Abstract
Exosomes are secreted vesicles of ~30 to 150 nm diameter that play important roles in human health and disease. To better understand how cells release these vesicles, we examined the biogenesis of the most highly enriched human exosome marker proteins, the exosomal tetraspanins CD81, CD9, and CD63. We show here that endocytosis inhibits their vesicular secretion and, in the case of CD9 and CD81, triggers their destruction. Furthermore, we show that syntenin, a previously described exosome biogenesis factor, drives the vesicular secretion of CD63 by blocking CD63 endocytosis and that other endocytosis inhibitors also induce the plasma membrane accumulation and vesicular secretion of CD63. Finally, we show that CD63 is an expression-dependent inhibitor of endocytosis that triggers the vesicular secretion of lysosomal proteins and the clathrin adaptor AP-2 mu2. These results suggest that the vesicular secretion of exosome marker proteins in exosome-sized vesicles occurs primarily by an endocytosis-independent pathway.
Targeting exosome marker proteins to the plasma membrane induces their budding from the cell.
INTRODUCTION
Exosomes are small extracellular vesicles (sEVs) of ~30 to 150 nm in diameter, have the same topology as the cell, and are highly enriched in a specific subset of exosome marker proteins, especially the exosomal tetraspanins (1–5). In mammals, exosomes are abundant in all biofluids, transmit signals and molecules between cells, and have been implicated in a wide array of physiological and disease processes (1, 2, 6–9). Moreover, the ubiquitous production of exosomes and other sEVs by all cell types allows their use as biosensors of tissue and organ health or disease, while the bionormal nature of exosomes makes them makes them an ideal nanovesicle for delivering vaccines, biologics, and other drugs (10–13). Thus, there is broad biomedical interest in understanding the biogenesis of exosome-sized, secreted vesicles.
It is widely assumed that exosomal proteins bud from cells by a multistep, endocytosis-dependent pathway that involves (i) the delivery of exosome marker proteins to endosomes, (ii) the loading of these proteins into nascent intralumenal vesicles (ILVs), and (iii) the subsequent release of ILVs as exosomes when ILV-containing endosomes [also known as multivesicular bodies (MVBs)] fuse with the plasma membrane (2, 8, 14–16). This model has also been extended to include CD63 and syndecans as recruiting factors for the scaffold protein syntenin, for syndecan-syntenin and CD63-syntenin complexes as recruiting factors for the protein Alix, and for Alix in recruiting the ESCRT (endosomal sorting complexes for transport) machinery to endosome membranes to drive ILV formation (14, 17–21). This model is based on well-established protein-protein interactions and is consistent with current models for how the ESCRT facilitate outward vesicle budding (outward = away from the cytoplasm) and membrane sealing in general (22, 23).
Given that other organelle biogenesis pathways were elucidated by studying their most highly enriched proteins (24–28), we study the biogenesis of exosomes by studying the most highly enriched exosomal proteins. In a recent side-by-side comparison of 24 human exosome marker proteins, we found that the exosomal tetraspanins CD81, CD9, and CD63 were more highly enriched in exosome-sized EVs than any of the 21 other marker proteins tested, including syntenin, Alix, and the ESCRT protein TSG101 (5). This result was gratifying at several levels, in part because CD81, CD9, and CD63 were the first three proteins shown to be enriched in exosomes (3, 4), and in part because CD81, CD9, and CD63 have for decades been used as exosome marker proteins (1, 29). However, our preliminary interrogation of CD81, CD9, and CD63 trafficking and vesicular secretion did not support the prevailing paradigm of exosome biogenesis. Specifically, we found that CD81 and CD9 reside at the plasma membrane and bud from cells at 15- and 5-fold higher efficiently than CD63, respectively, while CD63 is constitutively endocytosed from the plasma membrane and resides in endolysosomal compartments (5, 30). Moreover, we found that appending an endocytosis signal to CD9 blocked CD9 budding from the cell, while mutating CD63’s endocytosis signal induced its budding from the cell (5, 30). In short, these observations suggest that there is a large gap between the prevailing paradigm of exosome biogenesis and how cells actually bud exosome marker proteins from the cell.
To further investigate this gap, we examined how the vesicular secretion of CD81, CD9, and CD63 is affected by endocytosis signals, endocytosis inhibitors, and their level of expression. The results of our studies show that endocytosis strongly inhibits the exosomal secretion of all these highly enriched exosome marker proteins and, in the case of CD81 and CD9, triggers their destruction. We also show that six mechanistically distinct inhibitors of endocytosis all induced the vesicular secretion of CD63, which has an endocytosis signal, but have no effect on the vesicular secretion of CD81, which lacks an endocytosis signal. Finally, we show that high-level expression of CD63, which binds directly to the clathrin adaptor AP-2 subunit mu2, inhibits endocytosis, induces the plasma membrane accumulation and vesicular secretion of lysosomal proteins, and triggers the vesicular secretion and cellular depletion of the AP-2 subunit mu2. These and other results support the hypothesis that exosome marker proteins bud from cells along the spectrum of plasma and endosome membranes, and primarily by an endocytosis-independent pathway.
RESULTS
Mutational inhibition of AP-2 mu2 induces the vesicular secretion of CD63
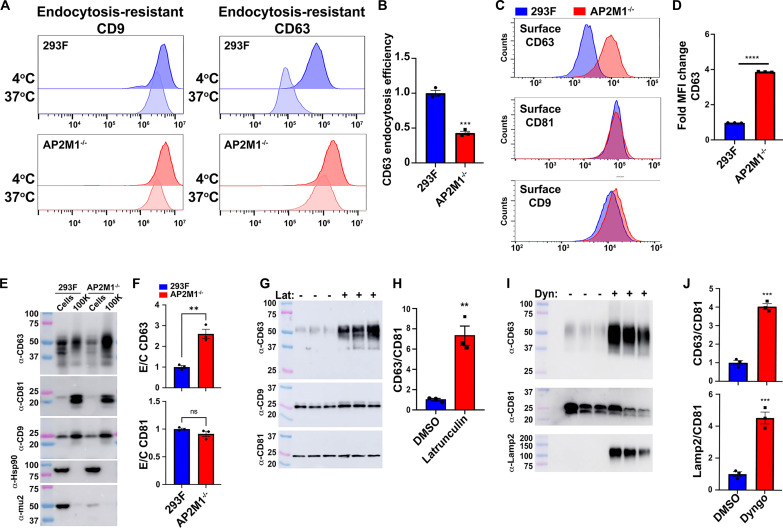
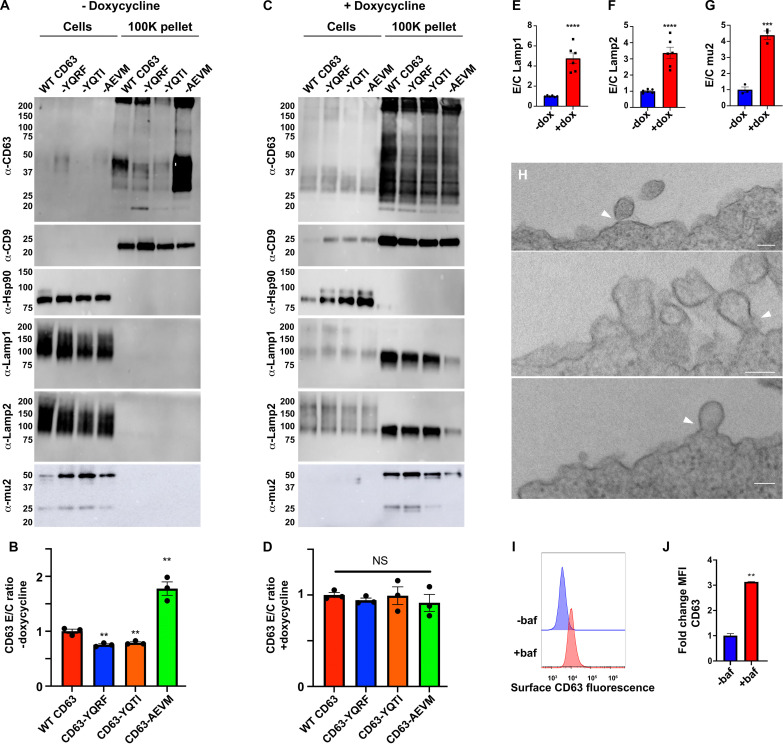
CD63 is a polytopic, integral membrane protein, synthesized in the endoplasmic reticulum, delivered to the plasma membrane by the canonical secretory pathway, and endocytosed to endosomes and lysosomes via the YxxΦ-type endocytosis signal located at its extreme C terminus, −YEVMcooh (31). To determine whether endocytosis promotes or inhibits the vesicular secretion of CD63, we generated a 293F cell line mutated in the gene (AP2M1) that encodes the mu2 subunit of the heterotetrameric clathrin adaptor protein complex AP-2, the protein that binds directly to YxxΦ-type endocytosis signals (32–34). This clonal AP2M1−/− cell line carries a large deletion on one AP2M1 allele and a one-codon deletion (ΔIle63) on its other AP2M1 allele (fig. S1) (cell lines used in this study are listed in table S1).
Given that this cell line still expresses a residual amount of the mu2/ΔIle63 protein, we measured CD63 endocytosis in 293F cells and AP2M1−/− cells to determine whether the AP2M1−/− cell line was actually defective in CD63 endocytosis. We did this using a five-step flow cytometry–based endocytosis assay in which we (i) chilled 293F and 293F/AP2M1−/− cells to 4°C to stop all vesicle traffic; (ii) incubated the two cell lines with unlabeled antibodies specific for either CD63 or the control protein CD9, separately, followed by washes to remove unbound antibody (Ab), all at 4°C; (iii) split each sample in half, leaving one at 4°C for 30 min, while the other was incubated at 37°C for 30 min to allow endocytosis to proceed; (iv) returned all samples to 4°C and incubated them at 4°C with fluorescently tagged anti-mouse immunoglobulin G (IgG); and (v) interrogated each cell population by flow cytometry to measure the relative levels of endocytosis-resistant CD63-mAb (monoclonal Ab) and CD9-mAb complexes that remained at the cell surface. In 293F cells, the extent of CD9 internalization was low (Fig. 1A, top left), while the extent of CD63 internalization was high (Fig. 1A, top right), consistent with the fact that CD63 has an endocytosis signal, whereas CD9 does not. However, in AP2M1−/− cells, the extent of CD9 internalization (Fig. 1A, bottom left) and CD63 internalization (Fig. 1A, bottom right) was low, indicating that the mutant AP2M1−/− cell line is defective in CD63 endocytosis. To quantify this effect, we calculated the CD63 endocytosis efficiency (EE) in these two cell lines from the mean fluorescence intensities (MFIs) of each sample, using the formula EE = [1 − (FICD63 @ 37oC/FICD63 @ 4oC)]/[1 − (FICD9 @ 37oC/FICD9 @ 4oC)]. Applying this formula to the data from three independent trials, we found that the CD63 endocytosis was 2.3-fold lower in AP2M1−/− cells than in 293F cells (P = 0.0003) (Fig. 1B), a result that is consistent with both AP-2’s key role in endocytosis and the hypomorphic nature of the AP2M1/ΔIle63 mutation. The endocytosis defect of AP2M1−/− cells was also apparent in a real-time immunofluorescence microscopy–based assay of CD63 and CD9 endocytosis (fig. S2).
Fig. 1. Inhibiting endocytosis induces a selective increase in the vesicular secretion of CD63.
(A) Flow cytometry histograms showing the fluorescence staining intensity for surface-exposed anti-CD63 and anti-CD9 mAbs following 30-min-long incubations at 4°C versus 37°C for (blue) 293F cells and (red) AP2M1−/− cells. (B) Bar graph of the CD63 EE in 293F and AP2M1−/− cells. Bar heights represent the mean, error bars show the SEM, and *** denotes a P value of <0.001. (C) Flow cytometry histograms display the levels of fluorescence brightness of (blue) 293F cells and (red) AP2M1−/− cells stained with fluorescently tagged antibodies specific for CD63, CD81, and CD9. (D) Bar graph showing the mean fluorescence intensities (MFI) of surface-stained CD63, with **** denoting a P value of <0.0001. (E) Immunoblots of cell and exosome-containing samples collected from 293F and AP2M1−/− cells. (F) Bar graph showing the mean secretion efficiency (amount in exosome-sized vesicles/amount in cells) for CD63 and CD81 in 293F and 293F/AP2M1−/− cells, with ** denoting a P value of <0.01. (G) Immunoblots of exosome-containing samples produced by 293F cells cultured in medium lacking or containing latrunculin A. (H) Bar graph showing the CD63/CD81 ratio in these exosome-containing samples, with ** denoting a value of 0.0028. (I) Immunoblots of exosome-containing fractions collected from 293F cells cultured in medium lacking or containing the dynamin inhibitor dyngo-4a. (J) Bar graphs showing the ratios of CD63/CD81 and Lamp2/CD81 in exosome-containing fractions, ± dyngo-4a, with *** denoting a P value of <0.001. Experiments were performed a minimum of three times.
The endocytosis defect of the AP2M1−/− cell line is expected to result in an accumulation of CD63 at the cell surface. To determine whether this occurred, we chilled 293F and AP2M1−/− cells to 4°C to stop all vesicle traffic, incubated them with fluorescently tagged antibodies specific for CD63, CD81, and CD9, again at 4°C, washed away the unbound antibodies, also at 4°C, and then measured bound antibodies by flow cytometry. This revealed that mutation of AP2M1 triggered a substantial increase in the cell surface staining for CD63, whereas the staining for CD81 and CD9 was relatively unchanged (Fig. 1C). To quantify this effect, we calculated the average MFI for CD63 staining in both cell lines across multiple independent trials, which revealed that staining for CD63 was 4.0-fold higher (P < 0.0001) on AP2M1−/− cells than on 293F cells (Fig. 1D), suggesting a significant increase in CD63 surface abundance.
These results allowed us to next test whether inhibiting CD63 endocytosis led to a decrease in its vesicular secretion from the cell, as predicted by the prevailing paradigm of exosome biogenesis, or alternatively, whether it led to an increase in CD63’s vesicular secretion from the cell. Specifically, we grew 293F and 293F/AP2M1−/− cells for 3 days in chemically defined medium, in shaker cultures, after which cells and exosome-sized vesicles were collected and examined by immunoblot. Using antibodies specific for CD63, CD81, CD9, Hsp90, and mu2, we found that inhibiting endocytosis led to a substantial increase in the vesicular secretion of CD63 (Fig. 1E). Quantitation of immunoblot (IB) signal intensities allowed us to calculate the efficiency of these proteins’ vesicular secretion by the formula E/C = [IB signal intensity in exosome-containing fraction]/[IB signal intensity in cell lysate], which showed that loss of mu2 resulted in a 2.6-fold increase in the vesicular secretion of CD63 (P < 0.0001) (Fig. 1F). In contrast, the vesicular secretion of CD81 was unaffected by mutational inhibition of endocytosis, as its E/C ratio was relatively unaffected (0.9-fold; P = 0.16) (Fig. 1F).
Inhibitors of actin polymerization or dynamin induce the vesicular secretion of CD63
If endocytosis is truly inhibitory to CD63’s vesicular secretion, other inhibitors of endocytosis should have similar effects, including acute, fast-acting drugs. One such drug is latrunculin A, which inhibits actin polymerization, a driving force for protein endocytosis (35–38). To determine whether latrunculin also induced the exosomal secretion of CD63, we grew 293F control cells in chemically defined medium ± latrunculin, followed by collection of exosome-containing fractions and interrogation of these samples by immunoblot. This revealed that latrunculin increased the amount of CD63 in exosome-containing fractions but had no effect on the vesicular secretion of CD81 (Fig. 1G). Quantitation of IB band intensities revealed that latrunculin increased the CD63/CD81 ratio by 7.3-fold (P = 0.0028) (Fig. 1H). Latrunculin also induced a slight increase in the CD9:CD81 ratio, but this was only ~1.4-fold (P < 0.05).
