Abstract
It is now appreciated that a group of lymphoid lineage cells, collectively called innate-like effector lymphocytes, have evolved to integrate information relayed by the innate sensory immune system about the state of the local tissue environment and to pass on this context to downstream effector innate and adaptive immune responses. Thereby, innate functions engrained into such innate-like lymphoid lineage cells during development can control the quality and magnitude of an immune response to a tissue-altering pathogen and facilitate the formation of memory engrams within the immune system. These goals are accomplished by the innate lymphoid cells that lack antigen-specific receptors, γδ T cell receptor (TCR)-expressing T cells, and several αβ TCR-expressing T cell subsets—such as natural killer T cells, mucosal-associated invariant T cells, et cetera. Whilst we briefly consider the commonalities in the origins and functions of these diverse lymphoid subsets to provide context, the primary topic of this review is to discuss how the semi-invariant natural killer T cells got this way in evolution through lineage commitment and onward ontogeny. What emerges from this discourse is the question: Has a “limbic immune system” emerged (screaming quietly in plain sight!) out of what has been dubbed “in-betweeners”?
Keywords: NKT cells, function, evolution, development
I. INTRODUCTION
The immune system is one constituent of the 10 physiologic systems of the body. It consists of two arms: the innate and adaptive immune systems. These two immune systems work in concert to sense alteration/s in the internal milieu—Claude Bernard’s milieu de l’intérieur or Walter Cannon’s homeostasis (see origins in1–3)—to process and integrate the perceived information, and actuate a response tailored to the altered state/s. Usually, the quick-to-act innate immune response is sufficient to return the host’s altered internal milieu back to normalcy; when it fails, however, the adaptive immune system is fully engaged. Even when the innate immune response is sufficient to ward off inflammatory intruders, an adaptive immune response is initiated so that memory engrams of the first encounter can be established in anticipation of future engagements. One end-product of the innate immune response is the display of fragments—breakdown products of chemical constituents derived from agents that incite alterations in the host’s internal milieu—on the surface of certain innate immune cells called dendritic cells (DCs). T lymphocytes recognize such fragmentary end-products—largely peptides, but also lipids, vitamin metabolites, and xenobiotics—to initiate an adaptive immune response. Whilst this initial recognition imparts specificity to the reaction, T lymphocytes require additional signals to elaborate a context-dependent response. This context is established by the nature of soluble mediators—cytokines and chemokines—secreted by the activated innate immune cells and the innate-like effector lymphocytes (ILELs).
The innate immune response occurs locally at barrier sites, such as the oral–respiratory, gastrointestinal, and urogenital mucosae, and the skin—the common ports of entry of noxious substances and microorganisms. The primary adaptive T cell response occurs in the local lymph nodes draining the various tissues of the host. The topologic barrier so imposed is solved by the migration of activated resident DCs from the barrier site to the local draining lymph node wherein processed antigens are presented to naïve, antigen-specific T cells. This DC-T cell interaction initiates an adaptive immune response, connecting innate immunity to the adaptive immune responses.
ILELs have evolved in vertebrates to bridge the innate and adaptive immune responses (Fig. 1). ILELs are a group of unconventional lymphoid lineage cells that either do not express antigen-specific receptors (innate lymphoid cells, ILCs) or express a defined repertoire of antigen-specific receptors generated through somatic recombination (innate-like T and B lymphocytes). Unlike conventional T and B cells, unconventional T & B lymphocytes exhibit innate-like recognition and functional characteristics (Fig. 2).4–7 Innate-like lymphocytes include both T [γδT and αβT cells such as natural killer T cells (NKT), mucosal-associated invariant T (MAIT) lymphocytes, and CD8αα-expressing intestinal intraepithelial lymphocytes] and B cells (B-1 and marginal zone B cells). Importantly, ILCs and innate-like T cells, like conventional T cells mediate type 1, type 2, and type 3 immunity (Fig. 2). As such, this group of immune cells can initiate, amplify, and fine-tune both the innate and adaptive immune responses.5,7,8 Engagement of multiple immune modules result in a context-dependent inflammatory response to maintain a stable milieu intérieur.9
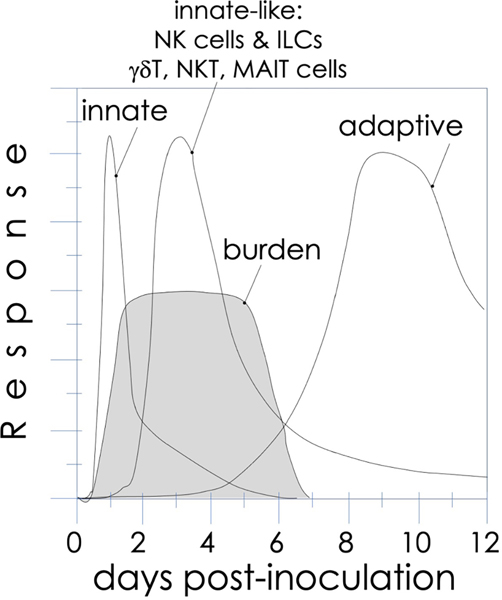
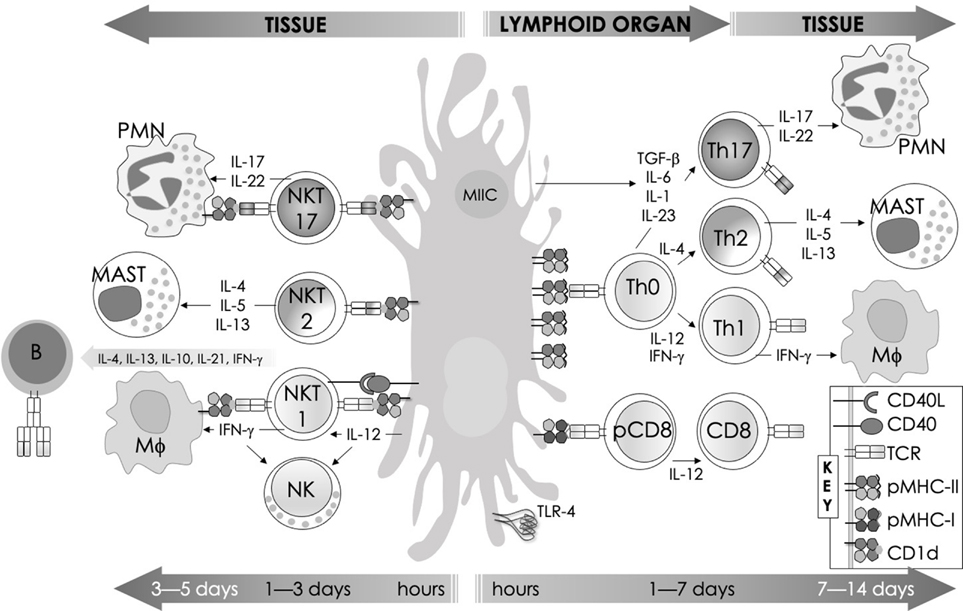
FIG. 1:
The kinetics of an immune response. The first peak signified by “innate” indicates the response of tissue macrophages and immature dendritic cells, which mature over the first few days following microbial (or antigenic) challenge to ferry antigen to local draining lymph nodes. Both also secrete innate cytokines and chemokines to recruit neutrophils. These sensory innate responses are integrated by innate-like effector lymphocytes, whose activities peak after the innate immune response, and relay context to the adaptive immune system T and B cells, impacting the antigenic burden.
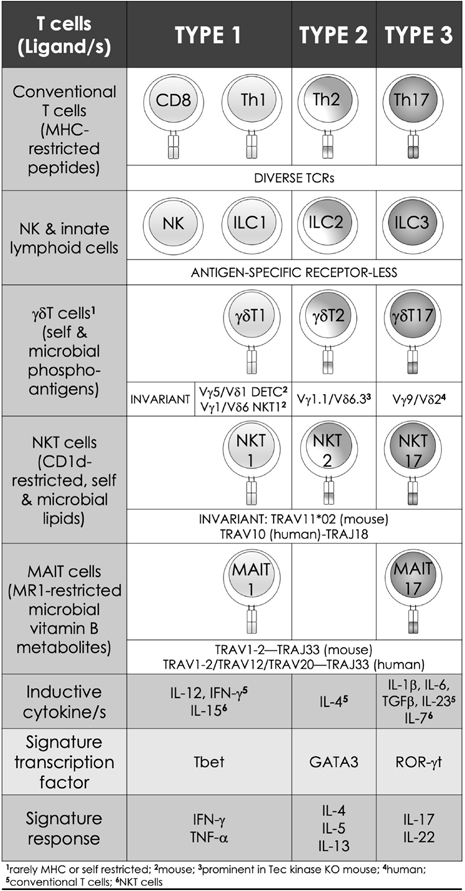
FIG. 2:
Features of innate-like effector lymphocytes mirror those of conventional T lymphocytes. Three types of effector immune responses are recognized: type 1, type 2, and type 3. They are characterized by the absence (ILCs) or presence of antigen-specific receptors on lymphoid lineage cells. Whilst conventional T cells express a diverse TCR (noted in the current IMGT nomenclature) repertoire, those of the innate-like effector lymphocytes express semi-invariant (NKT and MAIT cells) to invariant (γδT cells) TCRs. Type 1 effectors include both innate (NK) and adaptive cytotoxic (CD8+ T) cells and non-cytotoxic T helper (Th) 1 cells, as well as innate-like effector lymphocytes such as ILC1, NKT1, MAIT1, and γδT1 cells. They require IL-12 for induction, which is bolstered by IFN-γ. T-bet and related eomesodermin transcription factors control the differentiation of type 1 effector cells, which are essential for immunity against intracellular pathogens. Type 2 effector cells include Th2, ILC2, NKT2, and γδT cells. These cells are activated by IL-4 and require GATA3 for their effector differentiation. Type 2 effector cells secrete IL-4, IL-5, and IL-13, which are required for parasite expulsion. Their over activity results in allergic response and hypersensitivities. Type 3 effector cells include Th17, ILC3, NKT17, MAIT17, and γδT17 cells. These effector cells are induced by IL-6, TGF- β, IL-1 β, IL-23, and IL-7 (NKT17). RORγt is the lineage specific transcription factor. Type 3 effector cells secrete IL-17 and IL-22, which are important for immunity to extracellular bacteria and fungi.
NKT cells constitute one group of thymus-derived ILELs. NKT cell functions are controlled by CD1d restricted self and non-self lipid agonists.10 The majority of NKT cells (type I, invariant NKT cells—the protagonist of this review) express an invariant TCR α-chain generated by TRAV11*02 (mouse Vα14i) or TRAV10 (human Vα24i) to TRAJ18 (Jα18) rearrangement. The invariant α-chain largely pairs with mouse TRBV13–2*01 (Vβ8.2), TRBV29*02 (Vβ7), TRBV1 (Vβ2), or human TRBV25–1 (Vβ11) β-chain to form a functional semi-invariant TCR. At least in mice, a smaller NKT cell population—the type II NKT cells (reviewed elsewhere)—expresses a more diverse TCR repertoire and recognizes CD1d-restricted lipid antigens.11–16 Activated NKT cells rapidly secrete a variety of cytokines and chemokines, and upregulate costimulatory molecules. Similarly, MAIT cells and γδT cells also express a restricted TCR repertoire (Fig. 2).5,7 The ability of ILELs to respond quickly to specific agonists allows them to alert and steer downstream effector functions of myeloid and lymphoid cells. Hence, ILELs can control immunity to microbes and cancers, as well as autoimmune and inflammatory diseases.5,7,8,17–19
II. NKT CELL ACTIVATION AND FUNCTIONS—PARALLELS IN OTHER INNATE-LIKE EFFECTOR LYMPHOCYTES
“Nil ideo quoniam natumst in corpore ut uti possemus, sed quod natumst id procreat usum” (translated: “For nothing is born in the body in order that we may use it, but rather, having been born, it begets a use”20,21).
CD1d molecules, which bind to and present a variety of lipid ligands (Table 1 and references therein), control NKT cell functions.22 Pioneering studies from several research groups have shed light on how infectious agents or derived compounds activate NKT cells. Early studies demonstrated that DCs—by either directly presenting a microbial lipid/s or indirectly by inducing a self-lipid/s upon pathogen recognition—play an important role in NKT cell activation.22–25 Bacteria and certain fungi that biosynthesize glycosphinsphingolipids (GSLs) or glycoglycerolipids (GGLs), which when presented at the surface of DCs by CD1d, activate NKT cells.26–34 But, just as interestingly, microbes—both bacteria and viruses—that do not make NKT cell agonist/s themselves also activate NKT cells. This requires the expression of a pathogen-associated molecular pattern (PAMP) by the microbe and a PRR (pattern recognition receptor) by DCs.22,24,25 How both categories of microbes activate NKT cells is described below.
TABLE 1:
Synthetic, microbial, and self NKT cell agonists—structures and properties (adapted from refs 10,294)
| Lipid (class1) biologic vs. synthetic | Chain length2 | Structure | Response3 | Refs. |
|---|---|---|---|---|
| αGalCer (GSL) self |
C18 C24:l |
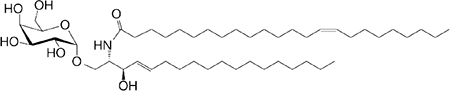
|
IFN-γ, IL-4 | 56 |
| Agel 9b (GSL) Agelas mauritianus |
C17 (C16-Me) phyto C24 |
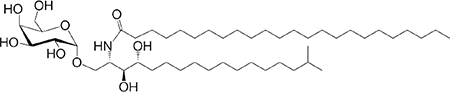
|
Anti-tumor; Agelas mauritianus | 286,287 |
| KRN7000 αGalCer (GSL) synthetic |
C18-phyto C26 |
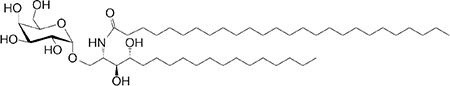
|
IFN-γ IL-4 and other cytokines | 236 |
| αCGal-Cer (GSL) synthetic |
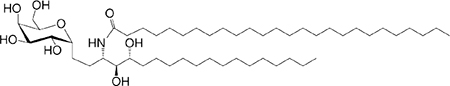
C18-phyto C26 |
|
Weak (mo4)-to-none (hu); IFN-γ | 288 |
| OCH (GSL) synthetic |
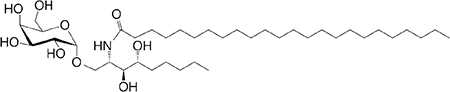
C9-phyto C24 |
|
Weak (mo)-to-none (hu); IL-4 (low-to-no IFN-γ) | 115 |
| C20-diene (GSL) synthetic |
C18-phyto C20:2 |
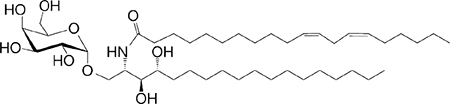
|
IL-4 (low-to-no IFN-γ) | 114 |
| αGalCer (GSL) Bacteroides fragilis |
CI7-C3OH C17 |
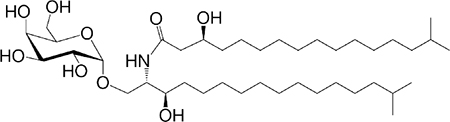
|
Stimulatory and inhibitory based on sphanganine branching | 27,33,289 |
| αGalU Cer (GSL) Sphingomonas spp. |
C18-phyto C14 |
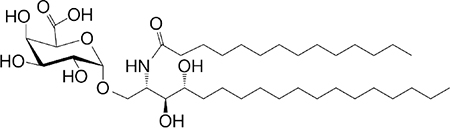
|
Weak IFN-γ | 30–32 |
| Asp B (GSL) Aspergillus fumigatus |
C20:2-C9 Me C16-C2 OH |
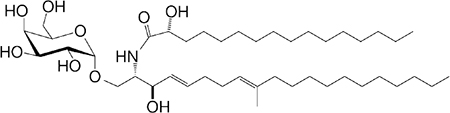
|
Weak agonist | 26 |
| αGlc-6-acyl-Chol Helicobacter pylori |
C14 |
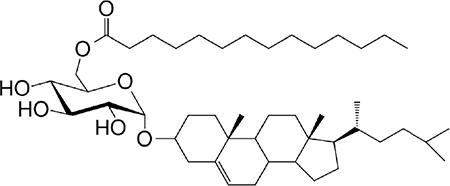
|
Activates a small NKT cell subset (mo) | 34 |
| βGalCer (GSL) self |
C18 C24:l |
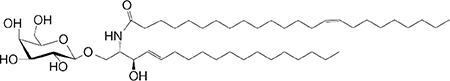
|
Weak agonist | 290,291 |
| iGb3 (GSL) self |
C18-C24 |
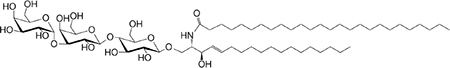
|
Weak (mo)-to-no (hu) response | 255 |
| αGal-DAG (GGL) Borrelia burgdorferi |
sn1-ClZA sn2-C16 |
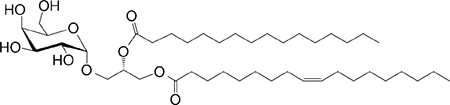
|
Weak agonist (mo) | 29 |
| αGlc-DAG (GGL) Streptococcus pneumoniae |
sn1-C18:1 sn2-C16 |
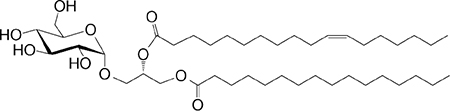
|
Weak agonist | 28 |
| PtdIno (GPL) self |
sn1-C18:1 sn2-C18:1 |
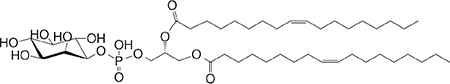
|
Week agonist (mo) | 59,292 |
| Plasma-logen (GPL) self |
sn1-C16 vinyl-ether sn2-lyso |
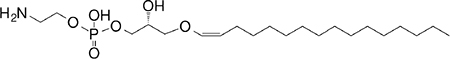
|
Agonist for positive selection (mo) | 293 |
| Lyso-PtdCho (GPL) self |
sn1-C16 sn2-lyso |
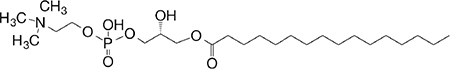
|
Weak (hu)-to-no (mo) GM-CSF (no IL-4, IFN-γ) response | 58 |
Agel, agelasphin; Asp B, asparamide B; Chol, cholesterol; DAG, diacylglycérol; GalCer, galactosylceramide; GalUCer, galacturonosylceramide; GlcCer, glucosylceramide; PtdCho, phosphatidylcholine; PtdIno, phosphatidylinositol; sn, stereo nomenclature for glycerolipids GGL, glycoglycerolipid; GPL, glycerophospholipid; GSL, glycosphingolipid
Sphingosine/phytosphingosine chain length indicated first and N-acyl chain length second
Agonist strength based on ref295
mo, mouse; hu, human
Our current understanding of NKT cell functions was gleaned from numerous in vitro and in vivo studies using the GSL, α-galactosylceramide (αGalCer, KRN7000), and its analogues (Table 1, and references therein). αGalCer/KRN7000—a potent NKT cell agonist with anti-cancer properties—is a natural compound isolated from the marine sponge, Agelas mauritianus, likely derived from its symbionts (Table 1, and references therein). NKT cell biology so gleaned came to question because marine sponges and associated symbionts are neither vertebrate parasites nor pathogens. Notwithstanding that, few Sphingomonas species, Bacteroides fragilis, Aspergillus fumigatus, and mammalian cells also generate αGalCers and/or related compounds (Table 1, and references therein). The prevalence of αGalCer and related compounds in nature lends credence to NKT cell biology learned from studies with αGalCer/KRN7000.
