Abstract
Mutations in the LMNA gene encoding lamins A/C cause an array of tissue-selective diseases, with the heart being the most commonly affected organ. Despite progress in understanding the perturbations emanating from LMNA mutations, an integrative understanding of the pathogenesis underlying cardiac dysfunction remains elusive. Using a novel conditional deletion model capable of translatome profiling, we observed that cardiomyocyte-specific Lmna deletion in adult mice led to rapid cardiomyopathy with pathological remodeling. Before cardiac dysfunction, Lmna-deleted cardiomyocytes displayed nuclear abnormalities, Golgi dilation/fragmentation, and CREB3-mediated stress activation. Translatome profiling identified MED25 activation, a transcriptional cofactor that regulates Golgi stress. Autophagy is disrupted in the hearts of these mice, which can be recapitulated by disrupting the Golgi. Systemic administration of modulators of autophagy or ER stress significantly delayed cardiac dysfunction and prolonged survival. These studies support a hypothesis wherein stress responses emanating from the perinuclear space contribute to the LMNA cardiomyopathy development.
Interplay of stress responses underlying the development of LMNA cardiomyopathy.
INTRODUCTION
Laminopathy is a collection of human diseases arising from mutations in the LMNA gene that encodes A-type lamins A and C (lamin A/C) (1). As a major component of the nuclear lamina, a fibrous meshwork lining the inner surface of the nuclear envelope (NE) (2, 3), lamin A/C play diverse cellular roles, such as providing protection from mechanical stress (4–7) and establishing scaffolding for binding chromatin (8, 9) as well as for cell signaling intermediaries (10–12). Despite ubiquitous expression in the nuclei of most differentiated mammalian somatic cells, LMNA incurs specific mutations that lead to tissue-selective syndromes affecting striated muscle, adipose tissue, and peripheral nerve, as well as multisystem disorders with features of accelerated aging. The most prevalent laminopathy is dilated cardiomyopathy (herein referred to as LMNA cardiomyopathy) with variable skeletal muscle involvement. Current therapies are limited to the treatment of congestive heart failure symptoms; hence, neither the underlying pathogenesis nor the eventual progression to heart failure is addressed. Targeted therapies based on mechanistic insights that prevent heart muscle deterioration are urgently needed.
Multiple established mouse models of LMNA cardiomyopathy that approximate the human disease currently exist. Germline homozygous Lmna knockout mice (Lmna−/−) develop rapid skeletal muscle dystrophy and cardiomyopathy and die within ~8 weeks of age (13, 14). Germline homozygous knock-in mice with point mutations corresponding to those identified in human patients (i.e., LmnaH222P/H222P and LmnaN195K/N195K) develop skeletal and cardiac muscle disease with slower kinetics, while heterozygotes are unaffected (15, 16). An important distinction between the mouse models of LMNA cardiomyopathy and the human disease is that in mice, mutations in both alleles of Lmna are required for the disease penetrance, whereas in humans, the vast majority of LMNA cardiomyopathy is autosomal dominant. Despite these differences, mouse models of LMNA cardiomyopathy have been instrumental in uncovering a wide array of cellular defects caused by Lmna mutations. For example, we and others showed that impaired macroautophagy (autophagy) in the heart underlies LMNA cardiomyopathy development in LmnaH222P/H222P and Lmna−/− mice (17, 18), but why and how this impairment contributes to disease pathogenesis is unknown.
Nuclear damage resulting from LMNA mutations is widely believed to be involved in the disease pathogenesis, but how it contributes is not well understood (19). Nuclear perturbations from defects in lamin A/C have been noted in cell culture models (5, 20, 21) and in the myocardium from murine models (15, 22, 23) and from humans (24), but how these alterations engender pathogenic mechanisms remain unclear. A detailed kinetic study using a temporally controlled, cardiomyocyte (CM)–specific deletion model of Lmna would enable accurate dissection of molecular pathogenesis emanating from nuclear fragility. To this end, we developed a novel Cre recombinase driver line referred to as CM-CreTRAP mice. It is a bi-cistronic transgenic line in which tamoxifen (Tam)–regulatable Cre recombinase (CreERT2) and EGFP-L10a fusion protein are coexpressed under the control of the myosin heavy chain 6 (Myh6) promoter, thereby directing their expression specifically in CMs. EGFP-L10a is enhanced green fluorescent protein fused to ribosomal protein L10a, a component of the 60S ribosomal protein, and the CM-specific driven expression of this fusion protein allows tagging of polysomes for immunoaffinity purification of translating mRNA [translating ribosome affinity purification (TRAP)] (25). Initially developed by the laboratory of N. Heintz for the purpose of characterizing neuronal subpopulations in mice (26, 27), the technique has been adapted to other model organisms including Drosophila melanogaster, Xenopus laevis, Danio rerio, and Arabidopsis thaliana (28–32). In addition, the method has evolved to enable Cre-directed EGFP-L10a tagging (33) and tetracycline operator–mediated temporal regulation (34).
Following intercross with the Lmnaflox/flox line, we induced CM-specific Lmna deletion in 12-week-old adult mice and detailed the development of LMNA cardiomyopathy. Using a combination of in vivo animal model, freshly isolated primary CMs, and in vitro culture models, we show that Lmna deletion in adult CMs causes substantial damage to the nucleus and the perinuclear space. The Golgi apparatus is particularly affected, leading to a CREB3-mediated Golgi stress response. Furthermore, in vitro disruption of Golgi recapitulates the disrupted autophagy phenotype observed in the germline mouse models of LMNA cardiomyopathy. Pharmacological modulators of autophagy or endoplasmic reticulum (ER) stress significantly improved the cardiac function and prolonged survival, suggesting that these pathways contribute to the pathogenesis and provide an additional avenue of therapeutic approaches.
RESULTS
Generation of a murine model with CM-specific Lmna deletion
To gain mechanistic insights into LMNA cardiomyopathy in a cell type–specific fashion, we engineered CM-CreTRAP mice that express a bi-cistronic construct consisting of CreERT2 and EGFP-L10a linked by an internal ribosomal entry site (IRES) (fig. S1A). The transgene’s expression is driven by the Myh6 promoter (35) and stabilized by the human growth hormone polyadenylation signal. We identified two founder lines (line 1 and line 2) with stable transgene transmission and robust expression of EGFP and CreERT2 in line 1 and lesser expression of these proteins in line 2 (fig. S1B). Direct fluorescence images of frozen heart sections confirmed the transgene protein expression in both founder lines (fig. S1C). The transgene mRNA expression in line 1 using primers that recognize CreERT2 sequences was specific to the heart with undetectable expression in other tissues tested (fig. S1D). Immunofluorescence staining on frozen heart sections from heterozygous transgenic positive CM-CreTRAP mice revealed a robust GFP signal that colocalized with sarcomeric actin, indicating CM-specific transgene expression (fig. S1E). We also noticed punctate signals within the nucleus (inset, white arrows), which is consistent with the established pattern of EGFP-L10a expression (27, 33).
Rapid myocardial dysfunction with robust fibrosis following Lmna deletion in adult CMs
We assessed the impact of Lmna deletion in CMs of CM-CreTRAP mice interbred with Lmnaflox/flox mice (36). All mice with Cre-driving transgenic allele (CM-CreTRAP) were studied as heterozygotes. Tam (100 mg/kg per day) was delivered intraperitoneally for five consecutive days followed by a 2-day rest (designate as “0 week”) in 12-week-old line 1 CM-CreTRAP:Lmnaflox/+, CM-CreTRAP:Lmnaflox/flox, mice, and corn oil for vehicle treated CM-CreTRAP:Lmnaflox/flox mice (Fig. 1A). To assess Lmna deletion efficiency, we isolated CMs from a subset of Tam and vehicle-treated CM-CreTRAP:Lmnaflox/flox mice for immunoblot analysis and observed ~96 and ~80% reduction in line 1 and line 2, respectively (fig. S2A). From this point on, most of our analyses were performed in line 1. With the confirmation of lamin A/C depletion in CMs, we then assessed its impact on the myocardium. The heart size was not significantly different at 2 weeks after Tam, but by 4 weeks, the enlargement was obvious (Fig. 1B). This enlargement was confirmed by heart weight/tibia length and heart weight/body weight ratio measurements (fig. S2B).
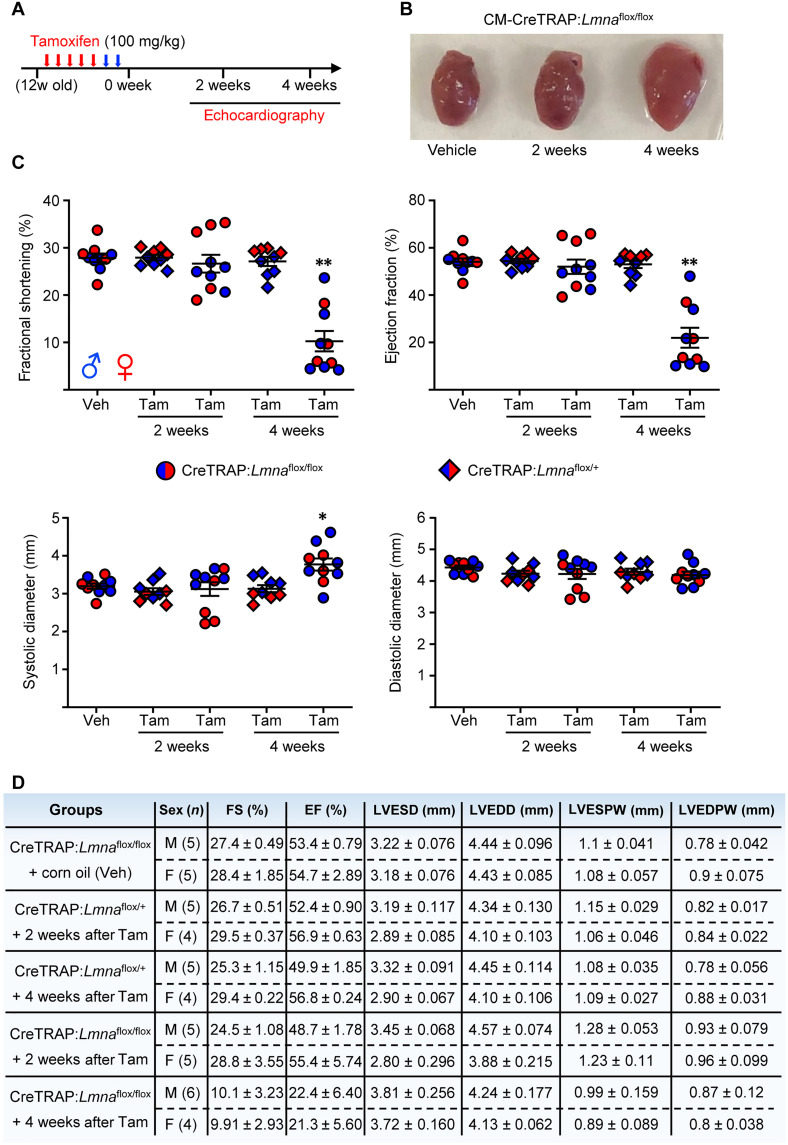
Fig. 1. Rapid cardiac performance deterioration after CM-specific Lmna deletion.
(A) Tam dosing schedule. Red arrows denote days of Tam injection and blue arrows show 2 days of rest. “12w old” denotes 12-week-old mice. (B) Hearts excised from CM-CreTRAP:Lmnaflox/flox mice treated with vehicle (Veh) or Tam for 2 and 4 weeks. (C) Echo on CM-CreTRAP:Lmnaflox/+ (diamond) and CM-CreTRAP:Lmnaflox/flox (circle) at 2 and 4 weeks Tam treatment. *P = 0.015 and **P < 0.0001 using one-way analysis of variance (ANOVA), followed by Dunnett’s multiple comparison correction with the Veh group serving as comparison control. Error bars = SEM. Blue = male and red = female mice. (D) Tabulation of echo data ± SEM. FS, fractional shortening; EF, ejection fraction; LVESD, left ventricular end systolic dimension; LVEDD, left ventricular end diastolic dimension; LVESPW, left ventricular end systolic posterior wall; LVEDPW, left ventricular end diastolic posterior wall. Veh-treated mice at 2 and 4 weeks were measured together with the Tam-treated group but pooled and presented as a single Veh control group because no differences were noted between them.
We then measured cardiac function by performing M-mode echocardiography (echo) at these time points to calculate fractional shortening (% reduction of left ventricular dimension at systole), ejection fraction (estimated calculation of % of blood ejected from left ventricle), and the systolic/diastolic left ventricular dimensions required to derive the first two parameters (Fig. 1C). Tam-treated CM-CreTRAP:Lmnaflox/flox mice revealed consistent and reproducible kinetics of cardiac dysfunction with a rapid decline in cardiac function by 4 weeks as determined by fractional shortening and ejection fraction (Fig. 1, C and D). Moreover, despite prior reports of Tam and Cre toxicity in the heart (37, 38), vehicle only–treated CM-CreTRAP:Lmnaflox/flox and Tam-treated CM-CreTRAP:Lmnaflox/+ mice revealed no cardiac dysfunction (Fig. 1, C and D), indicating that only the homozygous loss of Lmna in CMs caused rapid and severe cardiomyopathy in our experimental conditions.
