Abstract
Histone proteins are highly abundant and conserved among eukaryotes and play a large role in gene regulation as a result of structures known as posttranslational modifications (PTMs). Identifying the position and nature of each PTM or pattern of PTMs in reference to external or genetic factors allows this information to be statistically correlated with biological responses such as DNA transcription, replication, or repair. In the present work, a high-throughput analytical protocol for the detection of histone PTMs from biological samples is described. The use of complementary liquid chromatography, trapped ion mobility spectrometry, and time-of-flight mass spectrometry (LC-TIMS-ToF MS/MS) enables the separation and PTM assignment of the most biologically relevant modifications in a single analysis. The described approach takes advantage of recent developments in dependent data acquisition (DDA) using parallel accumulation in the mobility trap, followed by sequential fragmentation and collision-induced dissociation. Histone PTMs are confidently assigned based on their retention time, mobility, and fragmentation pattern.
SUMMARY:
An analytical workflow based on liquid chromatography, trapped ion mobility spectrometry, and time-of-flight mass spectrometry (LC-TIMS-ToF MS/MS) for high confidence and highly reproducible “bottom-up” analysis of histone modifications and identification based on principal parameters (retention time [RT], collision cross section [CCS], and accurate mass-to-charge [m/z] ratio).
INTRODUCTION:
In eukaryotic cells, DNA is packaged as chromatin into functional units called nucleosomes. These units are composed of an octamer of four core histones (two each of H2A, H2B, H3, and H4)1–4. Histones are amongst the most abundant and highly conserved proteins in eukaryotes, which are largely responsible for gene regulation5. Histone posttranslational modifications (PTMs) play a large role in the regulation of chromatin dynamics and rigger various biological processes such as DNA transcription, replication, and repair6. PTMs occur primarily on the accessible surface of the N-terminal regions of histones that are in contact with DNA3,7. However, tail and core modifications influence chromatin structure, altering inter-nucleosome interactions and recruiting specific proteins3,8.
A current challenge during liquid chromatography-mass spectrometry (LC-MS)-based proteomics is the potential co-elution of analytes of interest. In the case of data-dependent analyses (DDA), this translates into the potential loss of several precursor ions during the MS/MS acquisition process9. Time-of-flight (ToF) instruments acquire spectra at very high frequency9,10 (up to tens of kHz)11; this makes them capable of rapidly scanning the total precursor ions within a complex sample (MS1), thus promising optimal sensitivity and MS/MS sequencing rates (up to 100 Hz)9 and making them ideal for biological sample analysis10. Nevertheless, the sensitivity available at these high scan rates is limited by the MS/MS rate9. The addition of trapped ion mobility spectrometry (TIMS) in combination with an orthogonal quadrupole time-of-flight (qToF) mass spectrometer was used to mitigate these limitations. In TIMS, all precursor ions are accumulated in tandem and eluted as a function of their mobility, rather than selecting single precursor masses with a quadrupole9. Parallel accumulation–serial fragmentation (PASEF) allows for hundreds of MS/MS events per second without any loss of sensitivity9.
The principal aim of this work was to show the recent developments of DDA using parallel accumulation in the mobility trap followed by sequential fragmentation and collision-induced dissociation (CID). Histone PTMs were confidently assigned based on their retention times (RTs), mobilities, and fragmentation patterns.
PROTOCOL:
NOTE: Histone samples were extracted using a method adapted from Bhanu et al. (2020)12.
1. Sample preparation
1.1. Harvesting cultured cells
-
1.1.1. When cells are 80% confluent, ensure they are viable using trypan blue exclusion.
NOTE: A HeLa S3 cell line was used for these experiments, but this method can be applied to any cultured cells.
1.1.2. Aspirate the media, then apply 5 mL of 1x phosphate-buffered saline (PBS) to each plate.
1.1.3. Swirl the plate(s) to rinse all residual media, then aspirate PBS and apply 5 mL of 1x PBS.
1.1.4. Gently separate the cells from the plate by scraping them with a disposable cell lifter.
1.1.5. Transfer each cell suspension to a 15 mL conical tube.
1.1.6. Pellet the cells by centrifuging at 800 x g for 5 min.
1.1.7. Aspirate the PBS from the cell pellet.
-
1.1.8. Proceed to histone extraction.
NOTE: Flash-freeze the cell pellet in liquid nitrogen if it cannot be processed immediately. Store the pellets at −80 °C until ready to proceed.
1.2. Histone extraction
1.2.1. Estimate the volume of each cell pellet and mark the meniscus with a permanent marker.
-
1.2.2. Prepare enough nuclear isolation buffer (NIB; 15 mM Tris-HCl (pH 7.5), 15 mM NaCl, 60 mM KCl, 5 mM MgCl2, 1 mM CaCl2, and 250 mM sucrose) for all the samples. Alternatively, if many samples need to be processed over time, make the buffer in bulk and store at 2–8 °C for up to 6 months, or aliquot and freeze at −15 °C to −25 °C indefinitely by thawing only the amount necessary for each extraction.
NOTE: The buffer should remain clear during storage. If the buffer takes on a cloudy or otherwise abnormal appearance at any time, discard and prepare fresh buffer.
1.2.3. Prepare 50 times the volume of the cell pellets of wash buffer and add inhibitors as follows (approximately 10 mL of wash buffer per 2 samples).
1.2.3.1. To prepare 10 mL of wash buffer, mix 10 mL of NIB, 30 μL of 200 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride [AEBSF], 10 μL of 1 M dithiothreitol [DTT], 20 μL of 5 μM microcystin, 20 μL of 5 M sodium butyrate.
-
1.2.4. Remove 1/5 of the wash buffer to prepare the lysis buffer (1/5 volume from wash buffer, 0.3% NP-40 or NP-40 alternative).
NOTE: Do not use Triton-X 100 instead of NP-40 or NP-40 alternative, as it may be too abrasive for certain cell types.
1.2.5. Wash the cell pellet thoroughly by suspending it in 5 columns of wash buffer and centrifuging at 800 x g for 5 min at 4 °C. Complete this step twice, aspirating and discarding the supernatant between washes.
1.2.6. Ensure the volume of the cell pellet is still marked with a permanent marker. Resuspend in 10 volumes of lysis buffer.
1.2.7. Pipette-mix each pellet thoroughly to resuspend, then incubate for 15 min on ice.
1.2.8. After 15 min, centrifuge at 800 x g for 5 min at 4 °C.
-
1.2.9. Aspirate and discard the supernatant.
NOTE: The pellet should reduce to ≤ 1/2 the original pellet size (as indicated by the marker line). If the pellet has not reduced sufficiently, repeat the lysis procedure and include a gentle homogenization step using a pestle to break open the cells.
-
1.2.10. Once lysis is complete, resuspend the pellet in 500 μL of wash buffer, then centrifuge at 800 x g for 5 min at 4 °C. Aspirate and discard the supernatant, then repeat the wash step once more to remove all traces of NP-40.
NOTE: At this point, the pellet consists of chromatin, which contains histones.
1.2.11. Resuspend the pellet in 5 volumes (of the original cell pellet size) of 0.4 N H2SO4.
1.2.12. Incubate for 2 h in a cold room or refrigerator using an agitator.
1.2.13. After 2 h, centrifuge the sample(s) at 3400 x g for 5 min at 4 °C. Do not discard the supernatant.
1.2.14. Transfer the supernatant to new tubes, and spike 100% trichloroacetic acid (TCA) to 1/3 the volume of the contents (the final TCA concentration will be approximately 20%).
-
1.2.15. Gently invert the tube and observe that the clear, colorless solution turns white and/or cloudy, indicating protein precipitation.
NOTE: For solutions with low histone concentrations, the protein precipitation may not be immediately noticeable, but the precipitate should be visible after the overnight incubation.
1.2.16. Incubate without disturbance overnight (12–18 h) at 4 °C to completely precipitate the histone proteins.
1.2.17. The following day, centrifuge at 3400 x g for 5 min at 4 °C.
1.2.18. Aspirate the supernatant, being careful not to touch the sides of the tube with the pipette tip. At this stage, the histones are deposited primarily as a film around the sides of the tube(s).
1.2.19. Add 500 μL of ice-cold acetone + 0.1% HCl (acid acetone) to each tube, using a glass Pasteur pipette, then gently invert the tube(s) several times. Arrange the samples in order (1, 2, 3, etc.) while doing this, as any errant acetone may remove the markings on the tubes. Centrifuge at 3400 x g for 5 min at 4 °C and gently decant the supernatant.
1.2.20. Repeat this rinsing step with 500 μL of ice-cold 100% acetone, also with a glass Pasteur pipette. Centrifuge at 3400 x g for 5 min at 4 °C and gently decant the supernatant.
1.2.21. Leave the tubes open to dry at room temperature until the remaining acetone has evaporated.
1.2.22. When dry, add 100 μL of mass spectrometry (MS)-grade water to each tube. Use this droplet to swab all sides of the container to resuspend the entire histone film. Do this by pipetting the droplet onto the side of the tube and rotating it while pipetting up and down or dispensing half of the 100 μL and using the tip to stir it all around. A combination of both methods works best. Histones are readily soluble in water and will be in the solution.
1.2.23. After resuspending all samples, if there is any remaining white solid, sonicate in a bath at room temperature for 5 min.
1.2.24. Centrifuge at 800 x g for 5 min at 4 °C. Transfer the clear solution to fresh tubes. Discard any remaining insoluble pellet.
-
1.2.25. Run an SDS-PAGE under reducing conditions to verify that the extraction is clean.
NOTE: Gels can be run using any appropriate concentration of polyacrylamide as long as it can differentiate proteins in the range of 10–20 kDa. See the Table of Materials for the gels used in this protocol.
1.2.26. Perform a protein concentration assay (i.e., Bradford or BCA) to determine the total protein concentration.
1.3. Chemical derivatization (propionylation) of the lysine residues
-
1.3.1. Transfer 20 μg of histones (determined by the protein assay) to a clean tube. Dry this sample down to <5 μL using a vacuum concentrator, then resuspend using 20 μL of 100 mM ammonium bicarbonate (NH4CO3) (~1 μg/μL solution). Adjust the pH to ~8 using ammonium hydroxide if needed.
CAUTION: Do not use ammonium hydroxide (NH4OH) to resuspend, only to adjust the pH if necessary. Otherwise, proteins will denature and precipitate.
NOTE: To check the pH with minimal sample loss, use a pipette tip to dip in the sample and dab onto a pH strip. This testing procedure will be useful throughout the remaining sample preparation steps.
-
1.3.2. Prepare the propionylation reagent by adding propionic anhydride to acetonitrile (ACN) in a 1:3 (v/v) ratio (i.e., to make 40 μL of reagent, combine 10 μL of propionic anhydride with 30 μL of ACN).
NOTE: Traditionally, methanol or isopropanol have been used in preparation of a propionylation reagent. As propionylation is an amide formation reaction, a non-protonic solvent, like acetonitrile, is required to prevent unwanted side products and reactions, such as methyl propionyl ester, which results from using methanol. Only prepare enough propionylation reagent for up to 4 samples at a time so the reagent remains fresh. Use the reagent within 1–2 min of preparation. As the reagent sits, the propionic anhydride will react with any ambient moisture, and acetic acid will begin to form, which can change the effectiveness of the reagent and will change the pH of the histone solution once the reagent is added.
1.3.3. Add propionylation reagent to each sample in a 1:4 (v/v) (i.e., for 20 μL of histones, add 5 μL of propionylation reagent).
1.3.4. Quickly add 1:5 (v/v) NH4OH (i.e., add 4 μL for 20 μL of the histone solution) to re-establish the pH of the solution to ~8. If pH is still too low, add 1–2 μL of NH4OH at a time until a pH of 8 is achieved. Typically, a 1:5 (v/v) ratio is adequate.
1.3.5. Incubate the samples at room temperature for 15 min without disturbance.
1.3.6. Repeat the propionylation reaction for no more than 3–4 samples per batch of propionylation reagent to ensure minimal acid formation.
1.3.7. Repeat the propionylation procedure steps 1.3.2–1.3.5. A second round of propionylation ensures that >95% of available lysines are derivatized.
-
1.3.8. Dry the samples down to <5 μL using a vacuum concentrator. This will evaporate any unreacted propionylation reagent, acid products, and ammonia gas released from the NH4OH. If the samples dry out completely, this is fine, as no significant sample losses occur.
NOTE: Displace air in the propionic anhydride bottle with argon gas prior to storage to prevent the formation of acetic acid due to contact with ambient moisture remaining in the bottle.
1.4. Proteolytic digestion with trypsin
-
1.4.1. Resuspend histones in 100 mM NH4HCO3 to achieve a volume of 20 μL, achieving an optimal concentration of 1 μg/μL.
NOTE: Sample solutions with concentrations lower than 1 μg/μL will result in decreased Trypsin efficiency.
1.4.2. Add trypsin to histone samples at a 1:10 ratio (wt/wt) (i.e., add 2 μL of 1 μg/μL solution of trypsin to 20 μg of histones).
1.4.3. Incubate reactions at 37 °C for 6–8 h. Alternatively, incubate overnight (12–18 h) at room temperature.
-
1.4.4. Stop the digestion by freezing at −80 °C for at least 1 h.
NOTE: Do not use acid to quench the digestion reaction, as this will cause an unwanted drop in pH at this point in the procedure. The sample can be stored at −80 °C until ready to proceed (Interim stopping point).