A significant advance in MAIT cell biology was the discovery that the restricting MHC-related 1 (MR1) assembled with small molecules such as the folic acid (vitamin B9) metabolite 6-formylpterin (6-FP), a photodegradation product of B9. Although eluted from MR1, 6-FP is not a MAIT cell agonist.35 Further experiments revealed that bacterial species containing the yet to be identified rib gene for enzymatic production of the riboflavin biosynthetic intermediate 5-amino-6-d-ribitylaminouracil (5-A-RU),36 generated a MAIT cell agonist/s, whereas those bacterial species lacking this rib gene did not.35 It was then found that the vitamin B metabolite 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil (5-OP-RU) activated MAIT cells when presented by MR1 molecules.35 Much of what we know about MAIT cells owes to this seminal discovery (reviewed in refs. 5,7,37,38).
Some mouse and human γδT cells are CD1d-restricted. They recognize the GPL cardiolipin, or the GSLs αGalCer or sulfatide.39–42 Some mouse γδT cell clones recognize either nonclassical MHC class Ib molecules—such as the ligand-free T10 and T2243–48—or MHC class II molecules without peptide specificity.49,50 The specialized mouse dendritic epidermal T cells express Vγ9–Vδ2 TCR, home to the skin, and recognize Skint-1 expressed by keratinocytes.46,48,51,52 Most human γδT cells express the Vγ9–Vδ2 TCR and are specific for self or bacterial alkylamines, amino-bis-phosphonates, and phosphoantigens, which are presented by butyrophilins in a highly unconventional manner independent of a conventional antigen-binding groove as seen in MHC and related molecules.46,48,51 Other human γδT cells recognize stress-induced MIC, ULBP4, or endothelial protein C receptor.53 The recognition principles of some but not all agonistic γδTCRs have been elucidated.7
A. Multiple Mechanisms Activate NKT Cells: Parallels with Other Innate Effector Cells
1. Agonist-Dominated NKT Cell Activation
αGalCer, when presented by CD1d molecules, activates NKT cells in a TCR-dependent manner. Likewise, B. fragilis–derived αGalCer also directly activates NKT cells (Table 1, and references therein). Such an activation mechanism is considered agonist-dominated activation (Fig. 3, left panel).
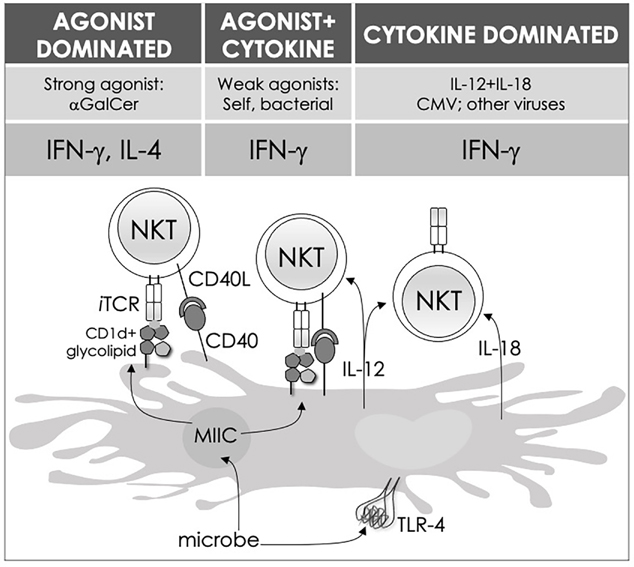
FIG. 3:
Three distinct strategies activate mouse NKT cells. Potent NKT cell agonists—such as αGalCer—directly activate NKT cells without the need for a second signal, in a TCR-signaling dominated fashion (left panel). Alternatively, microbes containing TLR ligands such as LPS activate NKT cells by inducing IL-12 production by DCs, which amplifies weak responses elicited upon recognition of CD1d bound with self-glycolipids by the NKT cell TCR. Several endogenous lipid agonists have been identified and characterized (see Table 1). Some microbes such as Sphingomonas capsulata, which are α-Proteobacteria, synthesize α-anomeric glycolipids for their cell walls. These glycolipids, when presented by CD1d, weakly activate NKT cells directly. In the presence of a second signal—generally a pro-inflammatory cytokine such as IL-12—such weak agonists strongly activate NKT cells (middle panel). Intriguingly, NKT cells can be activated solely by cytokines—mainly IL-12 plus IL-18—in a TCR-independent manner (right panel). Similar strategies activate MAIT cells as well. ILCs, owing to the lack of antigen-specific receptors, use cytokine-dependent mechanisms for initiation of an immune response. This diagram rendering the different strategies to NKT cell activation is an adaptation of past reviews10,101 and is based on works cited in the text.
2. Agonist-Dominated MAIT Cell Activation
MAIT cells share with NKT cells an agonist-dominated activation mechanism. As discussed above, MAIT cells recognize a limited number of agonistic small molecules. The best studied is the vitamin B metabolite 5-OP-RU.7,37,54 This agonist is biosynthesized by several Gram-positive and Gram-negative bacterial species that carry genes coding for enzymes that calalyze reactions in the riboflavin A biosynthetic pathway (reviewed in ref. 37).
3. Agonist- and Cytokine-Dominated NKT Cell Activation
Certain bacterial species generate weak NKT cell agonists; for example, Aspergilus fumigatus and Sphingomonas spp. biosynthesize asparamide B and α-galacturonosylceramide (αGalACer; Table 1; see references therein), respectively, which are αGalCer-related GSLs. Certain bacteria synthesise GGLs—e.g., α-galactosyldiacylglycerol by Borrelia burgdorferi and α-glucosyldiacylglycerol by Streptococcus pneumoniae—and cholesteryl-α-glycoside such cholesteryl-6-O-acyl α-glucoside by Helicobacter pylori (Table 1; see references therein). In the case of these weak agonists, NKT cell activation requires a second signal. Inflammatory cytokine/s elicited by DCs activated through their PRRs24,25,55 serve as a second signal. This form of NKT cell activation is considered agonist- and cytokine-dominated activation (Fig. 3, middle panel).
In many instances, the activation of antigen-presenting cells (APCs) by bacteria, fungi, and viruses induces self lipid biosynthesis and the generation of NKT cell self lipid agonists. Such self-agonists include glycerophospholipids (GPLs), mammalian αGalCer, and the GSL, iGb3 (isoglobotrihexosylceramide).31,56–59 NKT cells respond to CD1d molecules presenting self-lipids on host APCs in the presence of an inflammatory second signal.8,60 Notably, the inability of βGlcCer synthase-deficient cells to activate autoreactive NKT cell hybridomas61 and βGlcCer synthase-deficient thymocytes to support NKT cell development62 pointed to a cellular βGlcCer or a derived GSL as an endogenous mouse NKT cell agonist.61,62
As self-lipids are weak agonists, NKT cell activation requires a second signal such as IL-12 elicited upon activation via PRRs such as dectin-126,55 or toll-like receptor (TLR)-4.24,25 Type I interferon (IFN-I) produced by CpG-TLR9-activated DCs is also known to serve as a second signal for NKT cell activation by sialylated cellular glycolipids presented by CD1d molecules.63 Hence, this mode of activation is a variation of the agonist- and cytokine-dominated activation (Fig. 3, middle panel). Almost all virus infections and certain bacterial infections induce an IFN-I response.64–75 Hence, NKT cell activation by a self-agonist in conjunction with IFN-I notifies the host of a microbial infection even when an invading pathogen does not biosynthesize an NKT cell agonist.
4. Agonist- and Cytokine-Dominated MAIT Cell Activation
As virus and bacterial infections elicit innate cytokine responses, one study examined whether type I IFNs influence MAIT cell activation even when an agonist is presented. Akin to NKT cell activation, agonist-mediated human blood and liver MAIT cell activation was bolstered by the presence of type I IFNs.76,77
5. Cytokine-Dominated NKT Cell Activation
Under certain conditions, NKT cells are activated by the combined actions of IL-12 and IL-18, independent of a CD1d-restricted agonist.78–80 This mode of NKT cell activation, referred to as cytokine-dominated NKT cell activation (Fig. 3, right panel), is critical for immunity to cytomegalovirus80 and likely other viruses.
6. Cytokine-Dominated MAIT Cell Activation
MAIT cells were originally thought not to respond to viral infections.35,37 Subsequent studies demonstrated that viruses—influenza A, hepatitis, and dengue viruses—activated MAIT cells in a mechanism akin to cytokine-dominated activation of NKT cells, i.e., independent of the MAIT cell TCR. MAIT cell activation resulted in a MAIT1 cell-like activity characterized by IFN-γ release and granzyme B upregulation. MAIT cell activation by viruses depended on IL-18, which synergized with IL-12, IL-15 and type I IFNs.81,82 This form of activation protected mice from lethal influenza virus challenge.83
7. ILCs are Activated Solely by the Cytokine-Dominated Mechanism
ILCs lack antigen-specific B or T cell receptors. Their functions are induced in a cytokine-dominated mechanism. The inductive cytokines—which are quite similar to those that promote the differentiation of conventional CD4+ T cells (Fig. 2)—are produced by the local stromal cells and/or APCs, at the site of innate immune response to an injurious stimulus (reviewed in refs. 6,84).
Thus, ILELs have evolved common and distinct mechanisms to sense perturbations in milieu intérieur induced by microbial infections or derived products.
B. Self NKT Cell Agonist/s—One or Many and How are they Made?
Mouse and human NKT cells respond to several self-lipids, including the GSLs as well as the GPLs phosphatidylethanolamine, lysophosphatidylcholine, phosphatidylinositol, and plasmalogen (Table 1, and references therein). How cells biosynthesize GPLs is known, but how mouse and human APCs synthesize αGalCer or iGb3 is not. Current evidence suggests that humans lack the gene encoding iGb3 synthase (the α−3-galactosyltransferase-2), which adds the reducing galactosyl residue onto lactosylceramide to generate iGb3.85 Whilst mice carry the iGb3 synthase gene, it is expressed only in the dorsal root ganglion. Hence, iGb3 synthase deficiency does not have an impact on NKT cell development or function.86 These findings are further confounded by conflicting results from different groups as to the presence of iGb3 in mouse cells.85,87,88 An alternative route of iGb3 biosynthesis involving human A and mouse cis-AB enzyme has been suggested (schematized in Fig. 4A; see ref. 89) but remains unconfirmed.
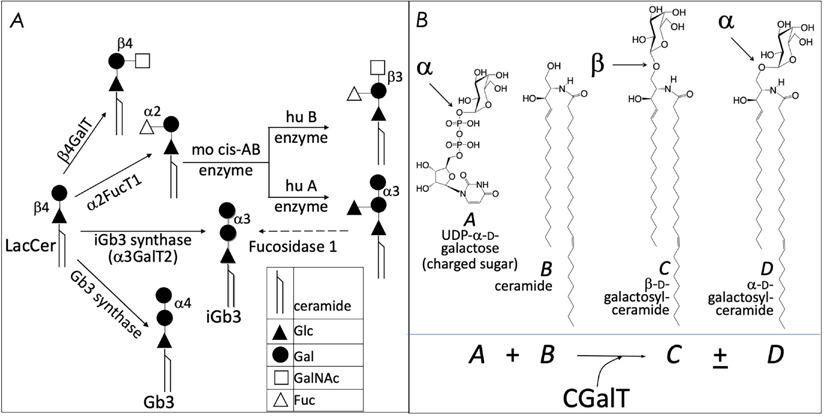
FIG. 4:
Alternate enzymatic strategies for the generation of cellular isoglobotriaosylceramide (A), and α GalCer (B). See text for details.
Another unsolved question pertains to the presence of αGalCer in mammalian cells even though αGalCer-like reactivity has been reported.56 Whilst β-glucosylceramide synthase (GCS)-deficient thymocytes—which are unable to biosynthesize βGclCer and higher order GSLs—do not promote NKT cell development,57 β-galactosylceramide synthase (CGT1)-deficient thymocytes do.61 These results suggested that a βGlcCer-derived GSL is a selecting ligand and implicated iGb3 as a potential ligand.57
β-glucosylceramide synthase-deficient cells do not activate NKT cell hybridomas.56,57,61 Hence, βGlcCer itself or a βGlcCer-derived GSL may be a self NKT cell agonist, which is supported by other evidence.90,91 One study found that αGalCer or an αGalCer-like compound could be a self NKT cell agonist, but a biosynthetic path to its generation is not established.56 One possible route to the biosynthesis of αGalCer and αGlcCer might respectively be CGT1 and CGS themselves. These enzymes may have an α-linkage retention property whereby the glycosytransferases use α-linked uridyldiphosphate-charged sugar donors to form β-linked monohexosylceramides by catalyzing α to β mutarotation prior to the condensation reaction. Nonetheless, the potential presence of αGlcCer/αGalCer in the absence of α-hexosylceramide synthase genes within mouse and human genomes poses a quandary.56,91 As a resolution, it is now recognized that the hexosylceramide synthases can retain the α-linkage of the charged sugar donor to generate α-linked monohexosylceramides (schematized in Fig. 4B). This notion is supported by biochemical evidence.92–95 Hence, in all likelihood, mammalian GCS and CGT have α-anomer retaining activity.
Cellular lipid content is stringently regulated. Hence, we hypothesized that the ER stress–induced unfolded protein response, which induces de novo lipid biosynthesis,96–99 may activate NKT cells.100,101 Recent findings have lent support to this hypothesis.102 Therefore, bacterial and viral infections, either by DC activation through TLR ligation or by ER stress induced by over-expression of virus-derived membrane glycoproteins, have the potential to alter cellular lipid content, both in variety and in concentration. Such changes in cellular self-lipid content can alter the quality and quantity of the ligands presented by CD1d.103 Because the NKT cell TCR exhibits co-operative ligand binding,104 it may recognize subtle qualitative and quantitative changes reflected in CD1d-associated lipid content. We hypothesize that sensitive antigen recognition at very early stages of infection, which may be initiated by very few infectious particles, is key to NKT cell function in vivo.
Thus, a model emerges in which CD1d detects alterations in cellular lipid content by virtue of its inherent affinity for such ligands. The presentation of self or non-self α-anomeric glycolipids on CD1d by DCs activates NKT cells. In vivo activation of NKT cells induces the prompt secretion of pro- and anti-inflammatory cytokines and chemokines, with the potential to jump start the immune system (Fig. 5). In doing so, the immune system is alerted by the entry of only a few intruders as occurs in natural infections.
FIG. 5:
Tissue distribution of innate-like effector lymphocytes. See text for details.
C. Steering Innate and Adaptive Immune Responses by Activated NKT Cells
NKT cells form immune synapse upon recognition of CD1d-bound lipid agonists displayed on APCs or planar membranes. The dynamics of the NKT cell TCR/ligand interactions determine the functional outcome.105 Positive co-operative interaction of NKT TCR with CD1d-lipid agonistic complexes enables NKT cells to sense and respond to subtle changes in cellular lipids.106 Upon activation, NKT cells rapidly polarize IFN-γ and lytic granules to the immune synapse to transmit an effector response.105,107,108 The transmission of effector molecules controls downstream innate and adaptive immune responses as described below.
Like the cells of the innate immune system (e.g., neutrophils, Mϕ, DCs, and NK cells), NKT cells respond within the first few hours upon agonist recognition in vivo and secrete inflammatory cytokines and chemokines (Fig. 5, left half). To accomplish this feat, NKT cells have epigenetically modified the Il4 and Ifng genes so as to constitutively express both transcripts.109–113 The nature of the activating NKT cell agonist determines the specific cytokine response (see Table 1). For example, the synthetic agonist αGalCer, within 30–90 minutes, elicits a wide variety of cytokines (Fig. 5, left half), while αGalCer variants containing different lipid chain length or unsaturation typically induce an IL-4 cytokine response.114,115 In contrast, αGalCer variants modified at distinct linkages induce an IFN-γ response (Table 1 and references therein). Thus, lipid agonists can be harnessed to direct immune responses that induce the desired therapeutic cytokine responses. This feature of αGalCer variants is further accentuated by the ability of activated NKT cells to transactivate cells of the innate and adaptive immune systems as narrated below (see Fig. 5).
DCs, especially CD8α+ DCs, which are major producers of IL-12,116 are critical for glycolipid agonist presentation, and subsequent NKT cell activation.117–123 In turn, activated NKT cells further stimulate and induce rapid maturity of the interacting DCs. The result is an upregulation of costimulatory molecules CD40, CD80, CD86, and several receptors necessary for antigen processing and presentation, such as DEC205 and MHC class II molecules, as well as an inflammatory cytokine (TNF-α and IL-12) response.23,124–127 Activated NKT cells also produce IFN-γ, which coupled with CD154 (CD40 ligand on NKT cells) and CD40 (on DCs) mediate reciprocal NKT-DC interactions.128,129 This NKT-DC dialogue directs various downstream immune responses, viz.: (1) IL-12 and IL-18 derived from NKT–DC interaction induce IFN-γ production by NK cells126; (2) NKT–DC crosstalk can induce IL-4, IL-6, IL-13, and IL-21 secretion, which cumulatively enhance B cell responses to protein antigens130–137; (3) NKT cell–DC interaction also sanctions DCs for antigen cross-presentation to CD8+ T cells,138–140 as well as the activation and differentiation of CD4 and CD8 T cells.124,139–141 Thus, the initial and sustained NKT cell–DC interactions amplify and steer downstream innate and adaptive immune responses.
Identification of the transactivation function of NKT cells through cytokine production and contact-dependent mechanisms (Fig. 5)8,142 led to the idea that these cells act as “cellular adjuvants.”8,143 NKT cell-DC crosstalk was recently shown to instruct inflammasome-independent IL-1β release during bacterial infection, especially by those pathogens that have devised ways to hide from the inflammasome pathway. Infected cells alter cellular lipid content, and NKT cells recognize such alterations when presented by CD1d. NKT cells, so activated, rapidly translocate the pre-existing intracellular pool of FasL to their cell surface, and transactivate Fas on APCs to induce the Fas-associated effector enzyme Caspase-8. Activated Caspase-8 cleave pro-IL-1β converting it to bioactive IL-1β, releasing it from infected APCs in a canonical Caspase-1 and non-canonical Caspase-11 inflammasome-independent manner.144 Caspase-8 also cleaves pore forming protein gasdermin D (GSDMD), resulting in GSDMD pore formation, K+ efflux and finally activation of the NLRP3-Caspase-1 inflammasome to further amplify IL-1β secretion.143 This two-cell back-up model for IL-1β release stymes pathogens leaving them no place to hide.145 Together, these findings provide a physiological context to NKT cell-APC crosstalk, bridging the topological barrier between the site of infection, sensory innate response, and T cell activation. Further, activated NKT cells provide context to downstream innate and adaptive effector responses.
