fig. S2A and F), leading to a 299 kb deletion of the rearranged host DNA.
Mapping the total RNA-seq reads (Fig. S2B through D) showed that the integrated HPV16 DNA expressed the E6E7 RNA using the viral early promoter P97 and a host 3ʹ splice site at Chr13 nt 73,456,962 (Fig. S2E) to reach a host PAS at Chr13 nt 73,452,999, ~60 kb downstream of the integration site (Fig. S2F). Through alternative RNA splicing, the integrated HPV16 DNA produces three spliced isoforms of viral E6E7 RNA (Fig. S2F, bottom) detectable both by RNA-seq and by Northern blot analyses of poly-A+ SiHa RNA (Fig. S2G). The transcript-2 and transcript-4 are two abundant isoforms, of which the transcript-2 is an E6*I RNA primarily used for translation of E7 (6, 31), and the transcript-4 presumably for an E6*I-host fusion protein. The less abundant transcript-3 should encode viral E6. Northern blots did not detect the less abundant unspliced precursor RNA transcript-1.
HPV18 E6E7 RNA in HeLa cells is mainly expressed from a single integrated DNA copy on chromosome 8
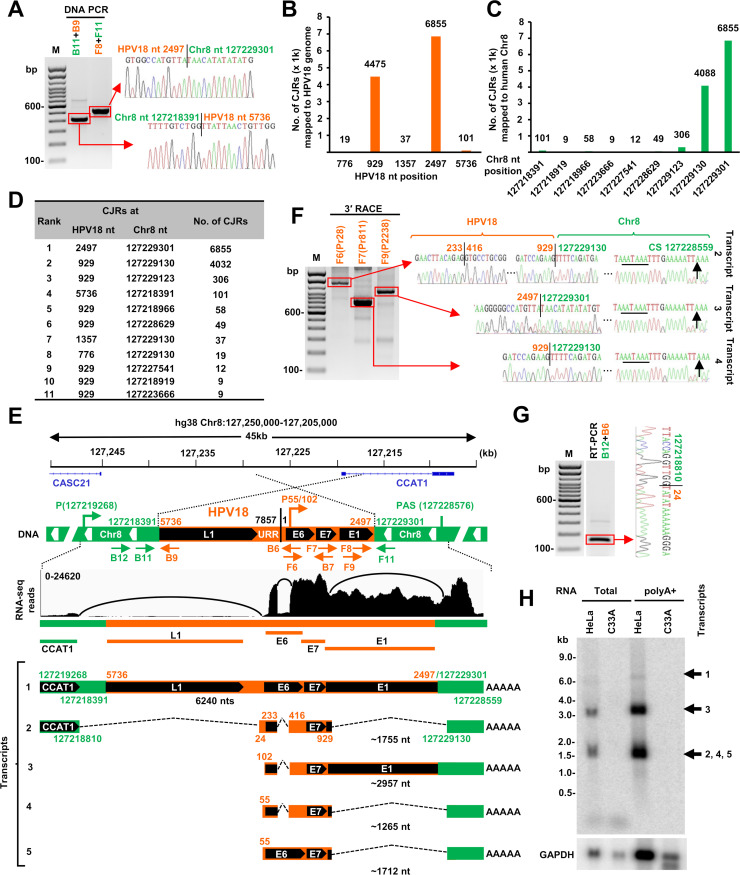
Each HeLa cell contains 62–68 chromosomes which have been reported to carry 20–50 copies of integrated HPV18 DNA (35, 43–45). Recent WGS identified multiple HPV18 DNA copies in an intergenic region on Chr8q24.21 (30, 40, 44, 45). We re-analyzed the published data (30) by searching host-virus DNA CJRs and concluded that the truncated HPV18 DNA is present in each of three copies of Chr8 (one with a Chr8p deletion) (37, 45) in HeLa cells. All three viral DNA copies are integrated in a rearranged region on Chr8 and share a common host-virus junction of which one end links HPV18 nt 5,736 in L1 to Chr8 nt 127,218,391. However, the other end of the integrated HPV18 DNA varies among the three Chr8 copies: one has HPV18 nt 2,497 in E1 linked to Chr8 nt 127,229,301, the second has HPV18 nt 25 in LCR linked to Chr8 nt 127,222,007, and the third has HPV18 nt 3,100 in E2 linked to Chr8 nt 127,221,122 (44, 45). The second copy of the integrated HPV18 DNA is not expressed because the second viral DNA copy does not have a viral early promoter P55/102 (32). The third copy does not produce stable RNA, presumably because there is no consensus downstream PAS near the integration site. Integration of the first copy of the truncated viral DNA (4,619 bp) occurred between the CASC21 gene and the lncRNA CCAT1 gene on Chr8 (46) and was accompanied by an ~11 kb deletion of host DNA and the loss of ~3.2 kb viral DNA. This is the only viral DNA copy that expresses a stable form of E6E7 RNA, which was confirmed by DNA PCR and sequencing (Fig. 4A). Intron 1 of CCAT1 was disrupted by HPV18 DNA integration, and thus, this integration prevents the expression of the CCAT1 on that copy of Chr8 due to the expression of viral E6E7 RNA.
Fig 4.
HPV18 transcription and RNA splicing from a single HPV18 DNA integration site in HeLa cells. (A) Verification of two integration junction sites in HeLa cells by PCR using two separate primer sets B11 + B9 and F8 + F11 shown in (E), with sequencing chromatograms on the gel right. (B and C) Mapping of the virus-host RNA CJRs to the HPV18 genome (B) and human Chr8 (C). (D) Distribution of top 11 virus-host CJR species identified by RNA-seq. (E) HPV18 DNA integration on Chr8 and its expression in HeLa cells. Diagrams showing the truncated HPV18 genome in size of 4,618 nt in a rearranged Chr8 region with one end of the viral DNA at nt 5,736 in L1 joined to Chr8 DNA at nt 127,218,391 and the other end of the viral DNA at nt 2,497 in E1 joined to Chr8 DNA at nt 127,229,301. Arrows below the chimeric host-virus genome show the primers used for validating the structure and expression of integrated viral DNA. RNA-seq reads-coverage (0–24,620 scale) and Sashimi plots for splice junctions in the Chr8 region with the integrated HPV18 DNA are illustrated by IGV, showing the transcription, splicing, and polyadenylation to produce five RNA isoforms, including the unspliced pre-mRNA transcript-1. The number in nt below each RNA isoform indicates the estimated length of the RNA without a pA tail. (F) HPV18 RNA splicing and polyadenylation cleavage sites (CS) were determined by 3′ RACE using HPV18-specific primers F6, F7, or F9. The RACE products were gel-purified and sequenced, with the mapped splice sites, integration junctions, and the PA cleavage site (black arrow) shown on the right chromatograms. (G) A viral-host fusion RNA splice junction was identified by RNA-seq and validated by RT-PCR using the primer set B12 + B6, with the sequencing chromatogram on the right. (H) Northern blot showing the major viral RNA isoforms transcribed from the identified integration site in HeLa cells. Total RNA (10 µg) and polyA + RNA purified from 100 µg of total RNA were analyzed by Northern blotting using an antisense HPV18 probe (B7). Corresponding RNA samples from an HPV-negative C33A cells served as negative controls. GAPDH RNA served as a sample loading control.
Mapping the total RNA-seq reads (Fig. 4B through E) showed that the truncated HPV18 DNA integrated at the Chr8 nt 127,229,301 site expresses E6E7 RNA using the viral early promoter P55/102 and a host 3ʹ splice site at Chr8 nt 127,229,130 to reach a host PAS at Chr8 nt 127,228,576, about 725 bp downstream of the HPV18 integration site. This integration site expresses five E6E7 RNA isoforms through alternative RNA splicing. Two of the RNAs are transcribed using a weak, cryptic host promoter within the CCAT1 intron upstream of the HPV18 integration site (Fig. 4E). We confirmed, by 3ʹ RACE, RT-PCR, and Sanger-sequencing, all of the RNA splice sites and the RNA cleavage site at Chr8 nt 127,228,559, 11 nt downstream of the mapped PAS, for RNA polyadenylation (Fig. 4F and G). Northern blot analysis of total HeLa cell RNA or polyA+ RNA detected transcripts of the predicted sizes (Fig. 4H). Three (transcript-2, -3, and -4) are E6* I which would be used for the translation of viral E7 (6, 8). Two (transcript-1 and -5), which are minor isoforms, would be used for the translation of E6 (Fig. 4E).
Inhibition of E6 and E7 expression using siRNAs targeting the host portion of specific virus-host fusion transcripts
Given that all of the examined cervical cancer tissues and the three established cervical cancer cell lines (CaSki, SiHa, and HeLa) express detectable E6E7-host fusion RNAs from a single chromosomal HPV integration site, we prepared siRNAs targeting the host portion of the E6E7-host fusion RNAs. If viral E6 and E7 are expressed from multiple integration sites, this target-specific siRNA approach should not block the expression of the viral oncogenes.
To confirm the expression of HPV16 E6E7 in CaSki cells from the viral DNA integrated at Chr6 nt 45,691,384 (Fig. 3A and B), we designed a CaSki-specific siRNA, si-C, to selectively target the host portion of the HPV16 E6E7-host fusion RNA (Fig. 5A). Western blot analysis confirmed that the expression of both HPV16 E6 and E7 proteins was almost completely abolished and that p53 expression was stabilized in CaSki cells treated with this target-specific siRNA compared to a non-targeting siRNA control (Fig. 5B). The si-C specific knockdown efficiency of E6 and E7 expression in CaSki cells was equal or even better than an HPV16 E7-specific siRNA and had no effect on the levels of the E6E7 fusion transcripts from HeLa or SiHa cells (Fig. S3A). The si-C-specific knockdown of viral E6 and E7 expression from the viral DNA integrated at this specific site also inhibited CaSki cell proliferation (Fig. 5E) and promoted CaSki cell senescence (16, 17) (Fig. 5F; Fig. S3B) but did not affect the growth or senescence of HPV18-positive HeLa, HPV16-positive SiHa, or the HPV-negative cervical cancer cell line C33A cells.