1.5. Chemical derivatization (propionylation) of peptide N-terminals
1.5.1. Dry the samples to <5 μL using a vacuum concentrator.
1.5.2. Resuspend the samples up to 20 μL (1 μg/μL) using 100 mM NH4HCO3.
-
1.5.3. Repeat propionylation as before (step 1.3).
NOTE: It is normal that the samples take longer to dry at this step due to a higher aqueous: organic phase ratio.
1.6. Sample desalting with stage-tips
1.6.1. Resuspend or dilute samples with 50 μL of MS-grade water + 0.1% TFA.
-
1.6.2. Using an 11-G sample corer, punch 5 disks of C18 material from a solid phase extraction disk (punch all 5 disks before transferring to the pipette tip). Insert and ensure the disks are securely and evenly wedged at the bottom of a 200 μL pipette tip (Figure 1).
NOTE: Use 15-G corer if desalting over 25 μg of sample through a single stage-tip.
-
1.6.3. Use a centrifuge adapter to hold the stage-tips in place in 1.5 mL or 2 mL microcentrifuge tubes.
NOTE: For the following centrifugation steps, use slow (400–500 x g) revolution at 4 °C, for 1–2 min at a time; the solvents normally pass through the resin in less than 1 min, depending on how tightly the C18 material is packed into the tips.
-
1.6.4. Rinse the resin by centrifuging with 50 μL of 100% acetonitrile to activate the C18 material and remove potential contaminants.
NOTE: It may be easier to load solutions onto the stage-tips using gel-loading pipette tips. Once the C18 material has been activated, it is important not to allow the resin to dry out for the duration of the desalting procedure.
1.6.5. Equilibrate the disk material with 80 μL of MS-grade water + 0.1% TFA by centrifugation.
1.6.6. Acidify the sample to pH 4 or lower using glacial acetic acid. Check the pH with pH strips as before to minimize sample loss.
1.6.7. Load the entirety of the sample onto the resin disk by slow centrifugation.
1.6.8. Wash the sample with 80 μL of MS-grade water + 0.1% TFA by centrifugation.
1.6.9. Elute the sample into a clean 1.5 mL tube by flushing 70 μL of 75% acetonitrile and 0.5% acetic acid by slow centrifugation. Additional centrifugation time may be used to ensure the full sample volume is eluted from the stage-tip. It is okay if the resin dries out with the additional centrifugation time, as it is no longer needed past the elution of the sample.
-
1.6.10. Dry each sample completely in a vacuum concentrator.
NOTE: Sample(s) can be stored at −80 °C until ready to proceed (Interim stopping point).
1.6.11. For LC-MS/MS analysis, reconstitute the samples in a volume of Solvent A (0.1% formic acid) from the liquid chromatography (LC) protocol that gives the final concentration of 0.4 μg/μL (i.e., dissolve 20 μg of histones in 50 μL of Solvent A).
Figure 1: Schematic representation and production of stage tips.
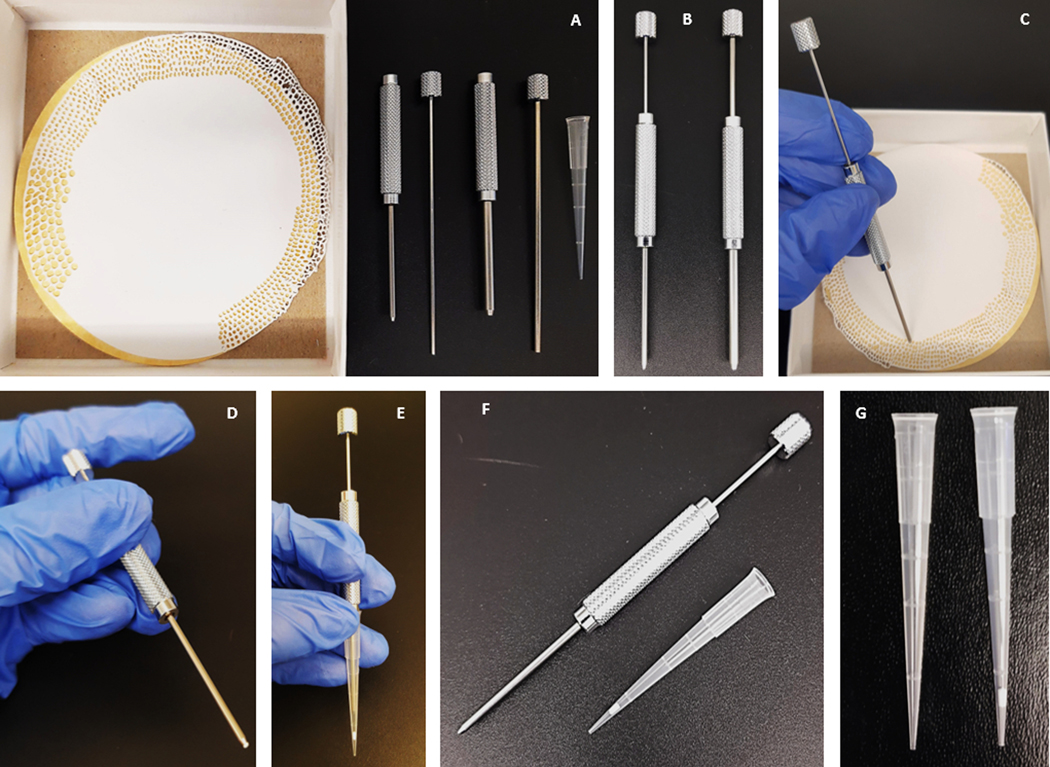
(A–G) Step-by-step guide on the manufacture of a C18-silica disk stage tip.
2. TIMS software interface
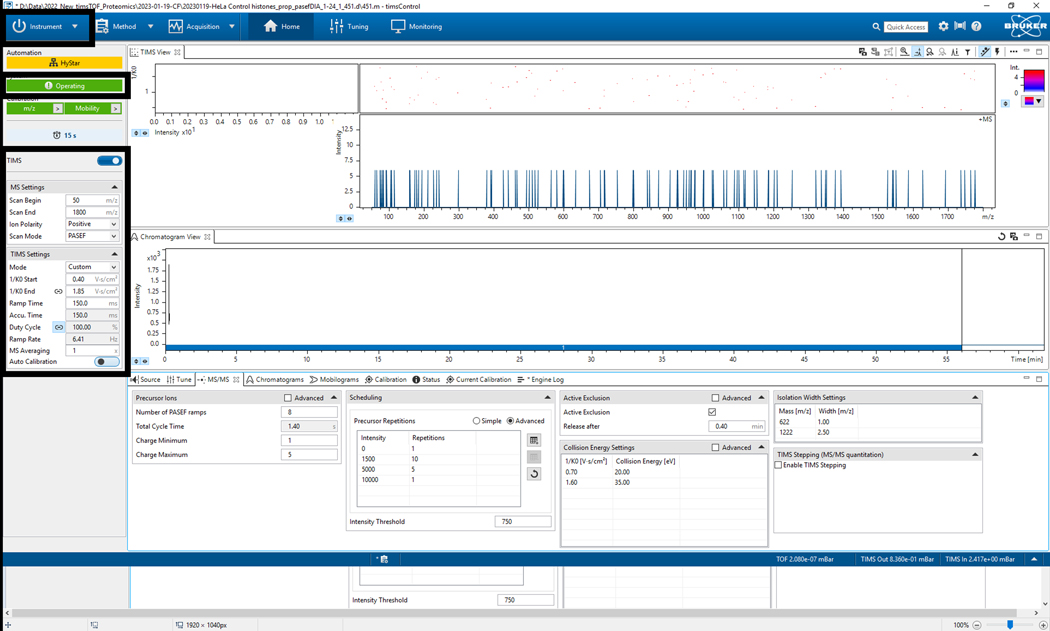
2.1. Select the Instrument tab and switch to Operate (verify that the instrument name becomes highlighted in green) (Figure 2).
2.2. Verify TIMS parameters (Figure 2).
2.3. Verify MS Settings (scan begin, scan end, ion polarity, scan mode) (Figure 2).
2.4. Verify TIMS settings (mode, mobility start, mobility end, ramp time, accumulation time, duty cycle, ramp rate, MS rate, MS averaging, and autocalibration) (Figure 2).
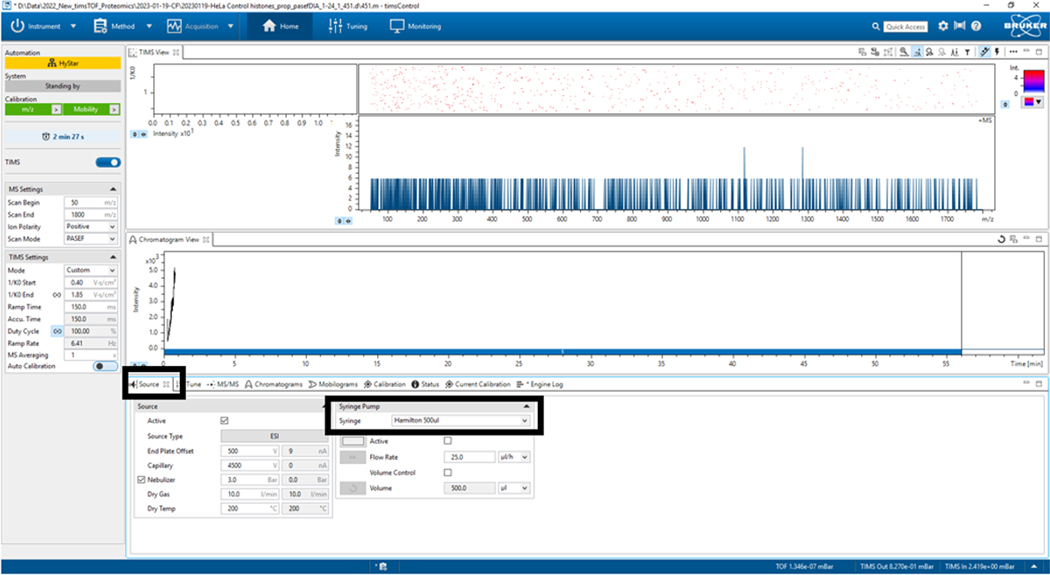
2.5. Go to the Source tab and activate the syringe option (Hamilton 500 μL) only for the TuneMix calibration step (Figure 3).13
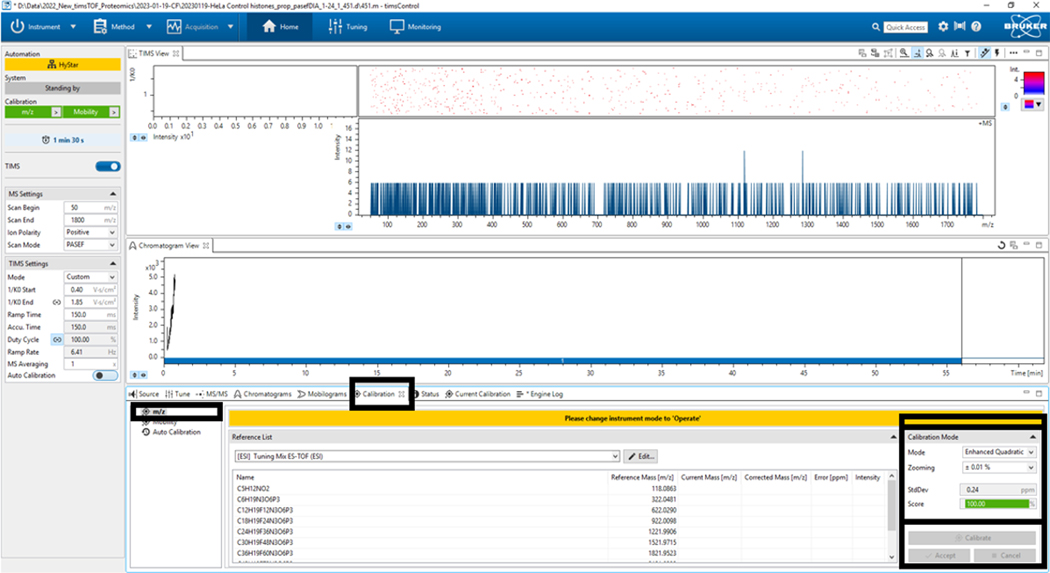
2.6. Go to the Calibration tab, click m/z, select Calibration Mode, and choose the mode (Enhanced Q, generally), zoom (+0.01%), and click Calibrate; when a score of 100% is achieved, accept (Figure 4).