D. Division of Labor by Innate-Like Effector Lymphocyte Subsets
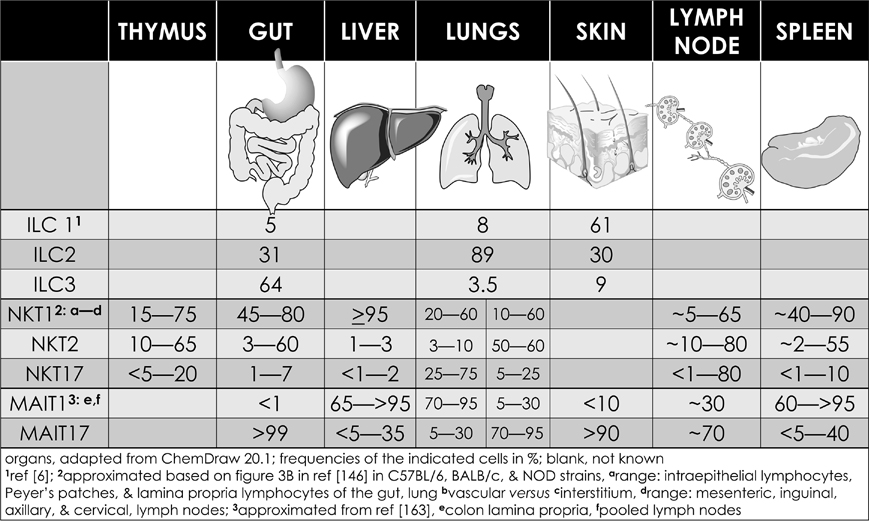
Innate-like effector lymphocyte activation results in rapid secretion of pro-inflammatory and regulatory cytokines and chemokines. Congruent with their ability to transactivate a range of innate and adaptive immune cells (see Fig. 5), ILELs control downstream immune responses. NKT cells are heterogeneous, consisting of at least three subsets—NKT1, NKT2, and NKT17146—as well as at least two induced subsets, NKT10 and NKTfh.133,134,136,147–150 Analagous subsets are also well characterized in ILCs and MAIT cells (although a MAIT2 cell subset remains as yet unidentified), and are emerging in γδT cells as well.5,7,151 Akin to conventional CD4+ T cell subsets, ILEL subsets are defined by subset-specific transcription factors and archetypal cytokine responses that, respectively, mediate type 1, type 2, and type 3 immunity (see Figs. 2 and 5). Each subset is represented in different proportions in different mouse strains, and varies within each strain at barrier tissues (Fig. 6).146–148,151–154
FIG. 6:
The effector functions of mouse NKT cells. The interactions between the invariant natural killer (NKT) cell receptor and its cognate antigen, as well as interactions between co-stimulatory molecules CD28 and CD40 and their cognate ligands CD80/86 (B7.1/7.2) and CD40L, respectively, activate NKT cells. Activated NKT cells participate in crosstalk with members of the innate and the adaptive immune systems by deploying cytokine and chemokine messengers. Upon activation in vivo, NKT cells rapidly secrete a variety of cytokines and chemokines, which influence the polarization of CD4+ T cells toward T helper (Th)1 or Th2 cells as well as the differentiation of precursor CD8+ T cells to effector lymphocytes, and B cells to antibody-secreting plasma cells. Some of these mediators facilitate the recruitment, activation, and differentiation of macrophages and DCs, which results in the production of IL-12 and possibly other factors. IL-12, in turn, stimulates NK cells to secrete IFN-γ. Thus, activated NKT cells have the potential to enhance as well as temper the immune response. This schematic rendition of NKT cell effector functions is an adaptation of past reviews8,10,101,296 and is based on works cited in the text.
TYPE 1 innate-like effector lymphocytes include NK cells, ILC1, γδT1 cells, NKT1 cells, and MAIT1 cells (Figs. 2 and 6). NKT1 cells activation results in a Th1-like cytokine response. Most NKT cells are of the NKT1 subset in C57BL/6 mouse thymus, spleen, and liver. NKT1 cell differentiation is dependent on T-bet (Tbx21) and IL-15, and to a limited extent, GATA3.146,152,153,155–157 Unlike in DP thymocytes, NKT cell lineage-specific depletion of HDAC3 selectively impairs NKT1 cell development as a result of diminished autophagy158–160—a cytoplasmic recycling process essential to both T and NKT cell development—as well as reduced expression of nutrient receptors GLUT1, CD71, and CD98.161 NKT1 cells mediate the anti-tumor effect of αGalCer,162 potentially through secreted IFN-γ and TNF-α.
TYPE 2 innate like effector lymphocytes include ILC2, γδT2 cells, and NKT2 cells (Figs. 2 and 6). MAIT2 cells have not been identified.5,151,163 γδT2 cells are enriched in Tec kinase deficient mice.164 NKT2 cells express the CD4 co-receptor, are enriched in mouse lungs and intestine, and their activation results in a type 2 cytokine and chemokine response (Fig. 6). This type 2 response may underlie airway hyperresponsiveness,152,153,165–168 a prominent feature of asthma, as a result of macrophage, eosinophil, neutrophil, and lymphocyte recruitment into the lungs and consequent tissue damage.165 Coincidently, NKT2 cells are overrepresented in BALB/c mice that are sensitive to airway hyperresponsiveness.153
TYPE 3 innate like effector lymphocytes include ILC3, γδT17 cells, NKT17 cells, and MAIT17 cells (Figs. 2 and 6). NKT17 cells are prominent in the peripheral lymph nodes, skin, and lungs, but scant in the liver and spleen (Fig. 6).169–171 NKT17 cells need IL-7 but not IL-15 for survival,152,172 and their development requires mTORC2 signaling and the transcription factors Runx1 and NKAP.173–177 Indeed, Runx1 depletion in NKT cells results in decreased IL-7Rα, BATF, and c-Maf expression, and increased Lef and Bcl11b expression.175 The role of NKAP in NKT17 cell development is not understood; it appears not to require mTOR, IL-7, or TGF-β signaling.173 NKT17 cells constitutively express RORγt,169 promote airway neutrophilia upon challenge with synthetic glycolipids or LPS, and rapidly secrete IL-17A in response to some bacterial infections.28,169,178 They may mediate ozone-induced airway hypersensitivity,179 experimental autoimmune encephalomyelitis,178 and acute hepatitis in mice.180
MOUSE NKT10 cells, the PLZF-independent subset,147 are found in low frequency in naive mice and in human peripheral blood. Upon re-activation, NKT10 cells that formerly responded to αGalCer in vivo secrete IL-10,148 which is thought to sustain immune-privileged sites. This NKT cell subset may also regulate TREG cell activities in adipose tissues.147
MOUSE NKT follicular helper (NKTFH) cells can provide cognate (when B cells present lipid antigens) or non-cognate (when B cells present protein antigens) help to B cells and regulate antibody responses.133,134,136,149,150 In vivo αGalCer induces a subset of NKT cells to take on a T follicular helper T (TFH) cell-like phenotype, forming NKTFH cells.150,181,182 NKTFH cells express CXCR5, PD1, BTLA, ICOS, and Bcl6. Their development depends on the same factors that induce TFH development.181 NKTFH cells–produced IL-21 rapidly drives germinal center formation, yielding appreciable levels of antigen-specific IgG antibodies.135,181,182 Nonetheless, NKTFH cell–induced antibody responses are ephemeral and inferior to TFH cell-driven antibody responses.135,181,182 NKTFH cells may control antibody responses against Borrelia hermsii, Streptococcus pneumoniae, and Plasmodium falciparum—all of which are human pathogens.135,181,182 NKTFH and TFH cells can synergize to promote robust antigen-specific antibody responses, thus highlighting the utility of αGalCer as a vaccine adjuvant.150
HUMAN NKT cell responses are as diverse in humans as in mice,183 yet subsets similar to those in mice were only recently described. Previous reports have demonstrated that human CD4+ and double-negative (DN) NKT cell subsets are functionally different. Activated human CD4+ NKT cells, in a pathologic role, accumulate in the lungs of chronic asthmatic patients; these NKT cells produce IL-4 and IL-13.184 Hence, human CD4+ NKT cells mirror the mouse NKT2 cell subset. Moreover, activated DN NKT cells produce IFN-γ and TNF-α and, thereby, mirror mouse NKT1 cells. Also, in the presence of inflammatory signals, both CD4+ and DN human NKT cell subsets induce perforin production. DN NKT cells also upregulate NKG2D expression, which combined with perforin may mediate cytotoxicity against infected cells and cancer cells.185,186 These human NKT cell functions parallel those of mouse NKT1 cells. Moreover, the presence of an NKT17-like subset is indicated by the finding that activated human NKT cells also secrete IL-17.183 Consistent with these findings, human liver perfusates contain both NKT, MAIT, and γδT cell subsets. Whilst NKT17, MAIT17, and γδT1 cells dominate the liver, proportionately low frequencies of NKT1, MAIT1, and γδT17 also are present.187 These subsets produce characteristic cytokines as do the mouse counterparts: the type 3 subsets produce IL-17, and the type I subsets, IFN-γ.187
Overall, ILELs divide labor among three distinct subsets. Specifically, global and single cell transcriptome analyses demonstrated that thymic NKT1, NKT2, and NKT17 cells represented definite subsets.187–190 While not formalized, human NKT cells potentially mirror mouse NKT cell subsets,187 hence requiring further enquiry. Differentiation of NKT cell subsets is influenced by tissue environment. Hence, NKT17 differentiation requires mammalian target of rapamycin (mTOR) complex-2177 and is inhibited by Tet enzymes that modify DNA 5-methylcytosine by controlling the expression of the Tbet and ThPOK transcription factors.191 A separate study using mice with monoclonal NKT cell populations showed that tissue homing pattern, but not NKT TCR avidity, dictated NKT cell subset differentiation.171 By contrast, other studies have found a significant role for TCR avidity in subset development.192–194
III. HOW NKT CELLS GOT THE WAY THEY ARE
A. Evolutionary Origins
“…the struggle against diseases, and especially infectious diseases, has been a very important evolutionary agent and that some of its results have been unlike those of the struggle for life…” (195 within collected papers in genetics by J.B.S. Haldane196). Nonetheless, “There is a natural and irrepressible tendency in the human mind to penetrate the mystery of the beginning of things…. But it is plainly denied to finite understandings to ascend to the very beginning, and to comprehend the nature of the operation of the First Cause of anything…. The ablest endeavours here to penetrate the beginning of things do but carry us, when most successful, a few steps nearer that beginning, and then leave us on the verge of a boundless ocean of the unknown truth, dividing the secondary or subordinate phenomena in the chain of causation from the great First Cause.”197
Sir Owen, a vocal critic of Darwin’s evolutionary theory by natural selection,198 may have been a bit too short-sighted with the above critique of the Origin. The exciting new biology of the 21st century knows no bounds. The essay below provides a glimpse on how one could “penetrate the mystery of the beginning of things” eons past!
Recent advances in whole genome sequencing (WGS) have ushered in comparative vertebrate genomics unveiling molecular signatures of selection upon genes that control various biologic functions, including immune responses. Pathobionts can apply inordinate selection pressure and significantly impact the evolution of immune response genes and cells. As early-life symbionts can impact health, microbial ecology may also control the evolution of immune response genes and cells.
The NKT cell and MAIT cell TCR engage their ligands with conserved germline-encoded residues in the complementarity-determining regions.199 Hence, phylogenetic analyses of the genes and gene segments that code for these molecules can inform their origins and whether selective pressures may have maintained them over evolutionary scales of time. Such analyses indicate that the MR1 and Cd1d genes, as well as the TRAV1 and TRAJ33, and TRAV10 and TRAJ18 gene segments that code for MAIT cell and NKT cell TCRs, respectively, evolved in a common ancestor/s of the therian mammals, before the metatherian (viviparous marsupial mammals) and eutherian (viviparous placental mammals) split but after the monotreme (oviparous mammals) therian split some 170 million years ago. Further, MR1, and TRAV1 and TRAJ33 genes may have co-evolved with each other. Curiously, however, not all eutherian (placental) mammals carry MR1 and Cd1d genes, as well as the TRAV1 and TRAJ33, and TRAV10 and TRAJ18 gene segments. Whilst the MR1 gene has evolved slowly, in comparison, Cd1 genes have evolved faster and diversified since their appearance in eutherian mammals. These findings are suggestive of selection pressure, presumably by riboflavin-producing microbial pathogens, in the maintenance of MR1, and TRAV1 and TRAJ33 genes.200
The above study did not find evidence for NKT cells or MAIT cells in other vertebrate animals.200 Nonetheless, there are reports of innate-like T cells in Xenopus laevis that look and function like NKT and MAIT cells.201,202 Notably, the search for MR1, and TRAV1 and TRAJ33 genes led to the discovery of co-evolving MR1-like, and TRAV1- and TRAJ33-like genes, TRAV41 and TRAJ38 genes in rabbits.200 Rabbits along with other lagomorphs, carnivores, and armadillos lack the MR1 gene. Thus, there are other potential ways to generate MAIT cells, lending credence to the idea that the Xenopus innate-like T cells may be another such example. This conjecture awaits formal evidence.
Cd1 and MR1 genes are relatively young when compared to the classical antigen-presenting MHC genes, Cd1 being older than MR1, the latter evolving around the same time as Cd1d. Cd1 was an amniote innovation, evolved in Mesozoic reptiles and sustained in the extant reptiles, such as the anapsid green anole lizard Anolis carolinensis and synapsid Siamese crocodile Crocodylus siamensis and Chinese alligator Alligator sinensis.203 Cd1 genes diversified in mammals, wherein evolved Cd1d.200 The reptilian Cd1 gene lacks orthology to avian or mammalian Cd1 genes,203 indicating that Cd1 genes may have risen several times in amniote evolution. Alternatively, Cd1 genes may have rapidly evolved and considerably diverged from the reptilian form within extinct synapsid and mammal-like reptiles prior to equilibration within eutherian species. The absence of Cd1 in oviparous monotremes such as platypuses, and the presence of a CD1d-like gene in a few metatherians such as the opossum, supports the alternative view to Cd1 evolution.
A phylogenetic analysis of TRAV10 (encoding the human Vα24 gene segment) or TRAV11 (encoding the mouse Vα14 gene segment) and TRAJ18 (encoding the Jα18 gene segment) indicated that gene elements related to TRAV10/11 and TRAJ18 are found solely in placental mammals.200 This discovery indicates that NKT cells are a eutherian contraption. As the host gut microbiota influences NKT cell terminal functional differentiation,27,33 and NKT cells can control gut microbial ecology,204 we predict that placental development, sudden perinatal exposure to maternal and environmental microbiota, and lactation may have contributed to the evolution of CD1d-restricted NKT cells. The same might be the case for MAIT cells200 and other ILELs, as they all recognize and/or respond to microbes, both symbiotic and pathogenic.
γδT cells are found in jawed (gnathan) vertebrates including cartilaginous (Chondrichthyes: rays, sharks, and skates) and bony (Osteichthyes: salmon, tuna, and zebra fish) fishes.51 γδT cells in these species use V(D)J recombined TCRs. Similarly, γδT-like cells are found in jawless (agnathan, e.g., lampreys and hagfish) vertebrates that use leucine-rich-repeat–based variable lymphocyte receptors, which also randomly rearrange in a Rag1/Rag2-independent mechanism to generate antigen-specific receptors.205,206 At the present time, it is not known whether these γδT cells are innate-like γδT or γδNKT cells.
Another means to penetrate the evolutionary beginnings of ILELs is the use of a comparative transcriptomics approach as previously described. This approach was applied to uncover the origins of NK cells—the natural cytotoxic ILCs (Fig. 2)—and T helper-like ILCs. Lineage specific inducers (soluble mediators), surface markers, and transcription factors trace back some 500 Mya, suggesting the origins of NK cells and ILC2s may have been conceived at the dawn of vertebrate evolution. ILC1 and ILC3 appeared more recently, but a clear point in time for their origins needs further study.207 Single cell transcriptomics190 shall illuminate their origins and firm those established. Further, such approaches could unveil any lymphoid lineage/s that may have preceded the likes of NKT and MAIT cells. Similarly, single cell transcriptomics can reveal the origins of γδT and γδT-like ILELs and whether the Xenopus NKT-like and MAIT-like cells are bona fide ILELs even though they use distinct restriction elements and TCRs.