To correlate organ function with tissue morphology, we performed histopathology assessing tissue structure, collagen deposition, and hypertrophy using hematoxylin and eosin (H&E), Picro-Sirius Red, and wheat germ agglutinin (WGA) staining, respectively (Fig. 2A). Because we did not observe sex-dependent differences in the cardiac function, we used both sexes for our experiments. H&E revealed obvious abnormalities at 2 weeks after Tam in the form of myocyte degeneration (blue arrows) and the presence of pyknotic nuclei (yellow arrows), with the inset image (black arrow) showing both abnormalities (Fig. 2A). By 4 weeks, substantial myocyte swelling and loss were obvious with abundant cytoplasmic vacuolation (Fig. 2A, green arrows). These results indicate that histological abnormalities in the myocardium are present at 2 weeks after Tam treatment despite the lack of cardiac functional changes. Furthermore, membrane staining with WGA revealed a progressive increase in the myocyte cross-sectional area, demonstrating myocyte swelling/hypertrophy (Fig. 2A). Picro-Sirius Red staining revealed a subtle increase in collagen deposition in the interstitial space as early as 1 week after Tam that progressively increased, with substantial fibrosis by 4 weeks (Fig. 2A). Masson’s Trichrome stain of the same hearts revealed a similar pattern (fig. S3A) with quantitation determined to be ~28% fibrosis at the 4-week time point (Fig. 2B). Furthermore, measurement of the myocyte cross-sectional area revealed a significant increase at 2 weeks after Tam (Fig. 2C), further demonstrating that, although cardiac function was normal at this time point, histological abnormalities have manifested. Together, these observations indicate a progressive pathological remodeling in response to CM-specific lamin A/C depletion. No abnormalities were observed in quadriceps from these mice, demonstrating specificity of the disease in the heart (fig. S3B). Moreover, hearts of Tam-treated CM-CreTRAP:Lmnaflox/+ mice displayed no obvious histological abnormalities (fig. S3C), further confirming that the myocardial pathogenesis requires the loss of both Lmna alleles, at least in the adult heart, and that Tam and the CreERT2 expression by themselves had no observable effect. Similar fibrotic responses were also observed in transgenic line 2 (fig. S3D).
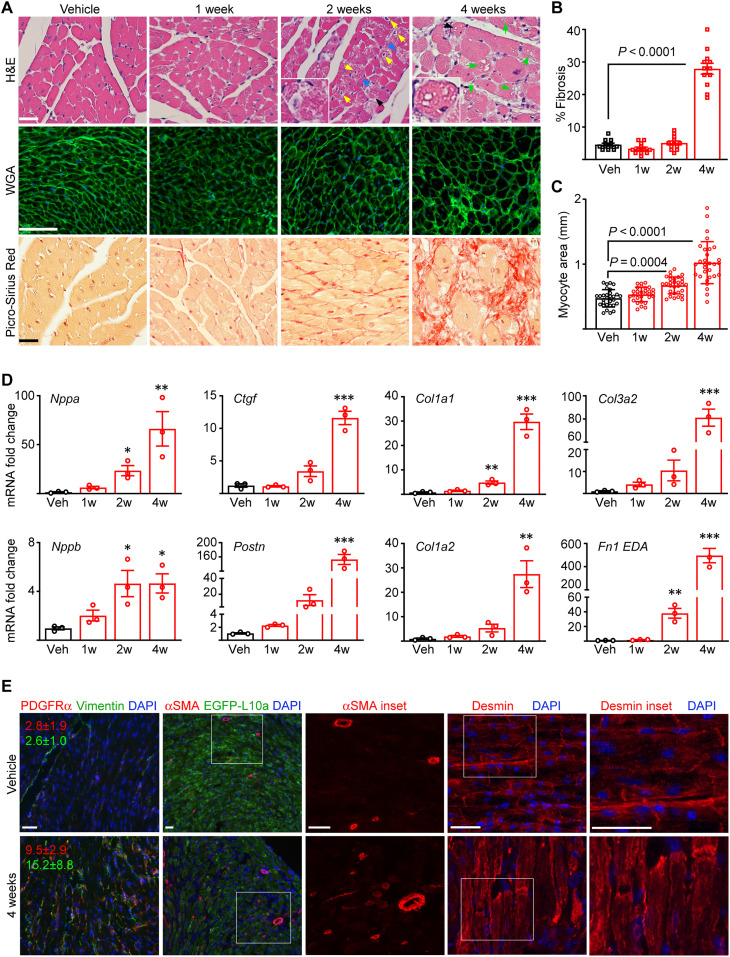
Fig. 2. Pathological cardiac remodeling accompanies cardiac dysfunction.
(A) Histological analyses of the hearts from CM-CreTRAP:Lmnaflox/flox mice at 1, 2, and 4 weeks after Tam or Veh treatment. Blue, yellow, and green arrows denote degenerating myocytes, pyknotic nuclei, and vacuolation, respectively. Black arrows denote insets. H&E, hematoxylin and eosin; WGA, wheat germ agglutinin. Scale bars, 50 μm. A representative image per group (n = 3) are shown. (B) Quantitation of average % fibrosis of Masson’s trichrome staining from 12 independent images from three mouse hearts per group. Error bars = SEM. % Fibrosis P value was determined using one-way ANOVA and Dunnett’s correction with Veh as control. (C) Quantitation of myocyte cross-sectional area based on WGA staining. Three images were taken per section, and average area of 30 individual CMs was measured in total per group across three experiments. Error bars = SD. P values were determined using one-way ANOVA and Dunnett’s correction with Veh as control. (D) qPCR of cardiac stress and profibrotic markers in hearts of Tam-treated CM-CreTRAP:Lmnaflox/flox mice. 1w to 4w denote weeks after final Tam dosing. Data are presented as fold change relative to Veh. *P < 0.05, **P <0.01, and ***P < 0.0001 using one-way ANOVA with Dunnett’s correction with Veh as control. Error bars = SEM. n = 3. (E) Immunofluorescence images of platelet-derived growth factor receptor α (PDGFRα), vimentin, α–smooth muscle actin (αSMA), EGFP-L10a, and desmin on mouse hearts 4 weeks after Veh or Tam treatment. The red and green numbers in line with the PDGFRα and vimentin micrographs denote their quantitation (% area per field ± SEM), respectively, calculated from 15 images from three mice per group across three replicate experiments. White boxes denote magnified insets shown to the right. Scale bars, 50 μm. Data from both sexes are shown in this figure.
The above histological changes were correlated with increased transcript expression of cardiac stress markers and markers of pathological fibrosis (Fig. 2D). Natriuretic peptide precursors Nppa and Nppb, which are produced in response to excessive CM stretch, were enhanced by 2 weeks after Tam treatment and increased further by 4 weeks. A similar pattern was observed for markers of activated cardiac fibroblasts (Postn and Ctgf) and extracellular matrix synthesis (Col1a1 and Col1a2) as well as those associated with pathological remodeling Col3a2 (39) and Fn1-extra domain A (40). These changes were absent in Tam-treated CM-CreTRAP:Lmnaflox/+ mice, demonstrating that again, in our system, Tam treatment or CreERT2 expression did not elicit pathological remodeling (fig. S3E). Indirect immunofluorescence revealed increased α–smooth muscle actin [expressed in myofibroblasts, stressed rodent CMs (41, 42), and pericytes (43)], vimentin (mesenchymal marker expressed in cardiac fibroblasts), and platelet-derived growth factor receptor α (also a cardiac fibroblast-selective marker) staining in the myocardial sections from CM-CreTRAP:Lmnaflox/flox mice at 4 weeks after Tam dosing, suggesting an increased number of cardiac fibroblasts, myofibroblasts, and capillaries within degenerated myocardium (Fig. 2E). Desmin staining further revealed disruption of typical CM alignment and distortions of CM shape owing to its degeneration (Fig. 2E).
NE abnormalities and partial UPR activation in Lmna-deleted CMs
A closer inspection of the higher magnification H&E-stained hearts revealed nuclear abnormalities uncharacteristic of those previously associated with LMNA cardiomyopathy models. Previous studies using germline knockout and knock-in mouse models revealed abnormal elongation of nuclei in CMs (23, 44). In the hearts of Tam-treated CreTRAP:Lmnaflox/flox mice, a greater variety of nuclear abnormalities at higher frequencies were noted (Fig. 3A). At 2 weeks after Tam, nuclear shapes were much more irregular, with many being swollen, pyknotic, or grossly misshapen (Fig. 3A). Even accounting for sectioning artifacts, abnormal nuclei were more prevalent in the Tam-treated hearts than those treated with corn oil. The severity and the frequency of these abnormalities became more pronounced at 4 weeks after Tam treatment (fig. S4A).
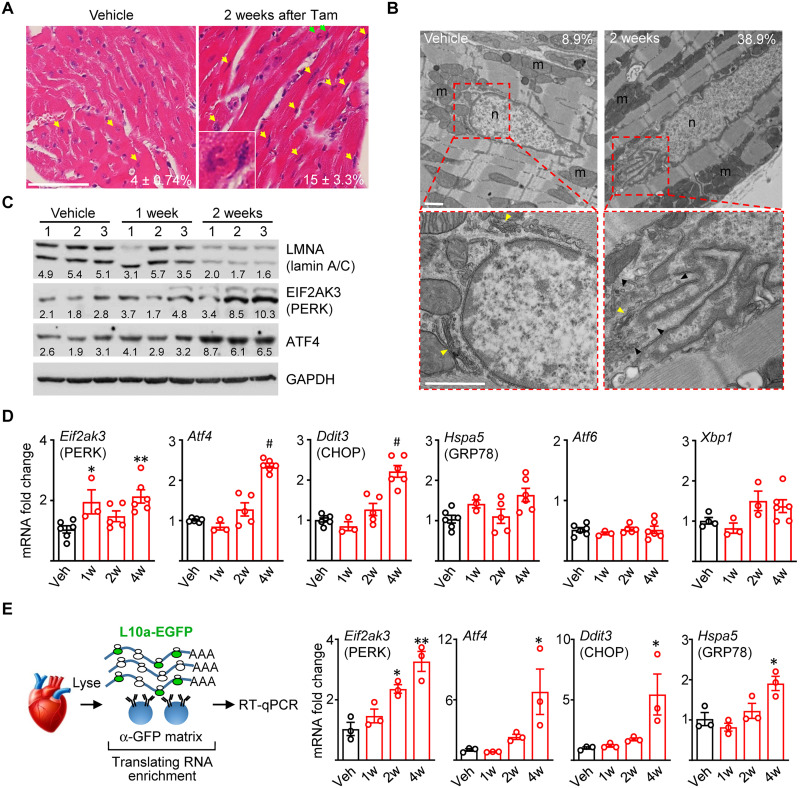
Fig. 3. Selective UPR activation in Lmna-deleted hearts.
(A) H&E staining of longitudinal heart sections from CM-CreTRAP:Lmnaflox/flox mice 2 weeks after Veh or Tam. Yellow arrows highlight abnormally shaped, ruptured, and stretched nuclei with green arrows showing inset. Average number ± SEM of abnormal nuclei per field counted from 15 fields from three mice per group from three experiments are shown. (B) TEM on hearts from CM-CreTRAP:Lmnaflox/flox mice 2 weeks after Veh or Tam. Red boxes show magnified perinuclear space. Yellow and black arrowheads denote Golgi and 100 nm vesicles, respectively. m, mitochondria; n, nucleus. Scale bars, 1 μm. Numbers in line with EM micrographs denote percent abnormal nuclei quantified in Veh (n = 45 total nuclei) and 2 weeks after Tam (n = 31) from two independent hearts per group. (C) Immunoblot of lamin A/C, PERK, ATF4, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in hearts from Veh or Tam-treated CM-CreTRAP:Lmnaflox/flox mice. Numbers on top denote individual hearts. One and 2 weeks denote time after final Tam treatment. Numbers in line with blots show quantification of protein bands normalized to GAPDH in arbitrary units. (D) qPCR analyses of mediators of unfolded protein response in hearts from CM-CreTRAP:Lmnaflox/flox mice treated with Veh or Tam. 1w, 2w, and 4w denote weeks after Tam. *P <0.05, **P <0.005, and #P <0.0001 using one-way ANOVA with Dunnett’s’s post hoc with Veh as control. n = 5 except for 1w (n = 3). All error bars in this figure = SEM. (E) Schema of experimental design using TRAP. Right panel shows qPCR on CM-specific translating mRNAs probed for Eif2ak3, Atf4, Ddit3, and Hspa5, respectively, from CM-CreTRAP:Lmnaflox/flox mice 1 to 4 weeks after Tam. *P < 0.05 and **P < 0.0005 using one-way ANOVA with Dunnett’s’s post hoc with Veh as control. n = 3.
We sought to obtain a more detailed view of the nuclear abnormalities, particularly on the perinuclear space, by using transmission electron microscopy (TEM) (Fig. 3B). Similar to H&E, we observed increased NE ruffling/breakdown from 2 weeks after Tam treatment (Fig. 3B and fig. S4, B and C), which is before detectable cardiac functional decline and obvious pathological remodeling. By 4 weeks after Tam, the frequency of NE undergoing breakdown increased further (fig. S4, B and C). Vacuoles of varying size (1 to 3 μm) adjacent to the NE and at the perinuclear space were present. Also, present in the perinuclear space was an abundance of small (~100 nm) vesicles that congest the ER-Golgi interface, particularly around areas of NE ruffling/breakdown (Fig. 3B and fig. S4B). These vesicles were abundant at both 2 and 4 weeks after Tam but far less so in vehicle-treated hearts, suggesting that the general abnormalities at the perinuclear region are associated with disease pathogenesis. Together, our data show that CM-specific Lmna deletion results in NE breakdown and perturbations at the perinuclear space, all before detectable cardiac functional decline.
