LETTER
Currently, the quantification of cytomegalovirus (CMV) DNA in blood samples (CMV-DNAemia) represents the gold standard for identifying active viral replication, preventing CMV-related disease, and monitoring response to drugs targeting CMV-DNA polymerase (1–5). The recent introduction of letermovir (LMV) showed that CMV-DNAemia may not be an accurate marker of active viral replication (6–9). Indeed, by blocking the terminase complex, LMV induces the release of free CMV-DNA fragments (abortive infection), which could lead to potential misinterpretation of molecular testing results (1, 6–9). Additional methods, such as CMV-viremia (shell vial method) and CMV-DNAemia post-DNase (DNase test), could be used to prove active viral replication. CMV viremia detects CMV infectious particles in cell culture, whereas the DNase test exploits the digestion activity of DNase I added to the sample before extraction to differentiate free naked DNA from the genome encapsidated into virions. Both methods, therefore, identify the presence of infectious virions in blood samples. However, the procedures are laborious and not standardized (8). This pilot study evaluates the clinical utility of a new CMV-RNAemia test to identify active viral replication and guide pre-emptive or prophylaxis strategies in transplant settings, especially in patients receiving LMV.
A total of 254 blood samples from 47 CMV-DNAemia-positive episodes that occurred in 44 transplant recipients (Table 1) were retrospectively tested for the detection and quantification of CMV-RNAemia, using the CMV RNA ELITe MGB kit on ELITe InGenius instrument (ELITechGroup). This is the first available commercial test targeting the virion-associated UL21.5 mRNA, a late transcript highly expressed during lytic infection (10–13). The assay was carried out on plasma samples stored at −80°C until processing. The results of CMV-RNAemia and CMV-DNAemia on the respective whole blood (WB) matrix, collected during virological routine monitoring, are reported in Table 1. Compared to CMV-DNAemia, the CMV-RNAemia showed sensitivity and specificity equal to 32.2% (73/227) and 100% (27/27), respectively; the percentage of agreement between the two methods was 39.3% (100/254, data not shown in Table 1). Specifically, 73 samples resulted positive by both tests (CMV-DNAemia versus CMV-RNAemia), and the correlation in the viral load measurement was high (r = 0.755, P < 0.001, Table 1). One hundred fifty-four samples were positive only for CMV-DNAemia. Out of these, 57.8% (89/154) were collected during the descending phase of positive CMV-DNAemia episodes (after reaching the peak) or were related to episodes with persistent low CMV-DNAemia levels (below 300 copies/mL). All the latter samples were from patients who were receiving LMV prophylaxis, suggesting that the type of anti-CMV drug administered should be considered during the viral load evaluations. In the analysis, the CMV-DNAemia episodes were stratified into three groups: LMV-prophylaxis, LMV off-label treatment, and pre-emptive therapy. Focusing on cases receiving LMV prophylaxis, CMV-RNAemia resulted positive in 6/25 (24%) episodes (Table 1). Among these, 4/6 (66.7%) were related to clinically significant CMV infections (based on both clinical and laboratory findings), treated by pre-emptive therapy. In 2/4 (50%) cases, genotypic LMV resistance (LMV-R) was documented. Considering the group receiving LMV off-label, six out of seven episodes (85.7%) showed positive CMV-RNAemia (Table 1), and in 3/6 cases (50%), LMV-R was found, leading to switching the treatment to other anti-CMV agents: maribavir, foscarnet, or immunoglobulins. In the remaining three cases, the LMV treatment was continued and combined with a reduced immunosuppressive therapy, until complete viral clearance (two consecutive CMV-DNAemia negative results). In the 12 out of 32 (37.5%) episodes in which CMV-RNAemia was detected during LMV administration, the active viral replication was documented by CMV-viremia and/or DNase tests. These additional methods also confirmed the CMV-RNAemia negative results in the remaining 20/32 (62.5%) episodes, suggesting that the positive CMV-DNAemia in these cases was possibly due to abortive infections. Finally, CMV-RNAemia was positive in all 15 episodes from 14 patients receiving pre-emptive therapy (Table 1).
TABLE 1.