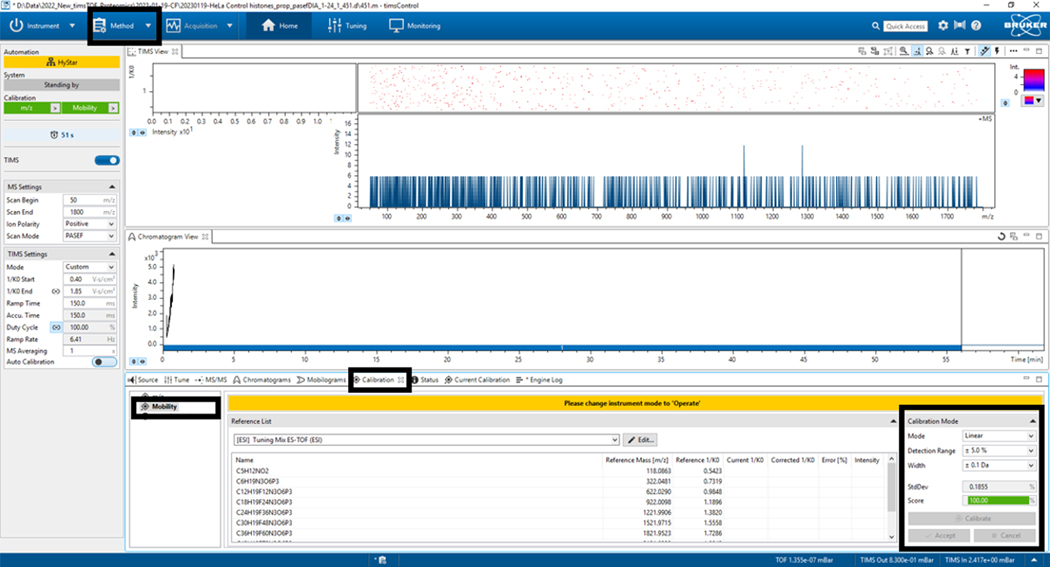
2.7. Go to the mobility tab and repeat the calibration process (Linear Mode, generally), detection range (+5%), width (0.1 Da), then click calibrate; when you get a score ≥ 98.5%, accept (Figure 5).
2.8. Go to the method and select the method to be used; for this example, Proteomic_/2023–01-19-CF/20230119-Hela Control histone_prep_pasefDIA_1–24_1_451.d/451.m-TimsControl was selected (Figure 5).
Figure 2: timsControl protocol steps 1–4.
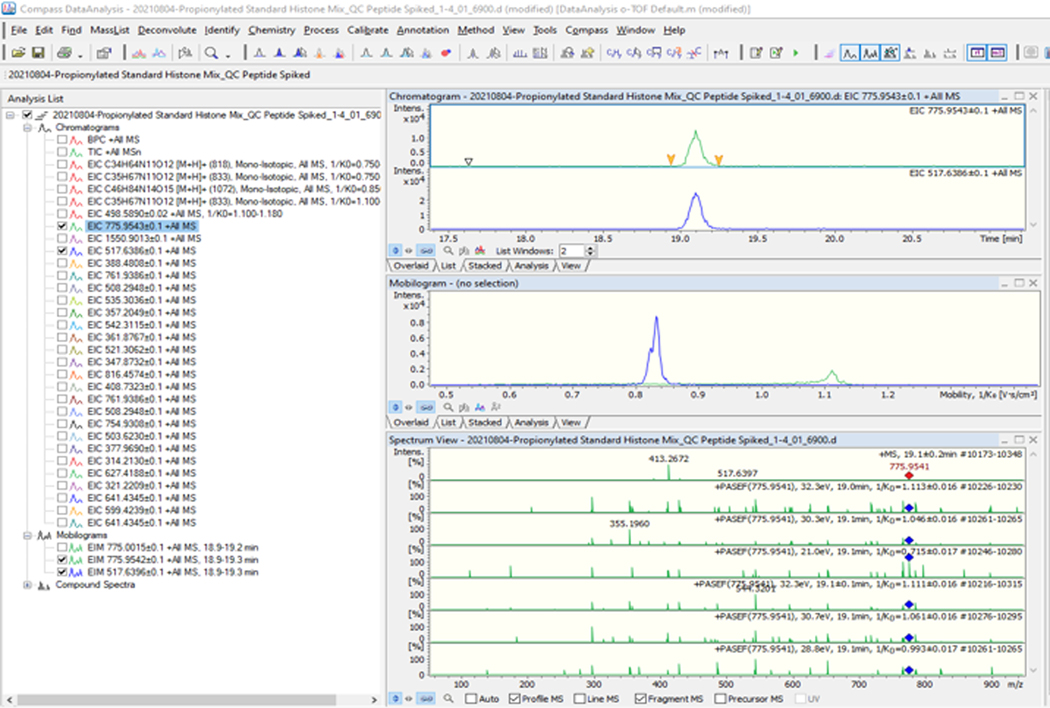
The figure shows the first four steps of the timsControl procedure. On the upper left-hand side, click the Instrument button to turn on and off the connection between the instrument and the software. Before executing any task, one must ensure that the software is in the operating mode. Finally, verify that the TIMS parameters are correct.
Figure 3: Source parameters.
In this case, Syringe Hamilton 500 μL was used only for TuneMix. Verify that the other parameters remain correct.
Figure 4: Mass-to-charge (m/z) calibration.
Select Calibrate until a score of 100% has been obtained in the bottom left panel Calibration Mode.
Figure 5: Mobility calibration.
Select Calibrate until a score of at least 98.5% has been obtained in the bottom left panel Calibration Mode.
3. LC-TIMS-PASEF-ToF MS/MS
3.1. Use the typical nESI operating conditions: 4500 V capillary voltage, 800 V endplate offset, 3.0 bar nebulizer pressure, 10.0 L/min dry gas, 200 °C dry heater, and 50 μL/min injection flow rate.
3.2. Use the typical MS settings: 6 eV collision energy, 1200 Vpp collision RF, 75 μs transfer time, 5 μs prepulse storage.
3.3. Determine the drift gas flow using the pressure difference from the entrance funnel P1 and the exit funnel P2. Parallel accumulation-serial fragmentation (PASEF) occurs in the TIMS cell, accumulating all precursor ions simultaneously rather than individually. Precursor ions are then released in narrow ion peaks versus the normally much wider peaks (about 50 times shorter), increasing the signal-to-noise ratio while still separating co-eluting peptides via mobility14.
-
3.4. Develop an LC-TIMS-ToF MS/MS method for analyzing proteolytic histone peptides. Couple a high-performance liquid chromatograph (HPLC) fitted with a C18 (300 Å, 5 μm, 4.6 mm x 250 mm) column with a commercial TIMS-TOF MS instrument with proprietary PASEF technology.
NOTE: This column size was determined to provide good separation at both high and low pH for peptide mixtures, based on previously published works15–17.
3.4.1. Set the injection volume to 20 μL (8 μg) of sample and a 0.4 mL/min flow rate.
3.4.2. Run a 60-min, non-linear LC gradient using water with 0.1% formic acid (Solvent A) and acetonitrile with 0.1% formic acid (Solvent B). Set the gradient: 10% B for 2.7 min, then to 20% B in 5.3 min, 28% B in 4 min, 35% B in another 18 min, to 40% B in 13 min, and 100% B in another 2 min. After holding 100% B for 5 min, lower the concentration to 10% B in 5 min and hold for the final 5 min.
3.5. Verify sample elution from the HPLC into the TIMS-TOF via nano-electrospray ionization (nESI) in positive ionization mode.
4. Data analysis
4.1. Identify the peptide sequences and modification sites.
4.1.1. Prepare a theoretical list of peptides using ProteinProspector [https://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msdigest] under the MS-digest tool.
4.1.1.1. Perform a theoretical digest while taking into consideration the conditions of the digest (enzyme used), types of PTMs being searched for (e.g., mono-, di-, or trimethylation), the size range of peptides being searched for, as well as mass detection range and the potential number of missed cleavages.
4.2. Manually analyze the acquired data based on theoretical peptides (Figure 6)12.
4.2.1. Search for the masses at several charge states (+1 to +4) for each theoretical peptide are searched for.
-
4.2.2. Following the initial identification of each m/z, select the peak and confirm the MS/MS using a theoretical list of fragmentation ions based on the peptide sequence, including PTMs.
NOTE: If the mobility of the identified peptide was known previously, this is also confirmed.
4.3. Calculate the relative abundances of various PTMs and report each modification as a percentage of the specified peptide sequence.
4.3.1. The relative abundance of each detected PTM is calculated using the following equation: Relative abundance = Area of PTM/Total area of unmodified and PTMs for a given peptide
Figure 6: Data processing: 20210804 Propionylated Standard Histones Mix_QC Peptide.
Before starting to process the data, make sure to prepare the theoretical list of possible charge states and their fragmentations (1550.9013; 775.9543; 517.6386; etc.) to extract those values from the base peak chromatogram (BPC +All MS). After extracting each peptide, make sure it looks like the analysis list shown in the figure. The peak 775.9543 was selected as an example. On the right side of the figure, three graphs are shown: the first corresponds to the chromatogram (intensity vs. time graph), the second to the mobilogram, and the third to the mass spectrum with PASEF fragmentation included.
REPRESENTATIVE RESULTS:
A bottom-up proteomic workflow (Figure 7) typically involves the following: extraction of the target protein(s) from a crude sample, followed by quantifying the concentration of the protein(s), and then fractionation, usually by gel electrophoresis or liquid chromatography. After fractionation, the proteins are digested using a proteolytic enzyme (often trypsin), and finally, mass spectrometric analysis of the resulting peptides and protein identification using an established database18. Sequence information is derived from precursor ions within the mass-to-charge (m/z) range indicated, which are subjected to collision-induced dissociation (CID), producing fragmentation patterns to be identified and sequenced using a database19 (Figure 8).
Figure 7: Typical bottom-up proteomics workflow.
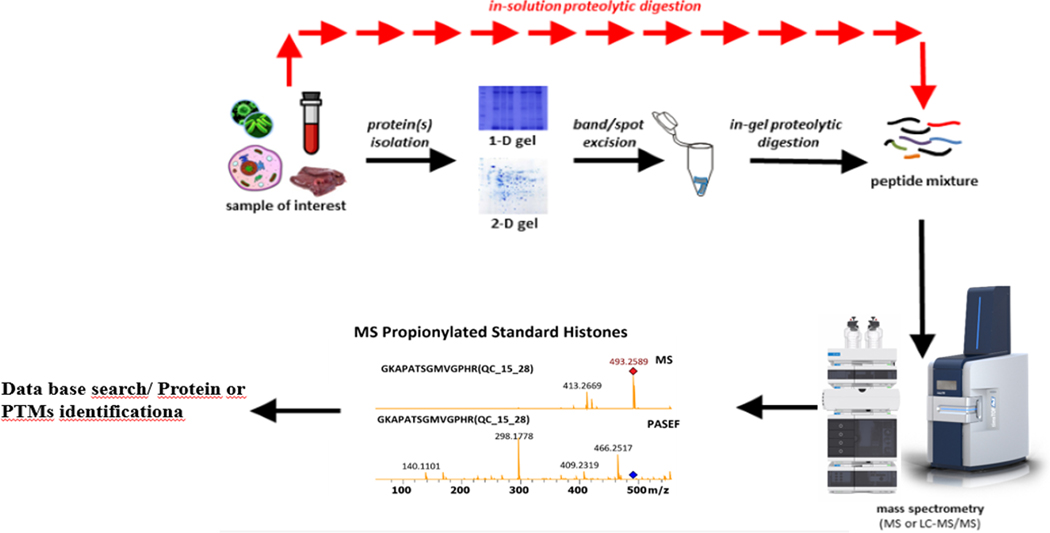
Step-by-step of the bottom-up procedures from sample preparation to identification9.
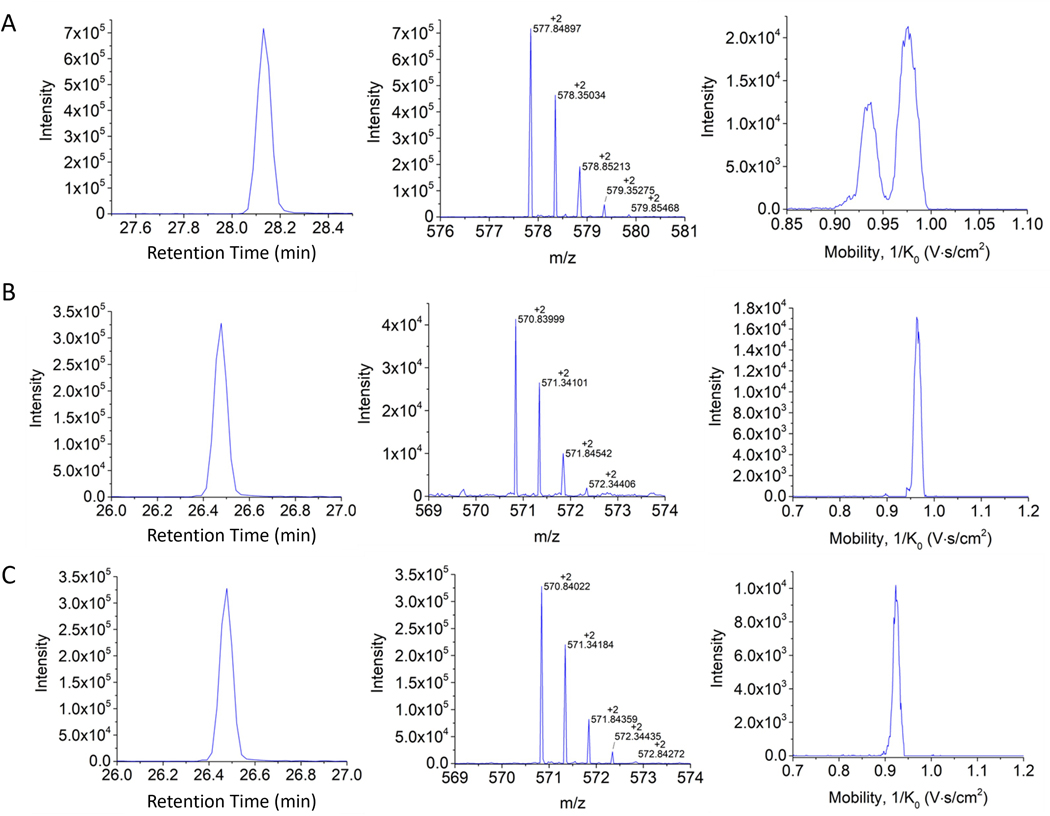
Figure 8: Retention time, isotopic pattern, and H3 18–26 mobility profiles.