In contrast to latrunculin, which inhibits a wide variety of actin-dependent processes, the drug dyngo-4a is a selective endocytosis inhibitor that targets dynamin (39), a large GTPase (guanosine triphosphatase) that catalyzes the scission of nascent endocytic vesicles from the plasma membrane (40). 293F cells were grown for 2 days in protein-free, chemically defined medium, in shaker cultures ± dyngo-4a, after which exosome-sized vesicles were collected from the medium and examined by immunoblot (dyngo-4a is somewhat toxic, hence our reduction of exosome collection by 1 day). Similar to what we observed for latrunculin, addition of dyngo-4a resulted in an increase in the vesicular secretion of CD63 (Fig. 1I). When we probed these samples using antibodies specific for Lamp2, which like CD63 is a lysosomal protein with a YxxΦ-type endocytosis signal (33), we found that dyngo-4a also induced its vesicular secretion from the cell. Quantitation of immunoblot band intensities revealed that dyngo-4a increased the CD63/CD81 and Lamp2/CD81 ratios by 4.0-fold (P = 0.0001) and 4.5-fold (P = 0.0009), respectively (Fig. 1J).
Syntenin, a known inhibitor of CD63 endocytosis, also induces the plasma membrane accumulation and vesicular secretion of CD63
Together, these results show that inhibiting endocytosis triggers a large and significant increase in the vesicular secretion of CD63, exactly the opposite of what the prevailing paradigm of exosome biogenesis would predict. Thus, it appears that CD63 buds from cells far better when it is localized to the plasma membrane than when it is endocytosed and delivered to endosomes, consistent with our earlier finding that mutating its endocytosis signal was all it took to increase its vesicular secretion by ~6-fold (5, 30). In light of these several observations, we took note of Latysheva et al. (41), which reported that the exosome biogenesis factor syntenin (17, 18) inhibits CD63 endocytosis by binding to the −EVMcooh region of CD63’s YxxΦ-type endocytosis signal, and that half of the syntenin molecules in the cell are bound to cell surface CD63 molecules (41). More specifically, we wondered whether syntenin drives the vesicular secretion of CD63 by inhibiting CD63 endocytosis.
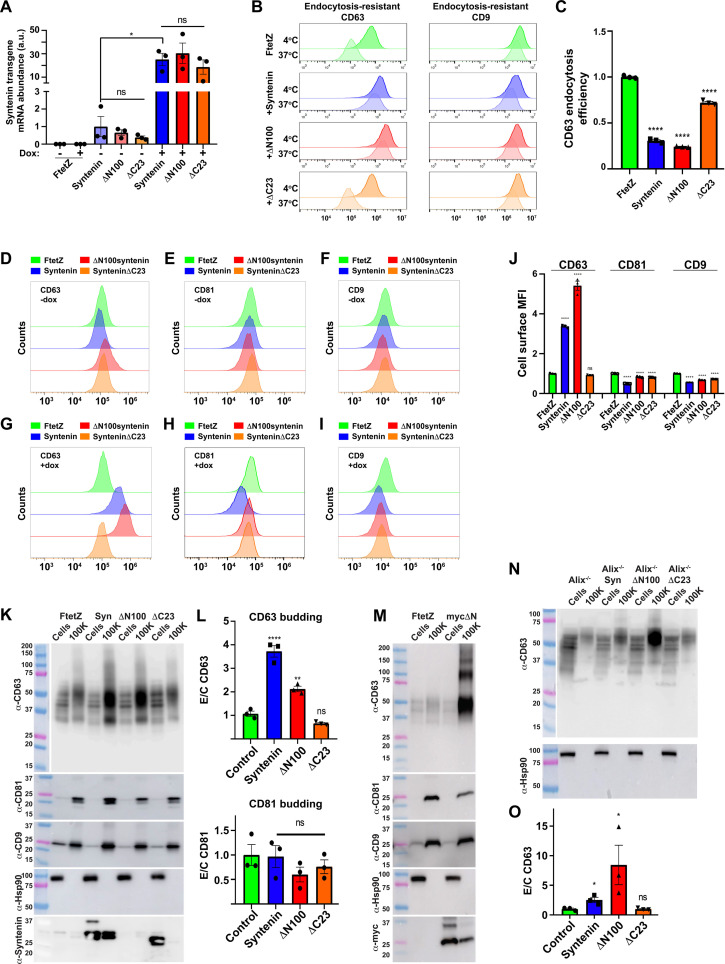
To test this hypothesis, we generated Tet-on 293F (FtetZ) cell lines that carry doxycycline (dox)–regulated, TRE3G-driven transgenes designed to inducibly express syntenin, the syntenin mutant ΔN100syntenin, or the syntenin mutant synteninΔC23 [ΔN100syntenin lacks the first 100 amino acids of syntenin, including its three Alix-binding YPLxL motifs, yet still binds CD63 and inhibits CD63 endocytosis (41), whereas synteninΔC23 retains syntenin’s Alix-binding sites but has greatly reduced CD63-binding activity (41)]. To ensure that these cell lines display the expected expression pattern for these syntenin transgenes, we grew these and control cells in the absence or presence of dox and then measured the expression of syntenin transgene mRNAs by reverse transcription–quantitative polymerase chain reaction (RT-qPCR). When grown in −dox medium, all three transgenes displayed a low but significant baseline level of transgene-encoded syntenin mRNA expression, while growth in +dox medium resulted in substantially higher levels of transgene-encoded syntenin mRNA (Fig. 2A).
Fig. 2. Syntenin drives the vesicular secretion of CD63 by blocking CD63 endocytosis.
(A) Bar graph of transgene-encoded syntenin mRNA levels in Tet-on 293F (FtetZ) cells and FtetZ cells carrying the TRE3G-regulated transgenes encoding syntenin (syntenin), ΔN100syntenin (ΔN100), or synteninΔC23 (ΔC23), grown ± dox (a.u., arbitrary units), with * denoting a P value of <0.05. (B) Flow cytometry histograms showing the fluorescence staining intensity for surface-exposed anti-CD63 and anti-CD9 mAbs following 30-min-long incubations at 4°C versus 37°C for dox-induced (green) control cells, (blue) syntenin-expressing cells, (red) ΔN100syntenin-expressing cells, or (orange) synteninΔC23-expressing cells. (C) Bar graph showing the EEs of CD63 in these cells, with **** denoting a P value of <0.0001. (D to I) Flow cytometry histograms of the same four cell lines, grown in the [(D) to (F)] absence of dox or [(G) to (I)] presence of dox, stained for surface-exposed [(D) and (G)] CD63, [(E) and (H)] CD81, or [(F) and (I)] CD9. (J) Bar graph of MFIs for these plasma membrane fluorescence brightness data, with **** denoting a P value of <0.0001. (K) Immunoblots of cells and exosome-containing fractions collected from dox-induced (FtetZ) control cells, (Syn) syntenin-expressing cells, (ΔN100) ΔN100syntenin-expressing cells, or (ΔC23) synteninΔC23-expressing cells. (L) Bar graphs showing the efficiency with which CD63 and CD81 were loaded into exosome-sized vesicles (E/C ratio) for, with ** and **** denoting P values of <0.01 and 0.0001, respectively. (M) Immunoblots of cell and exosome lysates collected from dox-induced FtetZ cells and FtetZ cells expressing mycΔN100syntenin (mycΔN). (N) Immunoblots of cell lysates and exosome-containing fractions of dox-induced Tet-on Alix−/− control cells and Alix−/− cells expressing syntenin (Syn), ΔN100syntenin (ΔN100), or synteninΔC23 (ΔC23). (O) Bar graph showing the vesicular secretion efficiency (E/C ratio) of CD63, with * denoting a Student’s t test P value of <0.05. All experiments were performed a minimum of three times.
The effect of syntenin, ΔN100syntenin, and synteninΔC23 on CD63 endocytosis was measured using the same five-step endocytosis assay described previously (see Fig. 1, A and B). The three syntenin transgenic cell lines and the control cell line (FtetZ) were grown in dox-containing medium overnight, after which the cells were chilled, incubated with unlabeled antibodies to either CD63 or CD9, maintained on ice or warmed to 37°C for 30 min, then returned to 4°C, probed with fluorescently tagged anti-mouse IgG, and examined by flow cytometry. Histograms of the flow cytometry data confirmed that high-level expression of either the syntenin or ΔN100syntenin transgenes induced a selective decrease in CD63 endocytosis (Fig. 2B). These flow cytometry data were used to calculate the MFI for each cell sample, and the MFI values were used to compute the EE for CD63 in each cell line by the formula EE = [1 − (FICD63 @ 37oC/FICD63 @ 4oC)]/[1 − (FICD9 @ 37oC/FICD9 @ 4oC)]. Application of this formula to the data from three independent trials revealed that high-level expression of syntenin expression resulted in a 3.3-fold decrease in CD63 endocytosis (P < 0.0001), that ΔN100syntenin caused a 4.1-fold decrease in CD63 endocytosis (P < 0.0001), but that synteninΔC23, which retains ~25% CD63-binding activity of syntenin (41), caused only a slight, 1.4-fold decrease in CD63 endocytosis (P < 0.0001) (Fig. 2C). The inhibition of CD63 endocytosis by expression of either syntenin or ΔN100syntenin was also observed using an immunofluorescence microscopy–based endocytosis assay (fig. S3).
To find out whether syntenin also affected CD63’s plasma membrane abundance, these same cell lines were grown overnight ± dox and then chilled and stained with a trio of fluorescently labeled antibodies specific for CD63, CD81, and CD9. In the absence of dox, surface staining for CD63 was similar in all cell lines, though slightly elevated in the ΔN100syntenin cell line, while cell surface staining for CD9 and CD81 was similar in all cell lines (Fig 2, D to F). In contrast, dox-induced expression of syntenin or ΔN100syntenin triggered a strong increase in cell surface staining for CD63 (Fig. 2G), while the cell surface staining for CD9 or CD81 was either unchanged or reduced (Fig. 2H). These flow data were used to calculate MFI for each marker, cell line, and condition. Normalization of these values to the FtetZ control line revealed that high-level expression of either the syntenin or ΔN100syntenin mRNAs triggered significant increases in cell surface MFI for CD63, whereas synteninΔC23 did not [3.4-fold for syntenin (P < 0.0001), 5.4-fold for ΔN100syntenin (P < 0.0001), and 0.9-fold for synteninΔC23 (P = 0.9)] (Fig. 2J). In contrast, we found that high-level expression of syntenin proteins caused a slight reduction in the cell surface staining MFI for CD9 and CD81. We also performed similar experiments in Tet-on Hela cell lines that inducibly express these same three syntenin proteins (fig. S4). The results from these Hela cell experiments revealed that high-level expression of syntenin or ΔN100syntenin once induced a sharp increase in cell surface staining for CD63, and that synteninΔC23 once again did not, demonstrating that the effect of syntenin on the cell surface staining for CD63 was not a peculiarity of 293 cells.
We next tested whether the syntenin-mediated inhibition of CD63 endocytosis resulted in an increase in CD63’s vesicular secretion. To do this, we grew the same four cell lines in dox-containing medium for 3 days in protein-free, chemically defined medium, in shaker cultures, after which cells and exosome-containing fractions were collected. These samples were interrogated by immunoblot using antibodies specific for CD63, CD81, CD9, Hsp90, and syntenin, which revealed that high-level expression of syntenin induced a 3.7-fold (P < 0.0001) increase in the vesicular secretion of CD63 but had little if any effect on the vesicular secretion of CD81 (Fig. 2, K and L). ΔN100syntenin also induced the vesicular secretion of CD63, by 2.1-fold (P = 0.0027), whereas synteninΔC23 had no detectable impact on CD63 budding from the cell.
To determine whether these results were peculiar to 293F cells, we repeated them using Tet-on Hela cells (StetZ) and Tet-on mouse NIH3T3 cells (3tetZ) engineered to inducibly express syntenin, ΔN100syntenin, or synteninΔC23 (fig. S5). These experiments confirmed that high-level expression of syntenin drove the vesicular secretion of CD63 from Hela and NIH3T3 cells, demonstrating that syntenin has this effect on three of the most commonly used cell lines in biomedical research. In addition to these studies, we also examined the effect of low-level syntenin expression on the vesicular secretion of CD63, CD9, and CD81, which revealed that ΔN100syntenin triggered a sharp increase in the vesicular secretion of CD63 even at low levels of expression (fig. S6).
One caveat to these results is that our anti-syntenin Ab failed to detect the ΔN100syntenin protein (Fig. 2K). This was somewhat surprising, since the transgene-encoded ΔN100syntenin mRNA was expressed to the same level as the transgene-encoded syntenin and synteninΔC23 mRNAs (see Fig. 2A). Given that dox-induced expression of ΔN100syntenin mRNA inhibited CD63 endocytosis (see Fig. 2, B and C), induced the cell surface accumulation of CD63 (see Fig. 2J), and triggered the vesicular secretion of CD63 (see Fig. 2, K and L), it is highly likely that the ΔN100syntenin protein had been expressed in these experiments. However, to demonstrate the expression of the ΔN100syntenin protein, we created a myc-tagged ΔN100syntenin (mycΔN100syntenin) transgene, and then a Tet-on 293F cell line designed to express mycΔN100syntenin in response to dox. Control and mycΔN100syntenin cells were grown in dox-containing medium as described above, followed by collection of cell lysates and exosome-containing fractions. Immunoblot of these samples confirmed that mycΔN100syntenin induced a selective increase in the vesicular secretion of CD63, with little to no effect on the vesicular secretion of CD81 or CD9, and was readily detected by anti-myc antibodies, confirming that the mycΔN100syntenin protein was expressed in these experiments (Fig. 2M). Additional experiments with cell lines designed to express myc-tagged forms of syntenin, ΔN100syntenin, and synteninΔC23 showed that mycΔN100syntenin expression is substantially lower than that of myc-tagged syntenin or myc-tagged synteninΔC23 (fig. S7), and also that none of the five different anti-syntenin antibodies that were raised against proteins or peptides contained within ΔN100syntenin were able to detect the mycΔN100syntenin protein.
Alix is not required for syntenin-induced CD63 budding from the cell
The observation that ΔN100syntenin and mycΔN100syntenin trigger the vesicular secretion of CD63 suggests that syntenin drives the vesicular secretion of CD63 independently of its binding partner Alix, as all three of syntenin’s Alix-binding sites are removed by the ΔN100 mutation. To determine whether syntenin is truly able to drive the exosomal secretion of CD63 independently of Alix, we created a Tet-on version of an Alix−/− human embryonic kidney (HEK) 293 cell line that we described previously (5) and then modified this Tet-on Alix−/− HEK293 cell line to carry the dox-inducible, TRE3G-regulated syntenin, ΔN100syntenin, and synteninΔC23 transgenes. The resulting cell lines were grown in dox-containing medium for 3 days, after which we collected cell lysates and exosome-containing fractions. Immunoblot of these fractions showed that dox-induced expression of syntenin triggered a 2.5-fold increase in the exosomal secretion of CD63 (P = 0.03) (Fig. 2, N and O), demonstrating that syntenin can drive the vesicular secretion of CD63 in the absence of Alix.