Characteristics of study population, infective episodes, and samples analyzed
| LMV-prophylaxis | LMV off-label treatmentb | Pre-emptive therapyc | Total | |
|---|---|---|---|---|
| No. of transplant recipients | 23 | 7 | 14 | 44 |
| Male sex | 13 | 5 | 10 | 28 |
| Age (mean ± SD) | 49 ± 13.2 | 34 ± 16.1 | 45 ± 21.2 | 45 ± 17 |
| Type of transplant | ||||
| Hematopoietic stem cells | 23 | 4 | 6 | 33 |
| Liver | 0 | 1 | 3 | 4 |
| Heart | 0 | 2 | 2 | 4 |
| Kidney | 0 | 0 | 3 | 3 |
| CMV serostatus D/Ra | ||||
| D+/R+ | 13 | 0 | 7 | 20 |
| D−/R+ | 10 | 4 | 3 | 17 |
| D+/R− | 0 | 3 | 4 | 7 |
| No. of CMV-DNAemia-positive episodesd | 25 | 7 | 15 | 47 |
| CMV-DNAemia-positive/total samples | 97/106 | 35/37 | 95/111 | 227/254 |
| Median CMV DNAemia levels in whole blood (copies/mLe, range) | 3.9 × 102 (3 × 102–1.9 × 105) | 1.4 × 103 (3 × 102–7.3 × 104) | 2.8 × 103 (3 × 102–2.4 × 106) | 9.8 × 102 (3 × 102–2.4 × 106) |
| No. of CMV-RNAemia-positive episodesf | 6 | 6 | 15 | 27 |
| CMV-RNAemia-positive/total samples | 14/106 | 9/37 | 50/111 | 73/254 |
| Median CMV-RNAemia levels in plasma (copies/mL, range) | 3 × 10 (3 × 10–1.9 × 103) | 5.1 × 10 (3 × 10–1.2 × 103) | 1.4 × 102 (3 × 10–9.1 × 103) | 8.1 × 10 (30–9.1 × 103) |
| Correlation coefficient between CMV DNAemia and CMV-RNAemia levels | 0.571g | 0.237g | 0.759h | 0.755h |
| Sensitivity–specificity of CMV-RNAemia vs CMV-DNAemia | 14.4%–100% | 25.7%–100% | 52.6%–100% | 32.2%–100%i |
D, donor; R, recipient; +, positive; −, negative.
LMV-off-label due to ganciclovir resistance (three patients receiving solid organ transplant), severe neutropenia (two pediatric hematopoietic stem cell transplant recipients), and positive CMV-DNAemia results when starting LMV (two adult hematopoietic stem cell transplant recipients).
The patients receiving therapy with (val)ganciclovir (n = 13) or foscarnet (n = 1).
CMV-DNAemia-positive episodes: at least two sequential positive CMV-DNAemia results in whole blood or one more than 300 copies/mL; a mean of 5.4 samples/episodes [±3.5 standard deviation (SD)] was analyzed.
Conversion factor from copies/mL to international units/mL is 0.46.
CMV-RNAemia-positive episodes: episodes with at least one positive result.
Spearman correlation, CMV-DNAemia and CMV RNAemia levels under the lower limit of quantification (300 and 30 copies/ml) were considered equal to 150 and 15 copies/ml, respectively.
Pearson correlation, CMV-DNAemia and CMV RNAemia levels under the lower limit of quantification (300 and 30 copies/ml) were considered equal to 150 and 15 copies/ml, respectively.
The specificity of 100% was also found in 100 additional plasma samples from transplant patients for whom CMV reactivation was ruled out by negative CMV-DNAemia results (data not shown).
Analyzing all the 227 positive samples (Table 1), the CMV-DNAemia levels were higher in specimens positive for CMV-RNAemia than in those negative (P < 0.001, Fig. 1). Considering the three groups of patients, lower median CMV-DNAemia values were found in cases receiving LMV, as prophylaxis or off-label, than in the pre-emptive group (P < 0.001, Mann–Whitney test). The results confirmed that detectable low DNAemia levels during LMV administration may reflect abortive rather than productive infection. These data also suggest that CMV-RNAemia could be useful for detecting active CMV replication in patients receiving antiviral therapy, especially with drugs that do not act on the viral DNA polymerase, such as LMV.
Fig 1.
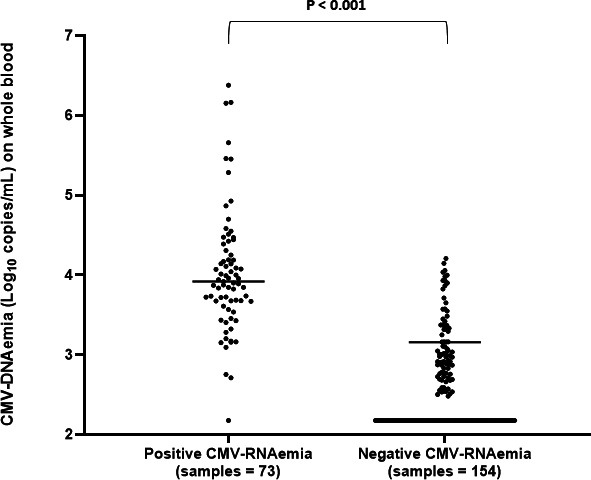
Comparison of CMV-DNAemia levels in samples positive and negative for CMV-RNAemia. Positive values under the lower limit of quantification (300 copies/mL) were reported as equal to 150 copies/mL. Higher median CMV-DNAemia values were observed in specimens positive for CMV-RNAemia than in the negatives: 8,289 copies/mL [interquartile range (IQR): 4,664–21,286.2] vs 373 copies/mL (IQR: 300–1,106.7), respectively; P < 0.001 (Mann–Whitney test).