The nuclear membranes are contiguous with the rough ER, representing distinct regions of a single membrane system (45). We reasoned that the nuclear abnormalities and the damage arising from them would also cause ER–nuclear membrane disruption, leading to ER stress. Moreover, evidence of ER stress activation has been documented in hearts of transgenic models of LMNA cardiomyopathy (22) and in patient-derived fibroblasts (46). We therefore assessed for ER stress activation in hearts of CreTRAP:Lmnaflox/flox mice following Lmna deletion. ER stress triggers a set of signaling pathways, collectively termed unfolded protein response (UPR), for which there are three canonical branches: protein kinase R-like endoplasmic reticulum kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme type 1 (IRE1) (47). Following CM-specific Lmna deletion, we observed elevated protein expression of PERK (EIF2AK3) and its downstream mediator ATF4 at 2 weeks after Tam dosing (Fig. 3C). Quantitative polymerase chain reaction (qPCR) analyses further indicated that Eif2ak3, Atf4, and Ddit3 (encoding CHOP, a downstream target of ATF4) expression were elevated at the transcript level (Fig. 3D). To assess the ATF6 branch of the UPR, we measured mRNA levels of genes induced by ATF6. Transcriptionally active ATF6 can transactivate Xbp1, Hspa5 (encoding BiP), and its own transcription to establish a positive feedback loop (48–50). Expression analysis revealed little changes in the mRNA levels of Xbp1, Hspa5, and Atf6 in the CM-specific Lmna-deleted hearts (Fig. 3D), suggesting that the ATF6 branch is not robustly activated. To probe the IRE1 branch, we measured Xbp1 splicing as a proxy for IRE1 activation (51) by using PCR primers that span the intron excised by activated IRE1 (52) in neonatal CMs (nCMs) that we used as a more relevant in vitro culture model. Although Xbp1 splicing was readily detectable in wild-type nCMs treated with brefeldin A (BFA), a reversible inhibitor of anterograde protein trafficking from the ER causing robust ER stress, this splicing was dampened in lamin A/C–depleted nCMs (fig. S5A) or absent in hearts from Tam-treated CreTRAP:Lmnaflox/flox mice (fig. S5B). Hearts from Tam-treated CM-CreTRAP:Lmnaflox/+ mice did not display increased expression of any of these ER stress markers, demonstrating that Tam or CreERT2, in and of themselves, did not trigger ER stress (fig. S5C). Our collective results suggest that CM-specific Lmna deletion elicits selective UPR activation toward the PERK axis. We observed comparable findings in the human disease, as the protein levels of ATF4 in hearts from patients with LMNA cardiomyopathy were significantly elevated (~2.2-fold), with PERK levels showing a similar trend (~1.6-fold) but not reaching statistical significance, relative to age- and sex-matched healthy controls (fig. S5D).
To remove the influence of non-myocytes in our analyses, we isolated CM-specific translating mRNA using TRAP from vehicle- and Tam-treated mice and performed reverse transcription qPCR (RT-qPCR) (Fig. 3E). To validate that the immunopurified mRNAs are CM specific, we interrogated TRAP mRNA with primers specific to markers of various cell types in the myocardium. The TRAP mRNAs were enriched with those encoding CM-specific markers with little to no cardiac fibroblast or endothelial cell–specific markers (fig. S5E), confirming that the isolated mRNAs are specifically from CMs. From 2 weeks after Tam treatment and onward, we observed increases in Eif2ak3, Atf4, Ddit3, and to a lesser degree Hspa5 expression (Fig. 3E), further supporting the notion that some form of UPR is activated in Lmna-deleted CMs.
Golgi stress in response to Lmna deletion
In adult CMs, the perinuclear ER and Golgi are tightly pack into a confined space around the perinuclear space. The area is also filled with perinuclear mitochondria in a fusiform shape (fig. S6A), which appear to have unique functions relative to their intrafibrillar counterparts (53). Given that partial UPR activation in response to Lmna deletion in CMs is characteristic of a Golgi stress response (54, 55), we reasoned that Golgi may also be perturbed. We analyzed our EM data with a particular focus on the Golgi and observed frequent Golgi stack dilation/fragmentation starting from 2 weeks after Tam that increased further in frequency at 4 weeks (Fig. 4A and fig. S6, B and C). Furthermore, the dilation of the Golgi stacks appeared to be mostly localized around areas adjacent to NE deterioration (fig. S6, B and C), indicating that Golgi swelling and NE damage may be pathologically linked.
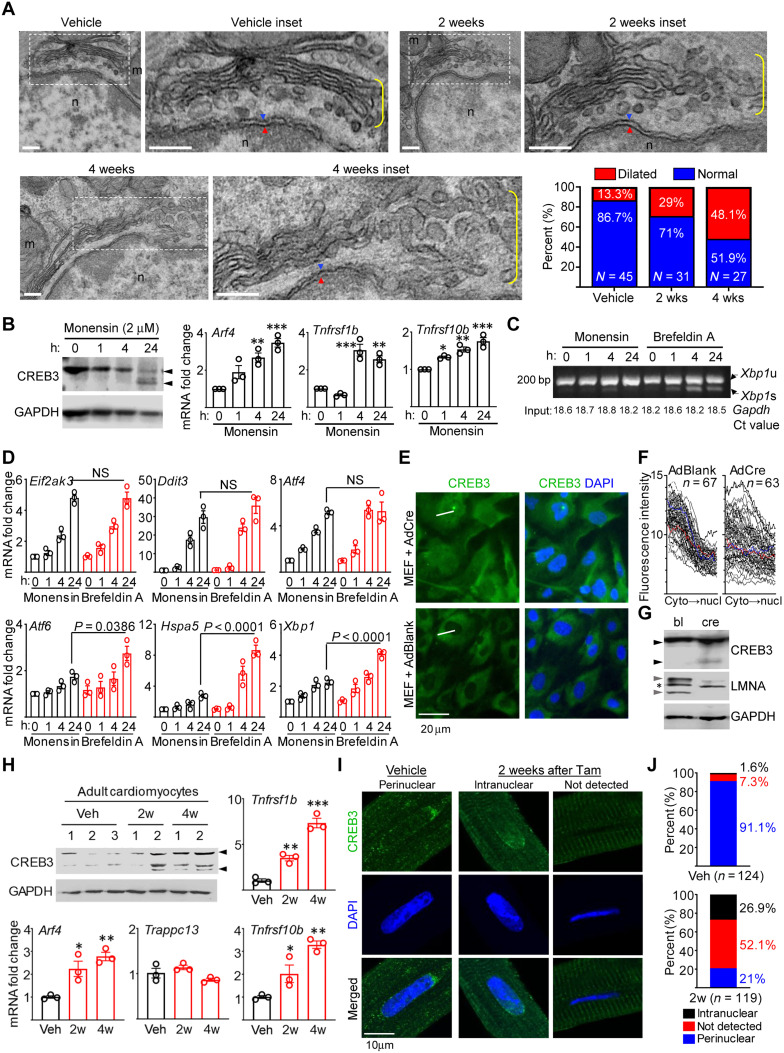
Fig. 4. Golgi stress in Lmna-deleted CMs.
(A) TEM of Lmna-deleted mouse hearts. Dotted boxes denotes inset. Yellow brackets denotes Golgi. Blue/red arrowheads denote outer/inner nuclear membranes. Scale bars, 200 nm. Quantitation of % dilated and normal Golgi in Veh (n = 45 nuclei), 2 weeks (n = 31), and 4 weeks (n = 27) after Tam. (B) CREB3/GAPDH immunoblots (left) and qPCR of CREB3-activated genes (right) in monensin-treated Lmnaflox/flox MEFs. Arrowheads denotes CREB3 cleavage. n = 3. (C) RT-PCR of Xbp1 mRNA splicing in monensin (2 μM)– or BFA (20 μM)–treated Lmnaflox/flox MEFs. Xbp1u and Xbp1s denote unspliced and spliced variants. Numbers on bottom denote Gapdh Ct values (input control). Representative image from three experiments are shown. (D) qPCR of UPR mediators in Lmnaflox/flox MEFs treated as in (C). n = 3. NS, not significant. (E) Representative CREB3 and DAPI images of AdBlank or AdCre-infected Lmnaflox/flox MEFs from three experiments. White lines denote linear fluorescence intensity measurement examples. Scale bar, 50 μm. (F) Fluorescence intensity profiles of CREB3 in AdBlank or AdCre-infected Lmnaflox/flox MEFs. Measurement directionality is from cytoplasm (cyto) to nucleus (nucl). (G) CREB3, LMNA, and GAPDH immunoblots in AdBlank (bl)– or AdCre (cre)–infected Lmnaflox/flox MEFs. Black and gray arrowheads denote CREB3 cleavage and lamin A/C, respectively. Asterisk denotes nonspecific band. Representative blot from n = 3 experiments. (H) Representative CREB3 and GAPDH immunoblot of adult CMs from Veh- or Tam-treated CM-CreTRAP:Lmnaflox/flox mice. n = 3 experiments. Numbers on top denote individual hearts. Arrowheads denote CREB3 and its fragments. Remaining graphs show qPCR on TRAP mRNA for Arf4, Trappc13, Tnfrsf1b, and Tnfrsf10b. n = 3 to 5. (I) Confocal CREB3 and DAPI images on adult CMs from Veh- or Tam-treated CM-CreTRAP:Lmnaflox/flox mice from three isolation per staining. Scale bar, 10 μm. (J) CREB3 quantification from (I). n = nuclei analyzed from three experiments. All error bars in this figure = SEM. All P = one-way ANOVA with Dunnett’s post hoc with *P < 0.05, **P < 0.005, and ***P < 0.0005.
The swollen Golgi stacks at the perinuclear space are reminiscent of a similar stress response induced by a ionophoric antibiotic monensin, which acts by collapsing Na+ and H+ gradients, causing Golgi dilation (56). As previously indicated, prior studies have shown that monensin-induced Golgi stress triggers the ATF4-CHOP axis of the UPR without the other two (XBP-1 and ATF6) (54, 55). Similar molecular expression profiles were observed in the hearts of our CM-Lmna–deleted mice, strongly indicating that Golgi stress response underlies the selective ATF4-CHOP activation. To confirm this and to establish causality, we used in vitro culture models to recapitulate the in vivo phenotype.
Golgi stress induced by monensin can activate CREB3, an ER-resident protein belonging to a family of transcription factors that includes ATF6 (57). In a similar fashion to ATF6, full-length CREB3 is translocated to the Golgi in response to stress where it undergoes proteolytic cleavage, producing a transcriptionally active transcription factor that enters the nucleus and transactivates the expression of ARF4, TRAPPC13, and TNFRSF10A (58). Following 2 μM monensin treatment, both mouse embryonic fibroblasts (MEFs) isolated from Lmnaflox/flox mice (Fig. 4B, left) and C2C12 myoblasts (fig. S6D) displayed CREB3 cleavage (from uncleaved ~46-kDa protein to ~38-kDa fragments) by 24 hours. Assessment of mRNA up-regulation of genes induced by CREB3 in MEFs revealed that although Arf4 levels were significantly elevated by 4 hour after monensin treatment (Fig. 4B, right), Trappc13 only showed a trend toward significance (fig. S6E). Moreover, we interestingly found that no murine homolog for TNFRSF10A exists. Therefore, we measured other members of the tumor necrosis factor (TNF) family of receptors Tnfrsf1b and Tnfrsf10b and observed that they were also significantly elevated by 4 hours after monensin treatment (Fig. 4B, right), indicating that CREB3 may mediate the broad transactivation of TNF receptor family members. Furthermore, consistent with the prior reports (54, 55), monensin robustly activated the ATF4-CHOP axis, without similar activation of Atf6, Hspa5, and Xbp1 (as well as its splicing), whereas BFA displayed no such selectivity (Fig. 4, C and D).
Having established the molecular phenotypes from Golgi stress induced by monensin, we then assessed whether Lmna deletion, in and of itself, can induce CREB3 activation. To achieve this, we deleted Lmna in Lmnaflox/flox MEFs using adenovirus carrying Cre recombinase (AdCre) and assessed CREB3 cleavage and nuclear entry. Relative to MEFs infected with a blank adenovirus (AdBlank), those infected with AdCre displayed increased CREB3 localization in the nucleus (Fig. 4, E and F, and fig. S7 for uncropped micrographs). Notably, the CREB3 localization appeared to be coincident with the emergence of misshapen nuclei after 4 days after AdCre transduction of MEFs cultured in regular tissue culture plastic. Also, concomitant with this time point (4 days after adenoviral transduction), we consistently detected the presence of CREB3 cleavage product in MEFs infected with AdCre but not in those infected with the blank virus (Fig. 4G). These results indicate that Lmna deletion alone can trigger CREB3 activation. To confirm that Golgi stress–induced CREB3 is activated in the primary tissue from mice, we isolated adult CMs from Tam-treated CM-CreTRAP:Lmnaflox/flox mice and assessed for CREB3 activation. Similar to our immunoblot data (Fig. 3C), we observed lamin A/C depletion in adult CMs 2 weeks after Tam relative to vehicle-treated controls (fig. S8A). Lamin A/C loss appears to coincide with the incipient phase of Golgi stress signaling as ~50% of 2-week adult CMs displayed CREB3 cleavage, whereas all 4-week samples showed CREB3 cleavage and significant up-regulation of translating RNA for Arf4, Tnfrsf1b, and Tnfrsf10b but not Trappc13 (Fig. 4H). Notably, the pattern of CREB3 regulation in primary CMs seems to be different from cultured models; total CREB3 levels appear to increase concomitant with their cleavage in primary CMs, whereas this is not the case in culture models. The relevance of this noted difference is currently unknown. We further probed for CREB3 localization by immunofluorescence (Fig. 4I). In adult CMs from vehicle-treated mice, CREB3 was predominantly perinuclear (91.1% of adult CM nuclei analyzed), which is consistent with ER localization (Fig. 4, I and J, and see fig. S8, B and C for uncropped and additional images, respectively). However, following Lmna deletion, we observed three distinct patterns; CREB3 remained perinuclear (21%), was intranuclear (26.9%), or not detected (52.1%), indicating that CREB3 was activated in a subset of CMs (Fig. 4, I and J). The significance of the differential localization pattern is currently unclear. Although other Golgi stress responses have been identified, such as those involving Golph3 (59), we did not observe its activation in monensin-treated MEFs and in the Lmna-deleted hearts (fig. S8D), indicating that either it is not involved or not as prevalent as CREB3-mediated responses. Our results show that Lmna deletion in CMs leads to CREB3-mediated Golgi stress and the activation of the ATF4-CHOP signaling axis.