Fig 5.
Inhibition of viral E6 and E7 expression and cell proliferation and promotion of cell senescence by specific siRNAs targeting to the host portion of virus-host fusion RNAs from a single HPV DNA integration site in HPV16+ CaSki and HPV18+ HeLa cells. (A and C) Diagram showing the specific targeting sites of siRNAs, si-C for CaSki (A) and si-H for HeLa RNAs (C). (B and D) Specific siRNAs targeting CaSki (si-C) or HeLa (si-H) host portion of the virus-host fusion transcripts inhibit the expression of viral E6 and E7 proteins but stabilizing p53 in HPV16+ CaSki (B) and HPV18+ HeLa cells (D). Cell lysates were prepared 48 h after transfection of si-C for CaSki and si-H for HeLa cells, along with non-specific siRNA control (si-NC). The relative HPV E6/E7 and p53 protein levels in each sample were immunoblotted by corresponding antibodies. The signal intensity of each viral protein band was quantified after normalizing with GAPDH serving as the protein loading control. (E and F) The siRNAs specific to CaSki (si-C) or HeLa (si-H) portion of the host-virus fusion RNAs inhibited cell proliferation (E) but promoted cell senescence (F). HPV16+ SiHa and HPV− cervical cancer cells C33A served as cell line controls and si-NC as a siRNA control. A CCK-8 cell proliferation assay was applied to examine the cell proliferation at the indicated time (H) after cell transfection once (time zero, vertical arrow) with 40 nM of individual siRNAs. The viable cell numbers (mean ± SE from three experimental repeats) in each group were measured by absorbance at 450 nm (E). Cell senescence was examined by β-Gal staining (F) at day 8 for the indicated cells transfected twice with 40 nM of siRNA in a 72-h interval after first siRNA transfection. The senescent cells with perinuclear blue staining beyond the background level from each experimental group were counted from ~100 cells from six random fields and averaged (mean ± SE) from three independent experiments (F). **P < 0.01 by Student’s t-test.
To test the reproducibility of the results we obtained with CaSki cells, we designed a HeLa-specific siRNA, si-H, which targets the host RNA portion of the HPV18 E6E7-host fusion RNA transcribed from the viral DNA integrated in Chr8 at nt 127,229,301 (Fig. 4E and 5C). Western blot analysis confirmed that the expression, in HeLa cells, of both the E6 and E7 proteins was almost completely abolished by the specific si-H siRNA, and that p53 was stabilized, when compared to a non-targeting siRNA control (Fig. 5D). This si-H specific knockdown of viral E6 and E7 expression and the stabilization of p53 protein only in HeLa, but not in CaSki and SiHa cells (Fig. S3A), led to a specific inhibition of cell proliferation and promotion of cell senescence (16, 17, 47) of HeLa, but not CaSki, SiHa, or C33A cells (Fig. 5E and F; Fig. S3B).
Effects of HPV DNA integration on host gene expression
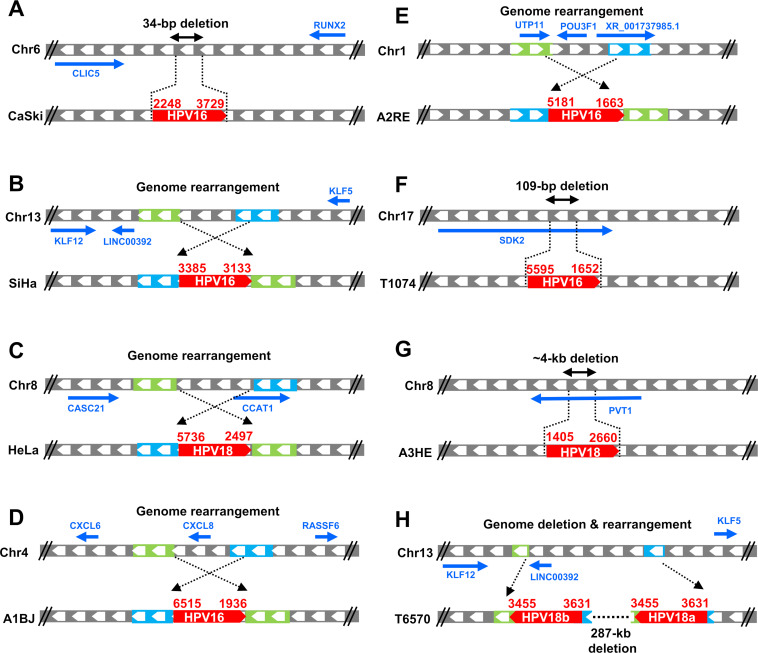
It appears that the all-integrated HPV DNAs (including the expressible and non-expressible) we identified from different chromosomes were linearized randomly and, in some cases, deleted (Fig. 6; Fig. S4A through G). Some, but not all, HPV DNA integration involves microhomology between the viral and host DNA sequences, as has been reported (30, 48). We found 2–6-bp microhomologies at some of the virus-host DNA junctions in cervical cancer tissues and derived-cell lines, SiHa and HeLa cells (Tables S1 and S2; Fig. S5). Most of the integrated viral DNAs in both cervical cancer tissues and cell lines were transcriptionally inactive or did not produce stable RNAs because the integrated HPV DNAs either lack a promoter region and/or an accessible downstream PAS (Fig. S4H through J). Distribution analyses of the virus-host DNA junctions from 525 expressible and non-expressible integrated HPV16 DNAs on multiple chromosomal sites in 17 cervical cancer tissues showed that there were break-points at many places in the viral genome (Fig. S4K; Table S3). However, the integrated HPV DNA fragments that were responsible for E6E7 expression in cervical cancer tissues or cell lines often had breaks in the viral genome in the E1 or E2 region as previously reported (30, 39, 49–52). Similarly, the integration sites in the host genome appeared to be widely distributed (Fig. S4K). However, we did find HPV DNA in the PVT1 gene on Chr8 in two tissues A3HE (HPV18, Fig. 2H) and A1M9 (HPV16, Fig. S1N) and ~1 Mb downstream of the PVT1 gene in HeLa (HPV18) cells (Fig. 4E). There are HPV DNA integrations in the region between KLF5 and KLF12 on Chr13 in SiHa cells (HPV16) (Fig. 6B; Fig. S2F) and in the tissues T6050 (HPV16) and T6570 (HPV18) (Fig. S1T and X).
Fig 6.
Simplified structures of HPV16 and HPV18 integration sites with viral E6 and E7 expression in three cervical cancer cell lines (A–C) and five cervical cancer tissues (D–H). The integrated viral DNA and its surrounding host genome segments and genes are indicated with arrows for transcription directions of host genes. Diagrams also show the viral DNA integration-induced host genome deletion and rearrangement.
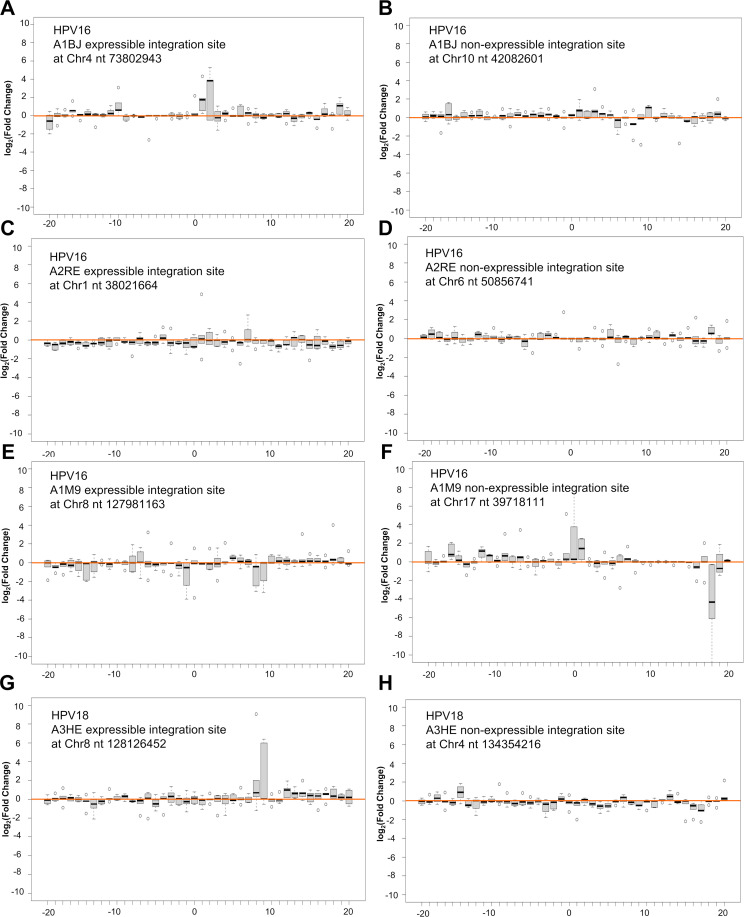
The integrations of viral DNA that led to the expression of virus-host fusion transcripts in both the cervical cancer tissues and three cell lines we examined were often associated with a deletion and/or rearrangement of the host DNA at the integration site (Fig. 6; Fig. S4A through G) which could affect the expression of host gene(s) from that copy of the chromosome. We found, in cases in which viral DNAs are integrated in an actively transcribed gene, that the viral DNAs were usually found to be inserted in an antisense orientation relative to the transcription of a host gene (Fig. 6; Fig. S4A through G). There is a recent report showing that integrated viral DNA can affect the expression of nearby host genes (53). We compared the expression of 100 genes upstream and 100 genes downstream of the integrated viral DNA that may or may not lead to the expression of E6E7 in cervical cancer tissues. To do so, we compared the expression of 200 genes from HPV16-positive A1BJ, A2RE, and A1M9, and HPV18-positive A3HE to the expression of the corresponding genes from HPV16-positive A3WB, A2LV, and A3HY or HPV18-positive A2RM, A1BF, and A2PK which contained only a single integration site on a different chromosome (Fig. 7; Table S1; Fig. S6A through H). Analysis of the eight integration sites (four each of E6E7 expressible and non-expressible) from cervical cancer tissues of A1BJ, A2RE, A1M9, and A3HE indicated that, in some cases, chromosomal HPV integration might have affected the expression of host genes both downstream and upstream of the integration site independent of viral E6E7 expression (Fig. 7; Fig. S6A through H).