Syntenin drives CD63 into CD81/CD9-containing vesicles
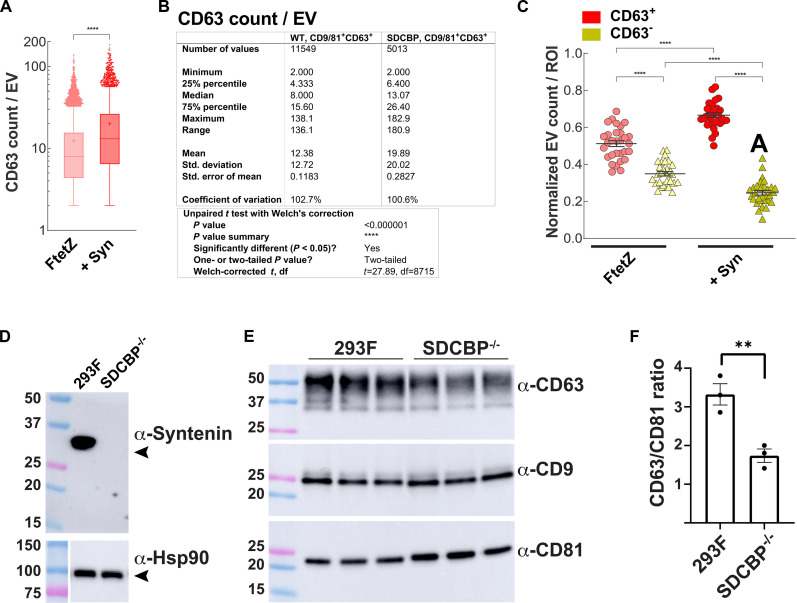
The preceding data raise the possibility that syntenin drives the vesicular secretion of CD63 by allowing it to remain at the cell surface, its preferred site of budding from the cell. Given that we previously established that plasma membrane–resident CD63 is loaded into the same exosome-sized vesicles as CD9 and CD81 (5), we expected that high-level expression of syntenin would also drive CD63 into exosome-sized vesicles together with CD81 and CD9. To explore this possibility, we grew control and syntenin-expressing 293F cells in dox-containing medium (3 days in protein-free, chemically defined medium, in shaker cultures), collected exosome-sized vesicles, and interrogated their immunophenotypes by quantitative single-molecule localization microscopy (qSMLM). Specifically, we immunopurified CD81- and CD9-containing vesicles on glass coverslips derivatized with anti-CD81 and anti-CD9 antibodies, stained the bound vesicles with fluorescently labeled anti-CD63 antibodies and fluorescently labeled anti-CD81/CD9 antibodies, and then interrogated >5000 CD81/CD9-containing, exosome-sized vesicles from each cell line by qSMLM. These experiments revealed that high-level expression of syntenin led to a 1.7-fold increase (P < 0.00001) in the number of detected CD63 molecules in CD81/CD9-positive vesicles, from an average of 12 ± 0.1 per vesicle to an average of 20 ± 0.3 per vesicle (Fig. 3A). This syntenin-induced increase in the amount of CD63 per CD81/CD9-positive vesicle was also reflected in an increase in the 25% to 75% range of detected CD63 molecules, which rose from 4 to 16 for control vesicles, to 6 to 26 for vesicles from syntenin-expressing cells (Fig. 3B). We also observed that two or more CD63 molecules were detected in most (51%) of CD81/CD9-positive vesicles collected from control cells, indicating that CD63 normally buds together with these plasma membrane–resident exosome marker proteins, and that this percentage rose to 67% for vesicles produced by syntenin-expressing cells, a 1.3-fold increase (P < 0.00001) (Fig. 3C). As for vesicle size, diameters of CD81/CD9-positive vesicles released by these two cell lines were within 10% of one another, 106 nm for 293F vesicles and 112 nm for vesicles released by syntenin-expressing cells.
Fig. 3. Syntenin drives CD63 into CD81/CD9-positive exosome-sized vesicles.
(A) Box and whisker plot showing the number of CD63 molecules detected on CD81/CD9-positive exosome-sized vesicles, as determined by qSMLM, from vesicles released by (WT) FtetZ cells and (SDCBP) FtetZ cells expressing high levels of syntenin. Note that the vesicle-to-vesicle variation spans two orders of magnitude in both samples. **** denotes P value of <0.000001from an unpaired t test with Welch’s correction. Data are from three independent trials. (B) Table of data from the same experiment as shown in (A). (C) Plot showing that high-level expression of syntenin increased the proportion of CD81/CD9-positive vesicles that contained two or more detected CD63 molecules. Each data point represents the mean values of all vesicles present within 30 regions of interest (ROIs) for each sample from six coverslips. Wide bars denote the average, and error bars denote the SEM. ****, ***, and * denote P values of <0.00005, <0.0005, and <0.05 from Dunnett’s T3 multiple comparisons test, respectively. Data are from three independent trials. (D) Anti-syntenin immunoblot of cell lysates prepared from 293F cells and the 293F/SDCBP−/− cell line that carries large indels at exons 2 and 3. Molecular weight (MW) markers are in kDa. (E) Immunoblot of exosome-containing fractions produced 293F cells and the 293F/SDCBP−/− cell line. (F) Bar graph of CD63/CD81 ratio in exosome-containing fractions produced by 293F cells and the 293F/SDCBP−/− cell line, in arbitrary units. ** refers to a Student’s t test P value of <0.01.
We also asked whether loss of syntenin causes the complementary phenotype, namely, a selective reduction in the vesicular secretion of CD63. We used Cas9-mediated gene editing to mutate the syntenin gene, SDCBP, in 293F cells, and this yielded an SDCBP−/− cell line with large insertion/deletion mutations that deleted the 3′ half of coding exon 2, all of intron 2, and the 5′ half of coding exon 3, on both alleles of the SDCBP gene. Not surprisingly, this cell line lacks syntenin protein (Fig. 3D). To determine the effect of syntenin loss on the vesicular secretion of CD63, we grew 293F and SDCBP−/− cells in suspension, in chemically defined medium for 3 days, after which we collected exosome fractions and interrogated them by immunoblot for CD63, CD9, and CD81. This revealed that loss of syntenin caused an ~50% drop (P < 0.01) in the CD63/CD81 ratio in exosome-containing fractions (Fig. 3, E and F), demonstrating that loss of syntenin caused a phenotype that was complementary to that of the syntenin-overexpressing cells. These results are not surprising, as they were reported previously by Zimmermann and colleagues (18) for syntenin knockout mice and for syntenin knockout cell lines.
To determine whether high-level expression of syntenin might affect the production of exosome-sized vesicles, we grew control cells, and the three syntenin transgenic cell lines were grown ± dox for 3 days in protein-free, chemically defined medium, in shaker cultures, after which exosome-containing fractions were collected by concentrating filtration and size exclusion chromatography. These exosome-containing fractions were then examined by nanoparticle tracking analysis (NTA), which measures the sizes and concentrations of exosome-sized vesicles and particles. This analysis revealed that there were no statistically significant differences in the overall number of exosome-sized particles released by any of these cell lines (fig. S8). These results complement those of Zimmermann and colleagues (18), who previously demonstrated a normal production of exosome-sized vesicles and particles in syntenin knockout cells.
CD63 is an expression-dependent inhibitor of mu2-mediated protein endocytosis
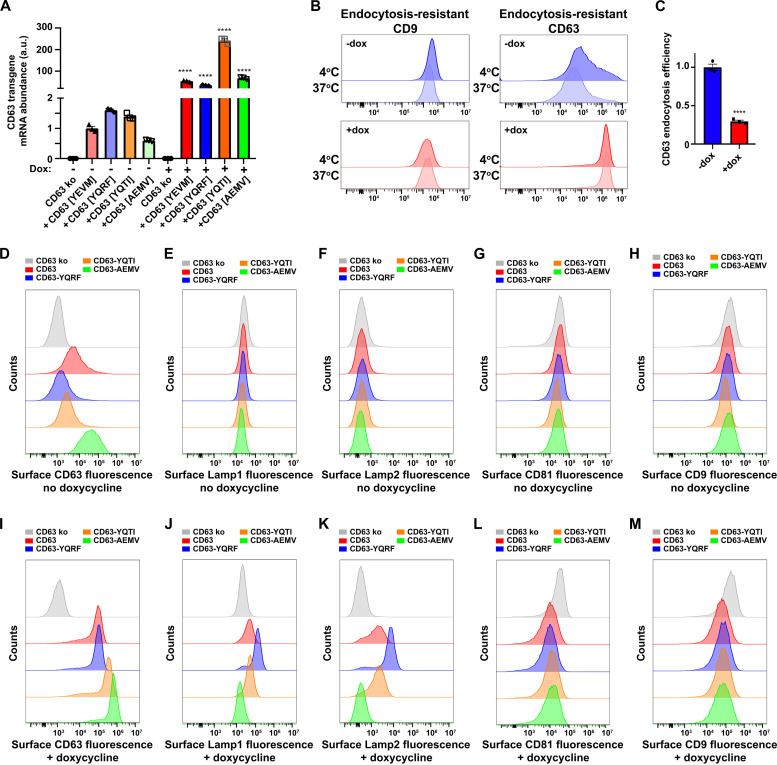
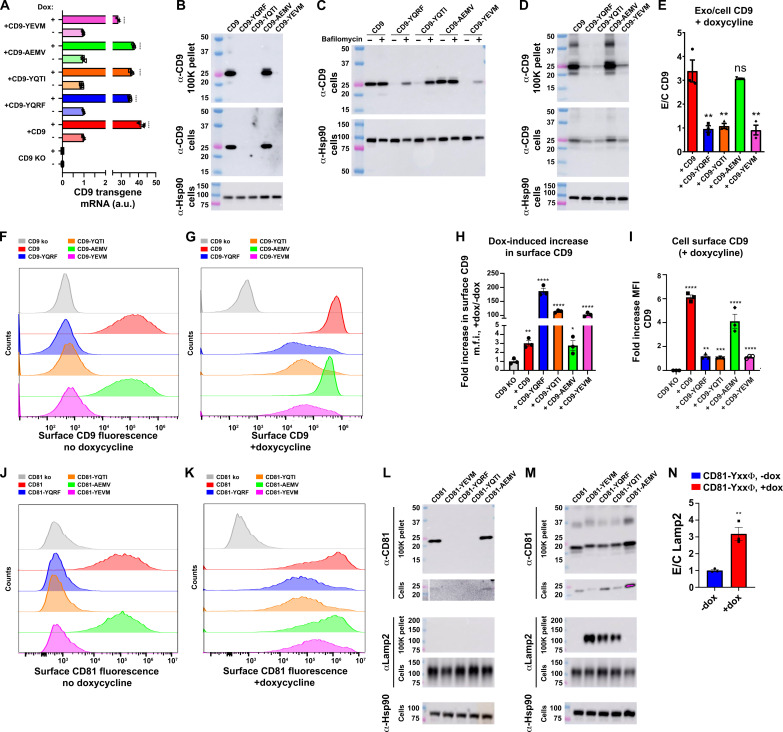
The model that emerges from these data is that endocytosis inhibits CD63’s vesicular secretion and that inhibitors of endocytosis, including syntenin, induce CD63’s budding from the cell. Because high-level expression of other endocytosis signal–containing protein is known to inhibit AP-2, through a combination of competition for limited amounts of mu2/AP-2 and protein crowding effects (32, 42–46), we next tested whether high-level expression of CD63 might do the same, resulting in the inhibition of endocytosis and the induction of CD63’s vesicualr secretion from the cell. To investigate this issue, we first created a Tet-on version of CD63−/− 293F cells (47) and then made derivatives of it that carry dox-inducible transgenes designed to express wild-type (WT) CD63, which has the endocytosis signal YEVM, CD63-YQRF [YQRF has a particularly high affinity for mu2 (32)], CD63-YQTI, or CD63-AEMV, which lacks the tyrosine at position −4 that is critical for binding mu2. Expression of these CD63 transgenes was tested by RT-qPCR, which confirmed that all four CD63 transgenic cell lines expressed low levels of their transgene-encoded CD63 mRNAs in the absence of dox, and much higher levels when the cells were grown in dox-containing medium (Fig. 4A).
Fig. 4. CD63 inhibits endocytosis and triggers the plasma membrane accumulation of itself, Lamp1, and Lamp2.
(A) Bar graph of CD63 transgene mRNA levels in Tet-on CD63−/− cells and CD63−/− cells carrying TRE3G-regulated CD63 transgenes. ****P < 0.0001. (B) Endocytosis assay flow cytometry histograms showing the cell surface fluorescence of endocytosis-resistant CD9 and CD63 following incubations at 4°C versus 37°C of CD63 transgenic cells from (blue) −dox or (red) +dox cultures. (C) Bar graph of CD63 EE values of CD63 transgenic cells grown ± dox. ****P < 0.0001. (D to M) Flow cytometry fluorescence brightness levels in FtetZ/CD63−/− cells carrying (gray) no transgene or the (red) WT CD63, (blue) CD63-YQRF, (orange) CD9-YQTI, or (green) CD63-AEMV transgenes, grown in the [(D) to (H)] absence of dox or [(I) to (M)] presence of dox, and stained for surface-exposed [(D) and (I)] CD63, [(E) and (J)], Lamp1, [(F) and (K)] Lamp2, [(G) and (L)] CD81, or [(H) and (M)] CD9.
To measure the effect of dox-induced expression of CD63 on the endocytosis of CD63, we used the same five-step endocytosis assay described previously. In brief, we grew the Tet-on CD63 knockout cell line that expresses WT CD63 (FtetZ/CD63−/−::CD63) ± dox and then (i) chilled the cells to 4°C; (ii) incubated them with unlabeled antibodies to either CD63 or CD9 followed by washing to remove unbound antibodies, also at 4°C; (iii) incubated the cells at 4°C or at 37°C for 120 min; (iv) then returned the cells to 4°C, incubated them with fluorescently tagged anti-mouse IgG, and washed them to remove unbound Ab; and (v) measured the amount of surface-labeled CD63-Ab and CD9-Ab complexes by flow cytometry. Cells grown in the absence of dox, which express low levels of CD63, displayed an internalization of surface CD63 (Fig. 4B, top right), whereas cells grown in the presence of dox failed to endocytose CD63 (Fig. 4B, bottom right). Using the MFI values from triplicate replicates of this experiment, and the formula EE = [1 − (FICD63 @ 37oC/FICD63 @ 4oC)]/[1 − (FICD9 @ 37oC/FICD9 @ 4oC)], we found that high-level expression of CD63 inhibited its own endocytosis by 3.4-fold (P < 0.0001) (Fig. 4C). This CD63 expression-induced inhibition of CD63 endocytosis was also observed using an independent, fluorescence microscopy–based CD63 endocytosis assay (fig. S9).
High-level expression of CD63 drives plasma membrane accumulation of YxxΦ proteins
Given that CD63 expression inhibits endocytosis (32, 42–46), we next examined the effect of the specific sequence of CD63’s endocytosis signal, and the level of CD63 expression, on the plasma membrane display of CD63 and two other mu2-binding, endocytosis signal–containing, lysosomal proteins, Lamp1 and Lamp2. To do so, we grew all four of the CD63 transgenic cell lines ± dox, chilled the cells to 4°C, stained them with fluorescently labeled antibodies specific for CD63, Lamp1, Lamp2, CD81, and CD9, and then examined them by flow cytometry.
In the absence of dox, the cell surface staining (MFI) for CD63 was highest for CD63-AEMV, which lacks a functional endocytosis signal, which was ~80-fold higher than background staining observed for CD63 knockout cells, whereas the cell surface staining for WT CD63 was ~4-fold lower, and the staining for CD63-YQRF was about half of that (Fig. 4D). Staining these same cells with antibodies specific for Lamp1, Lamp2, CD81, and CD9 showed that the low-level expression of these CD63 proteins had no effect on their cell surface staining, as the staining levels were in each case the same as in CD63 knockout cells (Fig. 4, E to H). Results were quite different for cells grown in dox-containing medium. Specifically, we found that staining (MFI) for all four forms of CD63 was now >100-fold above background, with the fold increase greatest for CD63-YQRF, which has the highest affinity for mu2 (Fig. 4I) (32). As for whether high-level expression of the three YxxΦ-containing forms of CD63 affected the cell surface staining of other YxxΦ-containing proteins from the plasma membrane, we found that the cell surface staining for Lamp1 and Lamp2 was substantially higher (Fig. 4, J and K). Quantification of these MFI changes across multiple trials revealed that high-level expression of WT CD63 increased the cell surface staining for Lamp1 and Lamp2 by 2.3-fold (P = 0.035) and 5.5-fold (P = 0.013), respectively, while high-level expression of CD63-YQRF increased the cell surface staining MFI for Lamp1 and Lamp2 by 5.9-fold (P = 0.011) and 19-fold (P = 0.0027), respectively. In contrast, cell surface staining for CD81 and CD9 was lower for all CD63-expressing cell lines (Fig. 4, L and M), highlighting the specificity of the Lamp1 and Lamp2 changes.
High-level expression of CD63 triggers the vesicular secretion of itself, Lamp1, Lamp2, and even AP-2 mu2
We next examined the effect of CD63’s endocytosis signal and expression level on the vesicular secretion of itself, the exosome marker CD9, the lysosomal proteins Lamp1 and Lamp2, the cytoplasmic protein Hsp90, and the CD63-binding endocytosis factor mu2. To do this, we grew the four CD63 transgenic cell lines 3 days in protein-free, chemically defined medium, in shaker cultures, in the absence dox, after which we collected cell lysates and exosome-containing fractions. Interrogation of these samples by immunoblot revealed that WT CD63 could be detected in the exosome-containing fractions but that CD63 lacking an endocytosis signal budded from the cell more efficiently, whereas CD63 proteins that retained an endocytosis signal but lacked an -EVMcooh syntenin-binding motif were secreted from the cell less efficiently (Fig. 5A). This was confirmed by quantitation of results from three independent trials, which revealed that the normalized vesicular secretion efficiency (E/C ratio) of CD63-YQRF and CD63-YQTI was reduced 0.8-fold relative to WT CD63 (P = 0.0042 and 0.0074, respectively), while the normalized E/C ratio of CD63-AEMV was increased significantly (1.8-fold, P = 0.0038) (Fig. 5B). Control immunoblots confirmed the presence of the exosome marker CD9 in the exosome-containing fractions and the presence of Hsp90, Lamp1, Lamp2, and mu2 in the cell lysates. It should be noted that all four CD63 proteins were detected upon longer exposure of the cell lysate immunoblots (fig. S10).