Considering the 27 episodes positive for both markers (Table 1), the CMV-DNAemia and CMV-RNAemia peaks were reached simultaneously, with median levels equal to 11,754 copies/mL (IQR: 7,386.5–26,174) and 81 copies/mL (IQR: positive <30–489.5), respectively. Interestingly, during the descending phase, negative results were obtained earlier with CMV-RNAemia than with CMV-DNAemia (mean time 7 days before, ±6.9 standard deviation), proving more rapidly an efficient viral clearance, as shown in a representative case detailed in Fig. 2. As reported in literature, during this phase, CMV-DNAemia could be detected due to the presence of free viral genome fragments that are released after infected cell degradation and are not necessarily associated with infectious viral particles (3, 4). These observations, together with the previous considerations, show that CMV-DNAemia alone may not be an accurate marker of active CMV replication, especially in patients undergoing LMV.
Fig 2.
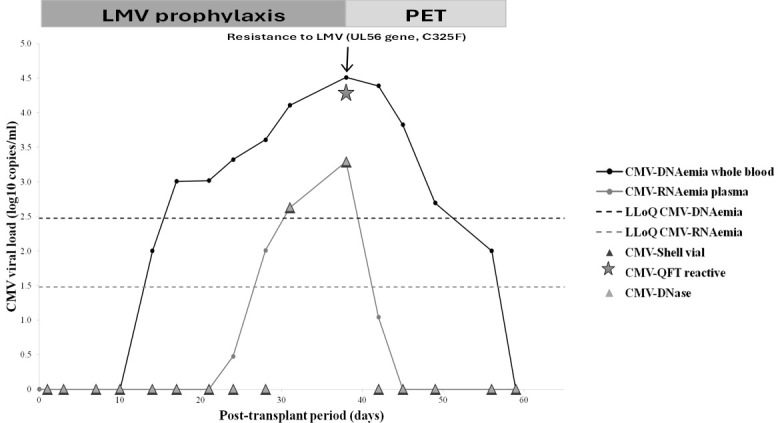
Virological monitoring by CMV-DNAemia and CMV-RNAemia in a hematopoietic stem cell transplant recipient managed by LMV prophylaxis during post-transplant. PET, pre-emptive therapy with foscarnet; LLoQ, lower limit of quantification; CMV-DNase, CMV-DNAemia detection in plasma samples after DNase digestion; CMV-QFT reactive, presence of CMV cell-mediated immune response measured by QuantiFERON-CMVhttps://www.qiagen.com/us/resources/download.aspx?id=42713b7e-2b8f-48b6-8198-44104d14aacb&lang=it-ITtest (QIAGEN). Adult hematopoietic stem cell transplant recipient receiving LMV prophylaxis. An episode of active viral replication was identified by positive results of CMV-RNAemia and of the additional routine methods (CMV-DNase and CMV-viremia). Pre-emptive therapy was started based on both clinical and laboratory findings, obtaining a complete viral clearance as shown by CMV-DNAemia and more rapidly by CMV-RNAemia.
In this study, CMV-DNAemia was measured in WB samples since previous investigations suggested this blood compartment as a preferable clinical sample for monitoring CMV infection post-transplantation (3, 4, 8). As RNA is an unstable and fragile molecule, plasma samples were immediately frozen at −80°C and analyzed within 3 weeks. At prospective and retrospective testing of CMV-RNAemia in a small group of samples, no significant differences were observed (data not shown), confirming the absence of degradation during storage.
In conclusion, CMV-RNAemia, detected by a standardized user-friendly assay and an automated and integrated system, together with CMV-DNAemia, could provide accurate information on viral load kinetics during post-transplant monitoring of CMV infection, especially in patients receiving LMV. Further studies with larger numbers of samples, including patients undergoing therapy with new anti-CMV drugs such as maribavir, are needed to confirm these preliminary data.
ACKNOWLEDGMENTS
We would like to thank Dr. Cristina Olivo and Dr. Guglielmo Stefanuto for their support and valuable feedback.
Contributor Information
Liliana Gabrielli, Email: liliana.gabrielli@aosp.bo.it.
Elitza S. Theel, Mayo Clinic Minnesota, Rochester, Minnesota, USA
ETHICS APPROVAL
The study was approved by the Ethics Committee of the Area Vasta Emilia Centro (CE-AVEC, approval number 985/2021/Disp/AOUBo).