Fig 7.
HPV DNA integration may or may not impact the expression of the nearby host genes. Host 100 genes from either side of the expressible (A, C, E, and G) and non-expressible (B, D, F, and H) HPV DNA integration sites (chromosome 0 position on x-axis) on the indicated chromosomal positions were compared for their expression of the genes in the corresponding chromosomal regions of other three separate cervical cancer tissues A3WB, A2LV, and A3HY with HPV16 (for A-F) or A2RM, A1BF, and A2PK with HPV18 (for G-H) single integration site on different chromosome regions. The non-expressible integration site with the highest HPV DNA CJRs in each sample was chosen for the paralleled analyses. Each bar in the panel’s x-axis represents the averaged expression levels of five binned genes (five genes in one bin, thus 10 = 50 and 20 = 100 genes) in each sample over the mean of all three controls. Box (interquartile range = IQR) and whisker plots illustrate the range of gene expression levels within each bin, with a black horizonal bar indicating the median value and small open circles for outliers. The fold change (FC) is log2 transformed. The solid pink line indicates the mean gene expression levels across the whole chromosomal regions. The genome range to the indicated chromosomal HPV DNA integration site for each panel comparison are: Chr4 nt 48,805,212–89,111,398 (A), Chr10 nt 13,438,484–67,918,390 (B), Chr1 nt 31,409,565–45,015,575 (C), Chr6 nt 41,789,896–85,615,234 (D), Chr8 nt 98,117,293–144,428,563 (E), Chr17 nt 34,285,668–41,966,503 (F), Chr8 nt 98,189,826–144,444,440 (G), and Chr4 nt 102,501,331–164,898,965 (H).
To confirm the results obtained from tissues, we compared the expression of 100 genes upstream and 100 genes downstream of the expressed and non-expressed viral DNA integration sites in CaSki cells to the expression of those genes in the corresponding chromosomal regions without HPV16 DNA integration in SiHa cells (Fig. S6I through N). We found differences in host RNA expression in the regions near viral DNA integration sites independent of E6E7 expression from the integrated viral DNA (Fig. S6I through K). However, when we compared the expression of 200 genes in the regions with no HPV16 DNA integration on Chr5, Chr8, and Chr22 from CaSki to the same chromosomal regions with no HPV16 DNA integration in SiHa cells, we found that there were differences in gene expression that were unrelated to HPV integration (Fig. S6L through N). We obtained similar results with CaSki cells when the expression of 200 host genes near expressed or non-expressed HPV DNA integration site were compared with a randomly selected region from a different chromosome without HPV DNA integration (Fig. S6O through W).
E6 and E7 point mutations in an expressed integrated viral DNA and production of predicted viral-host fusion proteins
By comparing the sequences of the integrated and expressed viral DNAs in cervical cancer tissues and in three cell lines CaSki, SiHa, and HeLa with the sequences of the corresponding reference HPV genomes, we identified a number of point mutations (Fig. S7 A through M) which are mostly consistent with what has been reported for the three cell lines (54). However, we did find, in CaSki cells, a new mutation, a G-to-T at nt 2,609 in the viral genome (Fig. S7A). We analyzed all 17 tissue samples with multiple HPV16 integration sites for the expressed HPV16 E6 and E7 from a single integrated HPV16 copy. Both a D32E mutation in E6 and an N29S mutation in E7 protein were found only in four Chinese samples (T1074, T6050, T6709, and T5350). A L90V mutation in E6 also seen in CaSki and SiHa cells, but no mutation in E7 protein was found in the samples A1BJ, A0VL, A3QD, and T3023. The other nine samples had no missense mutations in either E6 or E7.
Several of the virus-host fusion RNA transcripts identified in cervical cancer cell lines and tissues might encode virus-host fusion proteins. We identified four different virus-host fusion RNAs that are predicted to encode four putative virus-host fusion proteins (≥50 aa residues) in SiHa and HeLa cells (Fig. S7N through Q). We did not identify any RNA that might encode a virus-host fusion protein in CaSki cells. SiHa transcript-3 (Fig. S2F) encoding E6 protein might also express a E1^host fusion protein (54 aa residues, Fig. S7N). SiHa transcript-4 (Fig. S2F) is predicted to encode an E6*^host fusion protein (52 aa) of which 11 aa residues are from the host portion (Fig. S7O). HeLa transcript-1 from a weak host promoter would not express a truncated L1 protein because of an L1 frameshift from the fusion junction but could be spliced from Chr8 nt 127,218,810 to HPV18 nt 24 (Fig. 4E and G) to produce a spliced transcript-2 that is predicted to encode a putative protein of 64 aa residues (Fig. S7P). HeLa cells might also produce a truncated E1 protein (528 aa) from transcript-3 (Fig. 4E) using a host stop codon right at the integration junction (Fig. S7Q). However, transcript-3 in HeLa cells is an E6*I mRNA which functions primarily to produce E7 (6, 8). In theory, the truncated E1 should not be efficiently translated because its start codon is in a short distance (6 nt) from an E7 stop codon, which would be expected to prevent scanning ribosomes from re-initiating translation (6, 55, 56).
The transcript-4 in the tissue samples A1BJ (Fig. 2D) and A2RE (Fig. S1K) is predicted to encode potential E6*-host fusion proteins which would be 49-aa residues long in A1BJ and 105-aa residues in A2RE (Fig. S7R and S). RNAs (transcript-1 and -5) in tissue T1074 are predicted to encode two putative virus-host fusion proteins (Fig. S1Q): transcript-1 which encodes a 2560-aa SDK2-L1 fusion protein is derived from a weak host promoter (Fig. S7T) and transcript-5, which encodes an E6*-SDK2 fusion protein of 158-aa residues, is produced from the HPV16 early promoter P97 (Fig. S7U). In addition, transcript-4 in the tissue T5350 (Fig. S1S) is predicted to encode an E6*-host fusion protein of 105-aa residues (Fig. S7V). None of the viral transcripts detected in the other three tumor tissues, T6709, T6050, and T6570, are predicted to encode a virus-host fusion protein.
We investigated the function of the 52-aa E6 fusion protein 2 encoded by SiHa transcript-4 (Fig. S7O). The N-terminal 41-aa residues of the fusion are derived from HPV16 E6*I protein (31). Knockdown of transcript-4 expression in SiHa cells using a siRNA specifically targeting the splice junction of transcript-4 (Fig. S2H and I) led to decrease in the level of p53 protein by Western blot analysis (Fig. S2J) and to an increase in cell proliferation (Fig. S2K). This makes sense because viral E6*I modulates the E6-directed degradation of p53 and suppresses the growth of transformed cells (57, 58). Consistent with these results, cell cycle analysis (by flow cytometry) showed that siRNA knockdown of transcript-4 expression promoted cell entry into S phase, which was accompanied by a reduction in the fraction of the cells in G1 (Fig. S2L and M). The data suggest that, at least in some cases, the host-virus fusion proteins can be biologically active.
DISCUSSION
HPV integration can result in the expression of the oncogenes E6 and E7, which are critical for HPV carcinogenesis. Initial research predominantly focused on detecting integrated HPV DNA, pinpointing HPV integration sites, and estimating viral DNA copy numbers (22, 23, 59–62). Advanced techniques later facilitated the mapping of HPV oncogene transcripts from integration sites in cervical cancer cell lines and tissue samples using specific viral primers (62, 63). The introduction of whole-genome sequencing addressed earlier challenges in mapping the sites of HPV DNA integration (30, 39, 44). Nonetheless, questions remain. How is the integrated HPV DNA expressed, particularly when integration disrupts the viral E1/E2 regions in a way that prevents the bicistronic E6E7 RNA from using their normal viral early PAS? Does the E2/E6 DNA ratio, as determined by qPCR, truly reflect the integrated and episomal DNA ratio in clinical samples? Can viral E6 and E7 oncogenes be expressed from many integrated viral DNA copies? Is there any expression of virus-host fusion proteins from integrated HPV DNA? If the answer is yes, do any of the virus-host fusion proteins affect the growth and behavior of the cells?
In this report, we provide compelling evidence that in both cervical cancer tissues and cancer-derived cell lines, detectable levels of viral E6E7 RNA were derived from a single integrated copy of HPV DNA which had access to a downstream cellular PAS. This holds true for both cervical cancer tissues and cell lines that bear multiple HPV DNA copies integrated on several chromosomes. Our findings are consistent with an early report that, in CaSki and SiHa cells and 13 HPV cancers, only one HPV DNA integration site is transcriptionally active (24). Experiments using the inhibitor 5-azacytidine (24) in growing CaSki cells led to activate transcription from silent viral DNA centers, suggesting that methylation affected transcription of the integrated viral DNA. CpG methylation of HPV DNA was found to be important during carcinogenesis (64–66). We also showed that the absence of a viral early promoter and/or a functional viral/host PAS downstream from integrated HPV DNA results in transcriptionally dormant HPV DNA and/or in a failure to produce stable RNA (Fig. S4H through J). Consequently, for the expression of virus-host fusion transcripts and viral oncogenes, the integrated HPV DNA must have a viral early promoter and also must reside in a region of the host genome (which may have been rearranged during integration) that supplies the essential elements needed for the proper processing and polyadenylation of functional viral E6E7 RNA. Importantly, silencing the expression of individual virus-host fusion transcripts by siRNA targeting the host component of the fusion RNA resulted in a reduction of viral E6E7 expression, which inhibited the growth of the cancer cells and promoted cell senescence. These data further strengthen the idea that viral E6E7 expression from a single integrated HPV DNA copy drives proliferation and clonal expansion of the cancer cells and also show that development, in the future, of a precision medicine to treat individual cervical cancer cases is possible in addition to targeting the expression of viral E6 or E7.