Fig. 5. Endocytosis inhibits the vesicular secretion of CD63, while CD63 expression induces the vesicular secretion of Lamp1, Lamp2, and AP-2 mu2.
(A) Immunoblots of cell lysates and exosome-containing fractions (100K pellet) collected from uninduced FtetZ/CD63−/− cells carrying WT CD63, CD63-YQRF, CD9-YQTI, or CD63-AEMV transgenes. MW size markers in kDa. (B) Bar graph showing that strengthening the CD63 endocytosis signal inhibited its vesicular secretion, while mutationally inactivating the CD63 endocytosis signal increased its vesicular secretion. E/C ratios were normalized to 1 for the WT CD63 sample in this experiment. **P < 0.01. (C) Immunoblots of cell lysates and exosome-containing fractions (100K pellet) collected from dox-induced FtetZ/CD63−/− cells carrying WT CD63, CD63-YQRF, CD9-YQTI, or CD63-AEMV transgenes. MW size markers in kDa. (D) Bar graph showing that high-level expression led to all CD63 proteins displaying the same vesicular secretion (E/C ratio). E/C ratios were normalized to 1 for the WT CD63 sample in this experiment. denotes P value of >0.05. (E to G) Bar graphs of E/C ratios for (E) Lamp1, (F) Lamp2, and (G) mu2 in CD63 knockout cells carrying the WT CD63, CD63-YQRF, or CD9-YQTI transgenes, grown in the (blue) absence or (red) presence of dox. E/C ratios were in each case normalized to 1 for the -dox sample. ***P < 0.001, ****P < 0.0001. (H) Transmission electron micrographs of FtetZ/CD63−/− cells expressing a high level of CD63. Arrowheads point to the presumed neck of possible budding intermediates. Scale bar, 100 nm. (I) Flow cytometry histograms showing the surface-stained CD63 fluorescence of 293F cells grown overnight in the (blue) absence of bafilomycin or (red) presence of bafilomycin. (J) Bar graph showing the relative levels of cell surface CD63 staining (MFI) of 293F cells grown in the (blue) absence of bafilomycin or (red) presence of bafilomycin. **P < 0.01.
In contrast to these results, growth of these same four cell lines in dox-containing medium revealed that all four forms of CD63 displayed a robust vesicular secretion from the cell (Fig. 5C). Moreover, quantitation of their normalized vesicular secretion showed that all four forms of CD63 budded from the cell at approximately the same efficiency (Fig. 5D), consistent with our previous observation that these proteins all displayed a pronounced accumulation at the plasma membrane (see Fig. 4I). Interrogation of these same samples with antibodies specific for the YxxΦ motif–containing lysosome membrane proteins Lamp1 and Lamp2 showed that high-level expression of the YxxΦ motif–containing forms of CD63 (WT CD63, CD63-YQRF, or CD63-YQTI) led to a pronounced increase in their loading into exosome-sized EVs, with the E/C ratio of Lamp1 increasing by 4.8-fold (P < 0.0001) (Fig. 5E) and the E/C ratio of Lamp1 increasing by Lamp2 by 3.4-fold (P < 0.0001) (Fig. 5F), consistent with the effect of CD63 expression on their plasma membrane staining MFI levels (see Fig. 4, J and K). Furthermore, we found that high-level expression of YxxΦ motif–containing forms of CD63 also induced the vesicular secretion of the AP-2 subunit mu2 (which binds directly to YxxΦ motifs), reflected here in a 4.4-fold increase in its E/C ratio (P = 0.00040) (Fig. 5G). Thus, while other YxxΦ motif–containing proteins inhibit AP-2 by a combination of competitive inhibition and protein crowding effects (32, 42–46), high-level expression of CD63 also inhibits mu2 by driving its vesicular secretion and cellular depletion.
Our results show a strong positive correlation between a protein’s plasma membrane staining and its vesicular secretion, as well as a strong inhibitory effect of endocytosis on a protein’s vesicular secretion from the cell. This would suggest that CD63 and other exosome marker proteins bud directly from the plasma membrane. It also suggests that high-level expression of proteins like CD63 might allow us to detect the morphological signature of plasma membrane vesicle budding, namely, the presence of outward budding intermediates at the cell surface. To explore this possibility, we grew the CD63-expressing cell line in dox-containing medium overnight and then processed the cells for transmission electron microscopy (TEM). Specifically, the cells were fixed and dehydrated, a process that washes away all plasma membrane–derived vesicles, then embedded in Epon, sectioned, stained with uranyl acetate, and examined by TEM. Although the detection of a nascent budding intermediate in TEM thin sections is the topological equivalent of slicing through a speck of dust on the surface of a basketball, we detected plasma membrane structures that had the appearance expected of nascent budding intermediates (Fig. 5H). Some had very thin connections to the plasma membrane, some had the appearance expected for an earlier stage of budding in which constricted necks could be detected, and some had the appearance of structures arrested at an earlier stage of vesicle formation, a range of morphologies similar to what has been reported for retrovirus budding (48).
Bafilomycin induces a threefold increase in plasma membrane CD63 staining
The preceding results raise the question of whether lysosomotropic agents that are known to trigger endolysosomal exocytosis and induce the release of exosome-sized vesicles and exosome markers like CD63 might also increase the plasma membrane levels of CD63. To explore this possibility, we grew 293F cells in the presence or absence of bafilomycin, a vacuolar adenosine triphosphatase (V-ATPase) inhibitor known to trigger the release of CD63-containing, exosome-sized vesicles (20, 49–51). The cells were then chilled, stained with fluorescently labeled antibodies to CD63, and then examined them by flow cytometry. This experiment revealed that bafilomycin triggered a substantial increase in the cell surface staining for CD63 (Fig. 5I), 3.0-fold higher than that of control 293F cells (P = 0.0015) (Fig. 5J). Thus, the effect of bafilomycin and other inducers of protein budding from the cell might involve endocytosis-independent exosome budding from the plasma membrane, in addition to the release of intracellular stores of ILVs.
Endocytosis inhibits CD9 secretion and triggers its destruction
To determine whether endocytosis signals and expression level have similar effects on the cell surface accumulation and vesicular secretion of CD9, another highly enriched exosome marker protein, we created a Tet-on CD9−/−/− cell line (fig. S11) and then created derivatives of this cell line that carry dox-regulated transgenes encoding WT CD9 or CD9 proteins with the C-terminal peptides −RSGYEVMCOOH, −RSGYQRFCOOH, −RSGYQTICOOH, or −RSGAEMVCOOH. RT-qPCR confirmed that these dox-induced CD9 transgenes were expressed at low baseline levels in the absence of dox, and at higher levels when grown in the presence of dox (Fig. 6A). Next, we grew these five CD9 transgenic cell lines in medium lacking dox (3 days in protein-free, chemically defined medium, in shaker cultures), followed by collection of cell lysates and exosome-containing fractions. These were then examined by immunoblot, which revealed that WT CD9 and CD9-AEMV were readily detected in exosome-containing fractions, whereas CD9-YQRF, CD9-YQTI, and CD9-YEVM were not (Fig. 6B, top). We could only detect WT CD9 and CD9-AEMV in the cell lysates at much longer periods of blot exposure. Moreover, we were not able to detect CD9-YQRF, CD9-YQTI, or CD9-YEVM in the cell lysates, no matter how long we exposed the blot (Fig. 6B, middle panel), although their mRNAs were expressed to similar levels as the WT CD9 and CD9-AEMV mRNAs. These results confirm that CD9 buds from cells at high efficiency, confirm that endocytosis strongly inhibits the vesicular secretion of this exosome marker proteins, and show that endocytosis triggers an efficient and nearly complete degradation of CD9. CD9-YQRF, CD9-YQTI, and CD9-YEVM could be detected in cell lysates following incubation with the V-ATPase inhibitor bafilomycin (Fig. 5C), which is known to inhibit lysosomal proteolysis (52).
Fig. 6. Directed endocytosis of CD9 or CD81 inhibits their vesicular secretion and triggers their destruction.
(A) Bar graph of transgene-encoded CD9 mRNA levels ± dox, in Tet-on CD9−/−/− cells and matching cells carrying TRE3G-regulated CD9 transgenes, with **** denoting a P value of <0.0001. (B) Immunoblots of cells and exosome-containing fractions collected from uninduced Tet-on CD9−/−/− cells and matching cells expressing CD9, CD9-YQRF, CD9-YQTI, CD9-AEMV, or CD9-YEVM. (C) Immunoblots of cell lysates collected from the same cell lines grown overnight ± 30 nM bafilomycin A1. (D) Immunoblots of cells and exosome-containing fractions collected from dox-induced Tet-on CD9−/−/− cells expressing CD9, CD9-YQRF, CD9-YQTI, CD9-AEMV, or CD9-YEVM. (E) Bar graph showing the vesicular secretion efficiency (E/C ratio) for each form of CD9, with ** denoting a P value of <0.01. E/C ratios were normalized to 1 for the CD9-YQRF sample in this experiment. (F and G) Flow cytometry histograms showing the cell surface, anti-CD9 fluorescence staining intensity of (gray) Tet-on CD9−/−/− cells and cells expressing (red) CD9, (blue) CD9-YQRF, (orange) CD9-YQTI, (green) CD9-AEMV, or (purple) CD9-YEVM, which had been grown in the (F) absence of dox or (G) presence of dox. (H) Bar graph showing the fold increase in cell surface CD9 MFIs triggered by addition of dox, with **** denoting a P value of <0.0001. (I) Bar graph showing the MFIs of cell surface CD9 staining, with *** and **** denoting P values of <0.001 and <0.0001, respectively. (J and K) Immunoblots of cell lysates and exosome-containing fractions from (J) uninduced and (K) dox-induced Tet-on CD81−/− cells expressing CD81, CD81-YEVM, CD81-YQRF, CD81-YQTI, or CD81-AEMV. (L and M) Flow cytometry histograms showing the anti-CD81 surface-stained fluorescence staining intensities of (gray) Tet-on CD81−/− cells and matching cells expressing (red) CD81, (blue) CD81-YQRF, (orange) CD81-YQTI, (green) CD81-AEMV, or (purple) CD81-YEVM, grown in the (L) absence or (M) presence of dox. (N) Bar graph showing the vesicular secretion efficiency (E/C ratio) for Lamp2 in cells expressing endocytosed forms of CD81, normalized to 1 in the -dox sample, with ** denoting a P value of <0.01. Experiments were performed three times.
High-level expression of CD9-YxxΦ proteins stabilizes them and triggers their vesicular secretion
If high-level expression of AP-2 ligands like CD63 saturates protein endocytosis, and if endocytosis triggers CD9 destruction, then high-level expression should stabilize the CD9-YQRF, CD9-YQTI, and CD9-YEVM proteins and also induce their vesicular secretion from the cell. To test these predictions, we grew these same five cell lines in medium containing dox (3 days in protein-free, chemically defined medium, in shaker cultures), after which we collected cell lysates and exosome-containing samples. As predicted, immunoblot of these sample revealed that high-level expression led to the accumulation of CD9-YQRF, CD9-YQTI, and CD9-YEVM in cell lysates to a level nearly as high as WT CD9 and CD9-AEMV (Fig. 6D, middle panel). Furthermore, we found that all five forms of CD9 could now be detected in exosome-containing fractions (Fig. 6D, top), although the vesicular secretion of WT CD9 and CD9-AEMV (E/C ratio) was still significantly higher [threefold (P < 0.01)] than that of CD9-YQRF, CD9-YQTI, or CD9-YEVM (Fig. 6E) (note that blots of +dox samples were exposed for less time than blots of –dox samples).
High-level expression of CD9-YxxΦ proteins drives their plasma membrane accumulation
To examine the effect of expression level on the cell surface staining for CD9, we grew CD9 knockout cells and these five CD9 transgenic cell lines ± dox, then chilled the cells, stained them with fluorescently labeled CD9 antibodies, on ice, and examined them by flow cytometry. In the absence of dox, the cell surface staining for WT CD9 and CD9-AEMV was substantially higher than the background staining observed for CD9 knockout cells, whereas the staining for CD9-YQRF, CD9-YQTI, and CD9-YEVM was barely above background (Fig. 6F). These differences were quantified from MFI values across multiple independent trials, which revealed that the cell surface staining for WT CD9 and CD9-AEMV was 450-fold (P = 0.0001) and 330-fold (P = 0.0002) above background, respectively. In contrast, the cell surface staining for CD9-YQRF, CD9-YQTI, and CD9-YEVM was only ~2-fold above background [1.4-fold for CD9-YQRF (P = 0.008), 2.0-fold for CD9-YQTI (P = 0.0009), and 2.4-fold for CD9-YEVM (P = 0.001)]. Thus, the presence of an endocytosis signal reduced the cell surface staining of CD9 by >100-fold.
In contrast, high-level expression of these CD9 transgenes led to large increases in the cell surface staining for all forms of CD9, but especially for the YxxΦ-containing forms of CD9 (Fig. 6G). Specifically, we found that the cell surface staining (MFI) for WT CD9 and CD9-AEMV increased ~3-fold, to 1300-fold above background (P < 0.0001) for WT CD9 and 880-fold above background (P < 0.0001) for CD9-AEMV (Fig. 6H). In contrast, the cell surface staining (MFI) for CD9-YQRF was now 250-fold above background (P = 0.0005), a 180-fold increase relative to its −dox levels. Similar results were observed for CD9-YQTI and CD9-YEVM, as their cell surface staining MFI values were now 220-fold above background (P < 0.0001) and 240-fold above background (P < 0.0001), increases of 110-fold and 100-fold, respectively. Thus, the effect of high-level expression on the cell surface staining was evident for all forms of CD9, but was far larger, ~100- to 200-fold versus 3-fold, for the endocytosis signal–containing forms of CD9. That being said, the cell surface levels of WT CD9 and CD9-AEMV were still 6-fold and 4-fold higher than those of CD9-YQRF, CD9-YQTI, or CD9-YEVM (Fig. 6I), just as the efficiency of CD9’s vesicular secretion was ~3-fold higher for WT CD9 and CD9-AEMV than for CD9-YQRF, CD9-YQTI, or CD9-YEVM.
Endocytosis is a saturable inhibitor of CD81 plasma membrane expression
Together, these results indicate that exosome marker proteins bud from cells best when localized to the plasma membrane. To test this hypothesis in the context of CD81, the most highly enriched of all exosome marker proteins (3, 5), we created a CD81 knockout 293F cell line (fig. S11), converted it to a Tet-on line, and then generated derivatives of this Tet-on CD81−/− cell line that carry dox-regulated transgenes encoding WT CD81, CD81-RSGYQRF, CD81-RSGYQTI, CD81-RSGYEVM, or CD81-RSGAEMV. These cell lines were then grown overnight ± dox, chilled, incubated with fluorescently labeled anti-CD81 antibodies, and interrogated by flow cytometry. At the low, −dox level of CD81 transgene expression, we observed that the cell surface staining for WT CD81 and CD81-AEMV was far higher than the background staining of CD81 knockout cells, whereas the cell surface staining levels for CD81-YQRF, CD81-YQTI, or CD81-YEVM were barely above background (Fig. 6J). By comparison of MFI values across multiple trials, it appeared that the cell surface staining for WT CD81 and CD81-AEMV was 237-fold above background (P < 0.0001) and 210-fold above background (P < 0.0001), respectively. In contrast, the cell surface staining (MFI) across multiple trials for CD81-YQRF, CD81-YQTI, and CD81-YEVM was only 1.1× (P = 0.6), 1.1× (P = 0.4), and 1.5× (P = 0.014) above background, respectively.