B. Developmental Origins: Genomic Control of NKT Cell Ontogeny
“The child is the father of a man.” From “My Heart Leaps Up When I See a Rainbow in the Sky,” a poem by William Wordsworth
NKT cell, MAIT cell, and γδT cell development occurs in the thymus. As with all developmental processes, ILEL ontogeny depends on modular genome regulatory network/s composed of key apex transcription factors and downstream effectors. Thus, to become a T lineage ILEL versus an ILC rests upon the actions of a few apex transcription factors: NOTCH-TCF1-E2A and the E2A regulator Id2 (inhibitor of DNA-binding 2). NOTCH1 effectors TCF1 (encoded by Tcf7) and E2A (encoded by Tcf3) control Tcf12 (encoding HEB); HEB in turn regulates the expression and rearrangement of TCR gene loci, including TRA11*02 to TRAJ18 gene rearrangement. The expression of Id2 down regulates Tcf3 and, thereby, NOTCH1-TCF1 T cell apex transcription factors, begetting the ILC lineage.208,209
Commitment to the γδT innate-like cell lineage occurs prior to γδT innate-like lineage commitment. A key nuclear event that begets the ILEL-lineage and associated innate-like functions involves the activation of the zinc finger BTB domain-containing-16 (Zbtb16) gene that codes for promyelocytic leukemia zinc finger (PLZF). PLZF-mediated genomic control differentiates the unique NKT, MAIT, and γδT cell functions from those of other T lymphocytes. This section covers what we know of NKT cell development, much of which is similar to MAIT cell development, and perhaps γδT cell ontogeny as well.5 Developmental plans of other ILELs are reviewed elsewhere.5,6,51,209–211
Genetically modified mice whose thymocytes fail to develop beyond the (DN)2/DN3 stage also lack NKT cells (Fig. 7).54,212–216 Further, NKT cells fail to develop in mice with mutations in Myb (coding for c-Myb), Rorc (coding for RORγt), and Tcf12.217–220 As TRAV11*02 and TRAJ18 rearrangement occurs at a late DP stage,217,219 and NKT cells develop in Jα18-deficient mice that received highly purified tetramer-negative, DP-high thymocytes,221 commitment to the NKT cell lineage occurs at the DP stage.222 TRAV11*02 and TRAJ18 rearrangement, however, can occur within late DN thymocytes and the resulting precursors can differentiate into NKT1 cells.223
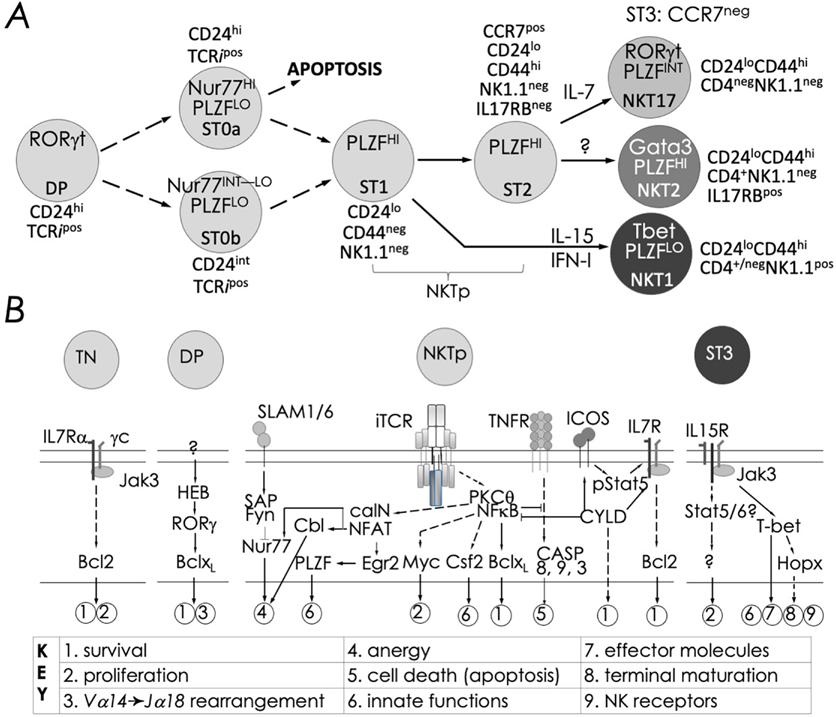
FIG. 7:
Schematic rendition of developmental stages and signaling events essential to NKT cell ontogeny: (A) Precursor ST0, immature ST1 & ST2, and mature ST3 represent distinct NKT cell developmental stages (ST) and NKT1, 2 and 17 are functional subsets. NKT cell ontogeny begins with the rearrangement of the TRAV11*02 to TRAJ18 TCR α-chain gene segments and proceeds after its interaction with the positively selecting CD1d-self lipid complex. Stage-specific NKT cell markers —e.g., CD24, CD44 and NK1.1—and subset-specific differentiation signals and transcription factors are indicated. Interleukin (IL)-7 and IL-15 are cytokines that mediate intercellular communication. (B) Thymocyte development to DP stage are essential for conventional CD4+ and CD8+ T as well as NKT cell development. Commitment to the NKT cell lineage occurs with the semi-invariant TCR expression at the DP stage under the influence of HEB (an E2A family transcription factor)- and RORγt. Positive selection at stage 0 induces the expression of PLZF, distinguishing NKT cells from other T lineages—e.g., CD4+ and CD8+ T cells. Onward development requires signals relayed via the lymphocyte activation molecule family member (SLAMF)-SLAM-associated protein (SAP)-Fyn (a Src kinase) module, which tempers agonistic signals relayed through the NKT cell TCR. NKT cell TCR signals are processed by protein kinase C (PKC)-θ and relayed to CBM (CARMA1-Bcl10-MALT1) complex for integration by the transcription factor nuclear factor-κB (NF-κB). Signals integrated by the SLAM-SAP-Fyn and the NKT cell TCR-PKC-θ-CBM-NF-κB modules culminate in Gata3 and Tbx21, encoding T-bet. The two transcription factors are essential for Il4 and Ifng transcription and lineage specific cytokine production. Calcineurin-NFAT-Egr2 and PKCθ-NF-κB signaling axes play essential roles in lineage proliferation (e.g., Myc), maintenance (e.g., Bcl-xL), and effector differentiation (e.g., Csf2), and maturation. Whilst the mechanism/s of subset differentiation are still debated, signals from IL-7 direct NKT17 cell differentiation and those from IL-15 and/or IFN-I (type I IFN) direct NKT1 cell differentiation. Transcription factors such as Egr-2, Ets-1, GATA3, Id2, Id3, MEF, Nur77, RORγt, and T-bet act at distinct stages shown to direct proper, functional NKT cell development. See text for details and references therein.
Commitment to the innate-like T cells occurs shortly after positive selection by thymic self agonists, and proceeds through defined stages and culminate in branched differentiation to three major subsets shown in Fig. 7. The ligation of the invariant NKT cell TCRα (iTCRa) by its cognate agonist on DP thymocytes induces Nr4a1 expression encoding Nur77, which lasts for about two days.224 Shutting down Nr4a1 expression is critical because Nur77 can induce cell death to anergy in developing NKT cells (discussed below; ref. 194). The mechanism of Nr4a1 shutdown, albeit unknown, perhaps results from dissociation from the agonist or by signal/s relayed by the SLAM-SAP-Fyn module, which is known to temper iTCRa signal intensity.193,224 Coincident with Nur77 depletion is the upregulation of Zbtb16 expression, encoding PLZF.224 It is not known whether positive selection results in two stage 0 (st0) subsets—one with low and the other with high Nur77, the latter destined to deletion by apoptosis. St0 cells with low Nur77 may then proceed to st1 as PLZF levels increase. Alternatively, SLAM-SAP-Fyn activation in st0 cells may bolster PLZF expression and repress Nr4a1 locus to prevent deletion and to permit onward development. Whichever be the mechanism, st1 cells committed to the lineage undergo proliferative burst and expand and upregulate CD44,187,190,224 becoming permissive to differentiation. NKT1 cells differentiate under the influence of IL-15 (and potentially type I IFN190) trans-presented by medullary stromal cells interacting with NKT cell precursors.155–157,225,226 Those that do not receive IL-15 and/or type I IFN signal/s or are not permissive to these signals beget st2 cells, which are precursors to NKT17 and NKT2 cell subsets. Whilst IL-7 receptor signaling programs NKT17 cell differentiation, signals that promote NKT2 cell differentiation is unknown. Alternatively, NKT2 cell subset could be default path that needs no specific signal/s. This NKT cell ontogenetic and differentiation pathway, extracted from recent reports,187,190,227 resembles the Waddington epigenetic landscape228,229 of binary cell fate decisions made by an immediate precursor until end-stage differentiation occurs. Furthermore, recent reports suggest that MAIT cell and γδT cell development and subset differentiation follow the same general principle as those described above.187,190
Mechanism/s of effector subset differentiation is incompletely understood. Evidence support a iTCR-agonist affinity-based model coupled with cytokine signaling,190,192,193 tissue homing site-specific,171 and sole cytokine-based differentiation.224 The induction of CD69 at the late stage of NKT cell differentiation190,224 has suggested a iTCR-agonist affinity-based differentiation model. Despite CD69 induction, which results from TCR activation, no induction of Nur77 —a proxy for TCR activation—was observed. IFN-I signaling can induce CD69, and IFN-I-regulated genes are induced in NKT1 cells.190,230 These findings support a cytokine-based differentiation model. It is possible that a low affinity iTCR ligand, which may not induce Nur77,231 sufficiently tickles the NKT cell TCR to make NKT cell precursors responsive to cytokine signaling. These competing models need further resolution.
Unlike conventional CD4+ and CD8+ T cells, which depend on thymic epithelial cells for positive and negative selection, NK1.1+ T cells rely on DP thymocytes for positive selection.232 NKT cell TCRs expressed by precursors interact with self lipid-bound CD1d on DP thymocytes233–237 and by homotypic signaling lymphocytic activation molecule (SLAM)-SLAM receptors on both cell types.238–240 These molecular interactions result in the activation of protein kinase Cθ-NF-κB and NFAT-Egr2 signaling modules, culminating in the induction of specific transcriptional programs crucial for NKT cell maturation (Fig. 7).238,239,241–246 PLZF-mediated transcriptional programs bestow upon NKT cells its unique functions through a differentiation process that results in NKT1, NKT2, and NKT17 cell subsets and the acquisition of cytokine secretion function.190,242,247,248 Whilst the NKT cell subsets are defined by the same subset-specific transcription factors expressed by the corresponding T helper cell subsets, NKT cells differ from T helper cells in the ability to quickly secrete cytokine/s in response to activation (Fig. 2).147,148,152–154,188
Gene regulatory networks (GRNs) control lineage-specific gene expression and unveil the developmental and evolutionary origins of cell lineages.249 PLZF is a lineage-specific master transcription factor, functioning as a critical node in the GRN that controls innate-like effector differentiation in developing NKT cells.247,248,250 Zbtb16 induction is in part regulated by acetylated Egr2251 induced downstream of NKT cell TCR signaling.244 A recent study revealed that the histone acetylase GCN (general control non-derepressible) 5 acetylates a critical lysine residue in Egr2, and DP thymocyte-specific depletion of GCN5 abrogated the progression of NKT cell development from st0 to st1 in a cell intrinsic manner. This st0 to st1 developmental block was secondary to the transcriptional downregulation of Zbtb16 and other essential NKT cell development genes such as Runx1, Tbx21, and Il2rb.251 GCN5 itself is an acetylated protein. Whether its function during NKT cell development depends on acetylation is currently unknown. In some models, the function of GCN5 depends on its deacetylation.252 Should GCN5 function in NKT cells depend on deacetylation, whether and which sirtuins (silent mating type information regulation 2 homologs 1–7; ref 252) play this role in NKT cells remains to be established.
While the mouse NKT cell iTCRa can pair with virtually all available TCR β-chains, the peripheral NKT cell repertoire consists of iTCRa paired with a restricted set of β-chains, viz., Vβ8, Vβ7, and Vβ2.253–256 It is generally thought that the semi-invariant NKT cell TCR repertoire is built solely by positive selection.257 This assumption is at odds with high affinity interactions between the NKT cell TCR and its cognate self-agonist, which results in self-reactive T cells. Hence, indirect evidence has implied a role for negative selection in sculpting the semi-invariant NKT cell TCR repertoire.258–262 Evidence in favor of both models has been discussed elsewhere.142
Nr4a1-encoded Nur77,263 a member of the orphan nuclear receptor transcription factor family,264,265 is expressed in both precursor and peripheral agonist-activated NKT cells.192,243,263 Nur77 expression is induced downstream of NKT cell TCR activation.192,243,263 Nur77 aids in negative selection of conventional T cells by converting the pro-survival factor Bcl-2 to a pro-apoptotic agent.264,266–269 Thus, a recent study found that Nur77 overexpression via a thymocyte-specific Nra4 transgene arrested NKT cell development at an early precursor stage shortly after positive selection, and induced negative selection and self-tolerance in thymic NKT cells, making these cells hyporesponsive to agonistic stimulation in the periphery.194 Moreover, homotypic SLAM-SLAM interactions between iNKT cell precursors and DP thymocytes are critical for the selection and differentiation of this lineage.232,238,241,270–276 Hence, this interaction is fine-tuned at the level of SLAM family member expression on the two cell types.277 NKT cells that develop in mice deficient in all six SLAM family receptors recapitulated the Nur77 overexpression phenotype.193 These findings together lend to a model in which strong and persistent NKT cell TCR signaling after positive selection induces high Nur77 expression. Because Nu77 induces apoptosis, precursors can become dead end cells as seen in NKT cells that overexpress Nur77 (Fig. 7; alternate paths at st0). Instead, the induction of SLAM family receptor signaling overrides Nur77-induced apoptosis and facilitates onward iNKT cell development. Consistent with this model, increased SLAM (SLAMF1) and Ly108 (SLAMF6) expression in st0 NKT cells could not temper Nu77 effects, perhaps because Nur77 overexpression was driven by the Lck proximal promotor and not the native Nr4a1 promotor. The next step would be to elucidate how the SLAM-SAP-Fyn signaling module controls Nr4a1 expression and function. Thus, both positive and negative selection events sculpt NKT cell TCR repertoire.
IV. END NOTES: A “LIMBIC IMMUNE SYSTEM” EMERGING IN PLAIN SIGHT
PLZF directs the distinct behaviors of a group of ILELs including γδT cells, NKT cells, MAIT cells, and ILCs.278–280 Also, PLZF is required for NK cell function, but not NK cell development.281 On the other hand, symbionts (gut and potentially other barriers such as skin and lungs) determine the development (MAIT cells, and potentially γδT cells) and functional differentiation (NKT cells, MAIT cells, and ILCs) of ILELs. We propose that these immune cells, all of lymphoid origin, be classified as the “limbic immune system” as they function at the edge (limbus in Latin) of the innate and adaptive immune systems. Remarkably, γδT, NK, and NKT cells localize to the interfollicular region of the lymph nodes, in between cells of that make up the innate and adaptive immune systems.282 Whilst not controlled by PLZF or the microbiota, by virtue of their physiologic functions, other tissue-restricted innate-like lymphocytes, such as CD8αα innate-type lymphocytes283 as well as B1 cells and NK cells,284 can be included in the limbic immune system. In its simplest form, the limbic immune system is anglicized Latin for the “in-betweeners”285 and, hence, synonymous with it.
In this proposal for a triune immune system we make no claims or assumption that the limbic immune system is an evolutionary transition between the innate and adaptive immune systems. But, instead, the limbic immune system is a conglomeration of independently acting modules, arising at different times in evolution, in many instances, repurposing loosely common genome regulatory circuits to accomplish a common task: to integrate information relayed by the innate sensory immune system about the local tissue environment and to provide context to downstream effector innate and adaptive immune responses. The multiple modules add robustness and evolvability to this limbic system to keep abreast of the ever-changing environment and the quick-evolving microbial cosmos, especially of those members of an otherwise symbiont community that turn pathobiont without much notice!
ACKNOWLEDGMENTS
We thank the contributions to the topics discussed here that were made by past members and collaborators of our laboratory. SJ is Research Career Scientist of the U.S. Department of Veterans Affairs (BX004595) and GDO is supported by the Vanderbilt University Medical Scientist Training Program. The authors and their works described herein were supported by a VA Merit Award (BX001444) as well as NIH Research Grants (AI042284, HL121139, and AI137082 to SJ, and DK104817 and AI139046 to LVK).
REFERENCES
- 1.Holmes FL. Claude Bernard, the milieu intérieur, and regulatory physiology. Hist Philos Life Sci. 1986;8(1):3–25. [PubMed] [Google Scholar]
- 2.Lanska DJ. Walter Bradford Cannon. Encyclopedia of neurological sciences. Elsevier, Inc; 2014. p. 580–583. [Google Scholar]
- 3.Holmes FL. Origins of the concept of the milieu intérieur. In Grande F, Visscher MB, editor. Claude Bernard and experimental medicine. Cambridge, MA: Shenkman; 1967. p. 179–191. [Google Scholar]
- 4.Lanier LL. Shades of grey—the blurring view of innate and adaptive immunity. Nat Rev Immunol. 2013. Feb 1;13(2):73. doi: 10.1038/nri3389. [DOI] [PubMed] [Google Scholar]
- 5.Pellicci DG, Koay HF, Berzins SP. Thymic development of unconventional T cells: How NKT cells, MAIT cells and γδ T cells emerge. Nat Rev Immunol. 2020. Dec;20(12):756–70. doi: 10.1038/s41577-020-0345-y. [DOI] [PubMed] [Google Scholar]
- 6.Eberl G, Powell N. Innate lymphoid cells. In: Principals of mucosal immunology. Boca Raton, FL: CRC Press; 2020. p. 115–122. [Google Scholar]
- 7.Mayassi T, Barreiro LB, Rossjohn J, Jabri B. A multilayered immune system through the lens of unconventional T cells. Nature. 2021. Jul;595(7868):501–10. doi: 10.1038/s41586-021-03578-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: An innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013. Feb;13(2):101–17. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 9.Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell. 2015. Feb 26;160(5):816–27. doi: 10.1016/j.cell.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill TM, Bezbradica JS, Van Kaer L, Joyce S. CD1d-restricted natural killer T cells. In eLS. Chichester, UK: John Wiley & Sons; 2001. [Google Scholar]
- 11.Marrero I, Ware R, Kumar V. Type II NKT cells in inflammation, autoimmunity, microbial immunity, and cancer. Front Immunol. 2015. Jun 17;6:316. doi: 10.3389/fimmu.2015.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhost S, Sedimbi S, Kadri N, Cardell SL. Immunomodulatory type II natural killer T lymphocytes in health and disease. Scand J Immunol. 2012. Sep;76(3):246–55. doi: 10.1111/j.1365-3083.2012.02750.x. [DOI] [PubMed] [Google Scholar]