Papillomavirus infection promotes host DNA repair activities and induces host genome instability due to the expression of viral E7 gene (48, 67–71). Unlike retrovirus integration, which is an obligatory step in viral replication (72–74) that is catalyzed by a viral integrase and is site-specific with respect to the viral genome, the integration of HPV DNA into host chromosomes is accidental and is a dead-end for the life cycle of the virus. We observed that almost all of the integrated viral DNA copies are truncated, perhaps because of host exonuclease digestion. When a circular HPV DNA is linearized in the infected cells, it immediately exposes both the 5′ and 3′ DNA ends to various host exonucleases for digestion and DNA repair. Thus, to be integrated, a piece of the linearized viral DNA must survive exonuclease digestion. Not surprisingly, the majority of the integrated viral DNA copies are truncated and either transcriptionally inactive or fail to produce stable RNA transcripts. Many of the integrated viral DNAs either lack a viral early promoter region and/or miss an accessible viral or host PAS downstream (Fig. S4H through J).
The distribution of integration sites that is seen in tumors and in tumor-derived cell lines is the result of both the initial distribution of the integration sites and any subsequent post-integration selections. In this study, the majority of the integrated HPV DNA copies that express E6E7 were often broken in the E1 or E2 region, which is consistent with previous reports (30, 39, 49–52). We think that it was likely due to a selection for the cells that express E6 and E7 because there is no obvious reason for the break points in the circular HPV genome to occur in specific places. This supposition is supported by an analysis of the break points in the integrated viral DNAs that do not express detectable transcripts. The breakpoints in the viral genome occur at many sites (Fig. S4K). Expression of E6E7 promotes growth of the cancer cells; blocking the expression of the E2 protein would allow the activation of the viral early promoter which is needed for the expression of E6E7 (15–17). As might be expected for a non-specific integration event, the integration sites in the host genome are widely distributed (Fig. 6; Fig. S4). HPV integration has been reported to prefer the host DNA fragile sites, intergenic regions, and transcriptionally active sites (23, 75–77). However, it is important to remember that these data come from the cells in which in vivo post-integration selection has had an opportunity to act. Viral DNA integration in the PVT1 region on Chr8 in the tissues A3HE (HPV18) and A1M9 (HPV16) and in HeLa cells and in the Chr3 region between KLF5 and KLF12 in the tissues T6050 (HPV16) and T6570 (HPV18) and in SiHa cells could be the result of preferential integration in fragile regions for HPV integration, as has suggested (77–82), but could also be either coincidental or the result of an as yet undefined post-integration selection.
In our study, most HPV integrations were located within host intergenic or intron regions, which is similar to what we observed for MmuPV1 integration (67). When viral DNA is integrated into a transcriptionally active site, it is usually found in an antisense orientation relative to the transcription of an adjacent host gene. As such orientation, the integrated viral DNA is less likely to interfere with the expression of the host gene. However, as noted earlier, it is likely that this is the result of post- integration selection, not a preference during integration, as has been suggested for HIV proviruses, where there is a strong selection against proviruses that are integrated in the introns of highly expressed genes in the same orientation as the gene (83). Integrated HPV DNA was found to be able to activate a host cryptic CCAT1 promoter in HeLa cells and a cryptic SDK2 promoter in the tissue T1074, neither of which are normally used. In both cases, the promoter activities were weak when compared with the viral early promoter. We did not find evidence that HPV integration broadly affects transcriptionally active promoters as has been proposed (23, 75, 76).
HPV integration can have significant effects on the host DNA, inducing deletions, duplications, and other rearrangements (75, 80, 84). We found that viral integration most commonly causes deletions in the host genome, which ranged in size from 22 bp in the tissue A1M9 to a 307 kb in the tissue T6570. Thus, the impact of chromosomal HPV DNA integration on the expression of nearby host genes depends on the specific characteristics of each integration event, as illustrated in Fig. 7 and Fig. S6. While previous research has indicated that HPV18 integration at Chr8q24 can influence the expression of host gene MYC (23, 85, 86), we did not find any HPV integrations that appeared to affect MYC gene expression in the cervical cancer tissues we analyzed. HPV integration has been shown to cause genome amplification/copy number alterations at the integration site in various cancers, including cervical precancer and invasive cancer (29, 87, 88), anal squamous cell carcinoma (89), head and neck cancer (84), and oropharyngeal cancers (90). In the cervical cancer tissue A3NI, we found that HPV integration induced host genome amplification of a 33.2 kb region that includes both the MAG and CD22 genes on Chr9.
While the exact mechanism underlying HPV integration-induced host genome amplification remains unclear, some aspects of HPV integration-induced duplications and deletions of viral or host genomic DNA can be partially explained. For instance, in CaSki cells, the 5ʹ end of the integrated HPV16 DNA contains a redundant 1,482 bp portion of E1-E2 region. Also, in A3HE, the 3ʹ end of the integrated HPV18 DNA bears a redundant 1,255 bp E1 region. We believe these duplications arose from two separate nicks on the two viral DNA strands during integration: one at or near HPV16 nt 2,248 on one strand and another at or near HPV16 nt 3,729 on the other strand. If each 3ʹ end strand of the viral DNA served as a primer for host DNA polymerases using distinct sequences on either strand of the host DNA in Chr6 as templates in the precursor to CaSki cells, the priming would have likely occurred at nt 45,691,417 of Chr6 for the E1 end and at the nt 45,691,384 for the E2 end. DNA repair would then remove host sequences and convert the single-stranded viral DNA into integrated dsDNA. A similar series of events could also explain the duplicated HPV18 DNA integrated in tissue A3HE. Because the viral DNA ends are used as primers, insertion often occurs where there is a short matching sequence of the host genome and the viral and host DNAs are joined at regions of microhomology (67, 91, 92). The identification of microhomology at some of the virus-host junctions (Fig. S5), although not all, supports the idea that integration occurred when viral DNA was used to prime DNA synthesis on the host genome (67, 91, 93–96).
HPV transcription from integrated viral DNA fragments is indicative of progression from low-grade cervical lesions to invasive cervical cancers. This has been monitored as a decrease in episome-derived transcripts in strong association with an increase of the integrated DNA-derived transcripts in a 3′ RACE-based APOT assay of the punch-biopsy samples, in parallel with cervical lesion progression (22, 28). Two cervical cell lines (HPV16+ W12 and HPV31+ CIN612) derived from a low-grade cervical lesion have been reported to contain heterogeneous populations of cells containing both episomal and integrated HPV16 or HPV31 DNA (97, 98) that could be separated into populations that contain either episomal or integrated forms of viral DNA (99, 100). However, recent reports have suggested that, in a small portion of cervical cancer cases, the growth of cancer cells could be induced by E6E7 expression purely from episomal HPV16 DNA. In these cases, the reported ratio of E2/E6 DNA (measured by qPCR) was approximately one (E2 was presumably not disrupted by integration) in the cervical cancer tissues (25–27). Although we think that these data are relevant, we are not persuaded that the PCR methods were sufficiently accurate to show that the ratio of E2/E6 DNA is exactly one. For example, there are issues pertaining to whether two primer sets will anneal with equal efficiency in the PCR detection. In addition (1), the integrated HPV DNA is inserted in multiple different chromosomal sites which have different GC to AT ratios (22/33 cases in this report). Some of the integrated viral DNA copies do have an intact E2 region but not express E6E7 (2); most of the integrated viral DNA copies that have a viral early promoter and express E6E7 have a disrupted E2, but there are the viral DNA copies expressing E6E7 that also have an intact E2 region. In the A3NI cancer tissue described in this report, we found that there were six chromosomal HPV integration sites, but we were not able to detect any virus-host fusion RNA (Table S1). The integrated DNA in the A3NI tissue had an intact E2 region and could express E6E7 using a viral early pA signal (Fig. S1A) (3); punch-biopsy tissues could also have been contaminated with low-grade lesions which contained high levels of replicating episomal HPV DNA.
To enable the growth and survival of a tumor cell, at least one integrated HPV DNA must express viral E6 and E7; these are critical for clonal cell selection and expansion. In some cases, novel virus-host fusion transcripts can arise, some of which encode fusion proteins, which might be able to enhance the oncogenic potential of the infected cells. Based on our examination of both cell lines and cervical cancer tissues, the expression of E6 and E7 from an integrated HPV DNA depends on the presence of both an intact viral early promoter and the appropriate surrounding host sequences. In most cases, the synthesis of E6E7 RNA was driven by a collaboration between a viral promoter and a host PAS. The use of a distant host PAS was often facilitated by the use of splice sites in the host DNA. Taken together, our results underscore the nuanced processes by which HPV integration leads to the production of viral-host fusion transcripts to express the viral oncogenes that play a pivotal role in the onset of HPV-associated cancers.
MATERIALS AND METHODS
Collection of cervical cancer tissues for HPV integration bioinformatics analysis
To analyze HPV16/18 integration and the expression of viral oncogenes from cervical cancer tissues, we first searched TCGA-cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) database and selected 67 cervical cancer cases with paired whole-genome sequencing (WGS, with sequencing reads-depth ≥200 million) and RNA-seq data (with sequencing reads-depth ≥100 million) from a total of 307 cases (29). We also identified two separate NCBI SRA data sets that have paired DNA-seq data (SRA189003) and the corresponding RNA-seq data (SRA189004) from seven Chinese cervical cancer tissues with high-quality in-depth reads (≥50 million in both DNA-seq and RNA-seq with respective 100 bp and 90 bp length pair-end reads per sample), of which six were infected with HPV16 and one with HPV18 and all displayed multiple copies of integrated HPV DNA (30). We analyzed both DNA-seq and RNA-seq reads against the combined chimeric human genome (GRCh38, hg38)/HPV18(gi|60975)/HPV16R (gi|333031) (https://pave.niaid.nih.gov), except G substitution of A at HPV16 nt 2,926 position, and focused on the CJRs because they reflect the virus integration site. The same analysis was conducted for the published genome sequences of CaSki (30, 39, 40), SiHa (39), and HeLa (30, 40, 44, 45) cells. CJR is a read where one part of the read aligned with the HPV16 or HPV18 genome and the other part aligned with the human genome at the integration junction (Fig. 1A). We only used CJR reads in which at least 20 nucleotides on each side of the junction were fully aligned with the virus and human genomes.