(A) Unmodified propionylated, (B) K23Ac peptide propionylated in the other two positions, and (C) K18Ac propionylated in the other two positions. Notice the advantages of the mobility separation for the case of the structural isomers shown in panels B and C.
For this work, the principal goal was to develop and apply an LC-TIMS-PASEF-ToF MS/MS DDA method following the steps described previously in the protocol section. Determining the positionality of posttranslational modifications on isomeric and isobaric peptides has presented a particular challenge regarding identification and spectrum interpretation. In this study, recombinant human histone standards and HeLa S3 cells were used as samples.
Histone PTM analysis of human histone standards via ESI-TIMS-PASEF-ToF MS/MS yielded mid- to large-sized peptides (3–30 amino acids in length) detected with as many as 5 charges per peptide. The propionylation procedure was successful in producing longer, more informative peptides than those commonly produced by Tryptic digestion. Upon data analysis, peptides were identified in variously modified states. As an advantage, the TIMS-based method differentiated some positional isomeric peptides carrying the same PTMs. For example, two isomeric species may overlap in retention time and m/z; however, the two signals could be separated in the mobility domain (Figure 9).
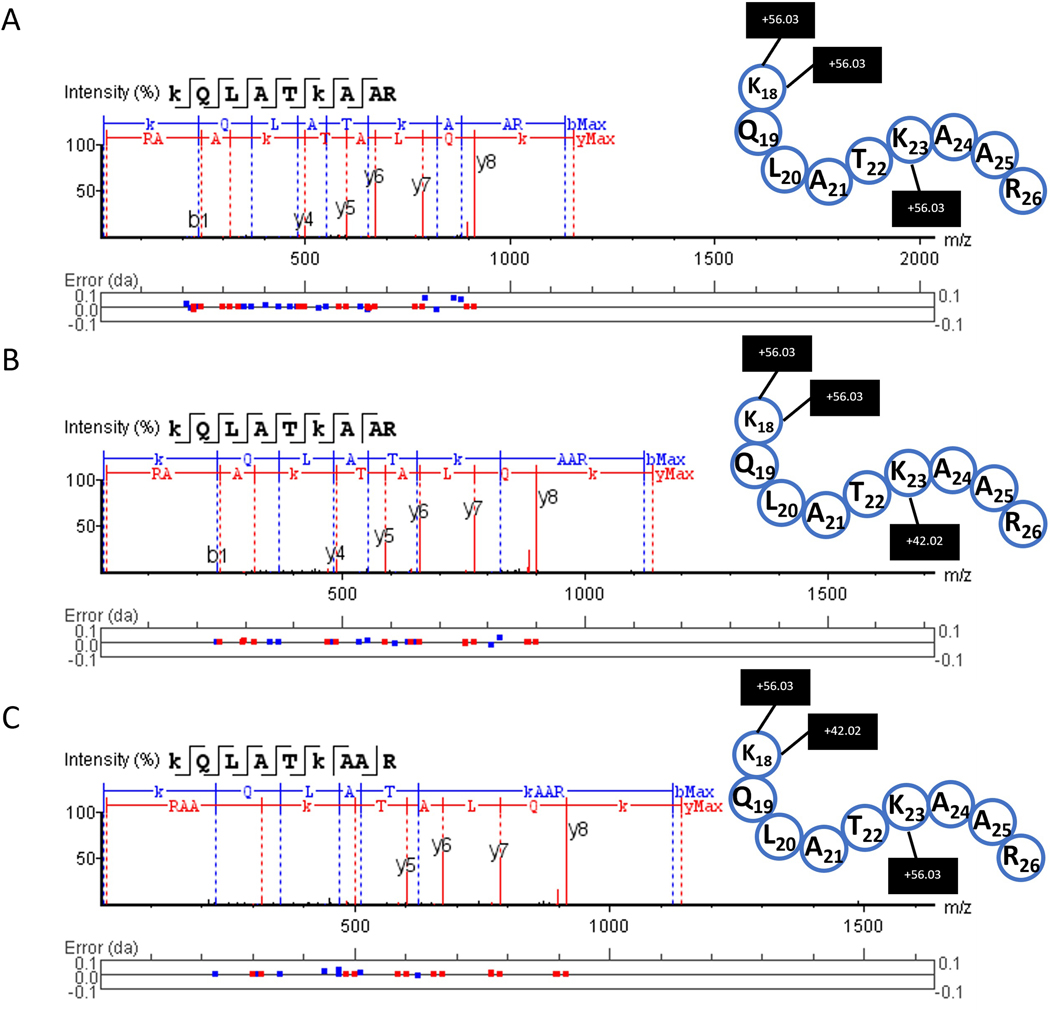
Figure 9: Example of MS/MS fragmentation peptide sequencing using PASEF.
Fragment spectra were obtained from proteomic analysis software for the H3 peptide with amino acid positions 18–26. (A) unmodified propionylated, (B) K23Ac peptide propionylated in the other two positions, and (C) K18Ac propionylated in the other two positions.
The corresponding fragmentation spectra for the peptides shown in Figure 9 were annotated by proteomic analysis software using the appropriate FASTA files. In Figure 9A, the unmodified peptide is seen with three propionyl (+56.03) groups (on the N-terminal, lysine 18, and lysine 23). In Figure 9B, the peptide is observed with an acetyl group (+42.02) on lysine 18 and two propionyl groups (one at the N-terminal and one on lysine 23). Finally, in Figure 9C the peptide is seen with an acetylation observed on lysine 23 and two propionyl groups (on the N-terminal and lysine 18). As published previously, the PASEF advantage could be used for increasing sequencing speed and sensitivity by targeting the same feature repeatedly9. This allows the user to obtain more structural information from biological samples. In this case, this is applied to the type and position of PTMs occurring on each histone.
Posttranslational modification analysis can also be represented visually as a sequence coverage plot, as seen in Figure 10. In Figure 10A, the histone H3 standard which has been propionylated prior to digestion, presents with longer peptides than would have otherwise resulted, denoted by the blue lines. Histones extracted from HeLa S3 cells were processed in the same fashion, as represented by Figure 10B. Several PTMs were indicated, including many different patterns at the same amino acid positions. This is to be expected from biological samples. Of note, the few gray lines in Figure 10B denote peptides that were identified ambiguously due to the lack of an MS2 resulting from the low intensity.
Figure 10: Example of a visual histone PTM analysis summary.

Results of observed peptides and PTMs from (A) an H3 standard and (B) H3 from HeLa S3 cells.
DISCUSSION:
Histones are basic proteins that regulate chromatin structure by interacting with DNA in the form of octamers consisting of the four core histones (two each of H2A, H2B, H3, and H4)20. Histones contain numerous lysine and arginine residues, which are readily modified, leading to extensive PTMs that alter the chromatin chemistry by influencing histone function or by binding to other cellular proteins21. PTMs can elicit biological responses by working in tandem, with specific groups of PTMs having been reported in several diseases, most notably, several types of cancer22.
When DNA damage is recognized at the cellular level, it is instantly followed by the action of a complex signaling cascade where lesions are marked, followed by the coordination of cell cycle progression and activation of the required repair pathways. In addition, DNA damage induces various modifications, such as acetyl and methyl adducts, which facilitate protein recruitment23. The great variety of PTMs that are involved in DNA lesions leads to the question of how these molecular mechanisms regulate their coexistence and what the functional importance of defending the integrity of the genome through an extremely complex integrated network is. For example, lysine 9 trimethylation of histone H3 (H3K9me3) has been linked to different pathologies in various diseases24. For reasons such as this, it is necessary to develop instrumental analytical methodologies that allow the complete characterization of these modifications at the cellular level23.
Analysis of the HeLa S3 histone extractions using manual data analysis software and proteomic analysis software revealed PTMs, including acetylation (+42.01 Da), methyl-propionylation (+70.04 Da), dimethylation (+28.03 Da), and trimethylation (+42.05) for several histone proteins. Additionally, the PASEF-based MS/MS method was able to differentiate some positional isomeric peptides carrying the same PTMs.
In the introduction, the advantages of coupling LC-TIMS-ToF MS/MS in the study of PTMs to show the recent developments of DDA using parallel accumulation in the mobility trap followed by sequential fragmentation and collision-induced dissociation are briefly described. The main idea is to establish a methodology that allows for the resolution of signals coming from different peptides and that, up until now, classical techniques have not been able to resolve. The derivatization process using propionic anhydride prevents the cleavage of lysine C-terminals by Trypsin, generating longer, more informative peptides. Peptides with the same m/z and retention time were able to be identified by their fragmentation patterns, but it was also seen that some of these species could be separated in the mobility domain using this LC-TIMS-PASEF-ToF MS/MS method.
To better understand this, Figure 8 represents three main characteristics of any molecule, thus allowing the identification of a compound, whether they are intact proteins, lipids, or peptides (in this case, histone H3 18–26), to name a few examples. These characteristics include the retention time (min) of a compound in the chromatographic column, the mass-charge ratio (m/z) of each compound, and the mobility (1/Ko) that these compounds present when they interact with the drift gas. In Figure 8A, the unmodified H3 peptide 18–26 is shown to have an RT of 28.15 min and that it presents two bands in its mobility spectrum, indicating that it has at least two conformations, a result that is suspected to be a result of the two lysines (18 and 23) that have been propionylated following the previously described protocol. The following spectra (Figure 8B,C) show the same peptide (H3 18–26) but varying the position of the acetylation group (42.02) between B, K18Ac and C, K23Ac. These two isomers (K18Ac and K23Ac) have been identified through the mobilogram, as they present with different spatial distributions, which results in different interactions with the gas in the TIMS cell. The importance of this method lies in the possibility of identifying and studying in more detail the different PTMs that have been associated with different diseases through, for example, DNA damage.
When fragmentation data are sparse, identifying a modification at a specific residue is challenging because two or more dissimilar modifications could occur simultaneously at (or near) the same residues and may be understood as a single modification25. This could be resolved by ensuring that the unmodified peptide has been identified, especially by using a standard to confirm or deny the presence of a single modification rather than multiple modifications (Table 1).
Table 1: Standard and HeLa S3 peptide LC-TIMS-ToF MS/MS characteristics.
Target and observed peptide list, including experimental properties (i.e., retention time, m/z, 1/Ko, and LC peak areas).