- 13.Terabe M, Berzofsky JA. The immunoregulatory role of type I and type II NKT cells in cancer and other diseases. Cancer Immunol Immunother. 2014. Mar 1;63(3):199–213. doi: 10.1007/s00262-013-1509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macho-Fernandez E, Brigl M. The extended family of CD1d-restricted NKT cells: Sifting through a mixed bag of TCRs, antigens, and functions. Front Immunol. 2015. Jul 28;6:362. doi: 10.3389/fimmu.2015.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasgupta S, Kumar V. Type II NKT cells: A distinct CD1d-restricted immune regulatory NKT cell subset. Immunogenetics. 2016. Aug;68(8):665–76. doi: 10.1007/s00251-016-0930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhodapkar MV, Kumar V. Type II NKT cells and their emerging role in health and disease. J Immunol. 2017. Feb 1;198(3):1015–21. doi: 10.4049/jimmunol.1601399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen NR, Garg S, Brenner MB. Antigen presentation by CD1: Lipids, T cells, and NKT cells in microbial immunity. Adv Immunol. 2009. Jan 1;102:1–94. doi: 10.1016/S0065-2776(09)01201-2. [DOI] [PubMed] [Google Scholar]
- 18.Kohlgruber AC, Donado CA, LaMarche NM, Brenner MB, Brennan PJ. Activation strategies for invariant natural killer T cells. Immunogenetics. 2016. Aug;68(8):649–63. doi: 10.1007/s00251-016-0944-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim EY, Lynch L, Brennan PJ, Cohen NR, Brenner MB. The transcriptional programs of iNKT cells. Semin Immunol. 2015. Feb 1;27(1):26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucretius. De Rerum Natura. Rome. Loeb Classical Library. Translated by Rouse WH. Revised by Ferguson Smith Martin. Cambridge, MA: Harvard University Press. [Google Scholar]
- 21.Darlington CD. The evolution of genetic systems (2nd ed). Edinburgh, UK: Oliver & Boyd; 1958. [Google Scholar]
- 22.Brigl M, Brenner MB. CD1: Antigen presentation and T cell function. Annu Rev Immunol. 2004. Apr 23;22:817–90. [DOI] [PubMed] [Google Scholar]
- 23.Vincent MS, Leslie DS, Gumperz JE, Xiong X, Grant EP, Brenner MB. CD1-dependent dendritic cell instruction. Nat Immunol. 2002. Dec;3(12):1163–8. [DOI] [PubMed] [Google Scholar]
- 24.Brigl M, Bry L, Kent SC, Gumperz JE, Brenner MB. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat Immunol. 2003. Dec;4(12):1230–7. [DOI] [PubMed] [Google Scholar]
- 25.Brigl M, Tatituri RV, Watts GF, Bhowruth V, Leadbetter EA, Barton N, Cohen NR, Hsu FF, Besra GS, Brenner MB. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med. 2011. Jun 6;208(6):1163–77. doi: 10.1084/jem.20102555. jem.20102555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albacker LA, Chaudhary V, Chang YJ, Kim HY, Chuang YT, Pichavant M, DeKruyff RH, Savage PB, Umetsu DT. Invariant natural killer T cells recognize a fungal glycosphingolipid that can induce airway hyperreactivity. Nat Med. 2013. Oct;19(10):1297–304. doi: 10.1038/nm.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014. Jan 16;156(1–2):123–33. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinjo Y, Illarionov P, Vela JL, Pei B, Girardi E, Li X, Li Y, Imamura M, Kaneko Y, Okawara A, Miyazaki Y, Gómez-Velasco A, Rogers P, Dahesh S, Uchiyama S, Khurana A, Kawahara K, Yesilkaya H, Andrew PW, Wong CH, Kawakami K, Nizet V, Besra GS, Tsuji M, Zajonc DM, Kronenberg M. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol. 2011. Sep 4;12(10):966–74. doi: 10.1038/ni.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, Khurana A. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006. Sep;7(9):978–86. [DOI] [PubMed] [Google Scholar]
- 30.Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong CH, Kronenberg M. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005. Mar;434(7032):520–5. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 31.Mattner J, DeBord KL, Ismail N, Goff RD, Cantu C, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, Hoebe K. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005. Mar;434(7032):525–9. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 32.Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol. 2005. Jun;35(6):1692–701. [DOI] [PubMed] [Google Scholar]
- 33.Wieland Brown LC, Penaranda C, Kashyap PC, Williams BB, Clardy J, Kronenberg M, Sonnenburg JL, Comstock LE, Bluestone JA, Fischbach MA. Production of α-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol. 2013. Jul 16;11(7):e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang YJ, Kim HY, Albacker LA, Lee HH, Baumgarth N, Akira S, Savage PB, Endo S, Yamamura T, Maaskant J, Kitano N. Influenza infection in suckling mice expands an NKT cell subset that protects against airway hyperreactivity. J Clin Invest. 2011. Jan 4;121(1):57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012. Nov;491(7426):717–23. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 36.Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, Chen Z, Reantragoon R, Meehan B, Cao H, Williamson NA, Strugnell RA, Van Sinderen D, Mak JY, Fairlie DP, Kjer-Nielsen L, Rossjohn J, McCluskey J. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014. May 15;509(7500):361–5. doi: 10.1038/nature13160. Epub 2014 Apr 2. [DOI] [PubMed] [Google Scholar]
- 37.Eckle SB, Corbett AJ, Keller AN, Chen Z, Godfrey DI, Liu L, Mak JY, Fairlie DP, Rossjohn J, McCluskey J. Recognition of vitamin B precursors and byproducts by mucosal associated invariant T cells. J Biol Chem. 2015. Dec 18;290(51):30204–11. doi: 10.1074/jbc.R115.685990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.López-Sagaseta J, Dulberger CL, Crooks JE, Parks CD, Luoma AM, McFedries A, Van Rhijn I, Saghatelian A, Adams EJ. The molecular basis for mucosal-associated invariant T cell recognition of MR1 proteins. Proc Natl Acad Sci U S A. 2013. May 7;110(19):E1771–8. doi: 10.1073/pnas.1222678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dieudé M, Striegl H, Tyznik AJ, Wang J, Behar SM, Piccirillo CA, Levine JS, Zajonc DM, Rauch J. Cardiolipin binds to CD1d and stimulates CD1d-restricted γδ T cells in the normal murine repertoire. J Immunol. 2011. Apr 15;186(8):4771–81. doi: 10.4049/jimmunol.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai L, Picard D, Anderson B, Chaudhary V, Luoma A, Jabri B, Adams EJ, Savage PB, Bendelac A. The majority of CD1d-sulfatide-specific T cells in human blood use a semiinvariant Vδ1 TCR. Eur J Immunol. 2012. Sep;42(9):2505–10. doi: 10.1002/eji.201242531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uldrich AP, Le Nours J, Pellicci DG, Gherardin NA, McPherson KG, Lim RT, Patel O, Beddoe T, Gras S, Rossjohn J, Godfrey DI. CD1d-lipid antigen recognition by the γδ TCR. Nat Immunol. 2013. Nov;14(11):1137–45. doi: 10.1038/ni.2713. [DOI] [PubMed] [Google Scholar]
- 42.Luoma AM, Castro CD, Mayassi T, Bembinster LA, Bai L, Picard D, Anderson B, Scharf L, Kung JE, Sibener LV, Savage PB. Crystal structure of Vδ1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human γδ T cells. Immunity. 2013. Dec 12;39(6):1032–42. doi: 10.1016/j.immuni.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bluestone JA, Cron RQ, Cotterman ME, Houlden BA, Matis LA. Structure and specificity of T cell receptor gamma/delta on major histocompatibility complex antigen-specific CD3+, CD4−, CD8-T lymphocytes. J Exp Med. 1988. Nov 1;168(5):1899–916. doi: 10.1084/jem.168.5.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crowley MP, Reich Z, Mavaddat N, Altman JD, Chien YH. The recognition of the nonclassical major histocompatibility complex (MHC) class I molecule, T10, by the γδ T cell, G8. J Exp Med. 1997. Apr 7;185(7):1223–30. doi: 10.1084/jem.185.7.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crowley MP, Fahrer AM, Baumgarth N, Hampl J, Gutgemann I, Teyton L, Chien YH. A population of murine γδ T cells that recognize an inducible MHC class Ib molecule. Science. 2000. Jan 14;287(5451):314–6. doi: 10.1126/science.287.5451.314. [DOI] [PubMed] [Google Scholar]
- 46.Shin S, El-Diwany R, Schaffert S, Adams EJ, Garcia KC, Pereira P, Chien YH. Antigen recognition determinants of γδ T cell receptors. Science. 2005. Apr 8;308(5719):252–5. doi: 10.1126/science.1106480. [DOI] [PubMed] [Google Scholar]
- 47.Adams EJ, Chien YH, Garcia KC. Structure of a γδ T cell receptor in complex with the nonclassical MHC T22. Science. 2005. Apr 8;308(5719):227–31. doi: 10.1126/science.1106885. [DOI] [PubMed] [Google Scholar]
- 48.Carding SR, Egan PJ. γδ T cells: Functional plasticity and heterogeneity. Nat Rev Immunol. 2002. May;2(5):336–45. doi: 10.1038/nri797. [DOI] [PubMed] [Google Scholar]
- 49.Matis LA, Fry AM, Cron RQ, Cotterman MM, Dick RF, Bluestone JA. Structure and specificity of a class II MHC alloreactive gamma delta T cell receptor heterodimer. Science. 1989. Aug 18;245(4919):746–9. doi: 10.1126/science.2528206. [DOI] [PubMed] [Google Scholar]
- 50.Schild H, Mavaddat N, Litzenberger C, Ehrich EW, Davis MM, Bluestone JA, Matis L, Draper RK, Chien YH. The nature of major histocompatibility complex recognition by γδ T cells. Cell. 1994. Jan 14;76(1):29–37. doi: 10.1016/0092-8674(94)90170-8. [DOI] [PubMed] [Google Scholar]
- 51.Chien YH, Meyer C, Bonneville M. γδ T cells: First line of defense and beyond. Annu Rev Immunol. 2014. Mar 21;32:121–55. doi: 10.1146/annurev-immunol-032713-120216. [DOI] [PubMed] [Google Scholar]
- 52.Turchinovich G, Hayday AC. Skint-1 identifies a common molecular mechanism for the development of interferon-γ-secreting versus interleukin-17-secreting γδ T cells. Immunity. 2011. Jul 22;35(1):59–68. doi: 10.1016/j.immuni.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 53.Murphy KM, Weaver C. Antigen presentation to T lymphocytes. In Janeway’s immunobiology (9th ed). New York: Garland Science; 2017. p. 213–256. [Google Scholar]
- 54.Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DI. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1. 1− CD4+ CD1d-dependent precursor stage. J Exp Med. 2002. Apr 1;195(7):835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohen NR, Tatituri RV, Rivera A, Watts GF, Kim EY, Chiba A, Fuchs BB, Mylonakis E, Besra GS, Levitz SM, Brigl M. Innate recognition of cell wall β-glucans drives invariant natural killer T cell responses against fungi. Cell Host Microbe. 2011. Nov 17;10(5):437–50. doi: 10.1016/j.chom.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kain L, Webb B, Anderson BL, Deng S, Holt M, Costanzo A, Zhao M, Self K, Teyton A, Everett C, Kronenberg M. The identification of the endogenous ligands of natural killer T cells reveals the presence of mammalian α-linked glycosylceramides. Immunity. 2014. Oct 16;41(4):543–54. doi: 10.1016/j.immuni.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou D, Mattner J, Cantu C, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, Teneberg S. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004. Dec 3;306(5702):1786–9. [DOI] [PubMed] [Google Scholar]
- 58.Fox LM, Cox DG, Lockridge JL, Wang X, Chen X, Scharf L, Trott DL, Ndonye RM, Veerapen N, Besra GS, Howell AR. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol. 2009. Oct 27;7(10):e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gumperz JE, Roy C, Makowska A, Lum D, Sugita M, Podrebarac T, Koezuka Y, Porcelli SA, Cardell S, Brenner MB, Behar SM. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000. Feb 1;12(2):211–21. [DOI] [PubMed] [Google Scholar]
- 60.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007. Apr 23;25:297–336. [DOI] [PubMed] [Google Scholar]
- 61.Stanic AK, De Silva AD, Park JJ, Sriram V, Ichikawa S, Hirabyashi Y, Hayakawa K, Van Kaer L, Brutkiewicz RR, Joyce S. Defective presentation of the CD1d1-restricted natural Va14Ja18 NKT lymphocyte antigen caused by β-D-glucosylceramide synthase deficiency. Proc Natl Acad Sci U S A. 2003. Feb 18;100(4):1849–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Popovic ZV, Rabionet M, Jennemann R, Krunic D, Sandhoff R, Gröne HJ, Porubsky S. Glucosylceramide synthase is involved in development of invariant natural killer T cells. Front Immunol. 2017. Jul 21;8:848. doi: 10.3389/fimmu.2017.00848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paget C, Mallevaey T, Speak AO, Torres D, Fontaine J, Sheehan KC, Capron M, Ryffel B, Faveeuw C, de Moraes ML, Platt F. Activation of invariant NKT cells by toll-like receptor 9-stimulated dendritic cells requires type I interferon and charged glycosphingolipids. Immunity. 2007. Oct 26;27(4):597–609. [DOI] [PubMed] [Google Scholar]
- 64.Grubor-Bauk B, Simmons A, Mayrhofer G, Speck PG. Impaired clearance of herpes simplex virus type 1 from mice lacking CD1d or NKT cells expressing the semivariant Vα14-Jα281 TCR. J Immunol. 2003. Feb 1;170(3):1430–4. [DOI] [PubMed] [Google Scholar]
- 65.Cornish AL, Keating R, Kyparissoudis K, Smyth MJ, Carbone FR, Godfrey DI. NKT cells are not critical for HSV-1 disease resolution. Immunol Cell Biol. 2006. Feb;84(1):13–9. doi: ICB[pii] 10.1111/j.1440-1711.2005.01396.x. [DOI] [PubMed] [Google Scholar]
- 66.Ashkar AA, Rosenthal KL. Interleukin-15 and natural killer and NKT cells play a critical role in innate protection against genital herpes simplex virus type 2 infection. J Virol. 2003. Sep 15;77(18):10168–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim EY, Battaile JT, Patel AC, You Y, Agapov E, Grayson MH, Benoit LA, Byers DE, Alevy Y, Tucker J, Swanson S. Persistent activation of an innate immune response translates respiratory viral infection into chronic lung disease. Nat Med. 2008. Jun;14(6):633–40. doi: 10.1038/nm1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson TR, Hong S, Van Kaer L, Koezuka Y, Graham BS. NK T cells contribute to expansion of CD8+ T cells and amplification of antiviral immune responses to respiratory syncytial virus. J Virol. 2002. May 1;76(9):4294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paget C, Ivanov S, Fontaine J, Renneson J, Blanc F, Pichavant M, Dumoutier L, Ryffel B, Renauld JC, Gosset P, Gosset P. Interleukin-22 is produced by invariant natural killer T lymphocytes during influenza A virus infection: Potential role in protection against lung epithelial damages. J Biol Chem. 2012. Mar 16;287(12):8816–29. doi: 10.1074/jbc.M111.304758.M111.304758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paget C, Ivanov S, Fontaine J, Blanc F, Pichavant M, Renneson J, Bialecki E, Pothlichet J, Vendeville C, Barba-Spaeth G, Huerre MR, Faveeuw C, Si-Tahar M, Trottein F. Potential role of invariant NKT cells in the control of pulmonary inflammation and CD8+ T cell response during acute influenza A virus H3N2 pneumonia. J Immunol. 2011. May 15;186(10):5590–602. doi: 10.4049/jimmunol.1002348. Epub 2011 Apr 13. Erratum in: J Immunol. 2011 Aug 1;187(3):1515. [DOI] [PubMed] [Google Scholar]
- 71.De Santo C, Salio M, Masri SH, Lee LY, Dong T, Speak AO, Porubsky S, Booth S, Veerapen N, Besra GS, Gröne HJ. Invariant NKT cells reduce the immunosuppressive activity of influenza A virus–induced myeloid-derived suppressor cells in mice and humans. J Clin Invest. 2008. Dec 1;118(12):4036–48. doi: 10.1172/JCI36264.36264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ho LP, Denney L, Luhn K, Teoh D, Clelland C, McMichael AJ. Activation of invariant NKT cells enhances the innate immune response and improves the disease course in influenza A virus infection. Eur J Immunol. 2008. Jul;38(7):1913–22. doi: 10.1002/eji.200738017. [DOI] [PubMed] [Google Scholar]
- 73.Kok WL, Denney L, Benam K, Cole S, Clelland C, McMichael AJ, Ho LP. Pivotal advance: Invariant NKT cells reduce accumulation of inflammatory monocytes in the lungs and decrease immune-pathology during severe influenza A virus infection. J Leukocyte Biol. 2012. Mar;91(3):357–68. doi: 10.1189/jlb.0411184.jlb.0411184. [DOI] [PubMed] [Google Scholar]
- 74.Zeissig S, Murata K, Sweet L, Publicover J, Hu Z, Kaser A, Bosse E, Iqbal J, Hussain MM, Balschun K, Röcken C, Arlt A, Günther R, Hampe J, Schreiber S, Baron JL, Moody DB, Liang TJ, Blumberg RS. Hepatitis B virus-induced lipid alterations contribute to natural killer T cell-dependent protective immunity. Nat Med. 2012. Jul;18(7):1060–8. doi: 10.1038/nm.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007. May 14;204(5):987–94. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lamichhane R, Munro F, Harrop TW, de la Harpe SM, Dearden PK, Vernall AJ, McCall JL, Ussher JE. Human liver-derived MAIT cells differ from blood MAIT cells in their metabolism and response to TCR-independent activation. Eur J Immunol. 2021. Apr;51(4):879–92. doi: 10.1002/eji.202048830. [DOI] [PubMed] [Google Scholar]
- 77.Lamichhane R, Galvin H, Hannaway RF, de la Harpe SM, Munro F, Tyndall JD, Vernall AJ, McCall JL, Husain M, Ussher JE. Type I interferons are important co-stimulatory signals during T cell receptor mediated human MAIT cell activation. Eur J Immunol. 2020. Feb;50(2):178–91. doi: 10.1002/eji.201948279. [DOI] [PubMed] [Google Scholar]
- 78.Nagarajan NA, Kronenberg M. Invariant NKT cells amplify the innate immune response to lipopolysaccharide. J Immunol. 2007. Mar 1;178(5):2706–13. [DOI] [PubMed] [Google Scholar]
- 79.Tyznik AJ, Tupin E, Nagarajan NA, Her MJ, Benedict CA, Kronenberg M. The mechanism of invariant NKT cell responses to viral danger signals. J Immunol. 181(7):4452–6. doi: 10.4049/jimmunol.181.7.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wesley JD, Tessmer MS, Chaukos D, Brossay L. NK cell–like behavior of Vα14i NK T cells during MCMV infection. PLoS Pathog. 2008. Jul 18;4(7):e1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Wilgenburg B, Scherwitzl I, Hutchinson EC, Leng T, Kurioka A, Kulicke C, de Lara C, Cole S, Vasanawathana S, Limpitikul W, Malasit P, Young D, Denney L; STOP-HCV consortium, Moore MD, Fabris P, Giordani MT, Oo YH, Laidlaw SM, Dustin LB, Ho LP, Thompson FM, Ramamurthy N, Mongkolsapaya J, Willberg CB, Screaton GR, Klenerman P. MAIT cells are activated during human viral infections. Nat Commun. 2016. Jun 23;7:11653. doi: 10.1038/ncomms11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Loh L, Wang Z, Sant S, Koutsakos M, Jegaskanda S, Corbett AJ, Liu L, Fairlie DP, Crowe J, Rossjohn J, Xu J. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity via IL-18–dependent activation. Proc Natl Acad Sci U S A. 2016. Sep 6;113(36):10133–8. doi: 10.1073/pnas.1610750113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Wilgenburg B, Loh L, Chen Z, Pediongco TJ, Wang H, Shi M, Zhao Z, Koutsakos M, Nüssing S, Sant S, Wang Z, D’Souza C, Jia X, Almeida CF, Kostenko L, Eckle SBG, Meehan BS, Kallies A, Godfrey DI, Reading PC, Corbett AJ, McCluskey J, Klenerman P, Kedzierska K, Hinks TSC. MAIT cells contribute to protection against lethal influenza infection in vivo. Nat Commun. 2018. Nov 9;9(1):4706. doi: 10.1038/s41467-018-07207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cortez VS, Robinette ML, Colonna M. Innate lymphoid cells: New insights into function and development. Curr Opin Immunol. 2015. Feb 1;32:71–7. doi: 10.1016/j.coi.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Speak AO, Salio M, Neville DC, Fontaine J, Priestman DA, Platt N, Heare T, Butters TD, Dwek RA, Trottein F, Exley MA. Implications for invariant natural killer T cell ligands due to the restricted presence of isoglobotrihexosylceramide in mammals. Proc Natl Acad Sci U S A. 2007. Apr 3;104(14):5971–6. doi: 10.1073/pnas.0607285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Porubsky S, Speak AO, Luckow B, Cerundolo V, Platt FM, Gröne HJ. Normal development and function of invariant natural killer T cells in mice with isoglobotrihexosylceramide (iGb3) deficiency. Proc Natl Acad Sci U S A. 2007. Apr 3;104(14):5977–82. doi: 10.1073/pnas.0611139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Y, Zhou D, Xia C, Wang PG, Levery SB. Sensitive quantitation of isoglobotriaosylceramide in the presence of isobaric components using electrospray ionization-ion trap mass spectrometry. Glycobiology. 2008. Feb 1;18(2):166–76. doi: 10.1093/glycob/cwm127. [DOI] [PubMed] [Google Scholar]