Cell lines and siRNAs
HPV16-positive cervical cancer cell lines, CaSki and SiHa cells, and HPV18-positive cervical cancer cell line, HeLa cells, were obtained from ATCC (Manassas, VA). HPV-negative cervical cancer cell line C33A cells (ATCC) with mutant p53 and mutant pRB were used as an HPV-negative cell control. All cell lines were maintained in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Waltham, MA) with 10% fetal bovine serum (GE Healthcare, Logan, UT) at 37°C under a 5% CO2 atmosphere.
To target the viral-host fusion RNA transcripts, a custom-designed synthetic siRNA specifically targeting the host portion of the identified chimeric virus-host RNAs in CaSki cells (si-C) or HeLa cells (si-H) or the virus-host junction of the fusion transcript-4 (si-Junction) in SiHa cells (Table S4) was purchased from Dharmacon (Lafayette, CO). Non-targeting control siRNA (Dharmacon, # D-001210-01) served as a negative control. SiHa/CaSki/HeLa cells at 24 h of cell passage were transfected with 40 nM of siRNA by LipoJet In Vitro Transfection Kit (Ver. II) (SignaGen Laboratories, Frederick, MD). Total protein and RNA extracts from one part of the transfected cells were prepared 48 h after the transfection and the other part of the transfected cells were used for cell proliferation and senescence analyses. The cells used for senescence assay by β-Gal staining were transfected twice by the indicated cell-specific siRNA at a 72 h interval.
Total RNA sequencing of cervical cancer cell lines and bioinformatics analysis
The total RNA extracted by TriPure reagent (Roche, Germany) was used for the construction of RNA-seq libraries followed by Illumina sequencing. Sequencing libraries were constructed following Illumina Stranded Total RNA protocol (Illumina, RS-122-2201). Paired-end 150 bp read length sequencing with a depth of 100 million reads per sample was performed on the NovaSeq platform according to the manufacturer’s instructions (Illumina) for CaSki cells. For SiHa cells, the length of the paired-end read was 75 bp with a depth of 75 million reads per sample. HeLa cells RNA-seq was performed on the NovaSeq platform with pair end reads 150-bp at a depth of 100 million reads. Adapters and low-quality bases were trimmed by CutAdapt v1.18 with the following parameters: cutadapt --pair-filter=any --nextseq-trim=2 --trim-n -n 5 -O 5 -q 10.10 m 35:35. Reference sequence of HPV16R used in this study was identical to HPV16 REF.1 (gi|333031) (https://pave.niaid.nih.gov), except G substitution of A at nt 2,926 position. HPV18REF.1 (gi|60975) (https://pave.niaid.nih.gov) and human genome (GRCh38, hg38) were also used to create chimeric HPV + hg38 genomes for reads mapping. Reads were aligned using STAR v 2.5.2b aligner with default setting (101). The paired reads fully aligned to the human or HPV16/18 genomes were removed, and the remaining “chimeric read pairs” were used in the follow-up analysis. Chimeric junction reads were the junction reads aligned to both HPV and human genomes with a threshold of minimal overhang ≥20 nts from the junction to each genome, with a corresponding mate fully aligned to HPV or human genome. STAR-generated “Chimeric.out.junction” files were used to calculate the number of chimeric reads. The uniquely mapped reads were then uploaded into the Integrative Genomic Viewer (IGV) program (https://software.broadinstitute.org/software/igv/) to visualize reads-coverage profile along with individual genomes. A Sashimi plot for splice junction visualization was generated by IGV. Raw data and analyzed RNA-seq data supporting the findings in this study have been deposited in the NCBI GEO database under accession numbers GSE158033 for CaSki, GSE174317 for SiHa, and GSE165475 for HeLa cells.
PCR, reverse transcription-PCR (RT-PCR), and quantitative RT-PCR (RT-qPCR)
To validate DNA break sites, total genomic DNA extracted by QIAamp DNA blood mini kit (Qiagen, Gaithersburg, MD) was used in PCR with the following primers: oLLY492/oLLY485 and oLLY489/oLLY495 for CaSki cell DNA, oLLY407/oLLY397 and oLLY402/oLLY409 for SiHa cell DNA, oLLY469/oXHW48 and oLLY474/oLLY490 for HeLa cell DNA (Table S4). For RT-PCR, total RNA was treated with Turbo DNase (Thermo Fisher Scientific), converted to cDNA by RT with random hexamers and Moloney murine leukemia virus (MuLV) reverse transcriptase (Thermo Fisher Scientific) and used in PCR with the following primers: oZMZ215/oLLY399 and oLLY405/oLLY399 for SiHa cell cDNA and oLLY473/oLLY480 for HeLa cell cDNA (Table S4).
RT-qPCR was carried out with total RNA isolated from the indicated cells using a TaqMan Gene Expression kit (Thermo Fisher Scientific, #4369016). Custom-designed primers and TaqMan probe to specifically detect the viral-host fusion transcripts are listed in Table S4. GAPDH TaqMan probe set (Thermo Fisher Scientific, assay #Hs02786624_g1) was used as an internal control.
5ʹ and 3ʹ rapid amplification of cDNA ends (RACE)
The 5′ and 3ʹ RACE assays were conducted with a Smart RACE cDNA amplification kit (Takara Bio, Ann Arbor, MI) as recommended with 1 µg/reaction of total RNA as a template. The following primers (Table S4) were used: HPV16-specific primer oZMZ260 was used for CaSki cell RNA and HPV18-specific primers oLLY478, oMA119, and oLLY475 were used for HeLa cell RNA.
Northern blotting
Northern blotting was performed as previously described (102). Briefly, 10 µg of total RNA or poly(A)+ mRNA selected from 50 or 100 µg of total RNA from each sample was separated in formaldehyde-containing 1% agarose gel in 1 × MOPS (morpholinepropanesulfonic acid). The separated RNAs were then capillary transferred onto a nylon membrane and cross-linked by UV light. After pre-hybridization, the membranes were probed with 32P-labeled oligos overnight at 42°C. The following probes (Table S4) were used for the hybridization: oZMZ302 for detection of HPV16 in CaSki cells; a mixture of oMA196, oZMZ302, and oZMZ220 for detection of HPV16 in SiHa cells; oXHW86 for HPV18 in HeLa cells and oZMZ270 for human GAPDH in all cell lines. The specific signal was captured by Typhoon biomolecular imager (GE Healthcare).
Cell proliferation and cell cycle distribution assays
The Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies, Rockville, MD) was used for the cell proliferation assay according to the manufacture’s protocol. The relative cell viability was calculated based on measured 450 nm absorbance in six biological repeats. For the cell cycle distribution analysis, 1.5 million cells were fixed with 70% ethanol at 4°C overnight, washed with PBS, followed by the incubation with PBS containing 200 µg/mL RNase A (Thermo Fisher Scientific), 20 µg/mL propidium iodide (Thermo Fisher Scientific), and 0.1% Triton X-100 (Promega) at room temperature for 30 min. Cell cycle distribution was determined by flow cytometry and analyzed by FlowJo software (BD Biosciences, Franklin Lakes, NJ).
Cell senescence assay
The cell senescence assay was assessed by the detection of β-Galactosidase (β-Gal) activity in cells. Senescence β-galactosidase Staining Kit (Millipore Sigma, # QIA117) was used according to the supplier’s protocol. The cells were transfected twice with 40 nM of siRNA before β-Gal staining. The cells were plated on day 1, transfected on day 2, and passed on day 4; the second siRNA transfection was on day 5; and the β-Gal staining was performed on day 8. For quantification, at least 100 cells from six random fields were observed and counted using a standard light microscopy.
Western blot and antibodies
For CaSki and HeLa cells, 5 million cells were collected in 350 µL of 1 × RIPA buffer with the addition of cocktail of protease inhibitors for chymotrypsin, thermolysin, papain, pronase, pancreatic extract, trypsin (Roche, #04693159001). Fifteen microliters of the lysate was then mixed with 5 µL 4 × SDS-loading buffer with the addition of 2-ME, heat denatured at 95°C for 10 min, and resolved in 4%–12% Bis-Tris NuPAGE gel (Thermo Fisher Scientific) in 1 × MOPS SDS buffer (Thermo Fisher Scientific). For SiHa cells, total proteins were prepared by direct lysis of 2 million cells in 350 µL of 4 × SDS-sample buffer containing 5% 2-mercaptoethanol (2-ME), and 15 µL of the sample was separated in 4%–12% Bis-Tris NuPAGE gel. After transferred onto a nitrocellulose membrane, the membrane was blocked for 1 h with 5% skimmed milk in 1 × TBS (Tris-buffered saline) containing 0.05% Tween-20 (TTBS) and incubated with a primary antibody diluted in TTBS overnight at 4°C. After three washes with 1 × TTBS buffer, the membrane was then incubated by a corresponding secondary peroxidase-conjugated antibody diluted in 2% milk/TTBS for 1 h at room temperature. The specific signal on the membrane was generated with SuperSignal West Pico (Thermo Fisher Scientific) and captured by the ChemiDoc Touch imaging system (Bio-Rad).