| Target Peptides | Histone Standard Mix | HeLa S3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid position | Peptide/mod | Charge State | m/z | Reporter Ions | RT (min) | m/z experimental | 1/k0 | LC peak area | RT (min) | m/z experimental | 1/k0 | LC peak area |
| H3_01_3_8 | TKQTAR | |||||||||||
| unmod | 1 | 816.4574 | 14.3 | 816.4552 | 1.314 | 92098 | 14.3 | 816.4555 | 1.338 | 14192 | ||
| 2 | 408.7323 | 659.38, 342.20 | 408.7326 | 0.746, .769 | 408.7325 | .771, .792 | ||||||
| K4me1 | 1 | 830.473 | ||||||||||
| 2 | 415.7402 | |||||||||||
| K4me2 | 1 | 788.4625 | ||||||||||
| 2 | 394.7349 | |||||||||||
| K4me3 | 1 | 802.4781 | ||||||||||
| 2 | 401.7427 | |||||||||||
| K4ac | 1 | 802.4417 | ||||||||||
| 2 | 401.7245 | |||||||||||
| H3_02_9_17 | KSTGGKAPR | |||||||||||
| unmod | 1 | 1069.6 | 16.7 | 1069.5965 | 1.585, 1.607, 1.616 | 171405 | 16.5 | 5083 | ||||
| 2 | 535.3036 | 829.45, 742.42, 641.37, 584.35, 527.33, 328.19, 429.23, 486.26, 543.28 | 535.3028 | 0.919 | 535.3033 | 0.921 | ||||||
| 3 | 357.2049 | |||||||||||
| K9me1 | 1 | 1083.6157 | 17.9 | 3056 | ||||||||
| 2 | 542.3115 | 542.3113 | 0.91 | |||||||||
| 3 | 361.8767 | |||||||||||
| K9me2 | 1 | 1041.6051 | 13.2 | 8285 | ||||||||
| 2 | 521.3062 | 829.45, 742.42, 641.37, 584.35, 527.33, 300.19, 401.24, 458.26, 515.28 | 521.3065 | 0.847 | ||||||||
| 3 | 347.8732 | |||||||||||
| K9me3 | 1 | 1055.6207 | 13.1 | 3468 | ||||||||
| 2 | 528.314 | 528.3144 | .856, .876 | |||||||||
| 3 | 352.5451 | |||||||||||
| K9ac | 1 | 1055.5844 | ||||||||||
| 2 | 528.2958 | 829.45, 742.42, 641.37, 584.35, 527.33, 314.17, 415.22, 472.24, 529.26 | ||||||||||
| 3 | 352.533 | |||||||||||
| K14ac | 1 | 1055.5844 | ||||||||||
| 2 | 528.2958 | 815.44, 728.41, 627.36, 570.34, 513.31, 328.19, 429.23, 486.26, 543.28 | ||||||||||
| 3 | 352.533 | |||||||||||
| K9me1K14ac | 1 | 1069.6 | ||||||||||
| 2 | 535.3036 | 815.44, 728.41, 627.36, 570.34, 513.31, 342.21, 443.25, 500.28, 557.30 | ||||||||||
| 3 | 357.2049 | |||||||||||
| K9me2K14ac | 1 | 1027.5894 | ||||||||||
| 2 | 514.2984 | |||||||||||
| 3 | 343.2013 | |||||||||||
| K9me3K14ac | 1 | 1041.6051 | ||||||||||
| 2 | 521.3062 | 815.44, 728.41, 627.36, 570.345, 513.31, 314.21, 415.26, 472.28, 529.30 | ||||||||||
| 3 | 347.8732 | |||||||||||
| K9acK14ac | 1 | 1041.5687 | ||||||||||
| 2 | 521.288 | |||||||||||
| 3 | 347.8611 | |||||||||||
| H3_03_18_26 | KQLATKAAR | |||||||||||
| unmod | 1 | 1154.6892 | 21.1 | 1154.6895 | 1.728 | 1749137 | 21 | 1154.6826 | 1.725 | 737276 | ||
| 2 | 577.8482 | 914.54, 786.48, 673.40, 602.36, 501.31, 369.21, 482.30, 553.33, 654.38 | 577.8482 | .918, .966 | 577.8478 | .919, .967 | ||||||
| 3 | 385.5679 | |||||||||||
| K23me1 | 1 | 1168.7048 | ||||||||||
| 2 | 584.856 | |||||||||||
| 3 | 390.2398 | |||||||||||
| K18me1 | 1 | 1168.7048 | ||||||||||
| 2 | 584.856 | |||||||||||
| 3 | 390.2398 | |||||||||||
| K18me1K23me1 | 1 | 1182.7205 | ||||||||||
| 2 | 591.8639 | |||||||||||
| 3 | 394.9117 | |||||||||||
| K18ac | 1 | 1140.6735 | ||||||||||
| 2 | 570.8404 | 914.54, 786.48, 673.40, 602.36, 501.31, 355.20, 468.28, 539.32, 640.37 | ||||||||||
| 3 | 380.896 | |||||||||||
| K23ac | 1 | 1140.6735 | 20.3 | 1140.672 | 1.709 | 204353 | ||||||
| 2 | 570.8404 | 900.53, 772.47, 659.38, 588.35, 487.30, 369.21, 482.30, 553.33, 654.38 | 570.8405 | .910, .957 | ||||||||
| 3 | 380.896 | |||||||||||
| K18acK23ac | 1 | 1126.6579 | ||||||||||
| 2 | 563.8326 | |||||||||||
| 3 | 376.2241 | |||||||||||
| H3_04_27_40 | KSAPATGGVKKPHR | |||||||||||
| unmod | 2 | 829.4728 | 17.7 | 829.4698 | 1.118 | 247198 | ||||||
| 3 | 553.3176 | 1417.79, 1330.76, 1259.72, 1162.67, 1091.63, 990.58, 933.56, 876.54, 777.47, 593.35, 328.19, 399.22, 496.28, 567.31, 668.36, 725.38, 782.40, 881.47 | 553.3176 | 0.891 | ||||||||
| 4 | 415.2401 | |||||||||||
| K36me1 | 2 | 836.4807 | ||||||||||
| 3 | 557.9895 | 1431.81, 1344.78, 1273.74, 1176.69, 1105.65, 1004.60, 947.58, 890.56, 791.49, 328.19, 399.22, 496.28, 567.31, 668.36, 725.38, 782.40, 881.47 | ||||||||||
| 4 | 418.744 | |||||||||||
| K27me1 | 2 | 836.4807 | 18.5 | 836.478 | 1.123, 1.141 | 7173 | ||||||
| 3 | 557.9895 | 1417.79, 1330.76, 1259.72, 1162.67, 1091.63, 990.58, 933.56, 876.54, 777.47, 342.21, 413.24, 510.30, 581.33, 682.38, 739.40, 796.42, 895.49 | 557.9894 | .890, .900, .909 | ||||||||
| 4 | 418.744 | |||||||||||
| K27me2 | 2 | 815.4754 | ||||||||||
| 3 | 543.986 | 1417.79, 1330.76, 1259.72, 1162.67, 1091.63, 990.58, 933.56, 876.54, 777.47, 300.19, 371.23, 468.28, 539.32, 640.37, 697.39, 754.41, 853.48 | ||||||||||
| 4 | 408.2413 | |||||||||||
| K36me2 | 2 | 815.4754 | ||||||||||
| 3 | 543.986 | 1389.80, 1302.76, 1231.73, 1134.67, 962.59, 905.57, 848.55, 749.48, 328.19, 399.22, 496.28, 496.28, 567.31, 668.36, 725.38, 782.40, 881.47 | ||||||||||
| 4 | 408.2413 | |||||||||||
| K27me3 | 2 | 822.4832 | 15.7 | 4310 | ||||||||
| 3 | 548.6579 | 1417.79, 1330.76, 1259.72, 1162.67, 1091.63, 990.58, 933.56, 876.54, 777.47, 314.21, 385.24, 482.30, 553.33, 654.38, 711.40, 768.43, 867.49 | 548.6568 | 0.896 | ||||||||
| 4 | 411.7452 | |||||||||||
| K36me3 | 2 | 822.4832 | ||||||||||
| 3 | 548.6579 | 1403.81, 1316.78, 1245.74, 1148.69, 1077.65, 976.61, 919.56, 862.56, 763.35, 328.19, 399.22, 496.28, 567.31, 668.36, 715.38, 782.40, 881.47 | ||||||||||
| 4 | 411.7452 | |||||||||||
| K27me2K36me1 | 2 | 822.4832 | ||||||||||
| 3 | 548.6579 | 1431.81, 1344.78, 1273.74, 1179.69, 1105.65, 1004.60, 947.58, 890.56, 791.49, 300.19, 371.23, 468.28, 539.32, 640.37, 697.39, 754.41, 853.62 | ||||||||||
| 4 | 411.7452 | |||||||||||
| K27me1K36me2 | 2 | 822.4832 | ||||||||||
| 3 | 548.6579 | 1389.80, 1302.76, 1231.73, 1134.67, 1063.64, 962.59, 905.55, 848.55, 749.48, 342.21, 413.24, 510.30, 581.33, 682.38, 739.40, 895.49 | ||||||||||
| 4 | 411.7452 | |||||||||||
| K27me1K36me1 | 2 | 843.4885 | ||||||||||
| 3 | 562.6614 | |||||||||||
| 4 | 422.2479 | |||||||||||
| K27me3K36me1 | 2 | 829.491 | ||||||||||
| 3 | 553.3298 | 1431.81, 1344.78, 1273.74, 1176.69, 1105.65, 1004.60, 947.58, 890.56, 791.49, 314.21, 385.24, 482.30, 553.33, 654.38, 711.40, 768.43, 867.49 | ||||||||||
| 4 | 415.2491 | |||||||||||
| K27me1K36me3 | 2 | 829.491 | ||||||||||
| 3 | 553.3298 | 1409.81, 1316.78, 1245.74, 1148.69, 1077.65, 976.61, 976.61, 919.58, 862.56, 763.49, 342.21, 413.24, 510.30, 581.33, 682.38, 739.40, 796.42, 896.49 | ||||||||||
| 4 | 415.2491 | |||||||||||
| K27me2K36me2 | 2 | 801.4779 | ||||||||||
| 3 | 534.6544 | |||||||||||
| 4 | 401.2426 | |||||||||||
| K27me3K36me2 | 2 | 808.4857 | ||||||||||
| 3 | 539.3263 | |||||||||||
| 4 | 404.7465 | |||||||||||
| K27me2K36me3 | 2 | 808.4857 | ||||||||||
| 3 | 539.3263 | |||||||||||
| 4 | 404.7465 | |||||||||||
| K27me3K36me3 | 2 | 815.4936 | ||||||||||
| 3 | 543.9981 | |||||||||||
| 4 | 408.2504 | |||||||||||
| K27ac | 2 | 822.465 | ||||||||||
| 3 | 548.6458 | |||||||||||
| 4 | 411.7361 | |||||||||||
| H3_04v3_27_40 | KSAPSTGGVKKPHR | |||||||||||
| unmod | 2 | 837.4703 | ||||||||||
| 3 | 558.6493 | |||||||||||
| 4 | 419.2388 | |||||||||||
| K36me1 | 2 | 844.4781 | ||||||||||
| 3 | 563.3212 | |||||||||||
| 4 | 422.7427 | |||||||||||
| K27me1 | 2 | 844.4781 | ||||||||||
| 3 | 563.3212 | |||||||||||
| 4 | 422.7427 | |||||||||||
| K27me2 | 2 | 823.4728 | ||||||||||
| 3 | 549.3176 | 1433.79, 1346.75, 1275.72, 1178.66, 1091.63, 990.58, 933.56, 876.54, 777.47, 593.35, 300.19, 371.23, 468.28, 555.28, 656.36, 713.38, 770.40, 869.47, 1053.59 | ||||||||||
| 4 | 412.2401 | |||||||||||
| K36me2 | 2 | 823.4728 | ||||||||||
| 3 | 549.3176 | 1405.79, 1318.76, 1247.72, 1150.67, 1063.64, 962.59, 905.57, 848.55, 749.48, 565.36, 328.19, 399.22, 496.28, 583.31, 684.36, 741.38, 798.40, 897.47, 1081.59 | ||||||||||
| 4 | 412.2401 | |||||||||||
| K27me3 | 2 | 830.4807 | ||||||||||
| 3 | 553.9895 | |||||||||||
| 4 | 415.744 | |||||||||||
| K36me3 | 2 | 830.4807 | ||||||||||
| 3 | 553.9895 | |||||||||||
| 4 | 415.744 | |||||||||||
| K27me2K36me1 | 2 | 830.4807 | ||||||||||
| 3 | 553.9895 | |||||||||||
| 4 | 415.744 | |||||||||||
| K27me1K36me2 | 2 | 830.4807 | ||||||||||
| 3 | 553.9895 | |||||||||||
| 4 | 415.744 | |||||||||||
| K27me1K36me1 | 2 | 851.4859 | ||||||||||
| 3 | 567.9931 | |||||||||||
| 4 | 426.2466 | |||||||||||
| K27me3K36me1 | 2 | 837.4885 | ||||||||||
| 3 | 558.6614 | |||||||||||
| 4 | 419.2479 | |||||||||||
| K27me1K36me3 | 2 | 837.4885 | ||||||||||
| 3 | 558.6614 | |||||||||||
| 4 | 419.2479 | |||||||||||
| K27me2K36me2 | 2 | 809.4754 | ||||||||||
| 3 | 539.986 | |||||||||||
| 4 | 405.2413 | |||||||||||
| K27me3K36me2 | 2 | 816.4832 | ||||||||||
| 3 | 544.6579 | |||||||||||
| 4 | 408.7452 | |||||||||||
| K27me2K36me3 | 2 | 816.4832 | ||||||||||
| 3 | 544.6579 | |||||||||||
| 4 | 408.7452 | |||||||||||
| K27me3K36me3 | 2 | 823.491 | ||||||||||
| 3 | 549.3298 | |||||||||||
| 4 | 412.2491 | |||||||||||
| K27ac | 2 | 830.4625 | ||||||||||
| 3 | 553.9774 | |||||||||||
| 4 | 415.7349 | |||||||||||
| H3_05_41_49 | YRPGTVALR | |||||||||||
| unmod | 1 | 1088.6211 | 18.9 | 2910608 | 18.8 | 232847 | ||||||
| 2 | 544.8142 | 544.8136 | 0.868 | 544.8148 | 0.87 | |||||||
| 3 | 363.5452 | |||||||||||
| Y41ph | 1 | 1168.5874 | ||||||||||
| 2 | 584.7973 | |||||||||||
| 3 | 390.2007 | |||||||||||
| H3_06_54_63 | YQKSTELLIR | |||||||||||
| unmod | 1 | 1362.7627 | 25.5 | 93690 | 25.4 | 63192 | ||||||
| 2 | 681.885 | 681.884 | 1.012 | 681.8846 | 1.013 | |||||||
| 3 | 454.9258 | |||||||||||
| 4 | 341.4461 | |||||||||||
| K56me1 | 1 | 1376.7784 | ||||||||||
| 2 | 688.8928 | |||||||||||
| 3 | 459.5976 | |||||||||||
| 4 | 344.9501 | |||||||||||
| K56me2 | 1 | 1334.7678 | ||||||||||
| 2 | 667.8875 | |||||||||||
| 3 | 445.5941 | |||||||||||
| 4 | 334.4474 | |||||||||||
| K56me3 | 1 | 1348.7835 | ||||||||||
| 2 | 674.8954 | |||||||||||
| 3 | 450.266 | |||||||||||
| 4 | 337.9513 | |||||||||||