- 88.Li Y, Teneberg S, Thapa P, Bendelac A, Levery SB, Zhou D. Sensitive detection of isoglobo and globo series tetraglycosylceramides in human thymus by ion trap mass spectrometry. Glycobiology. 2008. Feb 1;18(2):158–65. doi: 10.1093/glycob/cwm129. [DOI] [PubMed] [Google Scholar]
- 89.Zhou D, Xia C, Wang PG, Li Z, Zhang W, Ni G, Cheng J. Genetic studies of natural glycosphingolipid ligands for NKT cells. Methods Mol Biol. 2021;2388:13–25. doi: 10.1007/978-1-0716-1775-5_2. [DOI] [PubMed] [Google Scholar]
- 90.Brennan PJ, Tatituri RV, Brigl M, Kim EY, Tuli A, Sanderson JP, Gadola SD, Hsu FF, Besra GS, Brenner MB. Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat Immunol. 2011. Dec;12(12):1202–11. doi: 10.1038/ni.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brennan PJ, Tatituri RV, Heiss C, Watts GF, Hsu FF, Veerapen N, Cox LR, Azadi P, Besra GS, Brenner MB. Activation of iNKT cells by a distinct constituent of the endogenous glucosylceramide fraction. Proc Natl Acad Sci U S A. 2014. Sep 16;111(37):13433–8. doi: 10.1073/pnas.1415357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lairson LL, Chiu CP, Ly HD, He S, Wakarchuk WW, Strynadka NC, Withers SG. Intermediate trapping on a mutant retaining α-galactosyltransferase identifies an unexpected aspartate residue. J Biol Chem. 2004. Jul 2;279(27):28339–44. doi: 10.1074/jbc.M400451200. [DOI] [PubMed] [Google Scholar]
- 93.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: Structures, functions, and mechanisms. Annu Rev Biochem. 2008. Jul 7;77:521–55. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 94.Lee SS, Hong SY, Errey JC, Izumi A, Davies GJ, Davis BG. Mechanistic evidence for a front-side, SN i-type reaction in a retaining glycosyltransferase. Nat Chem Biol. 2011. Sep;7(9):631–8. doi: 10.1038/nchembio.628. [DOI] [PubMed] [Google Scholar]
- 95.Soya N, Fang Y, Palcic MM, Klassen JS. Trapping and characterization of covalent intermediates of mutant retaining glycosyltransferases. Glycobiology. 2011. May 1;21(5):547–52. doi: 10.1093/glycob/cwq190. [DOI] [PubMed] [Google Scholar]
- 96.Nunnari J, Walter P. Regulation of organelle biogenesis. Cell. 1996. Feb 9;84(3):389–94. doi: 10.1016/s0092-8674(00)81283-0. [DOI] [PubMed] [Google Scholar]
- 97.Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997. Sep;8(9):1805–14. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chapman R, Sidrauski C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu Rev Cell Biol. 1998;14:459–485. [DOI] [PubMed] [Google Scholar]
- 99.Brewer JW, Hendershot LM. Building an antibody factory: A job for the unfolded protein response. Nat Immunol. 2005. Jan;6(1):23–9. doi: 10.1038/ni1149. Erratum in: Nat Immunol. 2005 Feb;6(2):219. [DOI] [PubMed] [Google Scholar]
- 100.Joyce S CD1d and natural T cells: How their properties jump-start the immune system. Cell Mol Life Sci. 2001. Mar;58(3):442–69. doi: 10.1007/PL00000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Florence WC, Bhat RK, Joyce S. CD1d-restricted glycolipid antigens: Presentation principles, recognition logic and functional consequences. Expert Rev Mol Med. 2008. Jul;10:e20. doi: 10.1017/S1462399408000732. [DOI] [PubMed] [Google Scholar]
- 102.Govindarajan S, Verheugen E, Venken K, Gaublomme D, Maelegheer M, Cloots E, Gysens F, De Geest BG, Cheng TY, Moody DB, Janssens S. ER stress in antigen-presenting cells promotes NKT cell activation through endogenous neutral lipids. EMBO Rep. 2020. Jun 4;21(6):e48927. doi: 10.15252/embr.201948927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.De Libero G, Moran AP, Gober HJ, Rossy E, Shamshiev A, Chelnokova O, Mazorra Z, Vendetti S, Sacchi A, Prendergast MM, Sansano S. Bacterial infections promote T cell recognition of self-glycolipids. Immunity. 2005. Jun 1;22(6):763–72. doi: 10.1016/j.immuni.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 104.Stanic AK, Shashidharamurthy R, Bezbradica JS, Matsuki N, Yoshimura Y, Miyake S, Choi EY, Schell TD, Van Kaer L, Tevethia SS, Roopenian DC. Another view of T cell antigen recognition: Cooperative engagement of glycolipid antigens by Va14Ja18 natural TCR. J Immunol. 2003. Nov 1;171(9):4539–51. [DOI] [PubMed] [Google Scholar]
- 105.McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA, Bossi G, Salio M, Denkberg G, Reddington F, Tarlton A. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med. 2007. May 14;204(5):1131–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stanic AK, Shashidharamurthy R, Bezbradica JS, Matsuki N, Yoshimura Y, Miyake S, Choi EY, Schell TD, Van Kaer L, Tevethia SS, Roopenian DC, Yamamura T, Joyce S. Another view of T cell antigen recognition: Cooperative engagement of glycolipid antigens by Va14Ja18 natural T(iNKT) cell receptor [corrected]. J Immunol. 2003. Nov 1;171(9):4539–51. doi: 10.4049/jimmunol.171.9.4539. Erratum in: J Immunol. 2004 Jan 1;172(1):717. [DOI] [PubMed] [Google Scholar]
- 107.Bezbradica JS, Gordy LE, Stanic AK, Dragovic S, Hill T, Hawiger J, Unutmaz D, Van Kaer L, Joyce S. Granulocyte-macrophage colony-stimulating factor regulates effector differentiation of invariant natural killer T cells during thymic ontogeny. Immunity. 2006. Sep 1;25(3):487–97. [DOI] [PubMed] [Google Scholar]
- 108.Das R, Bassiri H, Guan P, Wiener S, Banerjee PP, Zhong MC, Veillette A, Orange JS, Nichols KE. The adaptor molecule SAP plays essential roles during invariant NKT cell cytotoxicity and lytic synapse formation. Blood. 2013. Apr 25;121(17):3386–95. doi: 10.1182/blood-2012-11-468868. Epub 2013 Feb 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang ZE, Gapin L, Kronenberg M, Locksley RM. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med. 2003. Oct 6;198(7):1069–76. doi: 10.1084/jem.20030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tanaka S, Tsukada J, Suzuki W, Hayashi K, Tanigaki K, Tsuji M, Inoue H, Honjo T, Kubo M. The interleukin-4 enhancer CNS-2 is regulated by Notch signals and controls initial expression in NKT cells and memory-type CD4 T cells. Immunity. 2006. Jun 1;24(6):689–701. doi: 10.1016/j.immuni.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 111.Collins PL, Chang S, Henderson M, Soutto M, Davis GM, McLoed AG, Townsend MJ, Glimcher LH, Mortlock DP, Aune TM. Distal regions of the human IFNG locus direct cell type-specific expression. J Immunol. 2010. Aug 1;185(3):1492–501. doi: 10.4049/jimmunol.1000124. Epub 2010 Jun 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Collins PL, Henderson MA, Aune TM. Lineage-specific adjacent IFNG and IL26 genes share a common distal enhancer element. Genes Immun. 2012. Sep;13(6):481–8. doi: 10.1038/gene.2012.22. Epub 2012 May 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang X, Bishop KA, Hegde S, Rodenkirch LA, Pike JW, Gumperz JE. Human invariant natural killer T cells acquire transient innate responsiveness via histone H4 acetylation induced by weak TCR stimulation. J Exp Med. 2012. May 7;209(5):987–1000. doi: 10.1084/jem.20111024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Karl OA, Im JS, Molano A, Dutronc Y, Illarionov PA, Forestier C, Fujiwara N, Arias I, Miyake S, Yamamura T, Chang YT. Modulation of CD1d-restricted NKT cell responses by using N-acyl variants of α-galactosylceramides. Proc Natl Acad Sci U S A. 2005. Mar 1;102(9):3383–8. doi: 10.1073/pnas.0407488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing Th 2 bias of natural killer T cells. Nature. 2001. Oct;413(6855):531–4. doi: 10.1038/35097097. [DOI] [PubMed] [Google Scholar]
- 116.Mashayekhi M, Sandau MM, Dunay IR, Frickel EM, Khan A, Goldszmid RS, Sher A, Ploegh HL, Murphy TL, Sibley LD, Murphy KM. CD8α+ dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity. 2011. Aug 26;35(2):249–59. doi: 10.1016/j.immuni.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bezbradica JS, Stanic AK, Matsuki N, Bour-Jordan H, Bluestone JA, Thomas JW, Unutmaz D, Van Kaer L, Joyce S. Distinct roles of dendritic cells and B cells in Va14Ja18 natural T cell activation in vivo. J Immunol. 2005. Apr 15;174(8):4696–705. [DOI] [PubMed] [Google Scholar]
- 118.Bai L, Constantinides MG, Thomas SY, Reboulet R, Meng F, Koentgen F, Teyton L, Savage PB, Bendelac A. Distinct APCs explain the cytokine bias of α-galactosylceramide variants in vivo. J Immunol. 2012. Apr 1;188(7):3053–61. doi: 10.4049/jimmunol.1102414jimmunol.1102414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fujii SI, Shimizu K, Kronenberg M, Steinman RM. Prolonged IFN-γ–producing NKT response induced with α-galactosylceramide–loaded DCs. Nat Immunol. 2002. Sep;3(9):867–74. [DOI] [PubMed] [Google Scholar]
- 120.Arora P, Baena A, Yu KO, Saini NK, Kharkwal SS, Goldberg MF, Kunnath-Velayudhan S, Carreño LJ, Venkataswamy MM, Kim J, Lazar-Molnar E, Lauvau G, Chang YT, Liu Z, Bittman R, Al-Shamkhani A, Cox LR, Jervis PJ, Veerapen N, Besra GS, Porcelli SA. A single subset of dendritic cells controls the cytokine bias of natural killer T cell responses to diverse glycolipid antigens. Immunity. 2014. Jan 16;40(1):105–16. doi: 10.1016/j.immuni.2013.12.004. Epub 2014 Jan 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bialecki E, Macho Fernandez E, Ivanov S, Paget C, Fontaine J, Rodriguez F, Lebeau L, Ehret C, Frisch B, Trottein F, Faveeuw C. Spleen-resident CD4+ and CD4− CD8α− dendritic cell subsets differ in their ability to prime invariant natural killer t lymphocytes. PLoS One. 2011. Oct 31;6(10):e26919. doi: 10.1371/journal.pone.0026919PONE-D-11-15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Macho-Fernandez E, Cruz LJ, Ghinnagow R, Fontaine J, Bialecki E, Frisch B, Trottein F, Faveeuw C. Targeted delivery of α-galactosylceramide to CD8α+ dendritic cells optimizes type I NKT cell-based antitumor responses. J Immunol. 2014. Jul 15;193(2):961–9. doi: 10.4049/jimmunol.1303029. Epub 2014 Jun 9. [DOI] [PubMed] [Google Scholar]
- 123.Zajonc DM, Cantu C, Mattner J, Zhou D, Savage PB, Bendelac A, Wilson IA, Teyton L. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol. 2005. Aug;6(8):810–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fujii SI, Shimizu K, Smith C, Bonifaz L, Steinman RM. Activation of natural killer T cells by α-galactosylceramide rapidly induces the full maturation of dendritic cells in vivo and thereby acts as an adjuvant for combined CD4 and CD8 T cell immunity to a coadministered protein. J Exp Med. 2003. Jul 21;198(2):267–79. doi: 10.1084/jem.20030324 jem.20030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fujii SI, Liu K, Smith C, Bonito AJ, Steinman RM. The linkage of innate to adaptive immunity via maturing dendritic cells in vivo requires CD40 ligation in addition to antigen presentation and CD80/86 costimulation. J Exp Med. 2004. Jun 21;199(12):1607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carnaud C, Lee D, Donnars O, Park SH, Beavis A, Koezuka Y, Bendelac A. Cutting edge: Cross-talk between cells of the innate immune system: NKT cells rapidly activate NK cells. J Immunol. 1999. Nov 1;163(9):4647–50. doi: ji_v163n9p4647. [PubMed] [Google Scholar]
- 127.Singh N, Hong S, Scherer DC, Serizawa I, Burdin N, Kronenberg M, Koezuka Y, Van Kaer L. Cutting edge: Activation of NK T cells by CD1d and α-galactosylceramide directs conventional T cells to the acquisition of a Th2 phenotype. J Immunol. 1999. Sep 1;163(5):2373–7. [PubMed] [Google Scholar]
- 128.Kitamura H, Iwakabe K, Yahata T, Nishimura S, Ohta A, Ohmi Y, Sato M, Takeda K, Okumura K, Van Kaer L, Kawano T, Taniguchi M, Nishimura T. The natural killer T (NKT) cell ligand alpha-galactosylceramide demonstrates its immunopotentiating effect by inducing interleukin (IL)-12 production by dendritic cells and IL-12 receptor expression on NKT cells. J Exp Med. 1999. Apr 5;189(7):1121–8. doi: 10.1084/jem.189.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vincent MS, Leslie DS, Gumperz JE, Xiong X, Grant EP, Brenner MB. CD1-dependent dendritic cell instruction. Nat Immunol. 2002. Dec;3(12):1163–8. [DOI] [PubMed] [Google Scholar]
- 130.Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, Berzins SP, Smyth MJ, Godfrey DI. Diverse cytokine production by NKT cell subsets and identification of an IL-17–producing CD4–NK1. 1–NKT cell population. Proc Natl Acad Sci U S A. 2008. Aug 12;105(32):11287–92. doi: 10.1073/pnas.08016311050801631105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chaudhry MS, Karadimitris A. Role and regulation of CD1d in normal and pathological B cells. J Immunol. 2014. Nov 15;193(10):4761–8. doi: 10.4049/jimmunol.1401805193/10/4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lang GA, Exley MA, Lang ML. The CD1d-binding glycolipid α-galactosylceramide enhances humoral immunity to T-dependent and T-independent antigen in a CD1d-dependent manner. Immunology. 2006. Sep;119(1):116–25. doi: IMM2413[pii] 10.1111/j.1365-2567.2006.02413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tonti E, Galli G, Malzone C, Abrignani S, Casorati G, Dellabona P. NKT-cell help to B lymphocytes can occur independently of cognate interaction. Blood. 2009. Jan 8;113 (2):370–6. doi: 10.1182/blood-2008-06-166249blood-2008-06-166249. [DOI] [PubMed] [Google Scholar]
- 134.Leadbetter EA, Brigl M, Illarionov P, Cohen N, Luteran MC, Pillai S, Besra GS, Brenner MB. NK T cells provide lipid antigen-specific cognate help for B cells. Proc Natl Acad Sci U S A. 2008. Jun 17;105(24):8339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.King IL, Fortier A, Tighe M, Dibble J, Watts GF, Veerapen N, Haberman AM, Besra GS, Mohrs M, Brenner MB, Leadbetter EA. Invariant natural killer T cells direct B cell responses to cognate lipid antigen in an IL-21-dependent manner. Nat Immunol. 2011. Nov 27;13(1):44–50. doi: 10.1038/ni.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Barral P, Eckl-Dorna J, Harwood NE, De Santo C, Salio M, Illarionov P, Besra GS, Cerundolo V, Batista FD. B cell receptor-mediated uptake of CD1d-restricted antigen augments antibody responses by recruiting invariant NKT cell help in vivo. Proc Natl Acad Sci U S A. 2008. Jun 17;105(24):8345–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chang PP, Barral P, Fitch J, Pratama A, Ma CS, Kallies A, Hogan JJ, Cerundolo V, Tangye SG, Bittman R, Nutt SL. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol. 2012. Jan;13(1):35–43. doi: 10.1038/ni.2166ni.2166. [DOI] [PubMed] [Google Scholar]
- 138.Semmling V, Lukacs-Kornek V, Thaiss CA, Quast T, Hochheiser K, Panzer U, Rossjohn J, Perlmutter P, Cao J, Godfrey DI, Savage PB. Alternative cross-priming through CCL17-CCR4-mediated attraction of CTLs toward NKT cell–licensed DCs. Nat Immunol. 2010. Apr;11(4):313–20. doi: 10.1038/ni.1848ni.1848. [DOI] [PubMed] [Google Scholar]
- 139.Gilchuk P, Hill TM, Guy C, McMaster SR, Boyd KL, Rabacal WA, Lu P, Shyr Y, Kohlmeier JE, Sebzda E, Green DR. A distinct lung-interstitium-resident memory CD8+ T cell subset confers enhanced protection to lower respiratory tract infection. Cell Rep. 2016. Aug 16;16(7):1800–9. doi: 10.1016/j.celrep.2016.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gilchuk P, Spencer CT, Conant SB, Hill T, Gray JJ, Niu X, Zheng M, Erickson JJ, Boyd KL, McAfee KJ, Oseroff C. Discovering naturally processed antigenic determinants that confer protective T cell immunity. J Clin Invest. 2013. May 1;123(5):1976–87. doi: 10.1172/JCI67388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hermans IF, Silk JD, Gileadi U, Salio M, Mathew B, Ritter G, Schmidt R, Harris AL, Old L, Cerundolo V. NKT cells enhance CD4+ and CD8+ T cell responses to soluble antigen in vivo through direct interaction with dendritic cells. J Immunol. 2003. Nov 15;171(10):5140–7. [DOI] [PubMed] [Google Scholar]
- 142.Kumar A, Suryadevara N, Hill TM, Bezbradica JS, Van Kaer L, Joyce S. Natural killer T cells: An ecological evolutionary developmental biology perspective. Front Immunol. 2017. Dec 22;8:1858. doi: 10.3389/fimmu.2017.01858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Donado CA, Cao AB, Simmons DP, Croker BA, Brennan PJ, Brenner MB. A two-cell model for IL-1β release mediated by death-receptor signaling. Cell Rep. 2020. Apr 7;31(1):107466. doi: 10.1016/j.celrep.2020.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bezbradica JS, Joyce S. NKT cells join the two step for inflammasome-independent IL-1beta release. Cell Rep. 2020;31(1):107481. doi: 10.1016/j.celrep.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shin S, Brodsky IE. The inflammasome: Learning from bacterial evasion strategies. Semin Immunol. 2015. Mar 1;27(2):102–10. doi: 10.1016/j.smim.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 146.Lee YJ, Wang H, Starrett GJ, Phuong V, Jameson SC, Hogquist KA. Tissue-specific distribution of iNKT cells impacts their cytokine response. Immunity. 2015. Sep 15;43(3):566–78. doi: 10.1016/j.immuni.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lynch L, Michelet X, Zhang S, Brennan PJ, Moseman A, Lester C, Besra G, Vomhof-Dekrey EE, Tighe M, Koay HF, Godfrey DI, Leadbetter EA, Sant’Angelo DB, von Andrian U, Brenner MB. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nat Immunol. 2015. Jan;16(1):85–95. doi: 10.1038/ni.3047. Epub 2014 Dec 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G. IL-10–producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest. 2014. Sep 2;124(9):3725–40. doi: 10.1172/jci72308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Galli G, Pittoni P, Tonti E, Malzone C, Uematsu Y, Tortoli M, Maione D, Volpini G, Finco O, Nuti S, Tavarini S. Invariant NKT cells sustain specific B cell responses and memory. Proc Natl Acad Sci U S A. 2007. Mar 6;104(10):3984–9. doi: 10.1073/pnas.0700191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tangye SG, Ma CS, Brink R, Deenick EK. The good, the bad and the ugly—T FH cells in human health and disease. Nat Rev Immunol. 2013. Jun;13(6):412–26. doi: 10.1038/nri3447. [DOI] [PubMed] [Google Scholar]