In contrast, high-level, dox-induced expression led to cell surface CD81 staining that was far above background for all five CD81 proteins (Fig. 6K). Cell surface staining MFI was highest for WT CD81 and CD81-AEMV, as these were 1200-fold above background for WT CD81 (P < 0.0001) and 1100-fold for CD81-AEMV (P < 0.0001), levels that are ~5-fold higher than in uninduced cells. In contrast, dox-induced expression led to cell surface staining for CD81-YQRF that was 380-fold above background (P < 0.0001), for CD81-YQTI that was 330-fold above background (P < 0.0001), and for CD81-YEVM that was 810-fold above background (P < 0.0001). These values represent increases in cell surface staining MFI that were 350-fold above their −dox staining for CD81-YQRF (P = 0.001), 290-fold above −dox staining for CD81-YQTI (P = 0.002), and 550-fold above −dox staining for CD81-YEVM (P = 0.0003). Thus, while endocytosis signals effectively remove CD81 from the plasma membrane at low levels of expression, high-level expression appears to saturate their endocytosis, resulting in >200-fold increases in the cell surface staining for YxxΦ motif–containing forms of CD81.
Endocytosis is a saturable inhibitor of CD81’s vesicular secretion
To determine whether endocytosis signals and expression level affected the vesicular secretion of CD81 the way they affected CD9 and CD63, we grew these five CD81 transgenic cell lines for 3 days, ± dox, in chemically defined medium, in shakers, and then collected cell lysates and exosome-containing fractions. Interrogation of the −dox samples revealed that WT CD81 and CD81-AEMV were readily detected in exosome-containing fractions, but CD81-YQRF, CD81-YQTI, and CD81-YEVM were not (Fig. 6L). Furthermore, while prolonged exposure of the cell lysates allowed us to detect faint bands for WT CD81 and CD81-AEMV in cell lysates, we were unable to detect the CD81-YQRF, CD81-YQTI, or CD81-YEVM, no matter how long we exposed the −dox cell lysate blots. These results are similar to what we observed for endocytosis signal–containing forms of CD9 and indicate that endocytosis also triggers the destruction of CD81. In addition, when we interrogated these cell and vesicle lysates for Lamp2, we detected it only in the cell lysates and not in the exosome-containing fractions.
These results changed substantially when these cells were grown in dox-containing medium, we found that high-level expression led to the cellular accumulation and vesicular secretion of all five CD81 proteins, regardless of whether they carried an endocytosis signal (Fig. 6M), consistent with the flow cytometry data and the idea that exosome markers bud from cells best when localized to the plasma membrane. In addition, we found that high-level expression of CD81-YQRF, CD81-YQTI, and CD81-YEVM triggered the vesicular secretion of Lamp2, leading to a 3.2-fold increase in its E/C ratio (P = 0.005) (Fig. 6N), results that are similar to what we observed earlier for cells treated with th dynamin inhibitor dyngo-4a or expressing high levels of CD63.
DISCUSSION
The biogenesis of intracellular organelles has been elucidated, in large part, by studying the biogenesis of their highly enriched proteins (24–28). This same approach can also be applied to elucidating the pathways of exosome biogenesis, as its been known for decades that these small extracellular vesicles are defined, in large part, by their high degree of enrichment in CD81, CD9, and CD63. Our analysis of their biogenesis demonstrates that these proteins bud from cells far better when they are localized to the plasma membrane, and far worse when they are endocytosed from the plasma membrane. These conclusions are based on a wide array of observations, summarized and interpreted below, and have significant implications for how how cells make exosomes.
Inhibitors of endocytosis induce the vesicular secretion of CD63
It is widely assumed that CD63 buds from cells by endocytosis from the plasma membrane, after which its loaded into nascent ILVs at the endosome membrane, and then secreted during the exocytosis of ILV-containing compartments (2, 8, 14–21). If this endocytosis-dependent model is correct, blocking CD63 endocytosis should inhibit CD63’s vesicular secretion from the cell. However, when we inhibited CD63 endocytosis, we observed that CD63 budding from the cell increased significantly. Moreover, we showed this using six mechanistically distinct inhibitors of CD63 endocytosis: (i) mutational inhibition of the AP-2 mu2 subunit gene AP2M1, (ii) inhibition of actin-dependent endocytosis with latrunculin, (iii) inhibition of dynamin-mediated endocytosis with dyngo-4a, (iv) syntenin-mediated inhibition of CD63 endocytosis, (v) mutational inactivation of CD63’s own endocytosis signal, and (vi) inhibition of endocytosis by high-level expression of CD63 itself. These results are also notable because they do not extend to CD81 of CD9, which are the most highly enriched of all human exosome marker proteins, lack endocytosis signals, and reside primarily at the plasma membrane (5, 30). Thus, the inhibitory effect of endocytosis on the vesicular secretion of CD63 appears to be indirect, manifested at the level of removing CD63 from the cell surface, and suggesting that exosomal CD63 arises primarily by the endocytosis-independent budding of CD63 from the plasma membrane.
Endocytosis inhibits the vesicular secretion of CD81 and CD9 and triggers their destruction
We showed here that endocytosis is also a potent inhibitor of CD81 and CD9 budding from the cell too. Specifically, we found that appending any of three different YxxΦ-type endocytosis signals to the C terminus of either CD81 or CD9 resulted in so strong a defect in their vesicular secretion that they could not be detected in secreted vesicles. These results add to the already considerable evidence that endocytosis inhibits the vesicular secretion of exosome marker proteins.
Our results also demonstrated that endocytosis also led to the destruction of these two widely-used exosome marker proteins, most likely by lysosomal proteolysis (52). This result is not particularly surprising, but it is revealing, as it suggests that the fate of endocytosed CD81 and CD9 is destruction rather than vesicular secretion. Furthermore, if these results are reflective of the fate of normally endocytosed CD81 and CD9 proteins, it would mean that ILVs containing CD81 or CD9 are likely destined for destruction, and that CD81/CD9-containing, exosome-sized EVs likely arose by direct budding from the plasma membrane.
Plasma membrane abundance positively correlates with a protein’s budding from the cell
Our data show that all three highly enriched exosome marker proteins budded from cells best when they were localized to the plasma membrane, and far worse when they were targeted to endosomes. Evidence for this is seen in nearly every experiment of this paper (Table 1). These findings also harken back to the original discovery of exosome marker proteins by Escola et al. (3), which was the first to show that CD81 and CD63 were highly enriched in exosomes, and that CD81 was ~18-fold more enriched in exosomes than CD63. We can now place the stark difference between CD81 and CD63 into its proper cell biological context, in part because we now know that CD81 resides at the plasma membrane, that CD63 has an endocytosis signal and is efficiently removed from the plasma membrane, and that redirecting CD63 from endosomes to the plasma membrane leads to a substantial and significant increase in its budding from the cell. The correlation between an exosome marker protein’s plasma membrane accumulation and its vesicular secretion is also reflected in previous studies that documented the budding of exosomes from endosome marker–containing domains of plasma membrane (53, 54), and from papers that showed that plasma membrane anchors are sufficient to induce the vesicular secretion of budding-competent proteins, whereas endosome membrane anchors are not (55, 56). Not surprisingly, TEM presented here shows the presence of what appear to be nascent vesicle budding intermediates at the plasma membrane, similar to those seen in retrovirus-infected cells (48).
Table 1. Comparison of experimental data with predictions of the endocytosis-independent and endocytosis-dependent pathways of exosome marker protein secretion.
This table lists 17 key observations of this report (left column), together with whether these observations are predicted by the endocytosis-independent pathway of exosome marker protein secretion (middle column), or by the prevailing hypothesis that exosomes are generated by an endocytosis-dependent pathway (right column). PM, plasma membrane.
| Observation | Endocytosis-independent pathway? | Endocytosis-dependent pathway? |
|---|---|---|
| Plasma membrane markers CD81 and CD9 budded from cells 15-fold to 5-fold better than the endocytosed marker CD63 | Yes | No |
| Knockout of AP2M1 induced the vesicular secretion of CD63 but not CD81 | Yes | No |
| Inhibiting actin-dependent endocytosis increased the vesicular secretion of CD63 but not CD81 | Yes | No |
| Inhibiting dynamin-dependent endocytosis increased the vesicular secretion of CD63 but not CD81 | Yes | No |
| Inhibiting dynamin-dependent endocytosis increased the vesicular secretion of Lamp2 but not CD81 | Yes | No |
| Syntenin induced the vesicular secretion of CD63 but not CD81 | Yes | No |
| ΔN100syntenin also induced the vesicular secretion of CD63 but not CD81 | Yes | No |
| Mutational inactivation of CD63’s YxxΦ endocytosis signal increased its vesicular secretion | Yes | No |
| Mutational strengthening of CD63’s YxxΦ endocytosis signal reduced its vesicular secretion | Yes | No |
| High-level expression of CD63 inhibited its own endocytosis and induced its vesicular secretion | Yes | No |
| High-level expression of CD63 induced the PM accumulation and vesicular secretion of Lamp1 and Lamp2 | Yes | No |
| High-level expression of CD63 induced the vesicular secretion and cellular depletion of AP-2 mu2 | Yes | No |
| Appending YxxΦ endocytosis signals to CD9 inhibited its vesicular secretion and triggered its destruction | Yes | No |
| High-level expression of CD9-YxxΦ proteins induced their PM accumulation and vesicular secretion, and inhibited their destruction | Yes | No |
| Appending YxxΦ endocytosis signals to CD81 inhibited its vesicular secretion and induced its destruction | Yes | No |
| High-level expression of CD81-YxxΦ proteins induced their PM accumulation and vesicular secretion, and inhibited their destruction | Yes | No |
| High-level expression of CD81-YxxΦ proteins induced the vesicular secretion of Lamp2 | Yes | No |
Exosome marker protein budding is mediated by a stochastic rather than self-assembly process
A hallmark of self-assembling particles is their high degree of uniformity in composition and stoichiometry. Examples of self-assembling particles include SV40 virions, which contain 72 pentamers of VP1 (57, 58), and ribosomes, which contain an invariable core of 72 proteins (59). In contrast, the immunopurified vesicles studied here, which have the same size as exosomes (~30 to 150 nm diameter), have the same topology as exosomes (outside out, inside in), and are loaded with exosome marker proteins (e.g. CD81, CD9, and CD63), show no evidence of self-assembly. Rather, they vary by >50-fold in the numbers of detected exosome marker proteins per vesicle even when produced by a single cell line (5, 30, 60, 61). These results were seen again in this study, as we interrogated thousands of 293F-derived, CD81/CD9-containing, exosome-sized vesicles by qSMLM, and found that the levels of CD63 varied by >50-fold. Moreover, half of these vesicles contained CD63, whereas half did not, thus, the data presented here suggest that cells bud exosome marker proteins by a stochastic mechanism, rather than by a self-assembly process.
Endocytosis-independent and endocytosis-dependent pathways of exosome biogenesis
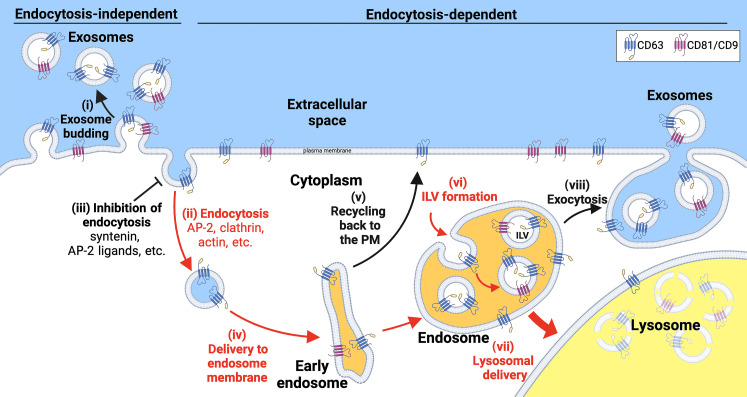
Together, our results support a model of exosome biogenesis in which highly enriched exosome marker proteins bud from cells by a single stochastic mechanism that operates along the spectrum of plasma and endosome membranes (Fig. 6). More specifically, this model proposes that the vesicular secretion of highly enriched exosome marker proteins (i) occurs primarily by an endocytosis-independent pathway at the plasma membrane, (ii) is strongly inhibited by their endocytosis, and (iii) is, for CD63 and perhaps other lysosomal proteins, strongly induced by inhibitors of endocytosis. As for what happens to endocytosed proteins, this likely involves their (iv) delivery to endosomes, (v) possible recycling back to the plasma membrane (directly or indirectly), and (vi) loading into ILVs, with the resultant ILVs retained indefinitely, (vii) destroyed in lysosomes, or (viii) secreted from the cell.
This model highlights the close interconnections between exosome biogenesis, protein endocytosis, and endolysosomal exocytosis. These interconnections are evident throughout our paper, from the effect of endocytosis inhibitors on the vesicular secretion of CD63, the effect of endocytosis signals on vesicular secretion and destruction of CD81 and CD9, and the effect of endolysosomal exocytosis inducer bafilomycin on the cell surface staining for CD63. It also draws attention to the fact that there is no way to know the origin membrane of any individual exosome-sized vesicle once it has left the cell, a problem noted more than 10 years ago (29), and affirmed by numerous other observations (1, 5, 29, 30, 53, 55, 56). The co-budding of CD63 along with CD81 and CD9 in half of all CD81/CD9 vesicles, and even more in syntenin-expressing cells, raises the possibility that much of the CD63 that is secreted from cells in exosome-sized EVs arises by direct budding from the plasma membrane.
In addition to its ability to predict and/or explain a wide array of probative empirical data (Table 1), the model presented here has significant conceptual advantages. First, by accepting the unitary nature of exosome protein budding from plasma and endosome membranes, this model adheres to the principle that “plurality should not be posited without necessity” [“pluralitas non est ponenda sine necessitate,” W. Occam, via Thorburn (62)] and thereby avoids the awkward use of two different names for vesicles that cannot be distinguished from one another by size, topology, or molecular content. Second, our model is a testable mechanistic hypothesis designed to facilitate experimental design, improve data interpretation, and invite experimental interrogation in ways that an inviolable definition cannot. Third, by adopting a traditional, empirically based definition of an exosome (i.e., small secreted vesicles of ~30 to 150 nm diameter that are enriched in exosome marker proteins and have the same topology as the cell), this model connects data from the long history of exosome research with modern principles of cell biology, one of which is that the mechanisms of exosome biogenesis can be elucidated by studying their most highly enriched proteins.
Although our use of the term “exosome” (i.e., small secreted vesicles of ~30 to 150 nm diameter that are enriched in exosome marker proteins and have the same topology as the cell) is acceptable to the flagship journal (Extracellular Vesicles) of the American Association of Extracellular Vesicles, and to several other peer-reviewed journals (1, 5, 29, 47, 53–56, 61, 63–66), it differs from the definition proposed by the International Society for Extracellular Vesicles (ISEV), which defines exosomes as secreted ILVs, and asserts that plasma membrane–derived vesicles are so different from exosomes that they have to be referred to by a different name (ectosome). We respect the right of ISEV and its members to adopt the definitions they prefer, and we ask the same in return.
Implications for syntenin, Alix, and the syntenin/Alix axis in the vesicular secretion of CD63
In addition to providing a simple yet coherent model for how cells bud exosome marker proteins, our data show that the exosome biogenesis factor syntenin contributes to the secretion of CD63 by inhibiting its endocytosis. Before our study, Latysheva et al. (41) found that syntenin binds the C-terminal tail of CD63 via its twin PDZ domains, that half of all syntenin is localized to the plasma membrane in complex with plasma membrane CD63 molecules, and that syntenin inhibits CD63 endocytosis by binding over the same peptide as the CD63 endocytosis signal. These observations, together with our discovery that redirecting CD63 to the plasma membrane induced its vesicular secretion by ~6-fold (5, 30), raised the possibility that syntenin drives the vesicular secretion of CD63 by inhibiting its endocytosis, inducing the plasma membrane accumulation of CD63, and thereby inducing its direct budding from the cell. We tested these predictions and found that they successfully predicted experimental outcomes, as high-level expression of syntenin inhibited CD63 endocytosis, induced the plasma membrane accumulation of CD63, triggered a fourfold increase in the vesicular secretion of CD63, had no effect on the production of exosome-sized vesicles, but increased the amount of CD63 present in CD81/CD9-positive vesicles. As a result, our model of exosome biogenesis places syntenin at the plasma membrane as an expression-dependent inhibitor of CD63 endocytosis (Fig. 7).