Anti-p53 (Proteintech, # 10442-1-AP) and anti-β tubulin (Sigma-Aldrich, #T5201), anti-HPV16 E6 (Arbor vita, # AVC-6), anti-HPV16 E7 (Gene Tex, # GTX133411), anti-GAPDH (Cell signaling, #2118), anti-HPV18 E6 (Arbor vita, # AVC-345), anti-HPV18 E7 (Gene Tex, # GTX133412) were used for Western blot.
ACKNOWLEDGMENTS
We thank Peter Johnson of NCI and his team in helping us for cell senescence assays, Paul Liu of NHGRI for his critical reading of our manuscript, and Johannes Schweizer from Arbor Vita company for providing us the HPV16/18 E6 antibodies. We also thank Frederick CCR/CIL Flow Cytometry Core for the cytometry analysis. The results in this report were partially deposited as a preprint in bioRxiv in 27 June 2020 and presented at the 34th International Papillomavirus Conference (IPVC), 2021 and the 35th IPVC, 2023.
This work was supported by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute, and the Center for Cancer Research (ZIASC010357 to Z.M.Z.).
L.Y. and Z.M.Z. designed the study and performed all data analyses and interpretations. L.Y. performed all the validation experiments. V.M. performed data analysis and interpretations. D.L. analyzed the results for interpretations and designed the cell functional analyses. A.L. and M.C. performed the RNA-seq data analysis. S.M. and V.B. performed SiHa cell RNA sequencing. H.L. performed CaSki cell RNA sequencing. S.H. analyzed the results and designed the breakpoint analysis. L.Y. and Z.M.Z. drafted the manuscript and finalized with input from all co-authors.
AFTER EPUB
[This article was published on 16 April 2024 with errors in Fig. 2 and S1. The figures were corrected in the current version, posted on 19 April 2024.]
Footnotes
This article is a direct contribution from Stephen H. Hughes, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Daniel DiMaio, Yale School of Medicine, and Louise Chow, University of Alabama at Birmingham.
Contributor Information
Zhi-Ming Zheng, Email: zhengt@exchange.nih.gov.
Thomas Shenk, Princeton University, Princeton, New Jersey, USA.
DATA AVAILABILITY
RNA-seq data are available under NCBI GEO accession numbers GSE158033 for CaSki, GSE174317 for SiHa, and GSE165475 for HeLa cells.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.00729-24.
Figures S1-S7.
Tables S1-S4.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Dürst M, Gissmann L, Ikenberg H, zur Hausen H. 1983. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci U S A 80:3812–3815. doi: 10.1073/pnas.80.12.3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. zur Hausen H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2:342–350. doi: 10.1038/nrc798 [DOI] [PubMed] [Google Scholar]
- 3. Schiffman M, Doorbar J, Wentzensen N, de Sanjosé S, Fakhry C, Monk BJ, Stanley MA, Franceschi S. 2016. Carcinogenic human papillomavirus infection. Nat Rev Dis Primers 2:16086. doi: 10.1038/nrdp.2016.86 [DOI] [PubMed] [Google Scholar]
- 4. Zheng ZM, Baker CC. 2006. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci 11:2286–2302. doi: 10.2741/1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu L, Majerciak V, Zheng ZM. 2022. HPV16 and HPV18 genome structure, expression, and post-transcriptional regulation. IJMS 23:4943. doi: 10.3390/ijms23094943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tang S, Tao M, McCoy JP, Zheng ZM. 2006. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J Virol 80:4249–4263. doi: 10.1128/JVI.80.9.4249-4263.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ajiro M, Tang S, Doorbar J, Zheng ZM. 2016. Serine/arginine-rich splicing factor 3 and heterogeneous nuclear ribonucleoprotein A1 regulate alternative RNA splicing and gene expression of human papillomavirus 18 through two functionally distinguishable cis elements. J Virol 90:9138–9152. doi: 10.1128/JVI.00965-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brant AC, Majerciak V, Moreira MAM, Zheng ZM. 2019. HPV18 utilizes two alternative branch sites for E6*I splicing to produce E7 protein. Virol Sin 34:211–221. doi: 10.1007/s12250-019-00098-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio L, Cibulskis K, Bertelsen B, et al. 2014. Landscape of genomic alterations in cervical carcinomas. Nature 506:371–375. doi: 10.1038/nature12881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kelley DZ, Flam EL, Izumchenko E, Danilova LV, Wulf HA, Guo T, Singman DA, Afsari B, Skaist AM, Considine M, Welch JA, Stavrovskaya E, Bishop JA, Westra WH, Khan Z, Koch WM, Sidransky D, Wheelan SJ, Califano JA, Favorov AV, Fertig EJ, Gaykalova DA. 2017. Integrated analysis of whole-genome ChIP-Seq and RNA-Seq data of primary head and neck tumor samples associates HPV integration sites with open chromatin marks. Cancer Res 77:6538–6550. doi: 10.1158/0008-5472.CAN-17-0833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao G, Wang J, Kasperbauer JL, Tombers NM, Teng F, Gou H, Zhao Y, Bao Z, Smith DI. 2019. Whole genome sequencing reveals complexity in both HPV sequences present and HPV integrations in HPV-positive oropharyngeal squamous cell carcinomas. BMC Cancer 19:352. doi: 10.1186/s12885-019-5536-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dürst M, Kleinheinz A, Hotz M, Gissmann L. 1985. The physical state of human papillomavirus type 16 DNA in benign and malignant genital tumours. J Gen Virol 66 (Pt 7):1515–1522. doi: 10.1099/0022-1317-66-7-1515 [DOI] [PubMed] [Google Scholar]
- 13. Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129–1136. doi: 10.1016/0092-8674(90)90409-8 [DOI] [PubMed] [Google Scholar]
- 14. Gonzalez SL, Stremlau M, He X, Basile JR, Münger K. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol 75:7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thierry F, Yaniv M. 1987. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J 6:3391–3397. doi: 10.1002/j.1460-2075.1987.tb02662.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodwin EC, Yang E, Lee CJ, Lee HW, DiMaio D, Hwang ES. 2000. Rapid induction of senescence in human cervical carcinoma cells. Proc Natl Acad Sci U S A 97:10978–10983. doi: 10.1073/pnas.97.20.10978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wells SI, Francis DA, Karpova AY, Dowhanick JJ, Benson JD, Howley PM. 2000. Papillomavirus E2 induces senescence in HPV-positive cells via pRB- and p21(CIP)-dependent pathways. EMBO J 19:5762–5771. doi: 10.1093/emboj/19.21.5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. el Awady MK, Kaplan JB, O’Brien SJ, Burk RD. 1987. Molecular analysis of integrated human papillomavirus 16 sequences in the cervical cancer cell line SiHa. Virology 159:389–398. doi: 10.1016/0042-6822(87)90478-8 [DOI] [PubMed] [Google Scholar]
- 19. Baker CC, Phelps WC, Lindgren V, Braun MJ, Gonda MA, Howley PM. 1987. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol 61:962–971. doi: 10.1128/JVI.61.4.962-971.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wagatsuma M, Hashimoto K, Matsukura T. 1990. Analysis of integrated human papillomavirus type 16 DNA in cervical cancers: amplification of viral sequences together with cellular flanking sequences. J Virol 64:813–821. doi: 10.1128/JVI.64.2.813-821.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smits HL, Cornelissen MT, Jebbink MF, van den Tweel JG, Struyk AP, Briët M, ter Schegget J. 1991. Human papillomavirus type 16 transcripts expressed from viral-cellular junctions and full-length viral copies in CaSki cells and in a cervical carcinoma. Virology 182:870–873. doi: 10.1016/0042-6822(91)90632-l [DOI] [PubMed] [Google Scholar]
- 22. Klaes R, Woerner SM, Ridder R, Wentzensen N, Duerst M, Schneider A, Lotz B, Melsheimer P, von Knebel Doeberitz M. 1999. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res 59:6132–6136. [PubMed] [Google Scholar]
- 23. Wentzensen N, Ridder R, Klaes R, Vinokurova S, Schaefer U, Doeberitz M von K. 2002. Characterization of viral-cellular fusion transcripts in a large series of HPV16 and 18 positive anogenital lesions. Oncogene 21:419–426. doi: 10.1038/sj.onc.1205104 [DOI] [PubMed] [Google Scholar]
- 24. Van Tine BA, Kappes JC, Banerjee NS, Knops J, Lai L, Steenbergen RDM, Meijer CLJM, Snijders PJF, Chatis P, Broker TR, Moen PT Jr, Chow LT. 2004. Clonal selection for transcriptionally active viral oncogenes during progression to cancer. J Virol 78:11172–11186. doi: 10.1128/JVI.78.20.11172-11186.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hong D, Liu J, Hu Y, Lu X, Li B, Li Y, Hu D, Lu W, Xie X, Cheng X. 2017. Viral E6 is overexpressed via high viral load in invasive cervical cancer with episomal HPV16. BMC Cancer 17:136. doi: 10.1186/s12885-017-3124-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheung JLK, Cheung T-H, Yu MY, Chan PKS. 2013. Virological characteristics of cervical cancers carrying pure episomal form of HPV16 genome. Gynecol Oncol 131:374–379. doi: 10.1016/j.ygyno.2013.08.026 [DOI] [PubMed] [Google Scholar]
- 27. Boulet GAV, Benoy IH, Depuydt CE, Horvath CAJ, Aerts M, Hens N, Vereecken AJ, Bogers JJ. 2009. Human papillomavirus 16 load and E2/E6 ratio in HPV16-positive women: biomarkers for cervical intraepithelial neoplasia >or=2 in a liquid-based cytology setting? Cancer Epidemiol Biomarkers Prev 18:2992–2999. doi: 10.1158/1055-9965.EPI-09-0025 [DOI] [PubMed] [Google Scholar]
- 28. Vinokurova S, Wentzensen N, Kraus I, Klaes R, Driesch C, Melsheimer P, Kisseljov F, Dürst M, Schneider A, von Knebel Doeberitz M. 2008. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res 68:307–313. doi: 10.1158/0008-5472.CAN-07-2754 [DOI] [PubMed] [Google Scholar]