- 151.Salou M, Legoux F, Lantz O. MAIT cell development in mice and humans. Mol Immunol. 2021. Feb 1;130:31–6. doi: 10.1016/j.molimm.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 152.Watarai H, Sekine-Kondo E, Shigeura T, Motomura Y, Yasuda T, Satoh R, Yoshida H, Kubo M, Kawamoto H, Koseki H, Taniguchi M. Development and function of invariant natural killer T cells producing Th2-and Th17-cytokines. PLoS Biol. 2012. Feb 7;10(2):e1001255. doi: 10.1371/journal.pbio.1001255PBIOLOGY-D-11-02362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of i NKT cells. Nat Immunol. 2013. Nov;14(11):1146–54. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hammond KJ, Pellicci DG, Poulton LD, Naidenko OV, Scalzo AA, Baxter AG, Godfrey DI. CD1d-restricted NKT cells: An interstrain comparison. J Immunol. 2001. Aug 1;167(3):1164–73. [DOI] [PubMed] [Google Scholar]
- 155.Castillo EF, Acero LF, Stonier SW, Zhou D, Schluns KS. Thymic and peripheral microenvironments differentially mediate development and maturation of iNKT cells by IL-15 transpresentation. Blood. 2010. Oct 7;116(14):2494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gordy LE, Bezbradica JS, Flyak AI, Spencer CT, Dunkle A, Sun J, Stanic AK, Boothby MR, He YW, Zhao Z, Van Kaer L, Joyce S. IL-15 regulates homeostasis and terminal maturation of NKT cells. J Immunol. 2011. Dec 15;187(12):6335–45. doi: 10.4049/jimmunol.1003965. Epub 2011 Nov 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Matsuda JL, Gapin L, Sidobre S, Kieper WC, Tan JT, Ceredig R, Surh CD, Kronenberg M. Homeostasis of V alpha 14i NKT cells. Nat Immunol. 2002. Oct;3(10):966–74. doi: 10.1038/ni837. Epub 2002 Sep 9. [DOI] [PubMed] [Google Scholar]
- 158.Parekh VV, Wu L, Boyd KL, Williams JA, Gaddy JA, Olivares-Villagómez D, Cover TL, Zong WX, Zhang J, Van Kaer L. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell–specific deletion of Vps34. J Immunol. 2013. May 15;190(10):5086–101. doi: 10.4049/jimmunol.1202071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pei B, Zhao M, Miller BC, Véla JL, Bruinsma MW, Virgin HW, Kronenberg M. Invariant NKT cells require autophagy to coordinate proliferation and survival signals during differentiation. J Immunol. 2015. Jun 15;194(12):5872–84. doi: 10.4049/jimmunol.1402154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Salio M, Puleston DJ, Mathan TS, Shepherd D, Stranks AJ, Adamopoulou E, Veerapen N, Besra GS, Hollander GA, Simon AK, Cerundolo V. Essential role for autophagy during invariant NKT cell development. Proc Natl Acad Sci U S A. 2014. Dec 30;111(52):E5678–87. doi: 10.1073/pnas.1413935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Thapa P, Arocha SR, Chung JY, Sant’Angelo DB, Shapiro VS. Histone deacetylase 3 is required for iNKT cell development. Sci Rep. 2017. Jul 19;7(1):1–3. doi: 10.1038/s41598-017-06102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Crowe NY, Coquet JM, Berzins SP, Kyparissoudis K, Keating R, Pellicci DG, Hayakawa Y, Godfrey DI, Smyth MJ. Differential antitumor immunity mediated by NKT cell subsets in vivo. J Exp Med. 2005. Nov 7;202(9):1279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, Richer W, Goubet AG, Daviaud C, Menger L, Procopio E. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J Exp Med. 2019. Jan 7;216(1):133–51. doi: 10.1084/jem.20181483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yin CC, Cho OH, Sylvia KE, Narayan K, Prince AL, Evans JW, Kang J, Berg LJ. The Tec kinase ITK regulates thymic expansion, emigration, and maturation of γδ NKT cells. J Immunol. 2013. Mar 15;190(6):2659–69. doi: 10.4049/jimmunol.1202531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Terashima A, Watarai H, Inoue S, Sekine E, Nakagawa R, Hase K, Iwamura C, Nakajima H, Nakayama T, Taniguchi M. A novel subset of mouse NKT cells bearing the IL-17 receptor B responds to IL-25 and contributes to airway hyperreactivity. J Exp Med. 2008. Nov 24;205(12):2727–33. doi: 10.1084/jem.20080698. Epub 2008 Nov 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Matangkasombut P, Marigowda G, Ervine A, Idris L, Pichavant M, Kim HY, Yasumi T, Wilson SB, DeKruyff RH, Faul JL, Israel E. Natural killer T cells in the lungs of patients with asthma. J Allergy Clin Immunol. 2009. May 1;123(5):1181–5. doi: 10.1016/j.jaci.2009.02.013S0091-6749(09)00337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim HY, Pichavant M, Matangkasombut P, Koh YI, Savage PB, DeKruyff RH, Umetsu DT. The development of airway hyperreactivity in T-bet-deficient mice requires CD1d-restricted NKT cells. J Immunol. 2009. Mar 1;182(5):3252–61. doi: 10.4049/jimmunol.0803339182/5/3252. [DOI] [PubMed] [Google Scholar]
- 168.Akbari O, Stock P, Meyer E, Kronenberg M, Sidobre S, Nakayama T, Taniguchi M, Grusby MJ, DeKruyff RH, Umetsu DT. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat Med. 2003. May;9(5):582–8. [DOI] [PubMed] [Google Scholar]
- 169.Michel ML, Keller AC, Paget C, Fujio M, Trottein F, Savage PB, Wong CH, Schneider E, Dy M, Leite-deMoraes MC. Identification of an IL-17–producing NK1. 1neg iNKT cell population involved in airway neutrophilia. J Exp Med. 2007. May 14;204(5):995–1001. doi: jem.20061551 [pii] 10.1084/jem.20061551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Doisne JM, Becourt C, Amniai L, Duarte N, Le Luduec JB, Eberl G, Benlagha K. Skin and peripheral lymph node invariant NKT cells are mainly retinoic acid receptor-related orphan receptor γt+ and respond preferentially under inflammatory conditions. J Immunol. 2009. Aug 1;183(3):2142–9. doi: 10.4049/jimmunol.0901059jimmunol.0901059. [DOI] [PubMed] [Google Scholar]
- 171.Clancy-Thompson E, Chen GZ, Tyler PM, Servos MM, Barisa M, Brennan PJ, Ploegh HL, Dougan SK. Monoclonal invariant NKT (iNKT) cell mice reveal a role for both tissue of origin and the TCR in development of iNKT functional subsets. J Immunol. 2017. Jul 1;199(1):159–71. doi: 10.4049/jimmunol.1700214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Webster KE, Kim HO, Kyparissoudis K, Corpuz TM, Pinget GV, Uldrich AP, Brink R, Belz GT, Cho JH, Godfrey DI, Sprent J. IL-17-producing NKT cells depend exclusively on IL-7 for homeostasis and survival. Mucosal Immunol. 2014. Sep;7(5):1058–67. doi: 10.1038/mi.2013.122mi2013122. [DOI] [PubMed] [Google Scholar]
- 173.Thapa P, Chen MW, McWilliams DC, Belmonte P, Constans M, Sant’Angelo DB, Shapiro VS. NKAP regulates invariant NKT cell proliferation and differentiation into ROR-γt–expressing NKT17 cells. J Immunol. 2016. Jun 15;196(12):4987–98. doi: 10.4049/jimmunol.1501653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Thapa P, Das J, McWilliams D, Shapiro M, Sundsbak R, Nelson-Holte M, Tangen S, Anderson J, Desiderio S, Hiebert S, Sant’Angelo DB. The transcriptional repressor NKAP is required for the development of iNKT cells. Nat Commun. 2013. Mar 12;4(1):1–1. doi: 10.1038/ncomms2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Thapa P, Manso B, Chung JY, Arocha SR, Xue HH, Sant’Angelo DB, Shapiro VS. The differentiation of ROR-γt expressing iNKT17 cells is orchestrated by Runx1. Sci Rep. 2017. Aug 1;7(1):1–3. doi: 10.1038/s41598-017-07365-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sklarz T, Guan P, Gohil M, Cotton RM, Ge MQ, Haczku A, Das R, Jordan MS. mTORC2 regulates multiple aspects of NKT-cell development and function. Eur J Immunol. 2017. Mar;47(3):516–26. doi: 10.1002/eji.201646343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wei J, Yang K, Chi H. Cutting edge: Discrete functions of mTOR signaling in invariant NKT cell development and NKT17 fate decision. J Immunol. 2014. Nov 1;193(9):4297–301. doi: 10.4049/jimmunol.1402042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Price AE, Reinhardt RL, Liang HE, Locksley RM. Marking and quantifying IL-17A-producing cells in vivo. PLoS One. 2012;7(6):e39750. doi: 10.1371/journal.pone.0039750PONE-D-12-02906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pichavant M, Goya S, Meyer EH, Johnston RA, Kim HY, Matangkasombut P, Zhu M, Iwakura Y, Savage PB, DeKruyff RH, Shore SA, Umetsu DT. Ozone exposure in a mouse model induces airway hyperreactivity that requires the presence of natural killer T cells and IL-17. J Exp Med. 2008. Feb 18;205(2):385–93. doi: 10.1084/jem.20071507. Epub 2008 Feb 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Milosavljevic N, Gazdic M, Simovic Markovic B, Arsenijevic A, Nurkovic J, Dolicanin Z, Djonov V, Lukic ML, Volarevic V. Mesenchymal stem cells attenuate acute liver injury by altering ratio between interleukin 17 producing and regulatory natural killer T cells. Liver Transpl. 2017. Aug;23(8):1040–50. doi: 10.1002/lt.24784. [DOI] [PubMed] [Google Scholar]
- 181.Chang PP, Barral P, Fitch J, Pratama A, Ma CS, Kallies A, Hogan JJ, Cerundolo V, Tangye SG, Bittman R, Nutt SL. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol. 2012. Jan;13(1):35–43. doi: 10.1038/ni.2166. [DOI] [PubMed] [Google Scholar]
- 182.Tonti E, Fedeli M, Napolitano A, Iannacone M, Von Andrian UH, Guidotti LG, Abrignani S, Casorati G, Dellabona P. Follicular helper NKT cells induce limited B cell responses and germinal center formation in the absence of CD4+ T cell help. J Immunol. 2012. Apr 1;188(7):3217–22. doi: 10.4049/jimmunol.1103501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chan AC, Leeansyah E, Cochrane A, D’Udekem d’Acoz Y, Mittag D, Harrison LC, Godfrey DI, Berzins SP. Exvivo analysis of human natural killer T cells demonstrates heterogeneity between tissues and within established CD4+ and CD4− subsets. Clin Exp Immunol. 2013. Apr;172(1):129–37. doi: 10.1111/cei.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Akbari O, Faul JL, Hoyte EG, Berry GJ, Wahlström J, Kronenberg M, DeKruyff RH, Umetsu DT. CD4+ invariant T-cell–receptor+ natural killer T cells in bronchial asthma. N Engl J Med. 2006. Mar 16;354(11):1117–29. doi: 10.1056/NEJMoa053614. [DOI] [PubMed] [Google Scholar]
- 185.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. 2002. Mar 4;195(5):625–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human Vα24 natural killer T cells. J Exp Med. 2002. Mar 4;195(5):637–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lee M, Lee E, Han SK, Choi YH, Kwon DI, Choi H, Lee K, Park ES, Rha MS, Joo DJ, Shin EC. Single-cell RNA sequencing identifies shared differentiation paths of mouse thymic innate T cells. Nat Commun. 2020. Aug 31;11(1):1–7. doi: 10.1038/s41467-020-18155-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, Kronenberg M. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol. 2016. Jun;17(6):728–39. doi: 10.1038/ni.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cohen NR, Brennan PJ, Shay T, Watts GF, Brigl M, Kang J, Brenner MB. Shared and distinct transcriptional programs underlie the hybrid nature of i NKT cells. Nat Immunol. 2013. Jan;14(1):90–9. doi: 10.1038/ni.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Krovi SH, Zhang J, Michaels-Foster MJ, Brunetti T, Loh L, Scott-Browne J, Gapin L. Thymic iNKT single cell analyses unmask the common developmental program of mouse innate T cells. Nat Commun. 2020. Dec 7;11(1):1–5. doi: 10.1038/s41467-020-20073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tsagaratou A, González-Avalos E, Rautio S, ScottBrowne JP, Togher S, Pastor WA, Rothenberg EV, Chavez L, Lähdesmäki H, Rao A. TET proteins regulate the lineage specification and TCR-mediated expansion of i NKT cells. Nat Immunol. 2017. Jan;18(1):45–53. doi: 10.1038/ni.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tuttle KD, Krovi SH, Zhang J, Bedel R, Harmacek L, Peterson LK, Dragone LL, Lefferts A, Halluszczak C, Riemondy K, Hesselberth JR. TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat Commun. 2018. Jul 9;9(1):1–3. doi: 10.1038/s41467-018-05026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lu Y, Zhong MC, Qian J, Calderon V, Tleugabulova MC, Mallevaey T, Veillette A. SLAM receptors foster iNKT cell development by reducing TCR signal strength after positive selection. Nat Immunol. 2019. Apr;20(4):447–57. doi: 10.1038/s41590-019-0334-0. [DOI] [PubMed] [Google Scholar]
- 194.Kumar A, Hill TM, Gordy LE, Suryadevara N, Wu L, Flyak AI, Bezbradica JS, Van Kaer L, Joyce S. Nur77 controls tolerance induction, terminal differentiation, and effector functions in semi-invariant natural killer T cells. Proc Natl Acad Sci U S A. 2020;117(29):17156–65. doi: 10.1073/pnas.2001665117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Haldane JBS. Disease and evolution. Ric Sci Suppl. 1949;19:2–11. [Google Scholar]
- 196.Dronamraju KR, editor. Selected genetic papers of J.B.S. Haldane. New York & London: Garland Publishing; 1990. [Google Scholar]
- 197.Owen R On parthenogenesis or the successive production of procreating individuals from a single ovum. London: John Van Voorst, Paternoster Row; 1849. [Google Scholar]
- 198.Owen R Darwin on the origin of species. Edinburgh Rev. 1860;3:487–532. [Google Scholar]
- 199.Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015. Mar 21;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 200.Boudinot P, Mondot S, Jouneau L, Teyton L, Lefranc MP, Lantz O. Restricting nonclassical MHC genes coevolve with TRAV genes used by innate-like T cells in mammals. Proc Natl Acad Sci U S A. 2016. May 24;113(21):E2983–92. doi: 10.1073/pnas.1600674113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Edholm ES, Banach M, Rhoo KH, Pavelka MS, Robert J. Distinct MHC class I-like interacting invariant T cell lineage at the forefront of mycobacterial immunity uncovered in Xenopus. Proc Natl Acad Sci U S A. 2018. Apr 24;115(17):E4023–31. doi: 10.1073/pnas.1722129115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hyoe RK, Robert J. A Xenopus tadpole alternative model to study innate-like T cell-mediated anti-mycobacterial immunity. Dev Comp Immunol. 2019. Mar 1;92:253–9. doi: 10.1016/j.dci.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yang Z, Wang C, Wang T, Bai J, Zhao Y, Liu X, Ma Q, Wu X, Guo Y, Zhao Y, Ren L. Analysis of the reptile CD1 genes: Evolutionary implications. Immunogenetics. 2015. Jun;67(5):337–46. doi: 10.1007/s00251-015-0837-2. [DOI] [PubMed] [Google Scholar]
- 204.Selvanantham T, Lin Q, Guo CX, Surendra A, Fieve S, Escalante NK, Guttman DS, Streutker CJ, Robertson SJ, Philpott DJ, Mallevaey T. NKT cell–deficient mice harbor an altered microbiota that fuels intestinal inflammation during chemically induced colitis. J Immunol. 2016. Dec 1;197(11):4464–72. doi: 10.4049/jimmunol.1601410. [DOI] [PubMed] [Google Scholar]
- 205.Boehm T, Hirano M, Holland SJ, Das S, Schorpp M, Cooper MD. Evolution of alternative adaptive immune systems in vertebrates. Annu Rev Immunol. 2018. Apr 26;36:19–42. doi: 10.1146/annurev-immunol-042617-053028. [DOI] [PubMed] [Google Scholar]
- 206.Boehm T, McCurley N, Sutoh Y, Schorpp M, Kasahara M, Cooper MD. VLR-based adaptive immunity. Annu Rev Immunol. 2012. Apr 23;30:203–20. doi: 10.1146/annurev-immunol-020711-075038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Vivier E, Van De Pavert SA, Cooper MD, Belz GT. The evolution of innate lymphoid cells. Nat Immunol. 2016. Jul;17(7):790–4. doi: 10.1038/ni.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Rothenberg EV. Transcriptional control of early T and B cell developmental choices. Annu Rev Immunol. 2014. Mar 21;32:283–321. doi: 10.1146/annurev-immunol-032712-100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kang J, Malhotra N. Transcription factor networks directing the development, function, and evolution of innate lymphoid effectors. Annu Rev Immunol. 2015. Mar 21;33:505–38. doi: 10.1146/annurev-immunol-032414-112025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Parham P. Chapter 3: Innate immunity. In The induced response to infection the immune system. New York: W.W. Norton & Company; 2021. [Google Scholar]
- 211.Kristin H, Hristo G. Recent advances in iNKT cell development. F1000Research. 2020;9. doi: 10.12688/f1000research.21378.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Boesteanu A, Silva AD, Nakajima H, Leonard WJ, Peschon JJ, Joyce S. Distinct roles for signals relayed through the common cytokine receptor γ chain and interleukin 7 receptor α chain in natural T cell development. J Exp Med. 1997. Jul 21;186(2):331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: What’s in a name?. Nat Rev Immunol. 2004. Mar;4(3):231–7. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 214.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen–specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000. Jun 5;191(11):1895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, Kronenberg M. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000. Sep 5;192(5):741–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002. Apr 19;296(5567):553–5. [DOI] [PubMed] [Google Scholar]
- 217.Bezbradica JS, Hill T, Stanic AK, Van Kaer L, Joyce S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4+ 8+ stage of thymic ontogeny. Proc Natl Acad Sci U S A. 2005. Apr 5;102(14):5114–9. doi: 10.1073/pnas.0408449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.D’Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol. 2010;11(3):240–9. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, Littman DR. Genetic evidence supporting selection of the Vα14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005. Jun 1;22(6):705–16. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 220.Hu T, Simmons A, Yuan J, Bender TP, Alberola-Ila J. The transcription factor c-Myb primes CD4+ CD8+ immature thymocytes for selection into the i NKT lineage. Nat Immunol. 2010. May;11(5):435–41. doi: 10.1038/ni.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001. Oct;2(10):971–8. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 222.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007. Jul;7(7):505–18. [DOI] [PubMed] [Google Scholar]
- 223.Dashtsoodol N, Shigeura T, Aihara M, Ozawa R, Kojo S, Harada M, Endo TA, Watanabe T, Ohara O, Taniguchi M. Alternative pathway for the development of V α 14+ NKT cells directly from CD4–CD8–thymocytes that bypasses the CD4+ CD8+ stage. Nat Immunol. 2017. Mar;18(3):274–82. doi: 10.1038/ni.3668. [DOI] [PubMed] [Google Scholar]