| K56ac | 1 | 1348.7471 | ||||||||||
| 2 | 674.8772 | |||||||||||
| 3 | 450.2539 | |||||||||||
| 4 | 337.9422 | |||||||||||
| H3_07_73_83 | EIAQDFKTDLR | |||||||||||
| unmod | 1 | 1447.7427 | 29.5 | 65430 | 29.3 | 119424 | ||||||
| 2 | 724.375 | 724.3749 | 1.022 | 724.3744 | 1.022 | |||||||
| 3 | 483.2524 | |||||||||||
| 4 | 362.6911 | |||||||||||
| K79me1 | 1 | 1461.7584 | ||||||||||
| 2 | 731.3828 | |||||||||||
| 3 | 487.9243 | |||||||||||
| 4 | 366.195 | |||||||||||
| K79me2 | 1 | 1419.7478 | ||||||||||
| 2 | 710.3775 | |||||||||||
| 3 | 473.9208 | |||||||||||
| 4 | 355.6924 | |||||||||||
| K79me3 | 1 | 1433.7634 | ||||||||||
| 2 | 717.3854 | |||||||||||
| 3 | 478.5927 | |||||||||||
| 4 | 359.1963 | |||||||||||
| K79ac | 1 | 1433.7271 | ||||||||||
| 2 | 717.3672 | |||||||||||
| 3 | 478.5805 | |||||||||||
| 4 | 359.1872 | |||||||||||
| H3_08_117_128 | VTIMPKDIQLAR | |||||||||||
| unmod | 1 | 1496.8505 | 30.5 | 100245 | 30.3 | 55421 | ||||||
| 2 | 748.9289 | 748.9275 | 1.021, 1.077 | 748.9276 | 1.021, 1.077 | |||||||
| 3 | 499.6217 | |||||||||||
| 4 | 374.9681 | |||||||||||
| K122ac | 1 | 1482.8348 | ||||||||||
| 2 | 741.9211 | |||||||||||
| 3 | 494.9498 | |||||||||||
| 4 | 371.4642 | |||||||||||
| H3_09u_64_135 | unmod | |||||||||||
| KLPFQR | 1 | 900.5301 | 23.7 | 900.528 | 1.406, 1.428 | 1024986 | 23.6 | 900.5261 | 1.404, 1.429 | 239181 | ||
| 2 | 450.7687 | 660.38, 547.30, 450.25, 354.24, 451.29, 598.36 | 450.7693 | 0.808 | 450.7688 | 0.807 | ||||||
| 3 | 300.8482 | |||||||||||
| RIRGER | 1 | 842.4955 | 16.2 | 2722 | ||||||||
| 2 | 421.7514 | 421.7505 | .779, .794 | |||||||||
| 3 | 281.5033 | |||||||||||
| IRGERA | 1 | 757.4315 | ||||||||||
| 2 | 379.2194 | |||||||||||
| 3 | 253.1487 | |||||||||||
| H4_01_4_17 | GKGGKGLGKGGAKR | |||||||||||
| unmod | 1 | 1550.9013 | 19.6 | 419558 | 19.5 | 250690 | ||||||
| 2 | 775.9543 | 775.9539 | 1.122, 1.140 | 775.9536 | 1.121, 1.140 | |||||||
| 3 | 517.6386 | 517.6376 | .799, .829 | 517.6387 | .815, .832 | |||||||
| 4 | 388.4808 | |||||||||||
| K5ac | 1 | 1536.8856 | ||||||||||
| 2 | 768.9464 | 1253.73, 1196.71, 1139.69, 284.16, 341.18, 398.20 | ||||||||||
| 3 | 512.9667 | |||||||||||
| 4 | 384.9769 | |||||||||||
| K8ac | 1 | 1536.8856 | ||||||||||
| 2 | 768.9464 | 955.57, 898.46, 785.46, 728.44, 298.17, 355.20, 412.22, 582.32, 639.35, 752.43, 809.45 | ||||||||||
| 3 | 512.9667 | |||||||||||
| 4 | 384.9769 | |||||||||||
| K12ac | 1 | 1536.8856 | ||||||||||
| 2 | 768.9464 | 941.55, 884.53, 771.45, 714.43, 544.32, 487.30, 430.28, 359.24, 596.34.653.36, 766.45, 823.47 | ||||||||||
| 3 | 512.9667 | |||||||||||
| 4 | 384.9769 | |||||||||||
| K16ac | 1 | 1536.8856 | 19 | 32497 | ||||||||
| 2 | 768.9464 | 530.30, 473.28, 416.26, 345.22, 1007.59, 1121.63, 1192.67 | 768.9464 | 1.112, 1.128 | ||||||||
| 3 | 512.9667 | 512.9662 | .805, .824, .837 | |||||||||
| 4 | 384.9769 | |||||||||||
| K5acK8ac | 1 | 1522.87 | ||||||||||
| 2 | 761.9386 | 1239.72, 1182.70, 1125.67, 955.57, 989.55, 785.46, 728.44, 544.32, 487.30, 359.24, 284.16, 341.18, 398.20, 568.31, 625.33, 738.41, 795.44, 979.56, 1036.58, 1093.6, 1164.64 | ||||||||||
| 3 | 508.2948 | |||||||||||
| 4 | 381.4729 | |||||||||||
| K5acK12ac | 1 | 1522.87 | ||||||||||
| 2 | 761.9386 | 1239.72, 1182.70, 1125.67, 941.55, 884.53, 771.45, 714.43, 544.32, 487.30, 430.28, 359.24, 284.16, 341.18, 398.20, 582.32, 639.35, 752.43, 809.45, 979.56, 1036.48, 1093.6, 1164.64 | ||||||||||
| 3 | 508.2948 | |||||||||||
| 4 | 381.4729 | |||||||||||
| K5acK16ac | 1 | 1522.87 | ||||||||||
| 2 | 761.9386 | 1239.72, 1182.70, 1125.67, 941.55, 884.53, 771.45, 714.43, 530.30, 473.28, 416.26, 345.22, 284.16, 341.18, 398.20, 582.32, 639.35, 752.43, 809.45, 993.57, 1050.59, 1107.62, 1178.65 | ||||||||||
| 3 | 508.2948 | |||||||||||
| 4 | 381.4729 | |||||||||||
| K8acK12ac | 1 | 1522.87 | ||||||||||
| 2 | 761.9386 | 941.55, 884.53, 771.45, 714.43, 544.32, 487.30, 430.28, 359.24, 298.18, 355.20, 412.22, 582.32, 639.35, 752.43, 809.45, 979.56, 1036.58, 1093.6, 1164.64 | ||||||||||
| 3 | 508.2948 | |||||||||||
| 4 | 381.4729 | |||||||||||
| K8acK16ac | 1 | 1522.87 | ||||||||||
| 2 | 761.9386 | 941.55, 884.53, 771.45, 714.43, 530.30, 473.28, 416.26, 345.22, 298.18, 355.20, 412.22, 582.32, 639.35, 752.43, 809.45, 993.57, 1050.59, 1107.62, 1178.65 | ||||||||||
| 3 | 508.2948 | |||||||||||
| 4 | 381.4729 | |||||||||||
| K12acK16ac | 1 | 1522.87 | ||||||||||
| 2 | 761.9386 | 927.54, 870.52, 757.43, 700.41, 530.30, 473.28, 416.26, 345.22, 298.18, 355.20, 412.22, 596.34, 653.36, 766.45, 823.47, 993.57, 1050.59, 1107.62, 1178.65 | ||||||||||
| 3 | 508.2948 | |||||||||||
| 4 | 381.4729 | |||||||||||
| K5acK8acK12ac | 1 | 1508.8543 | ||||||||||
| 2 | 754.9308 | 941.55, 884.53, 771.45, 714.43, 544.32, 487.30, 430.28, 359.24, 341.18, 568.31, 625.33, 738.41, 965.54, 1022.56, 1079.58, 1150.62 | ||||||||||
| 3 | 503.623 | |||||||||||
| 4 | 377.969 | |||||||||||
| K5acK8acK16ac | 1 | 1508.8543 | ||||||||||
| 2 | 754.9308 | 941.55, 884.53, 771.44, 714.43, 568.31, 625.33, 738.41, 795.44 | ||||||||||
| 3 | 503.623 | |||||||||||
| 4 | 377.969 | |||||||||||
| K5acK12acK16ac | 1 | 1508.8543 | ||||||||||
| 2 | 754.9308 | 927.54, 870.52, 757.43, 700.41, 582.32, 639.35, 752.43, 809.45 | ||||||||||
| 3 | 503.623 | |||||||||||
| 4 | 377.969 | |||||||||||
| K8acK12acK16ac | 1 | 1508.8543 | ||||||||||
| 2 | 754.9308 | 1211.69, 1154.66, 1097.64, 927.54, 870.52, 757.43, 700.41, 298.18, 355.20, 412.22, 582.32, 639.35, 752.43, 809.45 | ||||||||||
| 3 | 503.623 | |||||||||||
| 4 | 377.969 | |||||||||||
| K5acK8acK12acK16ac | 1 | 1494.8387 | ||||||||||
| 2 | 747.923 | |||||||||||
| 3 | 498.9511 | |||||||||||
| 4 | 374.4651 | |||||||||||
| H4_02_20_23 | KVLR | |||||||||||
| unmod | 1 | 627.4188 | 20.8 | 627.4178 | 1.215 | 835125 | 20.8 | 627.4204 | 1.213 | 2079 | ||
| 2 | 314.213 | 314.2138 | .720, .735 | |||||||||
| K20me1 | 1 | 641.4345 | 22.3 | 641.4339 | 1.23 | 35976 | ||||||
| 2 | 321.2209 | |||||||||||
| K20me2 | 1 | 599.4239 | 16.5 | 599.4245 | 1.172 | 283059 | ||||||
| 2 | 300.2156 | 300.2157 | 0.727 | |||||||||
| K20me3 | 1 | 613.4395 | 16.5 | 3838 | ||||||||
| 2 | 307.2234 | 307.223 | .692, .707, .723 | |||||||||
| K20ac | 1 | 613.4032 | ||||||||||
| 2 | 307.2052 | |||||||||||
| H4_03_24_35 | DNIQGITKPAIR | |||||||||||
| unmod | 1 | 1437.806 | 21.3 | 548144 | 21.2 | 337235 | ||||||
| 2 | 719.4066 | 719.4054 | .992, 1.013, 1.026 | 719.4055 | .994, 1.018, 1.031 | |||||||
| 3 | 479.9402 | 479.9387 | 0.818 | 479.9401 | .778, .797, .816 | |||||||
| 4 | 360.207 | |||||||||||
| H4_04_40_45 | RGGVKR | |||||||||||
| unmod | 1 | 784.4788 | 14 | 83319 | 13.7 | 12188 | ||||||
| 2 | 392.743 | 392.7429 | 0.758 | 392.7437 | 0.749 | |||||||
| K44ac | 1 | 770.4631 | ||||||||||
| 2 | 385.7352 | |||||||||||
| H4_05_79_92 | KTVTAMDVVYALKR | |||||||||||
| unmod | 1 | 1762.9771 | 43.9 | 27760 | ||||||||
| 2 | 881.9922 | 881.9916 | 1.234 | |||||||||
| 3 | 588.3306 | |||||||||||
| 4 | 441.4997 | |||||||||||
| H4_06u_46_102 | unmod | |||||||||||
| ISGLIYEETR | 1 | 1236.647 | 30.4 | 1236.648 | 1.723 | 630923 | 30.2 | 1236.6429 | 1.715 | 101001 | ||
| 2 | 618.8272 | 618.8266 | 0.951 | 618.826 | 0.952 | |||||||
| 3 | 412.8872 | |||||||||||
| 4 | 309.9172 | |||||||||||
| GVLKVFLENVIR | 1 | 1498.8991 | ||||||||||
| 2 | 749.9532 | |||||||||||
| 3 | 500.3046 | |||||||||||
| 4 | 375.4802 | |||||||||||
| DAVTYTEHAKR | 1 | 1402.6961 | 18.4 | 39543 | 18.3 | 1119 | ||||||
| 2 | 701.8517 | 701.8503 | 1.008 | 701.8525 | 1.01 | |||||||
| 3 | 468.2369 | 468.2373 | 0.758 | |||||||||
| 4 | 351.4295 | |||||||||||
| TLYGFGG | 1 | 770.3719 | 27.4 | 770.3708 | 1.283, 1.348 | 276638 | 27.1 | 770.3721 | 1.284, 1.303 | 17029 | ||
| 2 | 385.6896 | |||||||||||
| 3 | 257.4622 | |||||||||||
| 4 | 193.3484 | |||||||||||
| HH2A_01m1_36_42 | KGNYAER | |||||||||||
| unmod | 1 | 949.4738 | 17.1 | 949.4704 | 1.453 | 543640 | 17 | 949.4721 | 1.45 | 7694 | ||
| 2 | 475.2405 | 475.2401 | .812, .841 | 475.241 | 0.84 | |||||||
| K36ac | 1 | 935.4581 | ||||||||||
| 2 | 468.2327 | |||||||||||
| HH2A_01m3_36_42 | KGNYSER | |||||||||||
| unmod | 1 | 965.4687 | ||||||||||
| 2 | 483.238 | |||||||||||
| K36ac | 1 | 951.453 | ||||||||||
| 2 | 476.2302 | |||||||||||
| HH2A_01oX_36_42 | KGHYAER | |||||||||||
| unmod | 1 | 972.4897 | ||||||||||
| 2 | 486.7485 | |||||||||||
| K36ac | 1 | 958.4741 | ||||||||||
| 2 | 479.7407 | |||||||||||
| HH2A_02m1_4_11 | GKQGGKAR | |||||||||||
| unmod | 1 | 969.5476 | 15.5 | 969.5441 | 1.486 | 565276 | 15.4 | 969.5492 | 1.475, 1.486 | 51680 | ||
| 2 | 485.2774 | 672.38, 544.32, 487.30, 430.28, 298.18, 426.23, 483.26, 540.28 | 485.2762 | 0.875 | 485.2779 | 0.873 | ||||||
| K5ac | 1 | 955.5319 | ||||||||||
| 2 | 478.2696 | 284.16, 412.22, 469.24, 526.26, 672.28, 544.32, 487.30, 430.28 | ||||||||||
| K9ac | 1 | 955.5319 | ||||||||||
| 2 | 478.2696 | 658.36, 530.30, 473.28, 416.26, 298.18, 426.23, 483.26, 540.28 | ||||||||||
| K5acK9ac | 1 | 941.5163 | ||||||||||
| 2 | 471.2618 | |||||||||||
| K9me1 | 1 | 983.5632 | ||||||||||
| 2 | 492.2853 | 686.40, 558.34, 501.32, 444.30, 298.18, 426.23, 483.26, 543.28 | ||||||||||
| K5me1 | 1 | 983.5632 | ||||||||||
| 2 | 492.2853 | 672.38, 544.32, 487.30, 430.28, 312.20, 440.25, 497.28, 554.30 | ||||||||||
| HH2A_02oJ_4_11 | GKQGGKVR | |||||||||||
| unmod | 1 | 997.5789 | ||||||||||
| 2 | 499.2931 | |||||||||||
| K5ac | 1 | 983.5632 | ||||||||||
| 2 | 492.2853 | 700.41, 572.35, 515.33, 458.31, 284.16, 412.22, 469.24, 526.26 | ||||||||||
| K9ac | 1 | 983.5632 | ||||||||||
| 2 | 492.2853 | 686.39, 558.34, 501.31, 444.29, 298.18, 426.23, 483.26, 543.28 | ||||||||||
| K5acK9ac | 1 | 969.5476 | ||||||||||
| 2 | 485.2774 | 686.39, 558.34, 501.31, 444.29, 284.16, 412.22, 469.24, 526.26 | ||||||||||
| K9me1 | 1 | 1011.5945 | ||||||||||
| 2 | 506.3009 | 714.43, 586.37, 529.35, 472.33, 298.18, 426.23, 483.26, 543.28 | ||||||||||