- 29. The Cancer Genome Atlas Research Network . 2017. Integrated genomic and molecular characterization of cervical cancer. Nature 543:378–384. doi: 10.1038/nature21386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu Z, Zhu D, Wang W, Li W, Jia W, Zeng X, Ding W, Yu L, Wang X, Wang L, et al. 2015. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet 47:158–163. doi: 10.1038/ng.3178 [DOI] [PubMed] [Google Scholar]
- 31. Ajiro M, Jia R, Zhang L, Liu X, Zheng ZM. 2012. Intron definition and a branch site adenosine at nt 385 control RNA splicing of HPV16 E6*I and E7 expression. PLoS One 7:e46412. doi: 10.1371/journal.pone.0046412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang X, Meyers C, Wang HK, Chow LT, Zheng ZM. 2011. Construction of a full transcription map of human papillomavirus type 18 during productive viral infection. J Virol 85:8080–8092. doi: 10.1128/JVI.00670-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Greaves M, Maley CC. 2012. Clonal evolution in cancer. Nature 481:306–313. doi: 10.1038/nature10762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gisselsson D, Egnell R. 2017. Cancer - an insurgency of clones. Trends Cancer 3:73–75. doi: 10.1016/j.trecan.2016.11.010 [DOI] [PubMed] [Google Scholar]
- 35. Schwarz E, Freese UK, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. 1985. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 314:111–114. doi: 10.1038/314111a0 [DOI] [PubMed] [Google Scholar]
- 36. Mincheva A, Gissmann L, zur Hausen H. 1987. Chromosomal integration sites of human papillomavirus DNA in three cervical cancer cell lines mapped by in situ hybridization. Med Microbiol Immunol 176:245–256. doi: 10.1007/BF00190531 [DOI] [PubMed] [Google Scholar]
- 37. Macville M, Schröck E, Padilla-Nash H, Keck C, Ghadimi BM, Zimonjic D, Popescu N, Ried T. 1999. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res 59:141–150. [PubMed] [Google Scholar]
- 38. Yee C, Krishnan-Hewlett I, Baker CC, Schlegel R, Howley PM. 1985. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am J Pathol 119:361–366. [PMC free article] [PubMed] [Google Scholar]
- 39. Akagi K, Li J, Broutian TR, Padilla-Nash H, Xiao W, Jiang B, Rocco JW, Teknos TN, Kumar B, Wangsa D, He D, Ried T, Symer DE, Gillison ML. 2014. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res 24:185–199. doi: 10.1101/gr.164806.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lagström S, Umu SU, Lepistö M, Ellonen P, Meisal R, Christiansen IK, Ambur OH, Rounge TB. 2019. TaME-seq: an efficient sequencing approach for characterisation of HPV genomic variability and chromosomal integration. Sci Rep 9:524. doi: 10.1038/s41598-018-36669-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pett MR, Alazawi WOF, Roberts I, Dowen S, Smith DI, Stanley MA, Coleman N. 2004. Acquisition of high-level chromosomal instability is associated with integration of human papillomavirus type 16 in cervical keratinocytes. Cancer Res 64:1359–1368. doi: 10.1158/0008-5472.can-03-3214 [DOI] [PubMed] [Google Scholar]
- 42. Tang S, Tao M, McCoy JP, Zheng ZM. 2006. Short-term induction and long-term suppression of HPV16 oncogene silencing by RNA interference in cervical cancer cells. Oncogene 25:2094–2104. doi: 10.1038/sj.onc.1209244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lazo PA, DiPaolo JA, Popescu NC. 1989. Amplification of the integrated viral transforming genes of human papillomavirus 18 and its 5'-flanking cellular sequence located near the myc protooncogene in HeLa cells. Cancer Res 49:4305–4310. [PubMed] [Google Scholar]
- 44. Adey A, Burton JN, Kitzman JO, Hiatt JB, Lewis AP, Martin BK, Qiu R, Lee C, Shendure J. 2013. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature 500:207–211. doi: 10.1038/nature12064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Landry JJM, Pyl PT, Rausch T, Zichner T, Tekkedil MM, Stütz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, Gagneur J, Korbel JO, Huber W, Steinmetz LM. 2013. The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 3:1213–1224. doi: 10.1534/g3.113.005777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen L, Wang W, Cao L, Li Z, Wang X. 2016. Long non-coding RNA Ccat1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Molecules and Cells 39:330–336. doi: 10.14348/molcells.2016.2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goodwin EC, DiMaio D.. 2001. Induced senescence in HeLa cervical carcinoma cells containing elevated telomerase activity and extended telomeres. Cell Growth Differ 12:525-34. [PubMed] [Google Scholar]
- 48. Leeman JE, Li Y, Bell A, Hussain SS, Majumdar R, Rong-Mullins X, Blecua P, Damerla R, Narang H, Ravindran PT, Lee NY, Riaz N, Powell SN, Higginson DS. 2019. Human papillomavirus 16 promotes microhomology-mediated end-joining. Proc Natl Acad Sci U S A 116:21573–21579. doi: 10.1073/pnas.1906120116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vernon SD, Unger ER, Miller DL, Lee DR, Reeves WC. 1997. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer. Int J Cancer 74:50–56. doi: [DOI] [PubMed] [Google Scholar]
- 50. Xu B, Chotewutmontri S, Wolf S, Klos U, Schmitz M, Dürst M, Schwarz E. 2013. Multiplex identification of human papillomavirus 16 DNA integration sites in cervical carcinomas. PLoS One 8:e66693. doi: 10.1371/journal.pone.0066693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gagliardi A, Porter VL, Zong Z, Bowlby R, Titmuss E, Namirembe C, Griner NB, Petrello H, Bowen J, Chan SK, et al. 2020. Analysis of Ugandan cervical carcinomas identifies human papillomavirus clade-specific epigenome and transcriptome landscapes. Nat Genet 52:800–810. doi: 10.1038/s41588-020-0673-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou L, Qiu Q, Zhou Q, Li J, Yu M, Li K, Xu L, Ke X, Xu H, Lu B, Wang H, Lu W, Liu P, Lu Y. 2022. Long-read sequencing unveils high-resolution HPV integration and its oncogenic progression in cervical cancer. Nat Commun 13:2563. doi: 10.1038/s41467-022-30190-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Groves IJ, Drane ELA, Michalski M, Monahan JM, Scarpini CG, Smith SP, Bussotti G, Várnai C, Schoenfelder S, Fraser P, Enright AJ, Coleman N. 2021. Short- and long-range cis interactions between integrated HPV genomes and cellular chromatin dysregulate host gene expression in early cervical carcinogenesis. PLoS Pathog 17:e1009875. doi: 10.1371/journal.ppat.1009875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meissner JD. 1999. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol 80 ( Pt 7):1725–1733. doi: 10.1099/0022-1317-80-7-1725 [DOI] [PubMed] [Google Scholar]
- 55. Kozak M. 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299:1–34. doi: 10.1016/s0378-1119(02)01056-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grant CM, Miller PF, Hinnebusch AG. 1994. Requirements for intercistronic distance and level of eukaryotic initiation factor 2 activity in reinitiation on GCN4 mRNA vary with the downstream cistron. Mol Cell Biol 14:2616–2628. doi: 10.1128/mcb.14.4.2616-2628.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pim D, Massimi P, Banks L. 1997. Alternatively spliced HPV-18 E6* protein inhibits E6 mediated degradation of p53 and suppresses transformed cell growth. Oncogene 15:257–264. doi: 10.1038/sj.onc.1201202 [DOI] [PubMed] [Google Scholar]
- 58. Guccione E, Pim D, Banks L. 2004. HPV-18 E6*I modulates HPV-18 full-length E6 functions in a cell cycle dependent manner. Int J Cancer 110:928–933. doi: 10.1002/ijc.20184 [DOI] [PubMed] [Google Scholar]
- 59. Cannizzaro LA, Dürst M, Mendez MJ, Hecht BK, Hecht F. 1988. Regional chromosome localization of human papillomavirus integration sites near fragile sites, oncogenes, and cancer chromosome breakpoints. Cancer Genet Cytogenet 33:93–98. doi: 10.1016/0165-4608(88)90054-4 [DOI] [PubMed] [Google Scholar]
- 60. Luft F, Klaes R, Nees M, Dürst M, Heilmann V, Melsheimer P, von Knebel Doeberitz M. 2001. Detection of integrated papillomavirus sequences by ligation-mediated PCR (DIPS-PCR) and molecular characterization in cervical cancer cells. Int J Cancer 92:9–17. [PubMed] [Google Scholar]
- 61. Carmody MW, Jones M, Tarraza H, Vary CP. 1996. Use of the polymerase chain reaction to specifically amplify integrated HPV-16 DNA by virtue of its linkage to interspersed repetitive DNA. Mol Cell Probes 10:107–116. doi: 10.1006/mcpr.1996.0015 [DOI] [PubMed] [Google Scholar]
- 62. Ziegert C, Wentzensen N, Vinokurova S, Kisseljov F, Einenkel J, Hoeckel M, von Knebel Doeberitz M. 2003. A comprehensive analysis of HPV integration loci in anogenital lesions combining transcript and genome-based amplification techniques. Oncogene 22:3977–3984. doi: 10.1038/sj.onc.1206629 [DOI] [PubMed] [Google Scholar]
- 63. Vinokurova S, Wentzensen N, Einenkel J, Klaes R, Ziegert C, Melsheimer P, Sartor H, Horn L-C, Höckel M, von Knebel Doeberitz M. 2005. Clonal history of papillomavirus-induced dysplasia in the female lower genital tract. J Natl Cancer Inst 97:1816–1821. doi: 10.1093/jnci/dji428 [DOI] [PubMed] [Google Scholar]
- 64. Rösl F, Dürst M, zur Hausen H. 1988. Selective suppression of human papillomavirus transcription in non-tumorigenic cells by 5-azacytidine. EMBO J 7:1321–1328. doi: 10.1002/j.1460-2075.1988.tb02947.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rösl F, Arab A, Klevenz B, zur Hausen H. 1993. The effect of DNA methylation on gene regulation of human papillomaviruses. J Gen Virol 74 (Pt 5):791–801. doi: 10.1099/0022-1317-74-5-791 [DOI] [PubMed] [Google Scholar]