TRAP sequencing identifies up-regulation of MED25
Given the selective UPR activation signature specifically in CMs, we sought to obtain a broader snapshot of the gene expression profile. To achieve this, we performed RNA sequencing (RNA-seq) following TRAP. Translating mRNA isolated from ventricular tissue of mice 2 weeks after treatment with vehicle or Tam were profiled and compared (Fig. 5A). This time point was chosen on the basis of our results showing lamin A/C depletion in CMs but before detectable deterioration in cardiac function. The goal was to capture early perturbations elicited by lamin A/C depletion before large changes are brought on by cardiac functional decline. Data for sequencing depth, alignment rates, and complexity curves for the samples and PCA analysis demonstrate highly concordant behavior in replicates (fig. S9, A and B). However, we did notice large fold changes for some genes and a gap in the false discovery rate (FDR) distribution, which we surmise stem from lower complexity reads (fig. S9C). Raw sequencing results showed that a large proportion of the reads are dominated by mRNAs encoding sarcomeric, mitochondrial, and CM-specific isoform/enriched proteins (fig. S9D). This is expected from a CM-specific translatome and demonstrates that our TRAP worked as intended, which was to purify CM-specific translating mRNA from whole tissue. Nevertheless, to further ensure against false positives, we stringently filtered the genes by perfect markers, which is defined as all experimental replicates displaying same trends in expression over controls.
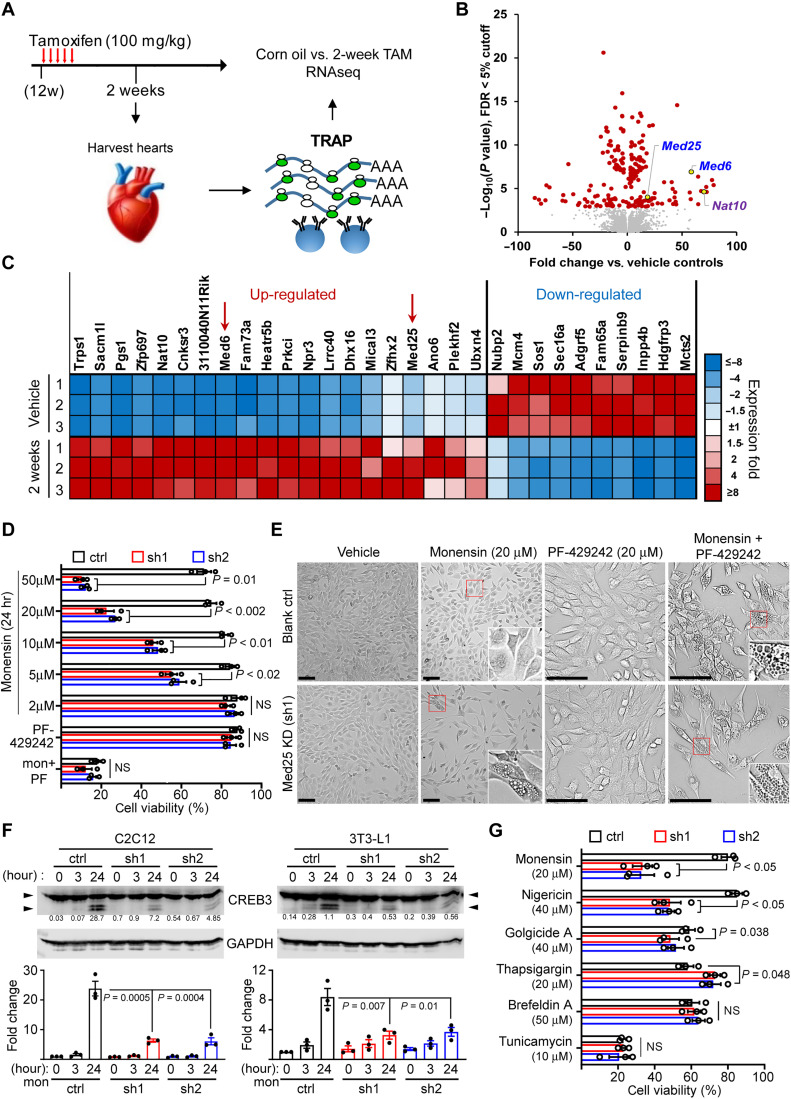
Fig. 5. MED25 depletion sensitizes cells to Golgi stress–induced vacuolar cell death.
(A) Experimental schema of TRAP/RNA-seq. (B) Volcano plot of differentially expressed genes sorted by –log10 of nominal P value with 5% FDR cutoff. (C) Heatmap of top 30 coding genes filtered by adjusted P values of perfect markers. Red arrows denote the position of Med6 and Med25. (D) Viability of MED25-depleted C2C12 treated with monensin, 20 μM PF-429242 (PF), and 20 μM PF-429242 + 2 μM monensin. The % cell viability was calculated relative to Veh controls. P values were calculated using two-way ANOVA with Dunnett’s correction. Error bars = SEM. n = 3 experiments. (E) Representative phase contrast images of blank control (ctrl) and MED25 KD C2C12 cells treated with Veh, monensin, PF-429242, and cotreatment. Insets, denoted by red boxes, show magnified cells with or without vacuoles. Scale bars, 50 μm. (F) CREB3 and GAPDH immunoblot of C2C12 (left) and 3T3-L1 (right) with MED25 KD (sh1 and sh2) treated with 2 μM monensin for 3 and 24 hours. Arrowheads denote CREB3 cleavage. Numbers in line with blots denote cleaved CREB3 quantitation normalized to GAPDH in arbitrary units. Bottom panels show cleaved CREB3 presented as fold change relative to ctrl at 0 hours (set to 1) from three experiments. P values were calculated using one-way ANOVA with Dunnett’s correction. (G) Viability of MED25-depleted C2C12 cells treated for 24 hours with chemical inducers of Golgi (monensin, nigericin, and golgicide A) and ER (BFA, thapsigargin, and tunicamycin) stress. The % cell viability was calculated relative to their respective cells treated with Veh. P values were calculated using two-way ANOVA with Dunnett’s correction. Error bars = SEM. n = 3. ns = not significant.
A volcano plot shows a pair of genes, Med6 and Med25, which were significantly up-regulated by 2 weeks (Fig. 5B). We also noted increased expression of Nat10, which was previously linked to misshapen nuclei in lamin A/C–depleted cells (60). A heatmap of top perfect markers showed that Med6 and Med25 are ranked 8th and 17th, respectively, as sorted by FDR-adjusted P values for up-regulated genes (Fig. 5C, red arrows). We confirmed their protein expression in hearts of both Tam-treated CM-CreTRAP:Lmnaflox/flox mouse lines 1 and line 2 (fig. S10A). MED25 and MED6 are members of the Mediator complex, an evolutionarily conserved transcriptional cofactors previously identified as regulators of UPR (61), which is consistent with the activation of some form of UPR in the hearts of Tam-treated CM-CreTRAP:Lmnaflox/flox mice. As we could not confirm the expression of MED6 in the myocardial tissue from human patients, as opposed to MED25 (62), we primarily focused our efforts on MED25.
Regulation of Golgi stress by MED25
Given that MED25 was shown to participate in UPR, we hypothesized that it may also play a role in Golgi stress regulation. Furthermore, it is unclear what role, if any, MED25 up-regulation plays in the context of LMNA cardiomyopathy. To begin to address this, we used muscle myoblast C2C12 culture model with short hairpin RNA (shRNA)–mediated silencing of Med25. Following MED25 depletion (fig. S10, B and C), C2C12 cells subjected to increasing concentration of Golgi stress inducer monensin (Fig. 5D). MED25 knockdown (KD) increased sensitivity to cell death in response to high concentrations (20 and 50 μM) of monensin, a phenotype that was evident starting from 5 μM (Fig. 5D). In contrast, control C2C12 cells (cells infected with lentivirus carrying an empty vector) were relatively resistant to high doses of monensin, indicating that MED25 confers protection against monensin-induced Golgi stress. These results suggest that MED25 may be up-regulated in the Lmna-deleted CMs to protect the cells from Golgi stress. Notably, the phenotypic features of death in the MED25 KD cells were notable; they resemble vacuolar cell death (VCD) as the cytoplasm accumulates with vacuoles (Fig. 5E, left, four panels), reminiscent of the H&E section images for Lmna-deleted hearts at 4 weeks after Tam.
Because various stress response mediators, including CREB3, require for their activation proteolytic cleavage by cis-Golgi–resident proteases such as site-1 proteases (S1P), we reasoned that perhaps MED25 KD impairs this process. To test this hypothesis, we cotreated cells with PF-429242, an S1P inhibitor, and monensin. While 20 μM PF-429242 alone had little to no effect on cell viability, its cotreatment with monensin sensitized control cells to VCD akin to those with MED25 KD (Fig. 5, D and E, right, four panels). MED25 KD cells displayed no differences in cell viability, further indicating that PF-429242 treatment and MED25 have overlapping effects. We also generated 3T3-L1 preadipocytes with MED25 KD (62), and they behave similarly to C2C12 cells in response to monensin and/or PF-429242 treatment (fig. S10D), suggesting that this may be a generalized effect and not specific to muscle myoblast lineage.
To further determine the impact of MED25 depletion on CREB3 activation, we assessed CREB3 cleavage in both C2C12 and 3T3-L1 cells with MED25 KD following 2 μM monensin for 3 and 24 hours (Fig. 5F). In both C2C12 and 3T3-L1 cells, MED25 KD significantly reduced the accumulation of cleaved CREB3 at 24 hours (Fig. 5F). We tested the most obvious potential mechanism; that MED25 is required for S1P transcription, but no differences were observed in the mRNA levels of Mbtps1 between control and MED25 KD cells (fig. S11A). These results strongly indicate that MED25 is a Golgi stress regulator that either facilitates the cleavage of CREB3 or its accumulation through unknown mechanisms. Because some ER stress inducers, such as BFA, can also induce CREB3 cleavage (63), we tested additional known chemical inducers of Golgi and ER stress for their ability to induce VCD in MED25 KD cells. Only monensin and nigericin were able to decrease cell viability selectively in MED25 KD cells relative to blank controls (Fig. 5G). Furthermore, MED25 KD cells dying from nigericin also displayed VCD that was less prominent then monensin (fig. S11B). Notably, monensin and nigericin are proton antiporters with selective binding preferences for Na+ and K+, respectively, suggesting that the collapse of ion gradients or lysosomal pH may be involved (64). Together, we conclude that MED25 depletion sensitizes cells to Golgi stress–induced VCD, and this phenotype is correlated with a reduction in cleaved CREB3.
Disruption of Golgi underlies autophagic abnormalities
We and others previously showed that autophagy, a stress-responsive lysosomal recycling mechanism that maintains cellular homeostasis, is impaired in germline models of LMNA cardiomyopathy (17, 18). To determine whether a similar impairment in autophagy is present in our inducible CM-specific Lmna deletion model, we assessed the levels of lipidated LC3B (LC3B-II) produced during autophagosome formation. These data were analyzed in the context of the expression level of p62, an autophagosome-associated protein that is degraded when autophagosomes are fused with lysosomes in a process described as autophagic flux. The combination of these two protein levels can be interpreted to distinguish whether a reduction in LC3B-II is caused by impaired autophagosome formation or its rapid degradation, and conversely, an increase in LC3B-II as a rapid expansion of a productive autophagic response or a blockade in autophagic flux. In the hearts of CM-Lmna–deleted mice, we observed a slight increase in both p62 and LC3B-II protein levels at 1 to 2 weeks after Tam treatment relative to vehicle controls (Fig. 6, A and B). By 4 weeks, obvious increases in p62 and LC3B-II were observed, which is indicative of autophagosome accumulation due to a blockade in autophagic flux.