Fig. 7. The endocytosis-independent and endocytosis-dependent pathways of exosome marker protein secretion.
The vesicular secretion of highly enriched exosome marker proteins (i) occurs primarily by the endocytosis-independent pathway and (ii) is strongly inhibited by their endocytosis, while (iii) inhibitors of endocytosis induce the vesicular secretion of CD63, Lamp2, and other constitutively-endocytosed exosomal proteins. Once a highly enriched exosome marker protein is internalized and (iv) delivered to endosomes, it can be (v) recycled to the plasma membrane or (vi) loaded into ILVs, which are then retained indefinitely, (vii) destroyed in lysosomes, or (viii) secreted as exosomes. In the case of CD81 and CD9, endocytosis triggers their nearly complete destruction, indicating that endocytosis results in their nearly stoichiometric delivery to lysosomes.
Our model does not, at this time, depict a role for Alix, although there is no doubt that syntenin binds directly to Alix. The omission of Alix from our model is based, in part, on our observation that high-level expression of syntenin induced the plasma membrane accumulation and vesicular secretion of CD63 in Alix−/− knockout cells. It is also based on our observation that ΔN100syntenin and mycΔN100syntenin induced the exosomal secretion of CD63, although these forms of syntenin lack all three of syntenin’s Alix-binding sites. It also reflects our previous observation that Alix knockout HEK293 cells failed to display a significant defect in the vesicular secretion of exosomal tetraspanins or the release of exosome-sized vesicles (5). These results do not, however, mean that Alix plays no role in the vesicular secretion of exosome marker proteins.
High-level expression of CD63 inhibits endocytosis, disrupts plasma membrane homeostasis, and alters exosome content
The endocytosis field has known for >20 years ago that high-level expression of endocytosis signal–containing proteins inhibits protein endocytosis (32, 42–46). In the case of proteins that carry YxxΦ-type endocytosis signals, ligand-mediated inhibition of AP-2/mu2-mediated endocytosis involves binding competition, which reflects the limiting amount of AP-2 in the cell, and protein crowding effects, which reflect the competition of ligands for packaging into clathrin-coated pits and vesicles. Thus, our discovery that high-level expression of CD63 inhibited protein endocytosis (3.0-fold) and drove the plasma membrane accumulation of itself, Lamp1, and Lamp2, is broadly consistent with our modern understanding of protein endocytosis (32, 42–46).
Nevertheless, the observation that CD63 inhibits endocytosis was unanticipated, as were the CD63-mediated induction of mu2 secretion and the resultant effects on exosome composition. Specifically, while previous examples of ligand-mediated inhibition of endocytosis have been ascribed to binding competition and protein crowding effects (32, 42–46), we demonstrated that high-level expression of CD63 led to the vesicular secretion and cellular depletion of mu2, the subunit of AP-2 that binds directly to YxxΦ motif–containing proteins (33). As a result, high-level expression of CD63 induced the vesicular secretion of Lamp1, Lamp2, and likely a broad array of other endocytosed proteins too, creating artifactual changes to exosome content. All told, there are >40 separate observations in this paper that support the conclusion that high-level expression of CD63 or another YxxΦ-containing exosome marker protein inhibited clathrin/AP-2–mediated endocytosis (table S2).
The dynamic interplay between YxxΦ motif–containing exosome marker proteins, the endocytosis machinery, and their vesicular secretion from the cell provide strong support for our model of exosome biogenesis (Fig. 7). However, for the exosome field, the CD63-induced changes to exosome content represent something of an inconvenient truth, as many studies have used CD63 overexpression to make broad conclusions about exosome biogenesis (15, 49, 67–71). Given that the TRE3G/dox-based expression system used in this report drives similar levels of transgene expression as the cytomegalovirus (CMV)/CAG-based expression systems (72) that were used in those earlier studies (15, 49, 67–71), it is likely that CD63-mediated inhibition of endocytosis has had unanticipated and potentially significant effects on the results of these prior studies. For example, Proteomic data from one of these studies shows that high-level expression of CD63 led to the vesicular secretion of Lamp1, Lamp2, and clathrin, whereas high-level expression of CD9 did not (15), a result that can now be ascribed to CD63-mediated inhibition of endocytosis. There is also clear evidence for CD63-induced artifacts in vivo, as high-level expression of CD63-GFP (green fluorescent protein) caused a highly penetrant neonatal lethality in transgenic rats, whereas a high but lower level of CD63-GFP expression allowed these transgenic rats to survive for up to 4 to 6 months (still an ~80% reduction in life span). In light of our results and our model (Fig. 7), it seems prudent to control for endocytosis effects, whenever CD63 is being expressed.
Implications for exosome engineering
Our data and model are also relevant to exosome engineering. After all, any attempt to engineer a protein into the endocytosis-dependent pathway is likely to result in the same effects that we found for CD63: an inhibition of protein endocytosis, disruption of plasma membrane homeostasis, corruption exosome content, and the budding of the protein from the plasma membrane anyway, or worse, its destruction. These inconvenient truths cannot easily be avoided, and as Feynman famously noted, “For a successful technology, reality must take precedence over public relations, for nature cannot be fooled” (73). Fortunately, there is no need to engineer the endocytosis-dependent pathway, as the endocytosis-independent pathway (Fig. 7) appears to be far more efficient, and easier to engineer. As we've shown recently, it requires nothing more than optimizing a protein’s expression, its delivery to the plasma membrane, and its loading into nascent budding vesicles (74).
MATERIALS AND METHODS
Plasmids
The plasmid pJM1463 is based on a pS series plasmid (65) and carries a bicistronic open reading frame (ORF) encoding rtTAv16-2a-BleoR downstream of the spleen focus-forming virus (SFFV) transcriptional control region. The plasmids pYA128, pYA129, pYA130, and pYA215 are Sleeping Beauty transposons based on pITRSB (65) that carry two genes: (i) an EFS-PuroR gene and (ii) a TRE3G regulated gene designed to express syntenin, ΔN100syntenin, synteninΔC23, or myc-tagged ΔN100syntenin, respectively. The plasmids pCG606, pCG732, pCG733, pCG734, and pCG607 are Sleeping Beauty transposons based on pITRSB that carry two genes: (i) an EFS-HygR gene and (ii) a TRE3G regulated gene designed to express codon-optimized CD9 ORFs that encode WT CD9, CD9-YQRF, CD9-YQTI, CD9-AEMV, and CD9-YEVM, respectively. The plasmids pCG602, pMG9, pCG604, and pMG10 are Sleeping Beauty transposons based on pITRSB that carry two genes: (i) an EFS-HygR gene and (ii) a TRE3G regulated gene designed to express codon-optimized ORFs that encode WT CD63, CD63-YQRF, CD63-YQTI, and CD63-AEMV, respectively. The plasmids pCG609, pLS14, pLS15, pLS16, and pCG610 are Sleeping Beauty transposons based on pITRSB that carry two genes: (i) an EFS-HygR gene and (ii) a TRE3G regulated gene designed to express codon-optimized CD81 ORFs that encode WT CD81, CD81-YQRF, CD81-YQTI, CD81-AEMV, and CD81-YEVM, respectively. All genes were synthesized in vitro, cloned into the appropriate expression vectors, and then sequenced in their entirety to ensure the absence of unwanted mutations.
The plasmid used for knockout of the SDCBP gene, pJM1087, was based on pFF (5) and contains three genes. The first of these consists of a CMV promoter driving expression of a single long quadricistronic ORF that encodes (i) Cas9-3xNLS, (ii) a viral 2a peptide, (iii) EGFP, (iv) another viral 2a peptide, (v) the thymidine kinase (tk) from herpes simplex virus (HSV), (vi) another viral 2a peptide, and (vii) the puromycin resistance protein PuroR (65), with the ORF flanked by a pair of loxP sites. The second of these genes consists of the PolIII-transcribed H1 promoter driving expression of a Cas9 gRNA with the target sequence of 5′-ATAAACCTACTTCCATCGTG-3′, which is complementary to a sequence in the second coding exon of the SDCBP gene. The third of these genes consists of the PolIII-transcribed 7sk promoter driving expression of a Cas9 gRNA with the target sequence of 5′-GGTTTCTGGTGCACCACTTC-3′, which is complementary to a sequence in the third coding exon of the SDCBP gene.
Plasmids carrying genomic DNA (gDNA) amplification products from mutant cell lines were generated by extracting gDNA from single-cell clones (SCCs), amplifying small fragments of the genome surrounding the target site, using Taq polymerase. The resulting PCR fragments were checked for proper size and then inserted into a bacterial cloning vector (A3600, Promega), and insert-containing clones were sequenced, with as many as 24 independent inserts sequenced for each cell line to ensure that all alleles were interrogated and that none carried the WT sequence of the gene. It should be noted that 293 cells carry three alleles of the CD9 gene, and thus, creating a 293 CD9 knockout cell line requires mutational inactivation of all three alleles.
Cell lines, transfections, and small molecules
Cells lines used in this study are summarized here (table S1). 293F cells were obtained from Thermo Fisher Scientific (A14528). NIH3T3 and Hela S3 cells were obtained from the American Type Culture Collection (CRL-1658 and CCL-2.2, respectively). The HEK293 Alix−/− cell line was described previously (5), as was the 293F/CD63−/− cell line (75). The 293F/AP2M1−/− cell line, 293F/CD9−/−/− cell line, and 293F/SDCBP−/− cell lines were generated in this report (see section below describing Cas9-mediated gene editing). Adherent cultures were grown in tissue culture plates in complete medium (CM) [Dulbecco’s modified Eagle’s medium (DMEM), 10% fetal bovine serum (FBS), 1% penicillin/streptomycin] at 37°C, 90% humidity, and 5% CO2. For suspension cultures of 293F and 293F-derived cell lines, cells were grown in Freestyle medium (Thermo Fisher Scientific) in Erlenmeyer shaker flasks at 110 rpm, 37°C, 90% humidity, and 8% CO2. DNA transfections were performed using Lipofectamine 3000 (L3000015, Thermo Fisher Scientific) according to the manufacturer’s instructions. Zeocin was used at 200 μg/ml. Puromycin was used at 3 μg/ml. Hygromycin was used at 200 μg/ml. Dox was used at 10 or 300 ng/ml. Latrunculin A was used at 1 μM. Bafilomycin A was used at 30 to 100 nM. Dyngo-4a was used at 30 μM.
To create the dox-inducible Tet-on cell lines (i.e., FtetZ, 3TetZ, StetZ, HtetZ/Alix−/−, FtetZ/CD63−/−, and FtetZ/CD9−/−/−), the parental cell lines (293F, NIH3T3, Hela-S, HEK293/Alix−/−, 293F/CD63−/−, and 293F/CD9−/−/−, respectively) were transfected with pJM1463 using Lipofectamine 3000. Two days later, the transfected cell populations were placed in zeocin-containing medium. The culture medium was changed every 3 to 4 days for 7 to 12 days to select for zeocin-resistant cells. The thousands of surviving SCCs from each transfection were then pooled to generate a single polyclonal Tet-on derivative of each parental cell line.
To create the cell lines that express the syntenin proteins, FtetZ, 3TetZ, StetZ, and HtetZ/Alix−/− cells were transfected with pYA128, YA129, pYA130, and pYA215. A day later, the cells were placed in puromycin-containing medium, followed by selection of puromycin-resistant clones and pooling of all clones to create polyclonal cell lines designed for the dox-induced expression of syntenin proteins. To create the cell lines that express CD9 proteins, FtetZ/CD9−/−/− cells were transfected with pCG606, pCG732, pCG733, pCG734, and pCG607. Two days later, the cells were placed in hygromycin-containing medium, followed by selection of hygromycin-resistant clones and pooling of all clones to create polyclonal cell lines designed for the dox-induced expression of CD9 proteins. To create the cell lines that express CD63 proteins, FtetZ/CD63−/− cells were transfected with pCG602, pMG9, pCG604, and pMG10. Two days later, the cells were placed in hygromycin-containing medium, followed by selection of hygromycin-resistant clones and pooling of all clones to create polyclonal cell lines designed for the dox-induced expression of CD63 proteins. To create the cell lines that express CD81 proteins, FtetZ/CD81−/− cells were transfected with pCG609, pCG610, pLS14, pLS15, and pLS16. Two days later, the cells were placed in hygromycin-containing medium, followed by selection of hygromycin-resistant clones and pooling of all clones to create polyclonal cell lines designed for the dox-induced expression of CD81 proteins.
Endocytosis assay by flow cytometry
Cells were trypsinized with TrypLE (Thermo Fisher Scientific), resuspended in ice-cold fluorescence-activated cell sorting (FACS) buffer, and then stained with unlabeled primary CD63 (1:20; NBP2-32830, Novus Biologicals) or CD9 (1:100; 312102, BioLegend) Ab for 30 min on ice. Cells were then washed three times with ice-cold FACS buffer to remove excess Ab. Cells were then split into two parallel populations, with one kept at 4°C while the other was incubated at 37°C in CM for 30 min. Cells were then returned to 4°C and then incubated with Alexa Fluor 488–labeled anti-mouse IgG Ab for 30 min at 4°C (1:800; 715-545-150, Jackson ImmunoResearch). Cells were washed three times with FACS buffer, then stained with 4′,6-diamidino-2-phenylindole (DAPI; 0.5 μg/ml), and interrogated for Alexa Fluor 488 and DAPI fluorescence using a CytoFLEX S flow cytometer (Beckman Coulter). Flow cytometry plots were generated using FlowJo (v10.8.1). EE was calculated using the following formula: EE = [1 − (FICD63 @ 37oC/FICD63 @ 4oC)]/[1 − (FICD9 @ 37oC/FICD9 @ 4oC)].
Endocytosis assay by confocal fluorescence microscopy
Cells were grown overnight on poly-d-lysine–coated coverglasses in CM and then transferred to prechilled 4°C CM. Cells were washed in 4°C phosphate-buffered saline (PBS) and then incubated for 30 min at 4°C with prechilled 4°C PBS (200 μl) containing 4 μl of fluorescein isothiocyanate (FITC)–conjugated anti-CD63 (clone H5C6, BioLegend) and 4 μl of allophycocyanin (APC)–conjugated anti-CD9 (clone H19a, BioLegend). Excess Ab was removed by two washes with 4°C PBS. Cells were then fixed immediately or transferred to CM at 37°C and incubated for 30 min at 37°C. Fixation was with 3.7% formaldehyde in PBS for 20 min. Cells were then incubated with DAPI to stain the nucleus and then examined by confocal fluorescence microscopy and imaged to assess the subcellular distribution of plasma membrane–labeled CD63 and CD9. Confocal fluorescence microscopy was performed using a Zeiss LSM880 microscope with gallium-arsenide phosphide (GaAsP) detectors and a 63×/1.4 NA (numerical aperture) Plan-Apochromat objective. Images were assembled into figures using ImageJ and Adobe Illustrator.
Preparation of exosome-containing fractions
For suspension cell cultures, cells were seeded into 30 ml of Freestyle medium at a density of 1 × 106 cells per milliliter and grown for 48 to 72 hours, with shaking. Culture medium was collected, and cells and cell debris were removed by centrifugation at 5000g at 4°C for 15 min, followed by passage of the resulting supernatant through a 200-nm pore size diameter filtration unit. To collect sEVs by size exclusion chromatography and filtration, the 200-nm filtrate was concentrated ~100-fold by centrifugal flow filtration across a 100-kDa pore size diameter filter (Centricon-70, MilliporeSigma), followed by purification by size exclusion chromatography using qEV nano columns (Izon Sciences) using 10-nm filtered PBS as buffer.
For adherent cell cultures (Alix−/− cell experiments only), 3 million cells were seeded onto 150-mm dishes in 30 ml of complete medium and allowed to adhere to the plates overnight, and then the medium was replaced and the cells were incubated for 3 days in complete medium. Culture medium was collected, and cells and cell debris were removed by centrifugation at 5000g at 4°C for 15 min, followed by passage of the resulting supernatant through a 200-nm pore size diameter filtration unit. The sEVs were then collected by differential centrifugation in which supernatants were spun for 30 min at 10,000g, spun a second time for 30 min at 10,000g, and then spun at 100,000g for 2 hours, all at 4°C. The supernatant was discarded, and the sEV pellet was resuspended in 10-nm filtered PBS for further analysis.