| K5me1 | 1 | 1011.5945 | ||||||||||
| 2 | 506.3009 | 700.41, 572.35, 515.33, 458.31, 312.20, 440.25, 497.28, 554.30 | ||||||||||
| HH2A_02oX_4_11 | GKTGGKAR | |||||||||||
| unmod | 1 | 942.5367 | ||||||||||
| 2 | 471.772 | |||||||||||
| K5ac | 1 | 928.521 | ||||||||||
| 2 | 464.7642 | 645.37, 544.32, 487.30, 430.28, 284.16, 385.21, 442.23, 499.25 | ||||||||||
| K9ac | 1 | 928.521 | ||||||||||
| 2 | 464.7642 | 631.35, 530.30, 473.28, 416.26, 298.18, 399.22, 456.25, 513.27 | ||||||||||
| K5acK9ac | 1 | 914.5054 | ||||||||||
| 2 | 457.7563 | |||||||||||
| K9me1 | 1 | 956.5523 | 16.6 | 11778 | ||||||||
| 2 | 478.7798 | 659.39, 558.34, 501.32, 444.30, 298.18, 399.22, 456.25, 513.27 | 478.779 | .843, .859 | ||||||||
| K5me1 | 1 | 956.5523 | ||||||||||
| 2 | 478.7798 | 645.37, 544.32, 487.30, 430.28, 312.20, 413.24, 470.27, 527.29 | ||||||||||
| HH2A_03m1_1_11 | SGRGKQGGKAR | |||||||||||
| unmod | 1 | 1269.7022 | 14 | 514 | ||||||||
| 2 | 635.3547 | 635.3556 | 0.95 | |||||||||
| 3 | 423.9056 | |||||||||||
| S1ac | 1 | 1311.7127 | ||||||||||
| 2 | 656.36 | |||||||||||
| 3 | 437.9091 | |||||||||||
| K5ac | 1 | 1255.6865 | ||||||||||
| 2 | 628.3469 | |||||||||||
| 3 | 419.2337 | |||||||||||
| HH2A_03oV_1_19 | AGGKAGKDSGKAKAKAVSR | |||||||||||
| unmod | 2 | 1062.0946 | ||||||||||
| 3 | 708.3988 | |||||||||||
| 4 | 531.5509 | |||||||||||
| 5 | 425.4422 | |||||||||||
| K4ac | 2 | 1055.0867 | ||||||||||
| 3 | 703.7269 | |||||||||||
| 4 | 528.047 | |||||||||||
| 5 | 422.6391 | |||||||||||
| K7ac | 2 | 1055.0867 | ||||||||||
| 3 | 703.7269 | |||||||||||
| 4 | 528.047 | |||||||||||
| 5 | 422.6391 | |||||||||||
| K11ac | 2 | 1055.0867 | ||||||||||
| 3 | 703.7269 | |||||||||||
| 4 | 528.047 | |||||||||||
| 5 | 422.6391 | |||||||||||
| K15ac | 2 | 1055.0867 | ||||||||||
| 3 | 703.7269 | |||||||||||
| 4 | 528.047 | |||||||||||
| 5 | 422.6391 | |||||||||||
| K4acK7ac | 2 | 1048.0789 | ||||||||||
| 3 | 699.055 | |||||||||||
| 4 | 524.5431 | |||||||||||
| 5 | 419.8359 | |||||||||||
| K4acK11ac | 2 | 1048.0789 | ||||||||||
| 3 | 699.055 | |||||||||||
| 4 | 524.5431 | |||||||||||
| 5 | 419.8359 | |||||||||||
| K4acK15ac | 2 | 1048.0789 | ||||||||||
| 3 | 699.055 | |||||||||||
| 4 | 524.5431 | |||||||||||
| 5 | 419.8359 | |||||||||||
| K7acK11ac | 2 | 1048.0789 | ||||||||||
| 3 | 699.055 | |||||||||||
| 4 | 524.5431 | |||||||||||
| 5 | 419.8359 | |||||||||||
| K7acK15ac | 2 | 1048.0789 | ||||||||||
| 3 | 699.055 | |||||||||||
| 4 | 524.5431 | |||||||||||
| 5 | 419.8359 | |||||||||||
| K11acK15ac | 2 | 1048.0789 | ||||||||||
| 3 | 699.055 | |||||||||||
| 4 | 524.5431 | |||||||||||
| 5 | 419.8359 | |||||||||||
| K7acK11acK15ac | 2 | 1041.0711 | ||||||||||
| 3 | 694.3831 | |||||||||||
| 4 | 521.0392 | |||||||||||
| 5 | 417.0328 | |||||||||||
| K4acK11acK15ac | 2 | 1041.0711 | ||||||||||
| 3 | 694.3831 | |||||||||||
| 4 | 521.0392 | |||||||||||
| 5 | 417.0328 | |||||||||||
| K4acK7acK15ac | 2 | 1041.0711 | ||||||||||
| 3 | 694.3831 | |||||||||||
| 4 | 521.0392 | |||||||||||
| 5 | 417.0328 | |||||||||||
| K4acK7acK11ac | 2 | 1041.0711 | ||||||||||
| 3 | 694.3831 | |||||||||||
| 4 | 521.0392 | |||||||||||
| 5 | 417.0328 | |||||||||||
| K4acK7acK11acK15ac | 2 | 1034.0633 | ||||||||||
| 3 | 689.7113 | |||||||||||
| 4 | 517.5353 | |||||||||||
| 5 | 414.2297 | |||||||||||
| HH2A_03oZ_1_19 | AGGKAGKDSGKAKTKAVSR | |||||||||||
| unmod | 2 | 1077.0998 | ||||||||||
| 3 | 718.4023 | |||||||||||
| 4 | 539.0536 | |||||||||||
| 5 | 431.4443 | |||||||||||
| K4ac | 2 | 1070.092 | ||||||||||
| 3 | 713.7304 | |||||||||||
| 4 | 535.5496 | |||||||||||
| 5 | 428.6412 | |||||||||||
| K7ac | 2 | 1070.092 | ||||||||||
| 3 | 713.7304 | |||||||||||
| 4 | 535.5496 | |||||||||||
| 5 | 428.6412 | |||||||||||
| K11ac | 2 | 1070.092 | ||||||||||
| 3 | 713.7304 | |||||||||||
| 4 | 535.5496 | |||||||||||
| 5 | 428.6412 | |||||||||||
| K15ac | 2 | 1070.092 | ||||||||||
| 3 | 713.7304 | |||||||||||
| 4 | 535.5496 | |||||||||||
| 5 | 428.6412 | |||||||||||
| K4acK7ac | 2 | 1063.0842 | ||||||||||
| 3 | 709.0586 | |||||||||||
| 4 | 532.0457 | |||||||||||
| 5 | 425.838 | |||||||||||
| K4acK11ac | 2 | 1063.0842 | ||||||||||
| 3 | 709.0586 | |||||||||||
| 4 | 532.0457 | |||||||||||
| 5 | 425.838 | |||||||||||
| K4acK15ac | 2 | 1063.0842 | ||||||||||
| 3 | 709.0586 | |||||||||||
| 4 | 532.0457 | |||||||||||
| 5 | 425.838 | |||||||||||
| K7acK11ac | 2 | 1063.0842 | ||||||||||
| 3 | 709.0586 | |||||||||||
| 4 | 532.0457 | |||||||||||
| 5 | 425.838 | |||||||||||
| K7acK15ac | 2 | 1063.0842 | ||||||||||
| 3 | 709.0586 | |||||||||||
| 4 | 532.0457 | |||||||||||
| 5 | 425.838 | |||||||||||
| K11acK15ac | 2 | 1063.0842 | ||||||||||
| 3 | 709.0586 | |||||||||||
| 4 | 532.0457 | |||||||||||
| 5 | 425.838 | |||||||||||
| K7acK11acK15ac | 2 | 1056.0764 | ||||||||||
| 3 | 704.3867 | |||||||||||
| 4 | 528.5418 | |||||||||||
| 5 | 423.0349 | |||||||||||
| K4acK11acK15ac | 2 | 1056.0764 | ||||||||||
| 3 | 704.3867 | |||||||||||
| 4 | 528.5418 | |||||||||||
| 5 | 423.0349 | |||||||||||
| K4acK7acK15ac | 2 | 1056.0764 | ||||||||||
| 3 | 704.3867 | |||||||||||
| 4 | 528.5418 | |||||||||||
| 5 | 423.0349 | |||||||||||
| K4acK7acK11ac | 2 | 1056.0764 | ||||||||||
| 3 | 704.3867 | |||||||||||
| 4 | 528.5418 | |||||||||||
| 5 | 423.0349 | |||||||||||
| K4acK7acK11acK15ac | 2 | 1049.0685 | ||||||||||
| 3 | 699.7148 | |||||||||||
| 4 | 525.0379 | |||||||||||
| 5 | 420.2318 | |||||||||||
| HH2A_04m1_12_17 | AKAKTR | |||||||||||
| unmod | 1 | 842.5094 | 17 | 842.5092 | 1.428 | 60315 | ||||||
| 2 | 421.7583 | 531.33, 460.29, 312.20, 383.23, 276.17 | 421.7597 | .772, .793 | ||||||||
| K13ac | 1 | 828.4938 | ||||||||||
| 2 | 414.7505 | |||||||||||
| K15ac | 1 | 828.4938 | ||||||||||
| 2 | 414.7505 | |||||||||||
| K13acK15ac | 1 | 814.4781 | ||||||||||
| 2 | 407.7427 | |||||||||||
| K15me1 | 1 | 856.5251 | ||||||||||
| 2 | 428.7662 | 531.33, 460.29, 326.21, 397.25 | ||||||||||
| K13me1 | 1 | 856.5251 | ||||||||||
| 2 | 428.7662 | 545.34, 474.31, 312.20, 383.23 | ||||||||||
| HH2A_04m3_12_17 | AKAKSR | |||||||||||
| unmod | 1 | 828.4938 | 16.9 | 828.49 | 1.411 | 1140381 | 16.8 | 828.4922 | 1.409 | 44577 | ||
| 2 | 414.7505 | 414.7502 | 0.788 | 414.7502 | 0.787 | |||||||
| K13ac | 1 | 814.4781 | ||||||||||
| 2 | 407.7427 | 517.31, 446.27, 298.18, 369.21 | ||||||||||
| K15ac | 1 | 814.4781 | ||||||||||
| 2 | 407.7427 | 503.29, 432.26, 312.20, 383.23 | ||||||||||
| K13acK15ac | 1 | 800.4625 | ||||||||||
| 2 | 400.7349 | |||||||||||
| K15me1 | 1 | 842.5094 | ||||||||||
| 2 | 421.7583 | 531.33, 460.29, 312.20, 383.23, 262.15 | ||||||||||
| K13me1 | 1 | 842.5094 | ||||||||||
| 2 | 421.7583 | 517.31, 446.27, 326.21, 397.25 | ||||||||||
| HH2A_05m1_72_77 | DNKKTR | |||||||||||
| unmod | 1 | 929.5051 | 16.2 | 929.5022 | 1.448 | 162881 | 16.1 | 929.5048 | 1.433, 1.446 | 18624 | ||
| 2 | 465.2562 | 465.2559 | 0.816 | 465.2567 | .805, .817 | |||||||
| K74ac | 1 | 915.4894 | ||||||||||
| 2 | 458.2483 | |||||||||||
| HH2A_06_82_88 | HLQLAIR | |||||||||||
| unmod | 1 | 906.5519 | 20.9 | 16685936 | 20.8 | 725428 | ||||||
| 2 | 453.7796 | 453.7796 | 0.823 | 453.7799 | 0.825 | |||||||
| 3 | 302.8555 | |||||||||||
| HH2A_07v_1_88 | unmod | |||||||||||
| H2AZ.AGGKAGKDSGKAKTKAVSR | 2 | 1077.0998 | ||||||||||
| 3 | 718.4023 | |||||||||||
| H2AY.SAKAGVIFPVGR | 1 | 1313.7576 | ||||||||||
| 2 | 657.3824 | |||||||||||
| 3 | 438.5907 | |||||||||||
| H2AX.GKTGGKAR | 1 | 942.5367 | ||||||||||
| 2 | 471.772 | |||||||||||
| 3 | 314.8504 | |||||||||||
| HH2A_08u_4_99 | unmod | |||||||||||
| HLQLAVR | 1 | 892.5363 | 19.8 | 19140 | ||||||||
| 2 | 446.7718 | 446.7721 | 0.815 | |||||||||
| 3 | 298.1836 | |||||||||||
| GGKKKSTKTSR | 1 | 1457.8322 | ||||||||||
| 2 | 729.4197 | |||||||||||
| 3 | 486.6156 | |||||||||||
| SGKKKMSKLSR | 1 | 1529.872 | ||||||||||
| 2 | 765.4396 | |||||||||||
| 3 | 510.6288 | |||||||||||
| IHRHLKTR | 1 | 1172.7011 | ||||||||||
| 2 | 586.8542 | |||||||||||
| 3 | 391.5719 | |||||||||||
| IHRHLKSR | 1 | 1158.6854 | ||||||||||
| 2 | 579.8463 | |||||||||||
| 3 | 386.9 | |||||||||||
| NDEELNKLLGR | 1 | 1412.738 | 27.5 | 12177 | ||||||||
| 2 | 706.8726 | 706.8716 | 0.996 | |||||||||
| 3 | 471.5842 | |||||||||||
| AGLQFPVGR | 1 | 1000.5574 | 23.4 | 1000.5555 | 1.444, 1.478, 1.510 | 14000051 | 23.2 | 1000.5538 | 1.475, 1.506 | 737499 | ||
| 2 | 500.7823 | 500.7814 | .836, .855 | 500.7828 | .837, .855 | |||||||
| 3 | 334.1907 | |||||||||||
| VHR | 1 | 467.2725 | 11.1 | 467.2726 | 1.009 | 68980 | ||||||
| 2 | 234.1362 | |||||||||||
| 3 | 156.4242 | |||||||||||
| LLR | 1 | 457.2832 | ||||||||||
| 2 | 229.1416 | |||||||||||
| 3 | 153.0944 | |||||||||||
| IHPELLAKKR | 1 | 1372.8311 | ||||||||||
| 2 | 686.9192 | |||||||||||
| 3 | 458.2819 | |||||||||||
| YIKKGHPKYR | 1 | 1513.8525 | ||||||||||
| 2 | 757.4299 | |||||||||||
| 3 | 505.289 | |||||||||||
| HH2B_01v_1_29 | unmod | |||||||||||
| 1C/1K.PEPAKSAPAPKKGSKKAVTKAQKKDGKKR | 3 | 1226.0397 | ||||||||||
| 4 | 919.7816 | |||||||||||
| 1H.PDPAKSAPAPKKGSKKAVTKAQKKDGKKR | 3 | 1221.3679 | ||||||||||
| 4 | 916.2777 | |||||||||||
| 2F.PDPAKSAPAPKKGSKKAVTKVQKKDGKKR | 3 | 1230.7116 | ||||||||||
| 4 | 923.2855 | |||||||||||
| 1B.PEPSKSAPAPKKGSKKAITKAQKKDGKKR | 3 | 1236.0433 | ||||||||||
| 4 | 927.2843 | |||||||||||
| 1N.PEPSKSAPAPKKGSKKAVTKAQKKDGKKR | 3 | 1231.3714 | ||||||||||
| 4 | 923.7804 | |||||||||||
| 1D.PEPTKSAPAPKKGSKKAVTKAQKKDGKKR | 3 | 1236.0433 | ||||||||||
| 4 | 927.2843 | |||||||||||
| 1M.PEPVKSAPVPKKGSKKAINKAQKKDGKKR | 3 | 1253.7309 | ||||||||||
| 4 | 940.55 | |||||||||||
| 1L.PELAKSAPAPKKGSKKAVTKAQKKDGKKR | 3 | 1231.3835 | ||||||||||
| 4 | 923.7895 | |||||||||||
| HH2B_02u_1_100 | unmod | |||||||||||
| LAHYNKR | ||||||||||||
| unmod | 1 | 1013.5527 | 19 | 815519 | 18.9 | 103031 | ||||||
| 2 | 507.28 | 507.2788 | 0.883 | 507.2803 | 0.883 | |||||||