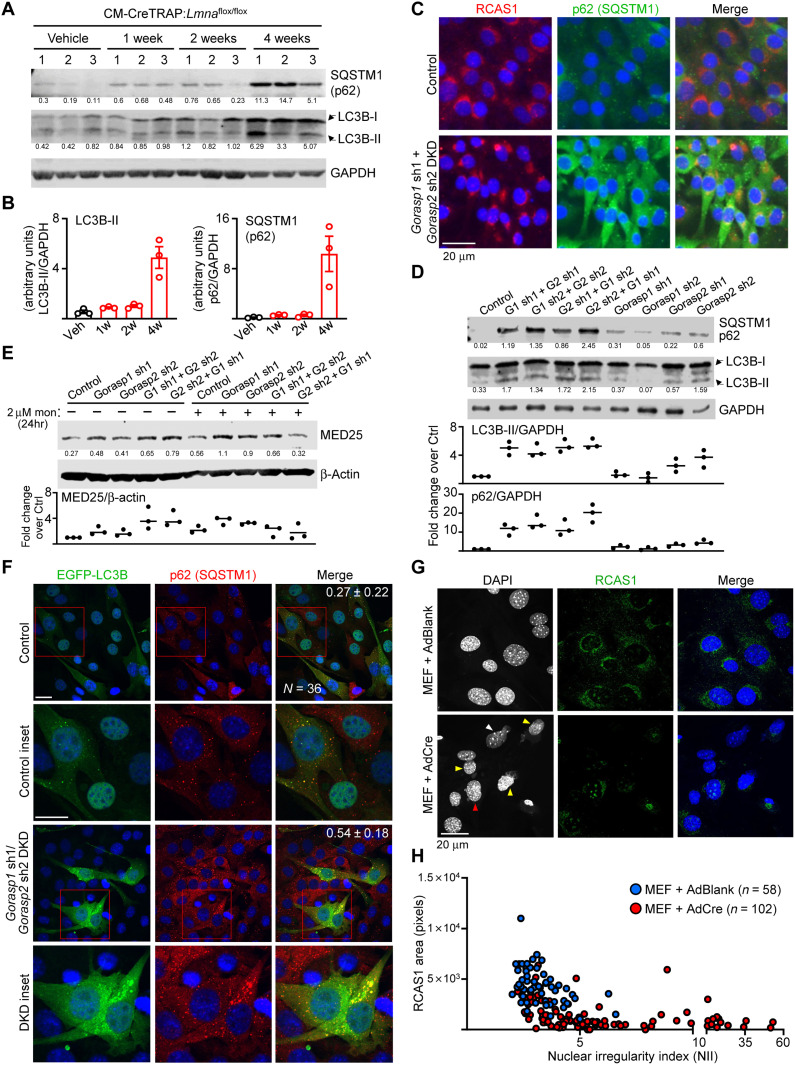
Fig. 6. Perinuclear damage underlies accumulation of autophagic markers.
(A) Immunoblot of p62, lipidated LC3B[LC3B-II], and GAPDH on Veh- or Tam-treated CM-CreTRAP:Lmnaflox/flox hearts at indicated times since last Tam dosing. n = 3 hearts per group. Numbers below blots denote quantified bands normalized to GAPDH in arbitrary units. (B) Compiled quantitation of LC3B-II and p62 blots in (A). (C) Representative immunofluorescence of C2C12 with Control or Gorasp1/Gorasp2 DKD stained for RCAS1 and p62 from three experiments. Control denotes C2C12 infected with lentivirus generated from blank plasmid. Scale bar, 20 μm. (D) Immunoblot of C2C12 with Gorasp1 or Gorasp2 KD alone or Gorasp1/Gorasp2 DKD probed for p62 and LC3B-II. GAPDH was assessed as loading control. Numbers below blots denote p62 and LC3B-II quantitation normalized to GAPDH in arbitrary units. Bottom shows LC3B-II and p62 fold change values relative to ctrl (set to 1) from n = 3 experiments. (E) Immunoblot/quantitation of monensin-treated C2C12 nuclear extracts with Gorasp1/2 KD probed for MED25 and β-actin as in (D). (F) Immunofluorescence of Control and Gorasp1/Gorasp2 DKD C2C12 transduced with EGFP-LC3B encoding retroviruses. After 1-week recovery, cells were stained for p62 and DAPI. Insets show magnified images of EGFP-LC3B puncta with p62. Numbers in line denote Mander’s coefficient ± SD (fraction of p62 overlapping with LC3B) from 36 random data points per group from three experiments. Scale bars, 10 μm. (G) Representative immunofluorescence images of Lmnaflox/flox MEFs 5 days after infection with AdBlank or AdCre stained for RCAS1 and DAPI from three experiments. Yellow arrowheads denote ruptured nuclei with extruding DNA. White arrow denotes nucleus with the highest nuclear irregularity index (NII). Higher NII denotes increased irregularities. Red arrow denotes nucleus with minor herniation but largely intact nuclear shape and Golgi. Scale bar, 20 μm. (H) NII plotted against perinuclear RCAS1 signal area from Lmnaflox/flox MEFs infected with AdBlank or AdCre. Data points were compiled from three experiments.
Golgi plays a direct and critical role for maintaining autophagy. For example, the delivery of degradative enzymes destined for lysosomes require conjugation with the mannose-6-phosphate residue in the cis-Golgi by N-acetylglucosamine–1 (GlcNAc-1)–phosphotransferase (65). The α and β subunits of GlcNAc-1-phosphotransferase are encoded by a single gene GNPTAB, which expresses as a precursor that requires proteolytic cleavage by S1P (66). Furthermore, Golgi also regulates the production and delivery of proteins and lipids required for autophagosome formation, such as ATG9A and PI(4)P, respectively (67). Given this critical role in autophagy, we sought to determine whether direct disruption of the Golgi apparatus will engender similar abnormalities in autophagy. To achieve this, we used shRNA-mediated KD of Golgi proteins GORASP1 and GORASP2 in C2C12 cells, as the depletion of both of these proteins was demonstrated to cause Golgi disruption and disassembly (68). We generated double KDs (DKDs) by a sequential delivery of shRNAs targeting Gorasp1 and Gorasp2 (fig. S11C) and then assessed for Golgi by staining for RCAS1, a protein highly enriched in Golgi (69). In C2C12 cells with a single KD (either Gorasp1 or Gorasp2) and in controls (C2C12 infected with lentivirus carrying a blank vector), RCAS1 staining exhibited crescent perinuclear staining indicative of the Golgi apparatus. In contrast, C2C12 with Gorasp1 and Gorasp2 DKD revealed a much smaller Golgi footprint with many cells displaying reduced signals (Fig. 6C; uncropped images are shown in fig. S11D). These results are indicative of Golgi disassembly and are consistent with the previously reported observations (68). We then assessed for p62 and LC3B-II and found that C2C12 cells with Golgi disruption (DKDs) displayed increased accumulation of both of these proteins relative to singly targeted cells even under basal conditions (Fig. 6D). This accumulation is posttranscriptionally mediated as no change in the mRNA levels of Sqstm1 (p62) and Map1lc3b (LC3B) was detected (fig. S11E). The Gorasp1/2 DKD also enhanced MED25 expression (Fig. 6E), further linking MED25 and Golgi stress. However, the MED25 levels decreased following monensin treatment (but not in the Gorasp1/2 single KD samples), suggesting that overt Golgi stress beyond a certain threshold may impede MED25 expression (Fig. 6E). We also probed for transcriptional activation of genes encoding ATF4-CHOP and observed that Golgi disruption did not lead to their increase (fig. S11E). These results demonstrate that p62 and LC3B-II accumulation results from Golgi disruption that is, in and of itself, insufficient to trigger the ATF4-CHOP axis and presumably require additional stressor such as damage to the nucleus.
To further demonstrate autophagy blockade in the Golgi-disrupted C2C12 cells, we coexpressed EGFP-LC3B in control and Gorasp1/Gorasp2 DKD cells by retroviral transduction. Following a 1-week recovery, we stained these cells for p62 to assess the extent of LC3B and p62 colocalization (Fig. 6F) and, after a 24 hours, 100 μM chloroquine treatment (fig. S12A) to establish a reference for LC3B-p62 colocalization in cells with chemically blocked autophagy. In controls, we observed mostly nuclear-localized EGFP-LC3B, which is an established pattern of its expression (70, 71), with few small puncta that colocalized with p62. However, in Gorasp1/Gorasp2 DKD cells, most of the EGFP-LC3B was extranuclear and formed larger aggregates (similar to but not as extensive as the chloroquine-treated cells; fig. S12A) that colocalized with p62 (Fig. 6F). This is further supported by the Mander’s coefficient, demonstrating a higher r value (r = 0.54) in the DKD cells relative to control cells (r = 0.27).
We then sought to determine whether lamin A/C depletion itself can disrupt the Golgi. Following infection of Lmnaflox/flox MEFs with either AdBlank or AdCre, we measured the protein expression of RCAS1 by immunoblot (fig. S12B) and immunofluorescence (Fig. 6G and fig. S12, C and D) as a surrogate for Golgi. We observed a ~50% reduction in the RCAS1 protein levels following lamin A/C depletion (fig. S12B). This reduction correlated with an increase in the number of nuclei with a higher nuclear irregularity index (NII) relative to Lmnaflox/flox MEFs with AdBlank infection (fig. S12, C and D). We confirm the reduced perinuclear RCAS1 signal in MEFs infected with AdCre relative to those infected with AdBlank (Fig. 6G and see fig. S13A for uncropped images). Upon closer inspection at the individual cells, there appeared to be a correlation between higher NII with lower Golgi footprint. To determine whether the severity of the nuclear abnormality is correlated with a reduced Golgi footprint, we measured the area of RCAS1 signal for each cell and plotted it against its respective NII (Fig. 6H). We observed a pattern in which a higher NII was correlated with lower RCAS1 signal area (Fig. 6H). By oversaturating the RCAS1 signal, the consequence of nuclear abnormalities resulting from lamin A/C depletion on the Golgi is better visualized (fig. S13B). In severely misshapen nuclei (fig. S13B, red arrowheads), the structure of the Golgi appeared to be convoluted and stretched, in a similar fashion to the nuclear damage. This suggests that the forces underlying the nuclear abnormalities in lamin A/C–depleted MEFs exert a similar physical stress on the Golgi that, consequently, damages the nucleus and the Golgi in tandem.
Enhancing autophagy or modulating ER stress preserves cardiac function
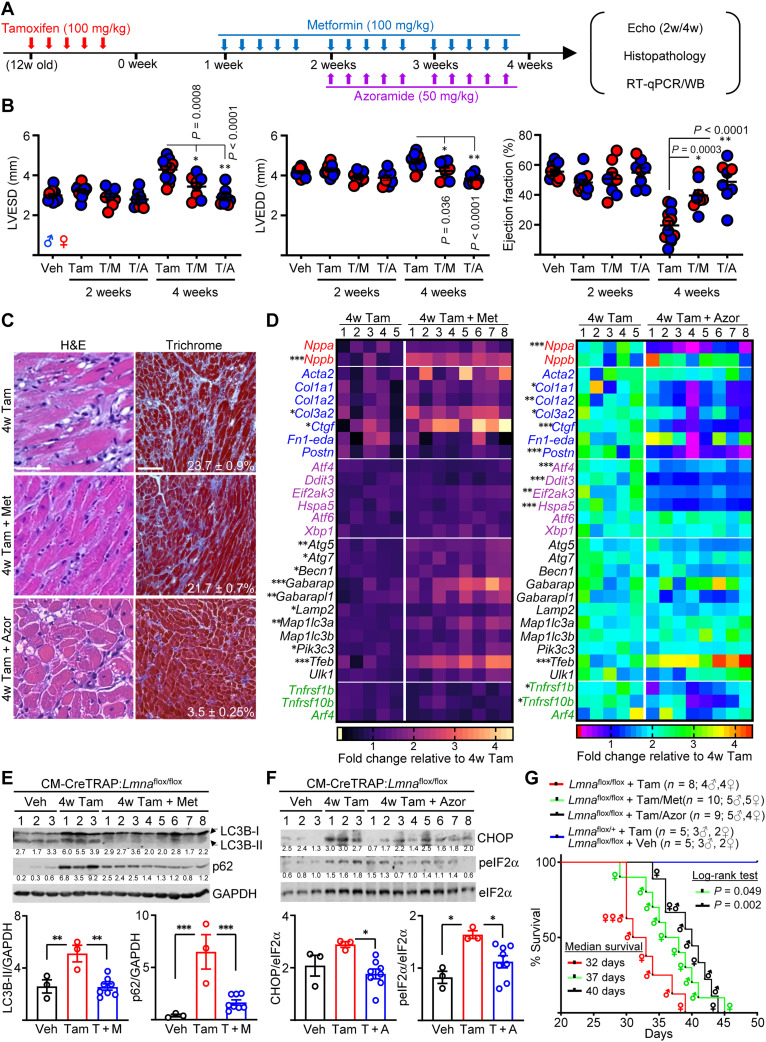
We and others previous demonstrated that reactivation of autophagy by rapamycin and its analog temsirolimus mitigates the disease in germline models of LMNA cardiomyopathy (17, 18), indicating a protective role of autophagy in this setting. Our current study suggests that damage in the perinuclear space emanating from nuclear abnormalities underlies the initial stressor that elicits a protective autophagy response, and when impaired, the damage to the CMs is exacerbated (72). Given the similar impairment in autophagy observed in our CM-specific model, we sought to perform pharmacological intervention studies aiming to mitigate stress signaling emanating from CM-specific lamin A/C depletion. Following CM-Lmna deletion in the adult heart, we administered either metformin (to stimulate autophagy) or azoramide (ER stress modulator) (73), as specific Golgi stress modulators have yet to be identified. Azoramide is a small molecule demonstrated to improve ER protein-folding ability by stimulating multiple chaperone expression in a dose-dependent fashion (73). Because high-dose azoramide administration (150 mg/kg) can enhance antidiabetic effects and glucose homeostasis (73), we decided on a far lower dose of 50 mg/kg (Fig. 7A). Given that the therapy was initiated before detectable cardiac function decline, our strategy represents a preventative therapy. Although no significant differences in cardiac function were observed at 2 weeks after Tam with or without metformin/azoramide relative to vehicle controls, these drugs significantly reduced both the systolic and the diastolic ventricular dimensions by 4 weeks, which resulted in preserved ejection fraction (Fig. 7B). Histopathology revealed that while the extent of cardiac fibrosis was similar between Tam versus Tam with metformin, the myocardial tissue disarray, cytoplasmic lysis, and vacuolation were less obvious (Fig. 7C and fig. S14A). Conversely, despite the similar preservation of cardiac function, hearts from azoramide-treated mice were phenotypically different. Although vacuolation was still obvious, we observed significantly reduced fibrosis (Fig. 7C and fig. S14A). These histological phenotypes were further supported by mRNA expression profiling analyses. We interrogated the expression of genes involved in several key cellular responses: cardiac stress adaptation (red), profibrotic (blue), UPR (purple), autophagy (black), and Golgi stress (green) related (Fig. 7D). Although several genes were enhanced by metformin relative to Tam alone, by far the largest number of significantly enhanced genes was in the autophagy response (left panel on Fig. 7D and fig. S14B). Alternatively, azoramide treatment predominantly reduced profibrotic genes and the UPR genes tested relative to Tam alone, although Atf6 and Xbp1 were not significantly altered (right panel on Fig. 7D and fig. S14B). Notably, Tnfrsf1b and Tnfrsf10b expressions were significantly reduced with the azoramide treatment but not with metformin despite the similar improvement in heart function, suggesting that azoramide may provide partial modulation of Golgi stress (Fig. 7D). At the protein level, metformin treatment reduced both LC3B-II and p62 levels, indicative of restored autophagy (Fig. 7E). Analogously, azoramide treatment reduced phospho-eIF2α (upstream activator of PERK) and CHOP expression (Fig. 7F), confirming the effects of azoramide treatment at the protein level. To determine whether these molecular changes associated with the improved cardiac function with metformin or azoramide therapy lead to increased survival, we performed survival analysis following treatment strategy outlined in Fig. 7A. Compared to the Tam-treated CreTRAP:Lmnaflox/flox mice, which showed a median survival of 32 days, both metformin and azoramide significantly enhanced survival by 15 and 25%, respectively (Fig. 7G). We observed no death events for our control group, which consisted of both Tam-treated CreTRAP:Lmnaflox/+ and vehicle-treated CreTRAP:Lmnaflox/flox mice. Together, these results indicate that CM-specific Lmna deletion leads to impaired autophagy in the heart, and either restoring autophagy or alleviating its need by dampening ER stress can delay LMNA cardiomyopathy and enhance survival in mice.