REFERENCES
- 1. Girmenia C, Lazzarotto T, Bonifazi F, Patriarca F, Irrera G, Ciceri F, Aversa F, Citterio F, Cillo U, Cozzi E, Gringeri E, Baldanti F, Cavallo R, Clerici P, Barosi G, Grossi P. 2019. Assessment and prevention of cytomegalovirus infection in allogeneic hematopoietic stem cell transplant and in solid organ transplant: a multidisciplinary consensus conference by the Italian GITMO, SITO, and AMCLI societies. Clin Transplant 33:e13666. doi: 10.1111/ctr.13666 [DOI] [PubMed] [Google Scholar]
- 2. Grossi PA, Baldanti F, Andreoni M, Perno CF. 2020. CMV infection management in transplant patients in Italy. J Clin Virol 123:104211. doi: 10.1016/j.jcv.2019.104211 [DOI] [PubMed] [Google Scholar]
- 3. Lazzarotto T, Chiereghin A, Piralla A, Piccirilli G, Girello A, Campanini G, Gabrielli L, Costa C, Prete A, Bonifazi F, Busca A, Cairoli R, Colombo AA, Zecca M, Sidoti F, Bianco G, Paba P, Perno CF, Cavallo R, Baldanti F, AMCLI-GLaIT working group . 2018. Cytomegalovirus and Epstein-Barr virus DNA kinetics in whole blood and plasma of allogeneic hematopoietic stem cell transplantation recipients. Biol Blood Marrow Transplant 24:1699–1706. doi: 10.1016/j.bbmt.2018.03.005 [DOI] [PubMed] [Google Scholar]
- 4. Lazzarotto T, Chiereghin A, Piralla A, Gibertoni D, Piccirilli G, Turello G, Campanini G, Gabrielli L, Costa C, Comai G, La Manna G, Biancone L, Rampino T, Gregorini M, Sidoti F, Bianco G, Mauro MV, Greco F, Cavallo R, Baldanti F, AMCLI-GLaIT . 2020. Kinetics of cytomegalovirus and Epstein-Barr virus DNA in whole blood and plasma of kidney transplant recipients: implications on management strategies. PLoS ONE 15:e0238062. doi: 10.1371/journal.pone.0238062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Razonable RR, Inoue N, Pinninti SG, Boppana SB, Lazzarotto T, Gabrielli L, Simonazzi G, Pellett PE, Schmid DS. 2020. Clinical diagnostic testing for human cytomegalovirus infections. J Infect Dis 221:S74–S85. doi: 10.1093/infdis/jiz601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shigle TL, Handy VW, Chemaly RF. 2020. Letermovir and its role in the prevention of cytomegalovirus infection in seropositive patients receiving an allogeneic hematopoietic cell transplant. Ther Adv Hematol 11:2040620720937150. doi: 10.1177/2040620720937150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nho D, Lee R, Cho SY, Lee DG, Kim EJ, Park S, Lee SE, Cho BS, Kim YJ, Lee S, Kim HJ. 2023. Cytomegalovirus infection after allogeneic hematopoietic cell transplantation under 100-day letermovir prophylaxis: a real-world 1-year follow-up study. Viruses 15:1884. doi: 10.3390/v15091884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cassaniti I, Colombo AA, Bernasconi P, Malagola M, Russo D, Iori AP, Girmenia C, Greco R, Peccatori J, Ciceri F, Bonifazi F, Percivalle E, Campanini G, Piccirilli G, Lazzarotto T, Baldanti F. 2021. Positive HCMV DNAemia in stem cell recipients undergoing letermovir prophylaxis is expression of abortive infection. Am J Transplant 21:1622–1628. doi: 10.1111/ajt.16450 [DOI] [PubMed] [Google Scholar]
- 9. Khawaja F, Spallone A, Kotton CN, Chemaly RF. 2023. Cytomegalovirus infection in transplant recipients: newly approved additions to our armamentarium. Clin Microbiol Infect 29:44–50. doi: 10.1016/j.cmi.2022.07.001 [DOI] [PubMed] [Google Scholar]
- 10. Terhune SS, Schröer J, Shenk T. 2004. RNAs are packaged into human cytomegalovirus virions in proportion to their intracellular concentration. J Virol 78:10390–10398. doi: 10.1128/JVI.78.19.10390-10398.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bresnahan WA, Shenk T. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288:2373–2376. doi: 10.1126/science.288.5475.2373 [DOI] [PubMed] [Google Scholar]
- 12. Huang ES, Johnson RA. 2000. Human cytomegalovirus - no longer just a DNA virus. Nat Med 6:863–864. doi: 10.1038/78612 [DOI] [PubMed] [Google Scholar]
- 13. Greijer AE, Dekkers CA, Middeldorp JM. 2000. Human cytomegalovirus virions differentially incorporate viral and host cell RNA during the assembly process. J Virol 74:9078–9082. doi: 10.1128/jvi.74.19.9078-9082.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]