| 3 | 338.5224 | |||||||||||
| LPHYNKR | 1 | 1039.5683 | ||||||||||
| 2 | 520.2878 | |||||||||||
| 3 | 347.1943 | |||||||||||
| EIQTAVR | 1 | 872.4836 | 18.6 | 872.4829 | 1.386 | 754301 | ||||||
| 2 | 436.7454 | 436.7443 | 0.797 | |||||||||
| 3 | 291.4994 | |||||||||||
| IAGEASR | 1 | 759.3995 | 17.5 | 759.3988 | 1.3 | 214007 | 17.4 | 759.3988 | 1.302 | 5806 | ||
| 2 | 380.2034 | 380.2035 | 0.746 | |||||||||
| 3 | 253.8047 | |||||||||||
| IASEASR | 1 | 789.4101 | ||||||||||
| 2 | 395.2087 | |||||||||||
| 3 | 263.8082 | |||||||||||
| LRTEVPRLPR | 1 | 1292.7797 | ||||||||||
| 2 | 646.8935 | |||||||||||
| 3 | 431.5981 | |||||||||||
To avoid excessive contamination or extractions of impure histones, it is important to check the quality of the reagents before use. For example, if the NIB buffer solution is stored and used in bulk, ensure that the solution is clear with no outward appearance of turbidity or abnormal presentation. Turbidity may be the result of bacterial growth, which would contaminate samples and could result in a mixture of histones and bacterial proteins. In addition, it is recommended to prepare fresh calibration curves for assays, such as the BCA or Bradford assay used to determine protein concentration, ensuring that the protein used for the calibration curve is not expired or degraded.
This method can be extended to other types of cells or organisms, for example, mosquitoes. In the case of whole or partial organisms, selecting an appropriate number of organisms is especially important to ensure that the final histone concentration is suitable for analysis.
Also, as a general guideline for mass spectrometer maintenance, the front end should be cleaned periodically to prevent buildup on the instrument and contamination between runs. This cleaning should include the curtain, orifice plate, and quadrupole, as required.
Generally, when an LC is used, it is necessary to take into consideration preparing fresh mobile phase(s) each week using MS-grade solvents. It is good practice to keep dedicated pipettes and glassware for mobile phase preparation and to purge the LC lines whenever new solutions are placed on the system. Guard and separation columns should usually be replaced every 100–200 injections and 500–1500 injections, respectively26. Be sure to inject blanks before and after running a batch of samples. If there are a large number of samples within a given batch, one may also consider running a blank at various intervals within the batch.
The protocol provides a PASEF-based DDA workflow for detecting histone PTMs and differentiation of isobaric and isomeric species based on ion mobility.
This protocol requires extensive sample preparation, and overall experimental sample preparation time should be accounted for. On average, the sample preparation protocol requires 2–3 business days to complete. Additionally, differences between laboratories and instrument versions can affect the overall sensitivity of the analysis.
Very few proteomic data analysis software have been deemed adequate for use in analyzing histones via bottom-up methods without manual adjustment or correction27–29. Results should (at least at first) be confirmed using manual analysis, which is also time-consuming. If analytical software is used, it should have MS/MS annotation capabilities, which are generally easy to confirm or reject.
It is also worth mentioning that it is impossible to separate isomers through mass spectrometry unless a TIMS cell is inserted and mobility values are used; for example, the positions of histone modifications can be determined using fragmentation patterns (PASEF).
ACKNOWLEDGMENTS:
The authors would like to acknowledge the initial support of Dr. Mario Gomez Hernandez during initial method developments.
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/65589.
DISCLOSURES:
Melvin A. Park and Matthew Willetts are employees of Bruker Daltonics Inc., the manufacturer of the timsTOF instrument.
REFERENCES:
- 1.Zhao Z, Shilatifard A. Epigenetic modifications of histones in cancer. Genome Biology. 20, 245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargen DF, Richmond TJ Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 389 (6648), 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Bentley GA, Lewit-Bentley A, Finch JT, Podjarny AD, Roth M. Crystal structure of the nucleosome core particle at 16 Å resolution. Journal of Molecular Biology. 176 (1), 55–75 (1984). [DOI] [PubMed] [Google Scholar]
- 4.Kornberg RD, Thomas JO Chromatin structure: Oligomers of the histones: The histones comprise an (F2A1)2(F3)2 tetramer, a different oligomer of F2A2 and F2B, and monomer of F1. Science. 184 (4139), 865–868 (1974). [DOI] [PubMed] [Google Scholar]
- 5.Borghini A, Cervelli T, Galli A, Andreassi MG DNA modifications in atherosclerosis: From the past to the future. Atherosclerosis. 230 (2), 202–209 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Jeanne Dit Fouque K. et al. Top- “Double-Down” mass spectrometry of histone H4 proteoforms: Tandem ultraviolet-photon and mobility/mass-selected electron capture dissociations. Analytical Chemistry. 94 (44), 15377–15385 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrence M, Daujat S, Schneider R. Lateral thinking: How histone modifications regulate gene expression. Trends in Genetics. 32 (1), 42–56 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Research. 21 (3), 381–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meier F. et al. Parallel accumulation-serial fragmentation (PASEF): Multiplying sequencing speed and sensitivity by synchronized scans in a trapped ion mobility device. Journal of Proteome Research. 14 (12), 5378–5387 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Chernushevich IV, Loboda AV, Thomson BA An introduction to quadrupole-time-of-flight mass spectrometry. Journal of Mass Spectrometry. 36 (8), 849–865 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Andrews GL, Simons BL, Young JB, Hawridge AM, Muddiman DC Performance characteristics of a new hybrid quadrupole time-of-flight tandem mass spectrometer (TripleTOF 5600). Analytical Chemistry. 83 (13), 5442–5446 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhanu NV, Sidoli S, Garcia B. A Workflow for ultra-rapid analysis of histone posttranslational modifications with direct-injection mass spectrometry. Bio-Protocol. 10 (18), e3756 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romson J, Emmer A. Mass calibration options for accurate electrospray ionization mass spectrometry. International Journal of Mass Spectrometry. 467, 11619 (2021). [Google Scholar]
- 14.Meier F, Park MA, Mann M. Trapped ion mobility spectrometry and parallel accumulation-serial fragmentation in proteomics. Molecular & Cellular Proteomics. 20, 100138 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.et al. Discovery of serum biomarkers for pancreatic adenocarcinoma using proteomic analysis. British Journal of Cancer. 103 (3), 391–400 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y. et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10S cells. Proteomics. 11 (10), 2019–2026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mertins P. et al. Integrated proteomic analysis of posttranslational modifications by serial enrichment. Nature Methods. 10 (7), 634–637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupree EJ, Jayathirtha M, Yorkie H, Mihasan M, Petre BA, Darie CC A critical review of bottom-up proteomics: The good, the bad, and the future of this field. Proteomes. 8 (3), 14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho CM et al. Bottom-up structural proteomics: cryoEM of protein complexes enriched from the cellular milieu. Nature Methods. 17 (1), 79–85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eickbush T, Moudrianakis E. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry. 17 (23), 4955–4964 (1978). [DOI] [PubMed] [Google Scholar]
- 21.Sadakierska-Chudy A, Filip M. A Comprehensive view of the epigenetic landscape. Part II: Histone posttranslational modification, nucleosome level, and chromatin regulation by ncRNAs. Neurotoxicity Research. 27 (2), 172–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan HT, Low J, Lim SG, Chung MCM Serum autoantibodies as biomarkers for early cancer detection. The FEBS Journal. 276 (23), 6880–6904 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Huang RX, Zhou PK DNA damage response pathways and targets for radiotherapy sensitization in cancer. Signal Transduction and Targeted Therapy. 5 (1), 60 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dantuma NP, van Attikum H. Spatiotemporal regulation of posttranslational modifications in the DNA damage response. The EMBO Journal. 35 (1), 6–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanner S, Pevzner PA, Bafna V. Unrestrictive identification of posttranslational modifications through peptide mass spectrometry. Nature Protocols. 1 (1), 67–72 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Dolan J. LC column problems everywhere 1. LCGC North America. 28 (9), 500–504 (2015). [Google Scholar]
- 27.Brusniak MK et al. An assessment of current bioinformatic solutions for analyzing LC-MS data acquired by selected reaction monitoring technology. Proteomics. 12 (8), 1176–1184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cham JA, Bianco L, Bessant C. Free computational resources for designing selected reaction monitoring transitions. Proteomics. 10 (6), 1106–1126 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Colangelo CM, Chung L, Bruce C, Cheung K. Review of software tools for design and analysis of large scale MRM proteomic datasets. Methods. 61, 287–298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]