Fig. 7. Modulating autophagy or ER stress attenuates cardiomyopathy development.
(A) Schema of therapy schedule. (B) Echo on CM-CreTRAP:Lmnaflox/flox mice at 2 and 4 weeks after Tam, Tam + metformin (T/M), or Tam + azoramide (T/A). LVESD and LVEDD denote left ventricular end systolic and diastolic diameters, respectively. P values were obtained using one-way ANOVA with Tukey’s correction. Error bars = SEM. (C) Representative H&E and Masson’s Trichrome staining of heart sections from CM-CreTRAP:Lmnaflox/flox mice at 4 weeks after treatments described in (A) from three mice per group. Scale bars, 100 μm. (D) Heatmap of mRNA expression profile on natriuretic peptide precursors (red), profibrotic (blue), ER stress–related (purple), autophagy (black), and Golgi stress (green)–related genes in hearts from CM-CreTRAP:Lmnaflox/flox mice at 4 weeks after treatments described in (A). Numbers on top of heatmap denote biological replicates. Color gradient scales and values on bottom denote fold change over mean value of 4w Tam from the five biological replicates. P values were obtained using unpaired, two-tailed Student’s t test. *P < 0.05, **P < 0.01, and ***P < 0.001. (E) p62, LC3B, and GAPDH immunoblots on CM-CreTRAP:Lmnaflox/flox mice hearts at 4 weeks after Veh, Tam, and Tam + metformin. Numbers on top denote biological replicates. Numbers below blots denote band quantitation normalized to GAPDH in arbitrary units. Bottom shows compiled quantitation from biological replicates. P values were obtained using one-way ANOVA with Tukey’s correction. *P < 0.05, **P < 0.01, and ***P < 0.001. Error bars = SEM. (F) Immunoblot similar to above but with azoramide therapy and probed for CHOP, phospho-eIF2α (peIF2α), and eIF2α. (G) Kaplan-Meier plot of CreTRAP:Lmnaflox/flox mice treated with Tam (n = 8), Tam + metformin (n = 10), and Tam + azoramide (n = 9). Tam-treated CreTRAP:Lmnaflox/+ (n = 5) and Veh-treated CreTRAP:Lmnaflox/flox (n = 5) mice are presented as a single group. P values were calculated using log-rank test with the survival curve of CreTRAP:Lmnaflox/flox mice + Tam as the comparison control. Sex of mice at death is shown within the curve. WB, Western blot.
DISCUSSION
It is well appreciated that lamin A/C maintain structural support of the nucleus, but how nuclear abnormalities resulting from Lmna mutations underlie pathology in a complex organ such as the heart is less well understood. In the current study, we found that adult CM-specific lamin A/C depletion caused rapid development of cardiomyopathy with accompanying pathological fibrosis. Before cardiac function decline, lamin A/C depletion caused nuclear abnormalities, activation of the CREB3-mediated Golgi stress response, and selective activation of the ATF4-CHOP axis. We further link MED25, a member of the mediator complex, which we identify as a Golgi stress regulator that confers protection from Golgi stress–induced VCD. Last, we show that Golgi disruption caused p62 and LC3B-II accumulation, and the administration of autophagy inducing drug metformin enhanced their clearance and ameliorated the disease. Likewise, a similar benefit to the cardiac function was achieved by UPR modulator azoramide.
To our knowledge, this is the first study demonstrating a connection between Golgi stress as a consequence of Lmna deletion in CMs. We posit that the interplay of stress responses emanating from nuclear damage and perinuclear organelle damage are at the nexus of the pathogenesis of LMNA cardiomyopathy. This assertion is consistent with recent studies demonstrating prolonged survival in CM-specific Lmna deletion mouse models, for which the NE and cytoskeleton were physically decoupled (74, 75). The NE damage from defects in lamin A/C is well established and has been noted as early as 1999 (14) and since then have been confirmed in cell culture models (20, 76–78) as well as in the myocardium from murine models (15, 22, 23) and from human patients (24). We expand on this foundation with our current study, in which nuclear abnormalities stemming from lamin A/C depletion, likely following repeated rupture/repair cycle, cause stress and damage at the adult CM perinuclear space tightly packed with ER, Golgi, and perinuclear mitochondria. As a part of the incipient stress response activation, we hypothesize that MED25 is activated as a compensatory response to confer cytoprotection. Over time, the NE damage worsens, leading to continued deterioration of ER/Golgi that is likely exacerbated by DNA damage (79, 80). Extending this line of reasoning, it is plausible that the progressive deterioration of the perinuclear space may eventually interfere with proper activation of various stress responses that depend on S1P-mediated cleavage in the cis-Golgi, such as CREB3 itself and its family members (81–83) and lipid metabolism regulator SREBP1/2 (84), among others. Although only a conjecture at this point, this may represent an evolution of disease progression, starting from nuclear fragility and rupture to ER/Golgi stress to eventually impaired stress response signaling that leads to VCD.
Golgi was first hypothesized to be implicated in laminopathies based on the observation that MEFs from Lmna−/− mice displayed Sun1 accumulation in the Golgi (85). Although it is plausible that Sun1 mislocalization contributes to the molecular pathogenesis, our data suggest that ionic/pH changes more likely underlie the dilation and the subsequent fragmentation of Golgi. This is based on (i) the observation that Golgi damage and the resulting stress signaling in the CMs of Lmna-deleted hearts are similar to those induced by monensin and nigericin, which acts by collapsing the Na+/K+ and H+ gradients (56), and (ii) the existence of tightly regulated Na+ and H+ gradients across the NE maintained by Na+- and K+-dependent adenosine triphosphatase (Na+-K+-ATPase) and vacuolar H+-ATPase, respectively (86). This damage to the Golgi may affect autophagy either directly or indirectly. As mentioned previously, the delivery of degradative enzymes destined for lysosomes require protein mediators that are processed by S1P (65, 66). Furthermore, the Golgi is a site for regulation of proteins and lipids required for autophagosome formation, such as ATG9A and PI(4)P, respectively (67). Notably, our results showing the accumulation of p62 and LC3B-II only after simultaneous KD of both Gorasp1 and Gorasp2 (Fig. 6D) are slightly different from previously reported study, in which the authors observed similar accumulation after GORASP2 depletion alone (87). Although we also observed a slight elevation of p62 and LC3B-II following Gorasp2 KD relative to blank controls and Gorasp1 KD, we observed an enhanced (possibly synergistic) effect with simultaneous Gorasp1 and Gorasp2 KD. While the reason is currently unclear, we surmise this may be due to the differences the cell type (C2C12 versus HeLa/HEK293) and the method of gene silencing (shRNA-mediated KD in a heterogeneous C2C12 population versus clonally derived HeLa/HEK293 following CRISPR-mediated deletion).
Another notable component of our study is the identification of MED25 as a Golgi stress regulator and its up-regulation in LMNA cardiomyopathy. The depletion of MED25 sensitizing cells specific to chemical inducers of Golgi stress (monensin and nigericin), but not to ER stress regulators, suggests that MED25 is up-regulated as a compensatory mechanism to protect Lmna-deleted CMs. However, the vacuolation observed in the late stage (4 weeks after Tam) hearts of our mouse model is reminiscent of VCD. If they are mediated by similar mechanisms, then this suggests that the cytoprotective mechanism conferred by MED25 up-regulation eventually collapses amid persistent Golgi stress. To our knowledge, the observed vacuolation is similar to two distinct forms of VCD described in the literature: methuosis (88, 89) and autosis (90). Methuosis is characterized by an accumulation of endocytosed fluorescent tracers in the vacuoles (91). Alternatively, cell death by autosis can be prevented by blocking autophagy by chloroquine (92). Future studies are required to delineate whether the VCD observed in our MED25-depleted cells subjected to Golgi stress is methuosis, autosis, or possibly another form of VCD.
Contrary to other mouse models of LMNA cardiomyopathy, such as the LmnaH222P/H222P model, we did not observe sex-dependent effects in our mice. We speculate that this is due to multiple reasons. First, we suspect that the aggressive nature and the rapid kinetic of the disease progression in our CM-specific Lmna deletion model may mask any differences due to sex. The LmnaH222P/H222P model comparatively is a more slowly progressing disease that is likely due to the nature of the mutation itself; H222P is a point mutant variant with comparable expression to the wild-type counterparts that also maintains proper localization of emerin (15). We speculate that these features of H222P model provide some degree of nuclear structural integrity and certainly more so relative to our CM-specific deletion model. Second, H222P is a germline mutation model, with abnormalities that are observed as early as during the embryonic development of heart (93). Coupled with the slower disease progression, we speculate that these conditions allow compensatory mechanisms to establish allostasis, which may be weakened by sex hormones, particularly androgen receptor nuclear accumulation (94).
Some limitations are worth noting. Although metformin can stimulate autophagy (95), it also exhibits pleiotropic effects in an organ-specific fashion. For example, metformin blocks hepatic gluconeogenesis, activates AMPK in striated muscle, and alters the gut microbiota in the intestine (96). Therefore, while the metformin therapy resulted in transcriptome changes indicative of increased autophagy in our study, the correlating cardiac protection observed may have benefitted from the aforementioned systemic effects of metformin. Same can be said for azoramide as its use at higher concentrations can have metabolic benefits as mentioned previously. Therefore, although the positive effects of these drugs on the heart function are only suggestive of autophagy blockade and ER stress involvement in the pathogenesis of LMNA cardiomyopathy, they still establish translational potential that can be realized into therapeutic strategies for a disease that, to date, has no effective targeted therapy. It is also worth noting the limitation of our two-dimensional (2D) culture model to recapitulate the in vivo condition. In the 2D culture, the nuclear damage likely results from compressive forces generated by cell flattening (21). However, forces applied to CMs in an intact heart would likely be more complex than a unidirectional compressive force. Nevertheless, given the recapitulation of nuclear abnormalities in 2D culture, we posit that the main driver of the observed nuclear damage is due to the lack of lamin A/C expression despite the noted differences in the forces felt by the CMs.
MATERIALS AND METHODS
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee of Thomas Jefferson University under protocol #01744-1. All methods adhered to the NIH Guide for the Care and Use of Laboratory. Lmnaflox/flox mice (36), obtained from the Jackson Laboratory on a mixed background, were backcrossed to C57BL/6J for at least eight generations and genotyped as instructed by the distributor. CM-CreTRAP transgenic mice were generated in a C57BL/6 background by Cyagen Biosciences. Sequences encoding EGFP and murine RPL10a was obtained and fused according to Heiman et al. (25) using In-Fusion HD (Takara Bio, catalog no. 011614). The EGFP-L10a fusion construct was incorporated downstream of CreERT2 (97) (Addgene, #14797) linked by an IRES (Addgene, #12254) while simultaneously being subcloned into pPSKII-MHC-GH (98) downstream of the Myh6 (α-myosin heavy chain) promoter and upstream of a human growth hormone polyadenylation signal using In-Fusion HD (Takara Bio, catalog no. 011614). The final transgenic targeting construct was verified by DNA sequencing. Genotyping was performed by standard PCR using genomic DNA isolated from tail clippings and primers indicated in the table S3. Mice were fed a chow diet and housed in a disease-free barrier facility with 12-hour light/12-hour cycles. Sample size estimations for in vivo analyses were calculated on the basis of initial pilot experimental results. Because our initial analyses did not reveal sex-dependent differences (Fig. 1), the both male and female mice were used in the rest of the study, unless indicated otherwise (Fig. 7, B and G). Tamoxifen (MilliporeSigma, catalog no. T5648) was dissolved in corn oil (MilliporeSigma, catalog no. C8267) and injected intraperitoneally. Intraperitoneal delivery of Metformin HCl at 100 mg/kg (MilliporeSigma, catalog no. PHR1084) was freshly prepared by dissolving in H2O and filter-sterilized before injection. Azoramide (Cayman Chemical, catalog no. 18045) was dissolved in dimethyl sulfoxide, filtered-sterilized, and injected at 50 mg/kg.