- 224.Bortoluzzi S, Dashtsoodol N, Engleitner T, Drees C, Helmrath S, Mir J, Toska A, Flossdorf M, Öllinger R, Solovey M, Colomé-Tatché M. Brief homogeneous TCR signals instruct common iNKT progenitors whose effector diversification is characterized by subsequent cytokine signaling. Immunity. 2021. Nov 9;54(11):2497–513. doi: 10.1016/j.immuni.2021.09.003. [DOI] [PubMed] [Google Scholar]
- 225.Kennedy MK. Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, Matsuki N, Charrier K, Sedger L, Willis CR, Brasel K, Morrissey PJ, Stocking K, Schuh JC, Joyce S, Peschon JJ. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin-15-deficient mice. J Exp Med. 2000;191:771–80. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.White AJ, Jenkinson WE, Cowan JE, Parnell SM, Bacon A, Jones ND, Jenkinson EJ, Anderson G. An essential role for medullary thymic epithelial cells during the intrathymic development of invariant NKT cells. J Immunol. 2014. Mar 15;192(6):2659–66. doi: 10.4049/jimmunol.1303057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang H, Hogquist KA. CCR7 defines a precursor for murine iNKT cells in thymus and periphery. Elife. 2018. Aug 13;7:e34793. doi: 10.7554/eLife.34793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Waddington CH. Chapter VII: The temporal course gene reaction. In Organisers and genes. Cambridge, UK: Cambridge University Press; 1947. [Google Scholar]
- 229.Waddington CH. Chapter 2: The cybernetics of development. In The strategy of genes: A discussion of some aspects of theoretical biology. George, Allen, & Unwin, Ltd. Oxfordshire, NY: Routledge; 1957. [Google Scholar]
- 230.Lin SJ, Chao HC, Kuo ML. The effect of interleukin-12 and interleukin-15 on CD69 expression of T-lymphocytes and natural killer cells from umbilical cord blood. Neonatology. 2000;78(3):181–5. doi: 10.1159/000014268. [DOI] [PubMed] [Google Scholar]
- 231.Holzapfel KL, Tyznik AJ, Kronenberg M, Hogquist KA. Antigen-dependent versus -independent activation of invariant NKT cells during infection. J Immunol. 2014. Jun 15;192(12):5490–8. doi: 10.4049/jimmunol.1400722. Epub 2014 May 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bendelac A Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182(6):2091–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Chen YH, Chiu NM, Mandal M, Wang N, Wang CR. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity. 1997. Apr;6(4):459–67. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 234.Mendiratta SK, Martin WD, Hong S, Boesteanu A, Joyce S, Van Kaer L. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity. 1997. Apr;6(4):469–77. doi: 10.1016/s1074-7613(00)80290-3. [DOI] [PubMed] [Google Scholar]
- 235.Smiley ST, Kaplan MH, Grusby MJ. Immunoglobulin E production in the absence of interleukin-4-secreting CD1-dependent cells. Science. 1997. Feb 14;275(5302):977–9. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 236.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997. Nov 28;278(5343):1626–9. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 237.Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997. Nov 28;278(5343):1623–6. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 238.Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, Mamchak AA, Terhorst C, Bendelac A. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007. Nov;27(5):751–62. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wang N, Satoskar A, Faubion W, Howie D, Okamoto S, Feske S, Gullo C, Clarke K, Sosa MR, Sharpe AH, Terhorst C. The cell surface receptor SLAM controls T cell and macrophage functions. J Exp Med. 2004. May 3;199(9):1255–64. doi: 10.1084/jem.20031835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Jordan MA, Fletcher JM, Jose R, Chowdhury S, Gerlach N, Allison J, Baxter AG. Role of SLAM in NKT cell development revealed by transgenic complementation in NOD mice. J Immunol. 2011. Apr 1;186(7):3953–65. doi: 10.4049/jimmunol.1003305.Epub 2011 Feb 25. [DOI] [PubMed] [Google Scholar]
- 241.Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, Stein PL. (2005). Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. 2005;11(3):340–5. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 2010;11(3):197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 243.Stanic AK, Bezbradica JS, Park JJ, Van Kaer L, Boothby MR, Joyce S. Cutting edge: The ontogeny and function of Va14Ja18 natural T lymphocytes require signal processing by protein kinase C theta and NF-kappa B. J Immunol. 2004. Apr 15;172(8):4667–71. doi: 10.4049/jimmunol.172.8.4667. [DOI] [PubMed] [Google Scholar]
- 244.Lazarevic V, Zullo AJ, Schweitzer MN, Staton TL, Gallo EM, Crabtree GR, Glimcher LH. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat Immunol. 2009. Mar;10(3):306–13. doi: 10.1038/ni.1696. Epub 2009 Jan 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Seiler MP, Mathew R, Liszewski MK, Spooner CJ, Barr K, Meng F, Singh H, Bendelac A. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol. 2012. Feb 5;13(3):264–71. doi: 10.1038/ni.2230. Erratum in: Nat Immunol. 2013 Apr;14(4):413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Qi Q, Kannan AK, August A. Tec family kinases: Itk signaling and the development of NKT αβ and γδ T cells. FEBS J. 2011. Jun;278(12):1970–9. doi: 10.1111/j.1742-4658.2011.08074.x. Epub 2011 Mar 29. [DOI] [PubMed] [Google Scholar]
- 247.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008. Sep 19;29(3):391–403. doi: 10.1016/j.immuni.2008.07.011. Epub 2008 Aug 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant’Angelo DB. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008. Sep;9(9):1055–64. doi: 10.1038/ni.1641. Epub 2008 Jul 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Peter I, Davidson E. Genomic control process: Development and evolution. London: Academic Press; 2015. [Google Scholar]
- 250.Mao AP, Constantinides MG, Mathew R, Zuo Z, Chen X, Weirauch MT, Bendelac A. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc Natl Acad Sci U S A. 2016. Jul 5;113(27):7602–7. doi: 10.1073/pnas.1601504113. Epub 2016 Jun 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wang Y, Yun C, Gao B, Xu Y, Zhang Y, Wang Y, Kong Q, Zhao F, Wang CR, Dent SYR, Wang J, Xu X, Li HB, Fang D. The lysine acetyltransferase GCN5 is required for iNKT cell development through EGR2 acetylation. Cell Rep. 2017. Jul 18;20(3):600–12. doi: 10.1016/j.celrep.2017.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Dominy JE Jr, Lee Y, Jedrychowski MP, Chim H, Jurczak MJ, Camporez JP, Ruan HB, Feldman J, Pierce K, Mostoslavsky R, Denu JM, Clish CB, Yang X, Shulman GI, Gygi SP, Puigserver P. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell. 2012. Dec 28;48(6):900–13. doi: 10.1016/j.molcel.2012.09.030. Epub 2012 Nov 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Mallevaey T, Scott-Browne JP, Matsuda JL, Young MH, Pellicci DG, Patel O, Thakur M, Kjer-Nielsen L, Richardson SK, Cerundolo V, Howell AR, McCluskey J, Godfrey DI, Rossjohn J, Marrack P, Gapin L. T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009. Jul 17;31(1):60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Chiu YH, Park SH, Benlagha K, Forestier C, Jayawardena-Wolf J, Savage PB, Teyton L, Bendelac A. Multiple defects in antigen presentation and T cell development by mice expressing cytoplasmic tail-truncated CD1d. Nat Immunol. 2002. Jan;3(1):55–60. doi: 10.1038/ni740. Epub 2001 Dec 3. [DOI] [PubMed] [Google Scholar]
- 255.Zhou D, Mattner J, Cantu C 3rd, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, Teneberg S, Wang D, Proia RL, Levery SB, Savage PB, Teyton L, Bendelac A. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004. Dec 3;306(5702):1786–9. doi: 10.1126/science.1103440. Epub 2004 Nov 11. [DOI] [PubMed] [Google Scholar]
- 256.Pajerowski AG, Nguyen C, Aghajanian H, Shapiro MJ, Shapiro VS. NKAP is a transcriptional repressor of notch signaling and is required for T cell development. Immunity. 2009. May;30(5):696–707. doi: 10.1016/j.immuni.2009.02.011. Epub 2009 Apr 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Wei DG, Curran SA, Savage PB, Teyton L, Bendelac A. Mechanisms imposing the Vbeta bias of Valpha14 natural killer T cells and consequences for microbial glycolipid recognition. J Exp Med. 2006. May 15;203(5):1197–207. doi: 10.1084/jem.20060418. Epub 2006 May 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Chun T, Page MJ, Gapin L, Matsuda JL, Xu H, Nguyen H, Kang HS, Stanic AK, Joyce S, Koltun WA, Chorney MJ. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J Exp Med. 2003. Apr 7;197(7):907–18. doi: 10.1084/jem.20021366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Pellicci DG, Uldrich AP, Kyparissoudis K, Crowe NY, Brooks AG, Hammond KJ, Sidobre S, Kronenberg M, Smyth MJ, Godfrey DI. Intrathymic NKT cell development is blocked by the presence of α-galactosylceramide. Eur J Immunol. 2003. Jul;33(7):1816–23. doi: 10.1002/eji.200323894. [DOI] [PubMed] [Google Scholar]
- 260.Schümann J, Pittoni P, Tonti E, MacDonald HR, Dellabona P, Casorati G. Targeted expression of human CD1d in transgenic mice reveals independent roles for thymocytes and thymic APCs in positive and negative selection of Vα14i NKT cells. J Immunol. 2005. Dec 1;175(11):7303–10. [DOI] [PubMed] [Google Scholar]
- 261.Stanic AK, Bezbradica JS, Park JJ, Matsuki N, Mora AL, Van Kaer L, Boothby MR, Joyce S. NF-κB controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes. J Immunol. 2004. Feb 15;172(4):2265–73. [DOI] [PubMed] [Google Scholar]
- 262.Napolitano A, Pittoni P, Beaudoin L, Lehuen A, Voehringer D, MacDonald HR, Dellabona P, Casorati G. Functional education of invariant NKT cells by dendritic cell tuning of SHP-1. J Immunol. 2013. Apr 1;190(7):3299–308. doi: 10.4049/jimmunol.1203466. Epub 2013 Feb 20. [DOI] [PubMed] [Google Scholar]
- 263.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011. Jun 6;208(6):1279–89. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Calnan BJ, Szychowski S, Chan FK, Cado D, Winoto A. A role for the orphan steroid receptor Nur77 in apoptosis accompanying antigen-induced negative selection. Immunity. 1995. Sep;3(3):273–82. doi: 10.1016/1074-7613(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 265.Winoto A Genes involved in T-cell receptor-mediated apoptosis of thymocytes and T-cell hybridomas. Semin Immunol. 1997. Feb 1;9(1): 51–8 doi: 10.1006/smim.1996.0053. [DOI] [PubMed] [Google Scholar]
- 266.Thompson J, Winoto A. During negative selection, Nur77 family proteins translocate to mitochondria where they associate with Bcl-2 and expose its proapoptotic BH3 domain. J Exp Med. 2008. May 12;205(5):1029–36. doi: 10.1084/jem.20080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Tao R, Hancock WW. Resistance of Foxp3+ regulatory T cells to Nur77-induced apoptosis promotes allograft survival. PLoS One. 2008. May 28;3(5):e2321. doi: 10.1371/journal.pone.0002321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Baldwin TA, Hogquist KA. Transcriptional analysis of clonal deletion in vivo. J Immunol. 2007. Jul 15;179(2):837–44. doi: 10.4049/jimmunol.179.2.837. [DOI] [PubMed] [Google Scholar]
- 269.Zhou T, Cheng J, Yang P, Wang Z, Liu C, Su X, Bluethmann H, Mountz JD. Inhibition of Nur77/Nurr1 leads to inefficient clonal deletion of self-reactive T cells. J Exp Med. 1996. Apr 1;183(4):1879–92. doi: 10.1084/jem.183.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.DeVault VL, Malagic M, Mei L, Dienz O, Lilley GW, Benoit P, Mistri SK, Musial SC, Ather JL, Poynter ME, Boyson JE. Regulation of invariant NKT cell development and function by a 0.14 Mbp locus on chromosome 1: A possible role for Fcgr3. Genes Immunity. 2019. Apr;20(4):261–72.doi: 10.1038/s41435-018-0031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Chen S, Cai C, Li Z, Liu G, Wang Y, Blonska M, Li D, Du J, Lin X, Yang M, Dong Z. Dissection of SAP-dependent and SAP-independent SLAM family signaling in NKT cell development and humoral immunity. J Exp Med. 2017. Feb;214(2):475–89. doi: 10.1084/jem.20161312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, Schwartzberg PL, Crotty S. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity. 2012. Jun 29;36(6):986–1002. doi: 10.1016/j.immuni.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Nunez-Cruz S, Yeo WJ, Rothman J, Ojha P, Bassiri H, Juntilla M, Davidson D, Veillette A, Koretzky GA, Nichols KE. Differential requirement for the SAP-Fyn interaction during NK T cell development and function. J Immunol. 2008. Aug 15;181(4):2311–20. doi: 10.4049/jimmunol.181.4.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Pasquier B, Yin L, Fondanèche MC, Relouzat F, Bloch-Queyrat C, Lambert N, Fischer A, de Saint-Basile G, Latour S. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Exp Med. 2005. Mar 7;201(5):695–701. doi: 10.1084/jem.20042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Wei DG, Lee H, Park SH, Beaudoin L, Teyton L, Lehuen A, Bendelac A. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J Exp Med. 2005. Jul 18;202(2):239–48. doi: 10.1084/jem.20050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Bedard M, Shrestha D, Priestman DA, Wang Y, Schneider F, Matute JD, Iyer SS, Gileadi U, Prota G, Kandasamy M, Veerapen N. Sterile activation of invariant natural killer T cells by ER-stressed antigen-presenting cells. Proc Natl Acad Sci U S A. 2019. Nov 19;116(47):23671–81. doi: 10.1073/pnas.1910097116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Jordan MA, Fletcher JM, Jose R, Chowdhury S, Gerlach N, Allison J, Baxter AG. Role of SLAM in NKT cell development revealed by transgenic complementation in NOD mice. J Immunol. 2011. Apr 1;186(7):3953–65. doi: 10.4049/jimmunol.1003305. [DOI] [PubMed] [Google Scholar]
- 278.Alonzo ES, Sant’Angelo DB. Development of PLZF-expressing innate T cells. Curr Opin Immunol. 2011. Apr 1;23(2):220–7. doi: 10.1016/j.coi.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol. 2013. Apr 1;25(2):161–7. doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Ishizuka IE, Constantinides MG, Gudjonson H, Bendelac A. The innate lymphoid cell precursor. Annu Rev Immunol. 2016. May 20;34:299–316. doi: 10.1146/annurev-immunol-041015-055549. [DOI] [PubMed] [Google Scholar]
- 281.Xu D, Holko M, Sadler AJ, Scott B, Higashiyama S, Berkofsky-Fessler W, McConnell MJ, Pandolfi PP, Licht JD, Williams BR. Promyelocytic leukemia zinc finger protein regulates interferon-mediated innate immunity. Immunity. 2009. Jun 19;30(6):802–16. doi: 10.1016/j.immuni.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Kastenmüller W, Torabi-Parizi P, Subramanian N, Lämmermann T, Germain RN. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell. 2012. Sep 14;150(6):1235–48. doi: 10.1016/j.cell.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Van Kaer L, Algood HM, Singh K, Parekh VV, Greer MJ, Piazuelo MB, Weitkamp JH, Matta P, Chaturvedi R, Wilson KT, Olivares-Villagómez D. CD8αα+ innate-type lymphocytes in the intestinal epithelium mediate mucosal immunity. Immunity. 2014. Sep 18;41(3):451–64. doi: 10.1016/j.immuni.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Bendelac A, Bonneville M, Kearney JF. Autoreactivity by design: Innate B and T lymphocytes. Nat Rev Immunol. 2001. Dec;1(3):177–86. doi: 10.1038/35105052. [DOI] [PubMed] [Google Scholar]
- 285.Borden Y, Bird L, Minton K, eds. Focus on: The inbetweeners: Innate-like lymphocytes. Nat Rev Immuol. 2013;13(February):73–149. [Google Scholar]
- 286.Morita M, Motoki K, Akimoto K, Natori T, Sakai T, Sawa E, Yamaji K, Koezuka Y, Kobayashi E, Fukushima H. Structure-activity relationship of. alpha.-Galactosylceramides against B16-bearing mice. J Med Chem. 1995. Jun;38(12):2176–87. [DOI] [PubMed] [Google Scholar]
- 287.Natori T, Koezuka Y, Higa T. Agelasphins, Novel a-galactosylceramides from the marine sponge Agelas mauritianus. Tetrahedron Lett. 1993;34:5591–2. [Google Scholar]
- 288.Schmieg J, Yang G, Franck RW, Tsuji M. Superior protection against malaria and melanoma metastases by a C-glycoside analogue of the natural killer T cell ligand α-galactosylceramide. J Exp Med. 2003. Dec 1;198(11):1631–41. doi: 10.1084/jem.20031192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Oh SF, Praveena T, Song H, Yoo JS, Jung DJ, Erturk-Hasdemir D, Hwang YS, Lee CC, Le Nours J, Kim H, Lee J. Host immunomodulatory lipids created by symbionts from dietary amino acids. Nature. 2021. Nov 10:1–6. doi: 10.1038/s41586-021-04083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Ortaldo JR, Young HA, Winkler-Pickett RT, Bere EW, Murphy WJ, Wiltrout RH. Dissociation of NKT stimulation, cytokine induction, and NK activation in vivo by the use of distinct TCR-binding ceramides. J Immunol. 2004. Jan 15;172(2):943–53. [DOI] [PubMed] [Google Scholar]
- 291.Parekh VV, Singh AK, Wilson MT, Olivares-Villagómez D, Bezbradica JS, Inazawa H, Ehara H, Sakai T, Serizawa I, Wu L, Wang CR. Quantitative and qualitative differences in the in vivo response of NKT cells to distinct α-and β-anomeric glycolipids. J Immunol. 2004. Sep 15;173(6):3693–706. [DOI] [PubMed] [Google Scholar]
- 292.Mallevaey T, Clarke AJ, Scott-Browne JP, Young MH, Roisman LC, Pellicci DG, Patel O, Vivian JP, Matsuda JL, McCluskey J, Godfrey DI. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity. 2011. Mar 25;34(3):315–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Facciotti F, Ramanjaneyulu GS, Lepore M, Sansano S, Cavallari M, Kistowska M, Forss-Petter S, Ni G, Colone A, Singhal A, Berger J. Peroxisome-derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat Immunol. 2012. May;13(5):474–80. doi: 10.1038/ni.2245. [DOI] [PubMed] [Google Scholar]
- 294.Kumar A, Bezbradica JS, Stanic AK, Joyce S. Characterization and functional analysis of mouse semi-invariant natural T cells. Curr Protoc Immunol. 2017. Apr;117(1):14–3. doi: 10.1002/cpim.22. [DOI] [PubMed] [Google Scholar]
- 295.Joyce S, Girardi E, Zajonc DM. NKT cell ligand recognition logic: Molecular basis for a synaptic duet and transmission of inflammatory effectors. J Immunol. 2011. Aug 1;187(3):1081–9. doi: 10.4049/jimmunol.1001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Van Kaer L α-Galactosylceramide therapy for autoimmune diseases: Prospects and obstacles. Nat Rev Immunol. 2005. Jan;5(1):31–42. [DOI] [PubMed] [Google Scholar]