Immunoblot
Cell samples and exosome-containing samples were lysed in Laemmli/SDS-PAGE (polyacrylamide gel electrophoresis) sample buffer lacking reducing agent. Samples were maintained in reducing agent-free sample buffer or adjusted to 5% β-mercaptoethanol, heated to 100°C for 10 min, spun at 13,000g for 2 min to eliminate insoluble material, and then separated by SDS-PAGE and processed for immunoblot as previously described (5). Cell lysates and vesicle lysates were loaded by proportion of total sample, using a constant 1:6 ratio of cell sample:100K pellet sample. Following separation by SDS-PAGE, proteins were transferred to Immobilon membranes (EMDMillipore), blocked for 2 hours at room temperature in 0.2% nonfat dry milk in tris-buffered saline with Tween 20 (TBST), and then incubated with primary antibodies for >2 hours, usually overnight, at 4°C. The next day, the membranes were washed five times with TBST, incubated for 1 to 2 hours with horseradish peroxidase (HRP) conjugates of secondary antibodies (Jackson ImmunoResearch), and then washed five more times. Membranes were incubated in ECL detection solution (MilliporeSigma), and antigens were detected using an Amersham Imager 600 gel imaging system (GE Healthcare Life Sciences). The resulting digitized IB images were processed in ImageJ (https://imagej.nih.gov/). In brief, each digital image was converted to an 8-bit grayscale file. Measurement parameter was set to integrated density, scale was set to pixel, images were inverted, bands were delineated using the freehand selection tool, and signal densities were converted to relative protein abundance by multiplying by the dilution factor for each sample. Relative vesicular secretion (E/C) was calculated by dividing the relative amount of the protein present in the exosome lysate by the sum of the protein abundance in the cell lysate ([amt in exos]/[amt in cells]).
We used antibodies specific for human CD63 (NBP2-32830, Novus Biologicals), mouse CD63 (NVG-2, BioLegend), CD9 (312102, BioLegend), CD81 (555675, BD Biosciences), Hsp90 (sc-13119, Santa Cruz Biotechnology), syntenin (PA5-76618, Thermo Fisher Scientific), syntenin (A5360, ABclonal), syntenin (A5497, ABclonal), syntenin (NBP1-33661, Novus Biologicals), syntenin (PA5-115875, Thermo Fisher Scientific), syntenin (PA5-28826, Thermo Fisher Scientific), Myc (clone 9E10), AP2M1 (68196, Cell Signaling Technology), Lamp1 (H4A3, Thermo Fisher Scientific), and Lamp2 (H4B4, Thermo Fisher Scientific). HRP-conjugated secondary antibodies were from Jackson ImmunoResearch and Cell Signaling Technology.
Reverse transcription qPCR
Total RNA was isolated using Quick-RNA Microprep Kit (Zymo Research). RNA was converted to single-stranded cDNA by reverse transcription using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems). qPCR analysis was performed using SYBR Green master mix (Bio-Rad) and the CFX96 Real-Time PCR Detection System (Bio-Rad), with gene-specific primers for our codon-optimized syntenin transgene (5′-GGCTCAAGTCTATTGATAATGGC-3′ and 5′-CCTTATCACTGGACCAACC-3′), our CD9 transgene (5′-GAA-ATGTATTAAATATCTTCTGTTCGGTTT-3′ and 5′-CCGGCAC-CGATAAGTATATAAAC-3′), and control primers for human 18S rRNA (5′-CGGCGACGACCCATTCGAAC-3′ and 5′-GAATCGA-ACCCTGATTCCCCGTC-3′). Data were analyzed with the ΔΔCT method.
Flow cytometry and FACS
Cells were released by trypsinization (TrypLE, Thermo Fisher Scientific), and cell clumps were removed using a cell strainer (Falcon, catalog no. 352235). Approximately 500,000 cells were then concentrated by a brief spin at 400g for 5 min and resuspended in 100 μl of 4°C FACS buffer (1% FBS in PBS) containing 2 μl of FITC-conjugated anti-CD63 (clone H5C6), 2 μl of APC-conjugated anti-CD9 (clone H19a), 2 μl of phycoerythrin (PE)–conjugated anti-CD81 (clone 5A6), or 2 μl of peridinin chlorophyll protein (PerCP)–conjugated anti-Lamp1 (clone H4A3) and 2 μl of PE-conjugated anti-Lamp2 (clone H4B4), all from BioLegend, for 30 min with gentle mixing every 10 min. Cells were washed three times with 1 ml of 4°C FACS buffer, with cells recovered by 400g spin for 5 min at 4°C. After the final wash, cells were resuspended in FACS buffer with DAPI (0.5 μg/ml) and analyzed using CytoFLEX S flow cytometer (Beckman Coulter). Flow cytometry histograms were generated using FlowJo (v10.8.1). For separation by FACS, labeled cells were sorted into single cells in a 96-well plate, on the basis of high or low cell surface labeling for CD63 or CD9.
Cas9-medited gene editing
To create the 293F/AP2M1−/− cell line, we transfected 293F cells with a mixture of Cas9 protein (A36498, Thermo Fisher Scientific) and an AP2M1-targeting single-guide RNA (sgRNA; target sequence of 5′-ACGTTAAGCGGTCCAACATT-3′) using Lipofectamine CRISPRMAX (CMAX00003, Thermo Fisher Scientific). Cells were cultured for several days and then seeded into 96-well plates at one cell per well. SCCs were expanded, gDNA was extracted from each clone, and each gDNA was interrogated by PCR using AP2M1 gene-specific primers. PCR products were ligated into the pGEM-T vector using a TA cloning kit (A3600, Promega) and transformed into Escherichia coli, and eight or more clones from each ligation were sequenced in their entirety. The 293F/AP2M1−/− cell line carried for further analysis carried one allele with a 198–base pair (bp) deletion and one allele with a = 1 codon deletion at a conserved position (Ile63Δ) of the AP2M1 protein (fig. S5).
A similar procedure was used to generate the 293F/CD9−/−/− cell line. 293F cells were transfected with a mixture of Cas9 protein (A36498, Thermo Fisher Scientific) and an sgRNA (target sequence of 5′-ATTCGCCATTGAAATAGCTG-3′) using Lipofectamine CRISPRMAX (Thermo Fisher Scientific). Cells were cultured for several days, trypsinized, and seeded into wells of 96-well plates at one cell per well. SCCs were expanded, gDNA was extracted from each clone, and each gDNA was interrogated by PCR using CD9 gene-specific primers. PCR products were ligated into the pGEM-T vector using a TA cloning kit (A3600, Promega) and transformed into E. coli, and 24 or more clones from each ligation were sequenced in their entirety. All gDNA amplification products from 293F/CD9−/−/− cell lines carried up to three different sequences, indicating that 293F cells carry three CD9 alleles. The cell line used for further experimentation carried one CD9 allele with a 4-bp deletion and two CD9 alleles with the same 8-bp deletion (fig. S8).
The 293F/SDCBP−/− cell line was created by transfecting 293F cells with the plasmid pJM1087 and then selecting for puromycin-resistant cell clones. After 7 days in selection, surviving cells were pooled, with EGFP-positive cells separated by FACS into individual wells of a 96-well plate. The emergent SCCs were expanded, and 10 were interrogated by immunoblot using antibodies specific for syntenin, revealing that all 10 lacked detectable level of syntenin protein. We then extracted gDNA from these clones and interrogated each gDNA by PCR using primers flanking both gRNA target sites in the SDCBP gene. The 293F/SDCBP−/− cell line selected carried deletions between the two target sites in coding exons 2 and 3 on both of its SDCBP alleles, rendering both functionally null. To delete the Cas9-EGFP-HSVtk-PuroR ORF, this cell line was transfected with a Cre recombinase expression vector, grown for 10 days in CM lacking antibiotics, and then seeded at various densities into CM containing ganciclovir to eliminate any HSVtk-expressing cells. After growth and selection for 2 weeks in ganciclovir-containing medium, the resulting cell clones were pooled and examined by flow cytometry, revealing that all cells in the population were EGFP-negative. Furthermore, exposure of these cells to puromycin confirmed that all of the cells in the ganciclovir-resistant population had also reverted to puromycin sensitivity and were therefore likely no longer expressing Cas9.
qSMLM analysis
Coverslips #1.5H (25 mm diameter) (Thermo Fisher Scientific, catalog no. NC9560650; Waltham, MA, USA) were functionalized with N-hydroxysuccinimide (NHS) groups, followed by covalent attachment of mAbs that bind to epitopes in the ectodomain of human CD81 and human CD9. FtetZ cells and FtetZ::syntenin cells were grown in Freestyle medium containing dox, followed by collection of their exosomes by concentrating filtration and size exclusion chromatography. The resulting exosome preparations were diluted in PBS containing 0.025% Tween 20 (1:50) to a final volume of 80 μl and placed on the surface of Ab-coated coverslips at room temperature overnight in a humidified chamber. Coverslips were then washed with PBS containing 0.025% Tween 20, and EVs were labeled with a cocktail of Alexa Fluor 647–labeled antibodies specific for human CD63 (Novus Biologicals, catalog no. NBP2-42225; Centennial, CO, USA) and CF568-labeled antibodies specific for human CD9 (BioLegend, catalog no. 312102; San Diego, CA, USA) and human CD81 (BioLegend, catalog no. 349502; San Diego, CA, USA). All antibodies were fluorescently labeled as described previously at a molar ratio of ~1 (60). Samples were fixed and stored as described previously (60).
For imaging, coverslips were placed in Attofluor cell chambers (Thermo Fisher Scientific, catalog no. A7816) loaded with direct stochastic optical reconstruction microscopy (dSTORM) imaging buffer. N-STORM super-resolution microscope (Nikon Instruments; Melville, NY, USA) was used for SMLM imaging using 561- and 640-nm lasers, respectively. Images were acquired using NIS-Elements software (Nikon Instruments). SMLM images were processed using N-STORM Offline Analysis Module of the NIS-Elements software to localize peaks. The localization data were analyzed with the Nanometrix software (version 1.0.4.61; Nanometrix Ltd., Oxford, UK) using a density-based spatial clustering of applications with noise (DBSCAN) algorithm. Before cluster analysis, the detected 640- and 561-nm channel localizations were aligned using the Uniform Dual Channel Alignment tool of Nanometrix. The DBSCAN-based cluster identification was performed at a neighbor search radius of 30 nm and minimum points per cluster of 30 as analysis conditions. Postprocessing of the detected cluster data, including concatenation and filtering, was performed using Matlab (version R2022a; MathWorks, Natick, MA, USA). Clusters were identified as EVs considering the following constraints. In the case of the 647-nm channel, the minimum and maximum number of localizations per cluster was set to 30 and 3000, and the minimum and maximum diameter was set to 20 and 400 nm, respectively. In the case of the 561-nm channel, the minimum and maximum number of localizations per cluster was set to 40 and 3200, and the minimum and maximum diameter was set to 30 and 400 nm, respectively. Colocalized (CD63+, CD81/CD9+) EVs were identified as overlapping clusters detected in the 640- and 561-nm channel. The number of tetraspanin molecules per exosome was calculated using an average of 15 (647-nm channel) and 16 (561-nm channel) localizations per single fluorescent tetraspanin Ab. Following EV identification, the EV count per region of interest (ROI), CD63 molecule count per EV, and EV diameter were further analyzed. To account for the difference in EV concentration across the samples, we normalized the number of detected CD63+, CD81/CD9+ and CD63−, CD81/CD9+ EVs by the total number of CD81/CD9+ EVs. Statistical significances in the normalized EV count per ROI data were determined using Brown-Forsythe and Welch analysis of variance (ANOVA) test. Statistical significance in the CD63 molecule count per EV and diameter data was assessed performing two-tailed Welch’s t test after logarithmic transformation. Statistical analysis and graph generation were performed in GraphPad Prism (version 9.5.1; GraphPad, San Diego, CA, USA).
NTA analysis
Exosome-containing fractions were diluted into 10-nm filtered PBS and examined for concentration of exosome-sized vesicles and particles by NTA using a Particle Metrix Zetaview Twin PMX-220, according to the manufacturer’s instructions.
Electron microscopy
FtetZ/CD63−/−::CD63 cells were grown in dox-containing CM overnight, after which they were fixed with formaldehyde/glutaraldehyde solution (in PBS), dehydrated in a graded ethanol series, embedded in Epon, sectioned, and stained with uranyl acetate. Sections were imaged on a Hitachi 7600 transmission electron microscope.
Acknowledgments
We thank J. Morrell for his outstanding technical assistance and the many colleagues who provided feedback over the course of this project, especially X. Breakefield, M. Caterina, C. Choi, S. Das, S. El-Andaloussi, D. Gupta, M. Pegtel, and A. Zickler.
Funding: This work was supported by grants from the NIH (UG3 CA241687, R35 HL150807, and UG3 TR002878) and the Johns Hopkins University.
Author contributions: Conceptualization: Y.A., C.G., M.G.-C., L.S.S.B., S.J.T., T.J.-T., and S.J.G. Methodology: Y.A., C.G., M.G.-C., L.S.S.B., A.S., T.J.-T., and S.J.G. Investigation: Y.A., C.G., M.G.-C., L.S.S.B., A.S., O.S., S.R., J.X., S.J.T., Y.D., and S.J.G. Visualization: Y.A., C.G., M.G.-C., L.S.S.B., A.S., and S.J.G. Funding acquisition: T.J.-T., R.L., and S.J.G. Project administration: T.J.-T., R.L., and S.J.G. Supervision: T.J.-T., R.L., and S.J.G. Writing—original draft: S.J.G. Writing—review and editing: Y.A., C.G., M.G.-C., L.S.S.B., S.J.T., A.S., T.J.-T., and S.J.G.