Human samples
Human heart tissue from patients with LMNA cardiomyopathy was obtained at the time of heart transplantation and myocardium from age- and sex-matched nonfailing hearts was obtained from brain-dead organ donors. Use of human heart tissue for research was approved by the University of Pennsylvania Institutional Review Board (protocol no. 848421), and the use of hearts from brain-dead organ donors for was approved by the Gift-of-Life donor Program in Philadelphia, PA. Written informed consent transplant for research use of heart tissues was obtained prospectively from transplant recipients and from next-of-kin in the case of organ donors. In all cases, hearts were arrested in situ with ice-cold, high-potassium cardioplegia; excised from the body; and transported to the laboratory in ice-cold Krebs-Henseleit buffer. Transmural samples taken from the left ventricular free wall were flash-frozen in liquid nitrogen and stored at −80°C until tissue analyses were performed.
Primary cell isolation and Lmna deletion
Adult murine CM isolation (from both male and female mice) was performed as described (99) with a constant pressure Langendorff perfusion system using Liberase TH (MilliporeSigma, catalog no. LIBTH-RO) at 25 μg/ml. Murine nCMs were isolated from the ventricles of 1- to 2-day-old wild-type C57BL/6 and Lmnaflox/flox mouse pups using magnetic-activated cell sorting neonatal heart dissociation kit according to the manufacturer (Miltenyi Biotec, catalog no. 130-098-373). Following digestion, the single-cell suspension was preplated for 30 min to selectively remove non-myocyte populations. The nonattached phase was recollected and replated in plates coated with laminin (10 μg/ml; Life Technologies, catalog no. 23017015) in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotic/antimycotic solution. To prevent the potential growth of non-myocyte cells, the culture medium was also supplemented with 100 μM 5-bromo-2-deoxyuridine and 10 μM cytosine arabinoside. The cells were maintained at 37°C in 95% humidity with 5% CO2 concentration. MEFs from Lmnaflox/flox mice were isolated using a previously described protocol (100) but with E9 embryos. Initial plating was designated passage 0 (p0) and used only up to p5. To delete Lmna in vitro, adenoviruses carrying mCherry (AdBlank) or mCherry-Cre (AdCre) (Vector Biolabs, catalog nos. 1767 and 1773, respectively) were used at 50 multiplicity of infection. Similar infection efficiencies were confirmed by mCherry.
In vitro culture, shRNA KD, and stress induction
C2C12 cells (American Type Culture Collection) were maintained in DMEM supplemented with 10% FBS (VWR, catalog no. 89510-186) at 37°C with 5% CO2 and subcultured at ~60 to 70% confluency. Viral packaging cell line 293T cells were maintained in the same media. For stable KD of Med25, we used two independent shRNAs in pLKO.1 lentiviral vector identified from murine Med25 shRNA (MilliporeSigma, catalog no. SHCLNG-NM_029365) with the sequences GCAGCTGTTCGATGACTTTAA (shRNA1) and TGCAGCTGTTCGATGACTTTA (shRNA2). For stable KD of Gorasp1 and Gorasp2, we used two independent shRNAs for each in pLKO.1 lentiviral vector with the following sequences: CTCTGAAGCTGATGGTGTATA (shRNA1) and ACCTCACAACTTACTGCCTTT (shRNA2) for Gorasp1 and GCTATGGTTATTTGCACCGAA (shRNA1) and CCCTGTCATGACTACTGCAAA (shRNA2) for Gorasp2. The lentiviral vectors were cotransfected into 293T cells with the packaging vectors pCMV-dR8.2 dvpr and pCMV-VSV-G (Addgene). Blank controls indicate C2C12 cells infected with viruses generated from an empty vector. Virus-infected cells were selected with puromycin (2 μg/ml). To stably overexpress EGFP-LC3B, blank control and Gorasp1/Gorasp2 DKD cells were infected with retroviruses generated from 293T transfected with pBABE-EFGP-LC3B (Addgene, catalog no. 22405) and pCL-Eco (Addgene, catalog no. 12371) plasmids. To induce stress, the cells were treated with indicated concentrations of thapsigargin (Cayman Chemical, catalog no. 10522), BFA (Cayman Chemical, catalog no. 11861), tunicamycin (Cayman Chemical, catalog no. 11445), monensin (Cayman Chemical, catalog no. 16488), golgicide A (Cayman Chemical, catalog no. 18430), nigericin (Cayman Chemical, catalog no. 11437), PF-429242 (Cayman Chemical, catalog no. 15140), and chloroquine (MilliporeSimga, catalog no. C6628). Cell viability was determined by trypan blue exclusion assay.
RNA isolation and RT-qPCR
Total RNA from tissues were isolated using a Direct-zol RNA kit (Zymo Research, catalog no. R2053) with a minor modification. Tissues were homogenized in TRIzol (Zymo Research, catalog no. R2050-1-200), and the aqueous phase containing the RNA fraction was separated by adding chloroform (20% vol of TRIzol). The aqueous fraction (being careful to avoid genomic DNA interface) was mixed with 100% molecular grade ethanol at 1:1 ratio and then further processed using the Direct-zol RNA kit according to the manufacturer’s instructions. cDNA were generated from RNA (100 ng for translating RNA and 1 μg for total RNA) primed with a 1:1 ratio of random hexameric primers and oligo(dT) using RevertAid RT kit (Thermo Fisher Scientific, catalog no. K1691). qPCR was performed on an QuantStudio5 qPCR system (Life Technologies) using PowerUP SYBR Green (Thermo Fisher Scientific, catalog no. A25743). Gapdh was assessed to ensure fidelity of enzymatic reactions and used as internal controls to normalize qPCR results. Fold changes in gene expression were determined by the ΔΔCt method (101) and presented as fold change over negative controls. For Xbp1 splicing, cDNA generated were amplified using standard thermocycler GeneAmp PCR system 9700 (Applied Biosystems), and the amplicons were run on 2% agarose gel. The transgene copy number (fig. S1D) was determined by qPCR analyses using a standard curve generated from known amounts (copy number) of transgenic targeting plasmid as a template. The transgene (CreERT2) copy number values were normalized to Rpl13a, within the respective tissue, by setting the median Rpl13a cycle of threshold (Ct) value as onefold. A complete list of primer sets used in the study is provided in the table S3.
Protein extraction and immunoblot analysis
Heart tissues were homogenized in ice-cold radioimmunoprecipitation assay buffer (MilliporeSigma, catalog no. R0278) with Pierce Protease Inhibitor cocktail (Thermo Fisher Scientific, catalog no. A32963) and 1 mM sodium vanadate (MilliporeSigma, catalog no. S6508). Following brief sonication (Dismembrator Model F60, Thermo Fisher Scientific), the samples were prepped in Laemmli buffer after which 15 to 30 μg of protein extract were loaded for SDS–polyacrylamide gel electrophoresis (SDS-PAGE). For immunoblots assessing CREB3 cleavage, cells were directly lysed in 2x Laemmli buffer and boiled for 5 min before loading in SDS-PAGE. Nuclear fractionation was done using NE-PER (Thermo Fisher Scientific, catalog no. 78833). Antibodies and dilutions used are provided in the table S4. Proper loading was initially confirmed by Ponceau S and then by probing with glyceraldehyde-3-phosphate dehydrogenase antibodies or otherwise indicated. Image capture was performed by Odyssey Fc Imaging System, and blot quantification was performed using Image Studio software (LI-COR Biosciences).
TRAP and RNA-seq
For TRAP, we processed the ventricular tissue exactly as described previously (25). Twelve-week-old mice were treated with either vehicle or Tam as described in Fig. 4A. Two weeks after Tam (or vehicle) dosing, ventricular tissue was harvested and translating mRNAs were purified from n = 3 biologically independent samples. Sequencing libraries were prepared from RNA samples using QuantSeq (Lexogen) and sequenced in a 75–base pair (bp) single end run on NextSeq 500 sequencer. RNA-seq data were aligned using STAR algorithm against mm10 mouse genome version and RSEM v1.2.12 software was used to estimate read counts and FPKM values using gene information from Ensembl transcriptome version GRCm38.89. Raw counts were used to estimate significance of differential expression difference between any two experimental groups using DESeq2 (ref3). Overall gene expression changes were considered significant if passed FDR < 5% threshold. Sequencing and data analysis were performed by Wistar Genomics and Bioinformatics Facilities. The data were submitted to National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database under the accession number GSE185620.
Transmission electron microscopy
Heparinized animals were euthanized, and the excised hearts were cannulated through the aorta on Langendorff apparatus. The hearts were initially perfused with Tyrode solution for 3 min [135 mM NaCl, 5.4 mM KCl, 5 mM MgCl2, 1 mM CaCl2, 0.33 mM NaH2PO4, 10 mM Hepes (pH 7.3)], followed by perfusion with calcium free Tyrode solution (same as above except for 0.02 CaCl2), and lastly by perfusion fixation with 2.5% glutaraldehyde in 0.15 M sodium cacodylate buffer (pH 7.4). Small pieces from left ventricle were cut (~1 to 3 mm3) and postfixed overnight in 4°C in 2% osmium tetroxide partially reduced by 0.8% K4Fe(CN)6 in 0.15 M Na-cacodylate buffer. Samples were contrasted en bloc with 1% uranylacetate in diH2O, dehydrated in graded series of acetone, and embedded in Spurr’s resin. Longitudinal, ultrathin sections (65 to 80 nm) were cut from the resin-embedded blocks with a diamond knife (Diatome-US, USA) using a Leica UCT ultramicrotome and caught on copper grid covered with formvar film. Images of longitudinally oriented CMs were obtained via an FEI Tecnai 12 TEM fitted with an AMT XR-111 10.5 Mpx charge-coupled device camera at ×3200 to ×15,000 magnification (80 kV). For quantitation of damaged nuclei (as determined by ruffled NE or those showing NE breakdown coupled with 100-nm vesicles present in the proximity) and Golgi dilation (determined as doubling/tripling of the Golgi stack width coupled with fragmentation), they were counted by a blinded observer unaware of the treatment conditions. For abnormal nuclei, the data are presented as a % of abnormal/normal nuclei from vehicle-treated (n = 45), 2 weeks after Tam-treat (n = 31), and 4 weeks after Tam-treated mice (n = 27), with n denoting the total number of unique nuclei identified from EM images from two independent hearts per group. For Golgi dilation, the data are presented as a % of unique nuclei with dilated Golgi in the perinuclear space.
Light microscopy and histopathological analysis
H&E, Masson’s Trichrome, and Picro-Sirius Red staining were performed by Translational Research and Pathology Shared Resources (Thomas Jefferson University, PA) using standard methods. For WGA staining, frozen heart sections (8 μm) were fixed in ice-cold 4% paraformaldehyde for 15 min, washed with 0.25% phosphate-buffered saline (PBS)–Tween, permeabilized with 0.2% Triton X-100 for 5 min, and blocked with 1% bovine serum albumin in PBS-Tween for 1 hour. The sections were stained with Alexa Fluor 488–conjugated WGA solution (Invitrogen, catalog no. W11261) at 5 μg/ml in Hanks’ balanced salt solution for 15 min at room temperature. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI), and the sections were mounted on coverslips with Prolong Diamond Antifade Mountant (Invitrogen, catalog no. P36961). For immunofluorescence, cells/tissue sections were fixed in ice-cold methanol:acetone (3:1) and processed using standard methods with antibodies at listed concentrations in table S4. Fluorescence microscopy images in Figs. 4I and 6 (E and F), and figs. S8 (A to C), S12A, and S13 are single plane images captured with the Nikon A1R confocal galvano (nonresonant) scanning microscope using a 40× oil immersion objective (Fluor 40× oil, numerical aperture of 1.30, working distance 0.20 mm) controlled by NIS Elements Imaging Software (version 5.11.02 64bit, Build 1369). All other fluorescence microscopy images were captured using the Life Technologies EVOS M7000 fluorescence and phase contrast microscope with 10× and 20× light-emitting diode Olympus objective controlled with the EVOS M7000 software (version 2.0.2094.0). All image analysis was performed using ImageJ 2.0 software (102). For myocyte cross-sectional area by WGA, the CMs were distinguished from one another by their distinct cell membranes stained with WGA. Partial or incomplete cells were excluded from the analysis. Three images were taken per section, and area of 30 individual CMs was measured in total. Values were expressed as the average of area/myocyte in microns. For quantification of fibrosis by Masson’s Trichrome staining, percent fibrotic area was calculated by dividing blue pixels in each image by total number of pixels taken up by the heart section in indicated number of independent images per condition. For quantifying abnormal nuclei on H&E sections, a blinded observer counted nuclei that were pyknotic, misshapen, elongated, and longitudinally collapsed. Those with multiple herniations were also considered abnormal. Extreme caution was taken to preclude from our analyses ostensibly abnormal nuclei that may result from sectioning artifacts. Furthermore, we only counted nuclei that were within the sarcolemma, indicative of bona fide CM nuclei, and avoid those that appear to be interstitial. Quantification of % abnormal nuclei was calculated by dividing the number abnormal nuclei identified in the CMs by the total number of CM nuclei across 15 fields from three mice per group. Fluorescence intensity profiles of CREB3 traversing the NE were determined using the ImageJ as previously described (103). Linear per-pixel fluorescence intensity of CREB3 was measured from the indicated number of cells from three separate experiments.