- 66. Mirabello L, Schiffman M, Ghosh A, Rodriguez AC, Vasiljevic N, Wentzensen N, Herrero R, Hildesheim A, Wacholder S, Scibior-Bentkowska D, Burk RD, Lorincz AT. 2013. Elevated methylation of HPV16 DNA is associated with the development of high grade cervical intraepithelial neoplasia. Int J Cancer 132:1412–1422. doi: 10.1002/ijc.27750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yu L, Majerciak V, Xue XY, Uberoi A, Lobanov A, Chen X, Cam M, Hughes SH, Lambert PF, Zheng ZM. 2021. Mouse papillomavirus type 1 (MmuPV1) DNA is frequently integrated in benign tumors by microhomology-mediated end-joining. PLoS Pathog 17:e1009812. doi: 10.1371/journal.ppat.1009812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Duensing S, Lee LY, Duensing A, Basile J, Piboonniyom S, Gonzalez S, Crum CP, Munger K. 2000. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc Natl Acad Sci U S A 97:10002–10007. doi: 10.1073/pnas.170093297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Duensing S, Münger K. 2002. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res 62:7075–7082. [PubMed] [Google Scholar]
- 70. Spardy N, Duensing A, Charles D, Haines N, Nakahara T, Lambert PF, Duensing S. 2007. The human papillomavirus type 16 E7 oncoprotein activates the Fanconi anemia (FA) pathway and causes accelerated chromosomal instability in FA cells. J Virol 81:13265–13270. doi: 10.1128/JVI.01121-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Moody CA, Laimins LA. 2009. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog 5:e1000605. doi: 10.1371/journal.ppat.1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Desfarges S, Ciuffi A. 2010. Retroviral integration site selection. Viruses 2:111–130. doi: 10.3390/v2010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH. 2014. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345:179–183. doi: 10.1126/science.1254194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hughes SH, Coffin JM. 2016. What integration sites tell us about HIV persistence. Cell Host Microbe 19:588–598. doi: 10.1016/j.chom.2016.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McBride AA, Warburton A. 2017. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog 13:e1006211. doi: 10.1371/journal.ppat.1006211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dooley KE, Warburton A, McBride AA. 2016. Tandemly integrated HPV16 can form a Brd4-dependent super-enhancer-like element that drives transcription of viral oncogenes. mBio 7:e01446-16. doi: 10.1128/mBio.01446-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Smith PP, Friedman CL, Bryant EM, McDougall JK. 1992. Viral integration and fragile sites in human papillomavirus-immortalized human keratinocyte cell lines. Genes Chromosomes Cancer 5:150–157. doi: 10.1002/gcc.2870050209 [DOI] [PubMed] [Google Scholar]
- 78. Thorland EC, Myers SL, Persing DH, Sarkar G, McGovern RM, Gostout BS, Smith DI. 2000. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res 60:5916–5921. [PubMed] [Google Scholar]
- 79. Thorland EC, Myers SL, Gostout BS, Smith DI. 2003. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene 22:1225–1237. doi: 10.1038/sj.onc.1206170 [DOI] [PubMed] [Google Scholar]
- 80. Wentzensen N, Vinokurova S, von Knebel Doeberitz M. 2004. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res 64:3878–3884. doi: 10.1158/0008-5472.CAN-04-0009 [DOI] [PubMed] [Google Scholar]
- 81. Gao G, Johnson SH, Vasmatzis G, Pauley CE, Tombers NM, Kasperbauer JL, Smith DI. 2017. Common fragile sites (CFS) and extremely large CFS genes are targets for human papillomavirus integrations and chromosome rearrangements in oropharyngeal squamous cell carcinoma. Genes Chromosomes Cancer 56:59–74. doi: 10.1002/gcc.22415 [DOI] [PubMed] [Google Scholar]
- 82. Jang MK, Shen K, McBride AA. 2014. Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome. PLoS Pathog 10:e1004117. doi: 10.1371/journal.ppat.1004117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Coffin JM, Bale MJ, Wells D, Guo S, Luke B, Zerbato JM, Sobolewski MD, Sia T, Shao W, Wu X, Maldarelli F, Kearney MF, Mellors JW, Hughes SH. 2021. Integration in oncogenes plays only a minor role in determining the in vivo distribution of HIV integration sites before or during suppressive antiretroviral therapy. PLoS Pathog 17:e1009141. doi: 10.1371/journal.ppat.1009141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Parfenov M, Pedamallu CS, Gehlenborg N, Freeman SS, Danilova L, Bristow CA, Lee S, Hadjipanayis AG, Ivanova EV, Wilkerson MD, et al. 2014. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci U S A 111:15544–15549. doi: 10.1073/pnas.1416074111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dürst M, Croce CM, Gissmann L, Schwarz E, Huebner K. 1987. Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proc Natl Acad Sci U S A 84:1070–1074. doi: 10.1073/pnas.84.4.1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ferber MJ, Thorland EC, Brink AATP, Rapp AK, Phillips LA, McGovern R, Gostout BS, Cheung TH, Chung TKH, Fu WY, Smith DI. 2003. Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene 22:7233–7242. doi: 10.1038/sj.onc.1207006 [DOI] [PubMed] [Google Scholar]
- 87. Thomas LK, Bermejo JL, Vinokurova S, Jensen K, Bierkens M, Steenbergen R, Bergmann M, von Knebel Doeberitz M, Reuschenbach M. 2014. Chromosomal gains and losses in human papillomavirus-associated neoplasia of the lower genital tract - a systematic review and meta-analysis. Eur J Cancer 50:85–98. doi: 10.1016/j.ejca.2013.08.022 [DOI] [PubMed] [Google Scholar]
- 88. Bodelon C, Vinokurova S, Sampson JN, den Boon JA, Walker JL, Horswill MA, Korthauer K, Schiffman M, Sherman ME, Zuna RE, Mitchell J, Zhang X, Boland JF, Chaturvedi AK, Dunn ST, Newton MA, Ahlquist P, Wang SS, Wentzensen N. 2016. Chromosomal copy number alterations and HPV integration in cervical precancer and invasive cancer. Carcinogenesis 37:188–196. doi: 10.1093/carcin/bgv171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Aldersley J, Lorenz DR, Mouw KW, D’Andrea AD, Gabuzda D. 2021. Genomic landscape of primary and recurrent anal squamous cell carcinomas in relation to HPV integration, copy-number variation, and DNA damage response genes. Mol Cancer Res 19:1308–1321. doi: 10.1158/1541-7786.MCR-20-0884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Symer DE, Akagi K, Geiger HM, Song Y, Li G, Emde AK, Xiao W, Jiang B, Corvelo A, Toussaint NC, Li J, Agrawal A, Ozer E, El-Naggar AK, Du Z, Shewale JB, Stache-Crain B, Zucker M, Robine N, Coombes KR, Gillison ML. 2022. Diverse tumorigenic consequences of human papillomavirus integration in primary oropharyngeal cancers. Genome Res 32:55–70. doi: 10.1101/gr.275911.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Oh J, Chang KW, Hughes SH. 2006. Mutations in the U5 sequences adjacent to the primer binding site do not affect tRNA cleavage by rous sarcoma virus RNase H but do cause aberrant integrations in vivo. J Virol 80:451–459. doi: 10.1128/JVI.80.1.451-459.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Varadarajan J, McWilliams MJ, Hughes SH. 2013. Treatment with suboptimal doses of raltegravir leads to aberrant HIV-1 integrations. Proc Natl Acad Sci U S A 110:14747–14752. doi: 10.1073/pnas.1305066110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Oh J, Chang KW, Alvord WG, Hughes SH. 2006. Alternate polypurine tracts affect rous sarcoma virus integration in vivo. J Virol 80:10281–10284. doi: 10.1128/JVI.00361-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Oh J, Chang KW, Wierzchoslawski R, Alvord WG, Hughes SH. 2008. Rous sarcoma virus (RSV) integration in vivo: a CA dinucleotide is not required in U3, and RSV linear DNA does not autointegrate. J Virol 82:503–512. doi: 10.1128/JVI.01441-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sfeir A, Symington LS. 2015. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem Sci 40:701–714. doi: 10.1016/j.tibs.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang H, Xu X. 2017. Microhomology-mediated end joining: new players join the team. Cell Biosci 7:6. doi: 10.1186/s13578-017-0136-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Stanley MA, Browne HM, Appleby M, Minson AC. 1989. Properties of a non-tumorigenic human cervical keratinocyte cell line. Int J Cancer 43:672–676. doi: 10.1002/ijc.2910430422 [DOI] [PubMed] [Google Scholar]
- 98. Bedell MA, Hudson JB, Golub TR, Turyk ME, Hosken M, Wilbanks GD, Laimins LA. 1991. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J Virol 65:2254–2260. doi: 10.1128/JVI.65.5.2254-2260.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jeon S, Allen-Hoffmann BL, Lambert PF. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol 69:2989–2997. doi: 10.1128/JVI.69.5.2989-2997.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hummel M, Hudson JB, Laimins LA. 1992. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J Virol 66:6070–6080. doi: 10.1128/JVI.66.10.6070-6080.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kang JG, Pripuzova N, Majerciak V, Kruhlak M, Le SY, Zheng ZM. 2011. Kaposi's sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from microRNA-mediated suppression. J Virol 85:2620–2630. doi: 10.1128/JVI.02144-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S7.
Tables S1-S4.
Data Availability Statement
RNA-seq data are available under NCBI GEO accession numbers GSE158033 for CaSki, GSE174317 for SiHa, and GSE165475 for HeLa cells.