Competing interests: Part of methodology used in this work is the subject of a provisional patent application. T.J.-T. and A.S. may be entitled to certain compensation through their institutions’ respective intellectual property policies in the event such intellectual property is licensed. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All nonproprietary materials are available upon request, provided that a material transfer agreement is completed with Johns Hopkins University.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Tables S1 and S2
REFERENCES AND NOTES
- 1.Pegtel D. M., Gould S. J., Exosomes. Annu. Rev. Biochem. 88, 487–514 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R., LeBleu V. S., The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Escola J. M., Kleijmeer M. J., Stoorvogel W., Griffith J. M., Yoshie O., Geuze H. J., Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273, 20121–20127 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Thery C., Regnault A., Garin J., Wolfers J., Zitvogel L., Ricciardi-Castagnoli P., Raposo G., Amigorena S., Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 147, 599–610 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fordjour F. K., Guo C., Ai Y., Daaboul G. G., Gould S. J., A shared, stochastic pathway mediates exosome protein budding along plasma and endosome membranes. J. Biol. Chem. 298, 102394 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixson A. C., Dawson T. R., Di Vizio D., Weaver A. M., Context-specific regulation of extracellular vesicle biogenesis and cargo selection. Nat. Rev. Mol. Cell Biol. 24, 454–476 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalluri R., McAndrews K. M., The role of extracellular vesicles in cancer. Cell 186, 1610–1626 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raposo G., Stoorvogel W., Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanez-Mo M., Siljander P. R., Andreu Z., Zavec A. B., Borras F. E., Buzas E. I., Buzas K., Casal E., Cappello F., Carvalho J., Colas E., Cordeiro-da Silva A., Fais S., Falcon-Perez J. M., Ghobrial I. M., Giebel B., Gimona M., Graner M., Gursel I., Gursel M., Heegaard N. H., Hendrix A., Kierulf P., Kokubun K., Kosanovic M., Kralj-Iglic V., Kramer-Albers E. M., Laitinen S., Lasser C., Lener T., Ligeti E., Line A., Lipps G., Llorente A., Lotvall J., Mancek-Keber M., Marcilla A., Mittelbrunn M., Nazarenko I., Nolte-'t Hoen E. N., Nyman T. A., O'Driscoll L., Olivan M., Oliveira C., Pallinger E., Del Portillo H. A., Reventos J., Rigau M., Rohde E., Sammar M., Sanchez-Madrid F., Santarem N., Schallmoser K., Ostenfeld M. S., Stoorvogel W., Stukelj R., Van der Grein S. G., Vasconcelos M. H., Wauben M. H., O., Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiklander O. P. B., Brennan M. A., Lotvall J., Breakefield X. O., El Andaloussi S., Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 11, eaav8521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li K., Rodosthenous R. S., Kashanchi F., Gingeras T., Gould S. J., Kuo L. S., Kurre P., Lee H., Leonard J. N., Liu H., Lombo T. B., Momma S., Nolan J. P., Ochocinska M. J., Pegtel D. M., Sadovsky Y., Sanchez-Madrid F., Valdes K. M., Vickers K. C., Weaver A. M., Witwer K. W., Zeng Y., Das S., Raffai R. L., Howcroft T. K., Advances, challenges, and opportunities in extracellular RNA biology: Insights from the NIH exRNA Strategic Workshop. JCI Insight 3, e98942 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mateescu B., Jones J. C., Alexander R. P., Alsop E., An J. Y., Asghari M., Boomgarden A., Bouchareychas L., Cayota A., Chang H. C., Charest A., Chiu D. T., Coffey R. J., Das S., De Hoff P., deMello A., D'Souza-Schorey C., Elashoff D., Eliato K. R., Franklin J. L., Galas D. J., Gerstein M. B., Ghiran I. H., Go D. B., Gould S., Grogan T. R., Higginbotham J. N., Hladik F., Huang T. J., Huo X., Hutchins E., Jeppesen D. K., Jovanovic-Talisman T., Kim B. Y. S., Kim S., Kim K. M., Kim Y., Kitchen R. R., Knouse V., LaPlante E. L., Lebrilla C. B., Lee L. J., Lennon K. M., Li G., Li F., Li T., Liu T., Liu Z., Maddox A. L., McCarthy K., Meechoovet B., Maniya N., Meng Y., Milosavljevic A., Min B. H., Morey A., Ng M., Nolan J., De Oliveira Junior G. P., Paulaitis M. E., Phu T. A., Raffai R. L., Reategui E., Roth M. E., Routenberg D. A., Rozowsky J., Rufo J., Senapati S., Shachar S., Sharma H., Sood A. K., Stavrakis S., Sturchler A., Tewari M., Tosar J. P., Tucker-Schwartz A. K., Turchinovich A., Valkov N., Van Keuren-Jensen K., Vickers K. C., Vojtech L., Vreeland W. N., Wang C., Wang K., Wang Z., Welsh J. A., Witwer K. W., Wong D. T. W., Xia J., Xie Y. H., Yang K., Zaborowski M. P., Zhang C., Zhang Q., Zivkovic A. M., Laurent L. C., Phase 2 of extracellular RNA communication consortium charts next-generation approaches for extracellular RNA research. iScience 25, 104653 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dooley K., McConnell R. E., Xu K., Lewis N. D., Haupt S., Youniss M. R., Martin S., Sia C. L., McCoy C., Moniz R. J., Burenkova O., Sanchez-Salazar J., Jang S. C., Choi B., Harrison R. A., Houde D., Burzyn D., Leng C., Kirwin K., Ross N. L., Finn J. D., Gaidukov L., Economides K. D., Estes S., Thornton J. E., Kulman J. D., Sathyanarayanan S., Williams D. E., A versatile platform for generating engineered extracellular vesicles with defined therapeutic properties. Mol. Ther. 29, 1729–1743 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoorvogel W., Resolving sorting mechanisms into exosomes. Cell Res. 25, 531–532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathieu M., Nevo N., Jouve M., Valenzuela J. I., Maurin M., Verweij F. J., Palmulli R., Lankar D., Dingli F., Loew D., Rubinstein E., Boncompain G., Perez F., Thery C., Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat. Commun. 12, 4389 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu D. A., Tao K., Wu B., Yu Z., Szczepaniak M., Rames M., Yang C., Svitkina T., Zhu Y., Xu F., Nan X., Guo W., A phosphoinositide switch mediates exocyst recruitment to multivesicular endosomes for exosome secretion. Nat. Commun. 14, 6883 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baietti M. F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E., Zimmermann P., David G., Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14, 677–685 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Kashyap R., Balzano M., Lechat B., Lambaerts K., Egea-Jimenez A. L., Lembo F., Fares J., Meeussen S., Kugler S., Roebroek A., David G., Zimmermann P., Syntenin-knock out reduces exosome turnover and viral transduction. Sci. Rep. 11, 4083 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larios J., Mercier V., Roux A., Gruenberg J., ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 219, e201904113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shelke G. V., Williamson C. D., Jarnik M., Bonifacino J. S., Inhibition of endolysosome fusion increases exosome secretion. J. Cell Biol. 222, e202209084 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghossoub R., Lembo F., Rubio A., Gaillard C. B., Bouchet J., Vitale N., Slavik J., Machala M., Zimmermann P., Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 5, 3477 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Vietri M., Radulovic M., Stenmark H., The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 21, 25–42 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Migliano S. M., Wenzel E. M., Stenmark H., Biophysical and molecular mechanisms of ESCRT functions, and their implications for disease. Curr. Opin. Cell Biol. 75, 102062 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Gould S. G., Keller G. A., Subramani S., Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J. Cell Biol. 105, 2923–2931 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurt E. C., Pesold-Hurt B., Schatz G., The amino-terminal region of an imported mitochondrial precursor polypeptide can direct cytoplasmic dihydrofolate reductase into the mitochondrial matrix. EMBO J. 3, 3149–3156 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalderon D., Roberts B. L., Richardson W. D., Smith A. E., A short amino acid sequence able to specify nuclear location. Cell 39, 499–509 (1984). [DOI] [PubMed] [Google Scholar]
- 27.Blobel Laboratory T., Günter Blobel: Pioneer of molecular cell biology (1936-2018). J. Cell Biol. 217, 1163–1167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.G. Blobel, D. D. Sabatini, Ribosome-membrane interaction in eukaryotic cells, in Biomembranes, L. A. Manson, Ed. (Springer, 1971), pp. 193–195. [Google Scholar]
- 29.Gould S. J., Raposo G., As we wait: Coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles 2, 20389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.F. K. D. Fordjour, G. G. Daaboul, S. J. Gould, A shared pathway of exosome biogenesis operates at plasma and endosome membranes. bioRxiv 545228 [Preprint] (2019). 10.1101/545228. [DOI]
- 31.Pols M. S., Klumperman J., Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 315, 1584–1592 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Ohno H., Stewart J., Fournier M. C., Bosshart H., Rhee I., Miyatake S., Saito T., Gallusser A., Kirchhausen T., Bonifacino J. S., Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269, 1872–1875 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Braulke T., Bonifacino J. S., Sorting of lysosomal proteins. Biochim. Biophys. Acta 1793, 605–614 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Mitsunari T., Nakatsu F., Shioda N., Love P. E., Grinberg A., Bonifacino J. S., Ohno H., Clathrin adaptor AP-2 is essential for early embryonal development. Mol. Cell. Biol. 25, 9318–9323 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akamatsu M., Vasan R., Serwas D., Ferrin M. A., Rangamani P., Drubin D. G., Principles of self-organization and load adaptation by the actin cytoskeleton during clathrin-mediated endocytosis. eLife 9, e49840 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin M., Shirazinejad C., Wang B., Yan A., Schoneberg J., Upadhyayula S., Xu K., Drubin D. G., Branched actin networks are organized for asymmetric force production during clathrin-mediated endocytosis in mammalian cells. Nat. Commun. 13, 3578 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamaze C., Fujimoto L. M., Yin H. L., Schmid S. L., The actin cytoskeleton is required for receptor-mediated endocytosis in mammalian cells. J. Biol. Chem. 272, 20332–20335 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Yarar D., Waterman-Storer C. M., Schmid S. L., A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol. Biol. Cell 16, 964–975 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCluskey A., Daniel J. A., Hadzic G., Chau N., Clayton E. L., Mariana A., Whiting A., Gorgani N. N., Lloyd J., Quan A., Moshkanbaryans L., Krishnan S., Perera S., Chircop M., von Kleist L., McGeachie A. B., Howes M. T., Parton R. G., Campbell M., Sakoff J. A., Wang X., Sun J. Y., Robertson M. J., Deane F. M., Nguyen T. H., Meunier F. A., Cousin M. A., Robinson P. J., Building a better dynasore: The dyngo compounds potently inhibit dynamin and endocytosis. Traffic 14, 1272–1289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mettlen M., Chen P. H., Srinivasan S., Danuser G., Schmid S. L., Regulation of clathrin-mediated endocytosis. Annu. Rev. Biochem. 87, 871–896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Latysheva N., Muratov G., Rajesh S., Padgett M., Hotchin N. A., Overduin M., Berditchevski F., Syntenin-1 is a new component of tetraspanin-enriched microdomains: Mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol. Cell. Biol. 26, 7707–7718 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marks M. S., Woodruff L., Ohno H., Bonifacino J. S., Protein targeting by tyrosine- and di-leucine-based signals: Evidence for distinct saturable components. J. Cell Biol. 135, 341–354 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warren R. A., Green F. A., Enns C. A., Saturation of the endocytic pathway for the transferrin receptor does not affect the endocytosis of the epidermal growth factor receptor. J. Biol. Chem. 272, 2116–2121 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Warren R. A., Green F. A., Stenberg P. E., Enns C. A., Distinct saturable pathways for the endocytosis of different tyrosine motifs. J. Biol. Chem. 273, 17056–17063 (1998). [DOI] [PubMed] [Google Scholar]
- 45.DeGroot A. C. M., Gollapudi S., Zhao C., LaMonica M. F., Stachowiak J. C., Weakly internalized receptors use coated vesicle heterogeneity to evade competition during endocytosis. Biochemistry 60, 2195–2205 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeGroot A. C. M., Zhao C., LaMonica M. F., Hayden C. C., Stachowiak J. C., Molecular thermodynamics of receptor competition for endocytic uptake. Soft Matter 15, 7448–7461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai S. J., Ai Y., Guo C., Gould S. J., Degron tagging of BleoR and other antibiotic-resistance genes selects for higher expression of linked transgenes and improved exosome engineering. J. Biol. Chem. 298, 101846 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Votteler J., Sundquist W. I., Virus budding and the ESCRT pathway. Cell Host Microbe 14, 232–241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verweij F. J., Bebelman M. P., Jimenez C. R., Garcia-Vallejo J. J., Janssen H., Neefjes J., Knol J. C., R. de Goeij-de Haas, Piersma S. R., Baglio S. R., Verhage M., Middeldorp J. M., Zomer A., van Rheenen J., Coppolino M. G., Hurbain I., Raposo G., Smit M. J., Toonen R. F. G., van Niel G., Pegtel D. M., Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J. Cell Biol. 217, 1129–1142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buratta S., Tancini B., Sagini K., Delo F., Chiaradia E., Urbanelli L., Emiliani C., Lysosomal exocytosis, exosome release and secretory autophagy: The autophagic- and endo-lysosomal systems go extracellular. Int. J. Mol. Sci. 21, 2576 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sagini K., Buratta S., Delo F., Pellegrino R. M., Giovagnoli S., Urbanelli L., Emiliani C., Drug-induced lysosomal impairment is associated with the release of extracellular vesicles carrying autophagy markers. Int. J. Mol. Sci. 22, 12922 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Futai M., Sun-Wada G. H., Wada Y., Matsumoto N., Nakanishi-Matsui M., Vacuolar-type ATPase: A proton pump to lysosomal trafficking. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 95, 261–277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Booth A. M., Fang Y., Fallon J. K., Yang J. M., Hildreth J. E., Gould S. J., Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J. Cell Biol. 172, 923–935 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen B., Fang Y., Wu N., Gould S. J., Biogenesis of the posterior pole is mediated by the exosome/microvesicle protein-sorting pathway. J. Biol. Chem. 286, 44162–44176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang Y., Wu N., Gan X., Yan W., Morrell J. C., Gould S. J., Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLOS Biol. 5, e158 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen B., Wu N., Yang J. M., Gould S. J., Protein targeting to exosomes/microvesicles by plasma membrane anchors. J. Biol. Chem. 286, 14383–14395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan Y., Stehle T., Liddington R. C., Zhao H., Harrison S. C., Structure determination of simian virus 40 and murine polyomavirus by a combination of 30-fold and 5-fold electron-density averaging. Structure 4, 157–164 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Liddington R. C., Yan Y., Moulai J., Sahli R., Benjamin T. L., Harrison S. C., Structure of simian virus 40 at 3.8-Å resolution. Nature 354, 278–284 (1991). [DOI] [PubMed] [Google Scholar]
- 59.Amirbeigiarab S., Kiani P., Velazquez Sanchez A., Krisp C., Kazantsev A., Fester L., Schluter H., Ignatova Z., Invariable stoichiometry of ribosomal proteins in mouse brain tissues with aging. Proc. Natl. Acad. Sci. U.S.A. 116, 22567–22572 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saftics A., Abuelreich S., Romano E., Ghaeli I., Jiang N., Spanos M., Lennon K. M., Singh G., Das S., Van Keuren-Jensen K., Jovanovic-Talisman T., Single extracellular VEsicle nanoscopy. J. Extracell. Vesicles 12, e12346 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fordjour F. K., Abuelreich S., Hong X., Chatterjee E., Lallai V., Ng M., Saftics A., Deng F., Carnel-Amar N., Wakimoto H., Shimizu K., Bautista M., Phu T. A., Vu N. K., Geiger P. C., Raffai R. L., Fowler C. D., Das S., Christenson L. K., Jovanovic-Talisman T., Gould S. J., Exomap1 mouse: A transgenic model for in vivo studies of exosome biology. Extracell. Vesicle 2, 100030 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thorburn W. M., Occam’s razor. Mind 24, 287–288 (1915). [Google Scholar]
- 63.Gan X., Gould S. J., Identification of an inhibitory budding signal that blocks the release of HIV particles and exosome/microvesicle proteins. Mol. Biol. Cell 22, 817–830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gould S. J., Booth A. M., Hildreth J. E., The Trojan exosome hypothesis. Proc. Natl. Acad. Sci. U.S.A. 100, 10592–10597 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo C., Fordjour F. K., Tsai S. J., Morrell J. C., Gould S. J., Choice of selectable marker affects recombinant protein expression in cells and exosomes. J. Biol. Chem. 297, 100838 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsai S. J., Atai N. A., Cacciottolo M., Nice J., Salehi A., Guo C., Sedgwick A., Kanagavelu S., Gould S. J., Exosome-mediated mRNA delivery in vivo is safe and can be used to induce SARS-CoV-2 immunity. J. Biol. Chem. 297, 101266 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sung B. H., Ketova T., Hoshino D., Zijlstra A., Weaver A. M., Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 6, 7164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sung B. H., von Lersner A., Guerrero J., Krystofiak E. S., Inman D., Pelletier R., Zijlstra A., Ponik S. M., Weaver A. M., A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nat. Commun. 11, 2092 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCann J. V., Bischoff S. R., Zhang Y., Cowley D. O., Sanchez-Gonzalez V., Daaboul G. D., Dudley A. C., Reporter mice for isolating and auditing cell type-specific extracellular vesicles in vivo. Genesis 58, e23369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verweij F. J., Revenu C., Arras G., Dingli F., Loew D., Pegtel D. M., Follain G., Allio G., Goetz J. G., Zimmermann P., Herbomel P., Del Bene F., Raposo G., van Niel G., Live tracking of inter-organ communication by endogenous exosomes in vivo. Dev. Cell 48, 573–589.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Yoshimura A., Kawamata M., Yoshioka Y., Katsuda T., Kikuchi H., Nagai Y., Adachi N., Numakawa T., Kunugi H., Ochiya T., Tamai Y., Generation of a novel transgenic rat model for tracing extracellular vesicles in body fluids. Sci. Rep. 6, 31172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qin J. Y., Zhang L., Clift K. L., Hulur I., Xiang A. P., Ren B. Z., Lahn B. T., Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLOS ONE 5, e10611 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.R. P. Feynman, Personal observations on reliability of shuttle (1986); https://www.nasa.gov/history/rogersrep/v2appf.htm.
- 74.C. Guo, J. Sachithanandham, W. Zhong, M. Craney, J. Villano, A. Pekosz, S. J. Gould, Antigen-display exosomes provide adjuvant-free protection against SARS-CoV-2 disease at nanogram levels of spike protein. bioRxiv 2024.01.04.574272 [Preprint] (2024). 10.1101/2024.01.04.574272. [DOI]
- 75.Tsai S. J., Ai Y., Guo C., Gould S. J., Degron-tagging of BleoR and other antibiotic-resistance genes selects for higher expression of linked transgenes and improved exosome engineering. J. Biol. Chem. 298, 101846 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S1 and S2