Cell-based image analysis
The colocalization analysis was performed blindly on randomly selected cells using ImageJ with JACOP plugin on a per cell basis. Briefly, individual EGFP-LC3B–positive cells were selected and traced to remove the background from analysis in ImageJ. The individual cell images were analyzed by ImageJ plugin JACOP (104). Both red (p62) and green (EGFP-LC3B) channel thresholds were set in and Mander’s coefficients (the proportion of p62 pixels that overlap EGFP-LC3B pixels) calculated under JACOP. For NII analysis, abnormal nuclei were assessed by the ImageJ plugin nuclear morphometric analysis (NMA) (105). Lmnaflox/flox MEFs infected with either AdBlank or AdCre were stained with antibodies against RCAS1 and DAPI. DAPI-stained nucleus images were fed into the NMA plugin with the “minimum nuclei area” set to 1000 pixel area and the “maximum nuclei area” set to maximum. “Nuclei intensity segmentation threshold” was adjusted to match the template nuclear shape, and the NII was calculated using the provided excel template according to the authors’ instructions. RCAS1 signal area was measured and calculated in ImageJ in a blinded fashion.
Cardiac function
Mice were anesthetized with 1 to 2% isoflurane and placed on a stereotactic heated scanning base (37°C) attached to an electrocardiographic monitor. Left ventricular function was determined using a Vevo 2100 imaging system (VisualSonics) equipped with the MS550D transducer (22 to 55 MHz). Parameters were measured for at least three cardiac cycles between the heart rate of 400 to 500 beats/min. A minimum of three separate M-mode images were used to derive the cardiac cycle parameters. An echocardiographer, blind to mouse genotype and/or treatment groups, performed the examinations and interpreted the results using AutoLV Analysis Software (VisualSonics).
Survival analysis
Survival analysis was performed until time of death or signs of distress requiring euthanasia. Mice were assigned to their treatment groups by simple randomization based on sex and littermate info. Signs of significant distress included (i) difficulty with normal ambulatory movement, (ii) failure to eat or drink, (iii) significant weight loss of more than 20%, (iv) depression, (v) unkempt hair coat, and (vi) significant respiratory distress. A staff veterinarian in the Office of Animal Resources at Thomas Jefferson University, blind to mouse genotype, determined if euthanasia was required. Those requiring euthanasia were euthanized according to the Office of Animal Resources consistent with American Veterinary Medical Association guidelines.
Statistical analysis
Graphpad (Prism 9) was used to perform statistical analyses. Statistical significance of binary variables was determined by a two-tailed, unpaired Student’s t test with a value of P < 0.05 considered significant. Statistical significance of three or more comparisons was determined by one-way analysis of variance (ANOVA) with post hoc Tukey’s error correction for multiple comparisons, in which mean values were compared to all other means. For comparisons in which three or more means were compared to a defined control, one-way ANOVA with Dunnett’s multiple comparison test was used. Statistical significance of three or more comparisons with two independent variables was calculated using two-way ANOVA with Dunnett’s correction. Median survival from the Kaplan-Meier plot was analyzed using a log-rank (Mantel-Cox) test. Survival analysis sample sizes are indicated in the figure legends. Values with error bars shown in figures are means ± SEM unless indicated otherwise. P < 0.05 was considered significant.
Acknowledgments
We thank R. Penn, S.-S. Sheu, T. Pera, and H. J. Worman for insightful discussions.
Funding: This work was supported by grants from the NIH/NHLBI R00HL118163 and R01HL150019 to J.C.C. and equipment grant S10 OD010408.
Author contributions: Conceptualization: K.S., E.P., and J.C.C. Methodology: K.S., E.P., D.M., J.S., N.W., Z.Z., Z.N. T.S., A.K., and J.C.C. Investigation: K.S., E.P., D.M., J.S., N.W., Z.Z., Z.N. T.S., A.K., and J.C.C. Supervision: G.C., K.B.M., and J.C.C. Writing—original draft: K.S., E.P., and J.C.C. Writing—review and editing: K.S., E.P., K.B.M., and J.C.C.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Heart tissue from human subjects with LMNA cardiomyopathy as well as the sex- and age-matched control samples were obtained through uniform biological materials transfer agreement (MTAs) with The Trustees of the University of Pennsylvania. The samples were collected de-identified and not specifically for the proposed research by interacting with living individuals. TRAP GFP antibodies were obtained through MTA with Bi-Institutional Antibody and Bioresource Core Facility. TRAP sequencing data are available in NCBI GEO database under accession number GSE185620. Newly generated materials are available upon request through the Thomas Jefferson University MTA office. The reporting of the data in preparation of this manuscript adhered to ICMJE guidelines.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Tables S1 to S4
REFERENCES AND NOTES
- 1.Dauer W. T., Worman H. J., The nuclear envelope as a signaling node in development and disease. Dev. Cell 17, 626–638 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Aebi U., Cohn J., Buhle L., Gerace L., The nuclear lamina is a meshwork of intermediate-type filaments. Nature 323, 560–564 (1986). [DOI] [PubMed] [Google Scholar]
- 3.Turgay Y., Eibauer M., Goldman A. E., Shimi T., Khayat M., Ben-Harush K., Dubrovsky-Gaupp A., Sapra K. T., Goldman R. D., Medalia O., The molecular architecture of lamins in somatic cells. Nature 543, 261–264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broers J. L., Peeters E. A., Kuijpers H. J., Endert J., Bouten C. V., Oomens C. W., Baaijens F. P., Ramaekers F. C., Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: Implications for the development of laminopathies. Hum. Mol. Genet. 13, 2567–2580 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Denais C. M., Gilbert R. M., Isermann P., McGregor A. L., te Lindert M., Weigelin B., Davidson P. M., Friedl P., Wolf K., Lammerding J., Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lammerding J., Schulze P. C., Takahashi T., Kozlov S., Sullivan T., Kamm R. D., Stewart C. L., Lee R. T., Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 113, 370–378 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swift J., Ivanovska I. L., Buxboim A., Harada T., Dingal P. C., Pinter J., Pajerowski J. D., Spinler K. R., Shin J. W., Tewari M., Rehfeldt F., Speicher D. W., Discher D. E., Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341, 1240104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Sandoval A., Towbin B. D., Kalck V., Cabianca D. S., Gaidatzis D., Hauer M. H., Geng L., Wang L., Yang T., Wang X., Zhao K., Gasser S. M., Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell 163, 1333–1347 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Solovei I., Wang A. S., Thanisch K., Schmidt C. S., Krebs S., Zwerger M., Cohen T. V., Devys D., Foisner R., Peichl L., Herrmann H., Blum H., Engelkamp D., Stewart C. L., Leonhardt H., Joffe B., LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Choi J. C., Worman H. J., Nuclear envelope regulation of signaling cascades. Adv. Exp. Med. Biol. 773, 187–206 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez J. M., Navarro-Puche A., Casar B., Crespo P., Andres V., Fast regulation of AP-1 activity through interaction of lamin A/C, ERK1/2, and c-Fos at the nuclear envelope. J. Cell Biol. 183, 653–666 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Berlo J. H., Voncken J. W., Kubben N., Broers J. L., Duisters R., van Leeuwen R. E., Crijns H. J., Ramaekers F. C., Hutchison C. J., Pinto Y. M., A-type lamins are essential for TGF-β1 induced PP2A to dephosphorylate transcription factors. Hum. Mol. Genet. 14, 2839–2849 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Kubben N., Voncken J. W., Konings G., van Weeghel M., van den Hoogenhof M. M., Gijbels M., van Erk A., Schoonderwoerd K., van den Bosch B., Dahlmans V., Calis C., Houten S. M., Misteli T., Pinto Y. M., Post-natal myogenic and adipogenic developmental: Defects and metabolic impairment upon loss of A-type lamins. Nucleus 2, 195–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C. L., Burke B., Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147, 913–920 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arimura T., Helbling-Leclerc A., Massart C., Varnous S., Niel F., Lacene E., Fromes Y., Toussaint M., Mura A. M., Keller D. I., Amthor H., Isnard R., Malissen M., Schwartz K., Bonne G., Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum. Mol. Genet. 14, 155–169 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Mounkes L. C., Kozlov S. V., Rottman J. N., Stewart C. L., Expression of an LMNA-N195K variant of A-type lamins results in cardiac conduction defects and death in mice. Hum. Mol. Genet. 14, 2167–2180 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Choi J. C., Muchir A., Wu W., Iwata S., Homma S., Morrow J. P., Worman H. J., Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci. Transl. Med. 4, 144ra102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos F. J., Chen S. C., Garelick M. G., Dai D. F., Liao C. Y., Schreiber K. H., MacKay V. L., An E. H., Strong R., Ladiges W. C., Rabinovitch P. S., Kaeberlein M., Kennedy B. K., Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci. Transl. Med. 4, 144ra103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen N. Y., Kim P. H., Fong L. G., Young S. G., Nuclear membrane ruptures, cell death, and tissue damage in the setting of nuclear lamin deficiencies. Nucleus 11, 237–249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Vos W. H., Houben F., Kamps M., Malhas A., Verheyen F., Cox J., Manders E. M., Verstraeten V. L., van Steensel M. A., Marcelis C. L., van den Wijngaard A., Vaux D. J., Ramaekers F. C., Broers J. L., Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum. Mol. Genet. 20, 4175–4186 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Hatch E. M., Hetzer M. W., Nuclear envelope rupture is induced by actin-based nucleus confinement. J. Cell Biol. 215, 27–36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu D., Lian H., Zhang X., Shao H., Huang L., Qin C., Zhang L., LMNA E82K mutation activates FAS and mitochondrial pathways of apoptosis in heart tissue specific transgenic mice. PLOS ONE 5, e15167 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolova V., Leimena C., McMahon A. C., Tan J. C., Chandar S., Jogia D., Kesteven S. H., Michalicek J., Otway R., Verheyen F., Rainer S., Stewart C. L., Martin D., Feneley M. P., Fatkin D., Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J. Clin. Invest. 113, 357–369 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park Y. E., Hayashi Y. K., Bonne G., Arimura T., Noguchi S., Nonaka I., Nishino I., Autophagic degradation of nuclear components in mammalian cells. Autophagy 5, 795–804 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Heiman M., Kulicke R., Fenster R. J., Greengard P., Heintz N., Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protoc. 9, 1282–1291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyle J. P., Dougherty J. D., Heiman M., Schmidt E. F., Stevens T. R., Ma G., Bupp S., Shrestha P., Shah R. D., Doughty M. L., Gong S., Greengard P., Heintz N., Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heiman M., Schaefer A., Gong S., Peterson J. D., Day M., Ramsey K. E., Suarez-Farinas M., Schwarz C., Stephan D. A., Surmeier D. J., Greengard P., Heintz N., A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang Y., Gupta V., Karra R., Holdway J. E., Kikuchi K., Poss K. D., Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc. Natl. Acad. Sci. U.S.A. 110, 13416–13421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas A., Lee P. J., Dalton J. E., Nomie K. J., Stoica L., Costa-Mattioli M., Chang P., Nuzhdin S., Arbeitman M. N., Dierick H. A., A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PLOS ONE 7, e40276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tryon R. C., Pisat N., Johnson S. L., Dougherty J. D., Development of translating ribosome affinity purification for zebrafish. Genesis 51, 187–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Jiao Y., Translating ribosome affinity purification (TRAP) for cell-specific translation profiling in developing flowers. Methods Mol. Biol. 1110, 323–328 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Watson F. L., Mills E. A., Wang X., Guo C., Chen D. F., Marsh-Armstrong N., Cell type-specific translational profiling in the Xenopus laevis retina. Dev. Dyn. 241, 1960–1972 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou P., Zhang Y., Ma Q., Gu F., Day D. S., He A., Zhou B., Li J., Stevens S. M., Romo D., Pu W. T., Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proc. Natl. Acad. Sci. U.S.A. 110, 15395–15400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drane L., Ainsley J. A., Mayford M. R., Reijmers L. G., A transgenic mouse line for collecting ribosome-bound mRNA using the tetracycline transactivator system. Front. Mol. Neurosci. 7, 82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramaniam A., Jones W. K., Gulick J., Wert S., Neumann J., Robbins J., Tissue-specific regulation of the alpha-myosin heavy chain gene promoter in transgenic mice. J. Biol. Chem. 266, 24613–24620 (1991). [PubMed] [Google Scholar]
- 36.Kim Y., Zheng Y., Generation and characterization of a conditional deletion allele for Lmna in mice. Biochem. Biophys. Res. Commun. 440, 8–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bersell K., Choudhury S., Mollova M., Polizzotti B. D., Ganapathy B., Walsh S., Wadugu B., Arab S., Kuhn B., Moderate and high amounts of tamoxifen in αMHC-MerCreMer mice induce a DNA damage response, leading to heart failure and death. Dis. Model. Mech. 6, 1459–1469 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lexow J., Poggioli T., Sarathchandra P., Santini M. P., Rosenthal N., Cardiac fibrosis in mice expressing an inducible myocardial-specific Cre driver. Dis. Model. Mech. 6, 1470–1476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuivaniemi H., Tromp G., Type III collagen (COL3A1): Gene and protein structure, tissue distribution, and associated diseases. Gene 707, 151–171 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.W. S. To, Midwood K. S., Plasma and cellular fibronectin: Distinct and independent functions during tissue repair. Fibrogenesis Tissue Repair 4, 21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eppenberger-Eberhardt M., Flamme I., Kurer V., Eppenberger H. M., Reexpression of alpha-smooth muscle actin isoform in cultured adult rat cardiomyocytes. Dev. Biol. 139, 269–278 (1990). [DOI] [PubMed] [Google Scholar]
- 42.Kern S., Feng H. Z., Wei H., Cala S., Jin J. P., Up-regulation of alpha-smooth muscle actin in cardiomyocytes from non-hypertrophic and non-failing transgenic mouse hearts expressing N-terminal truncated cardiac troponin I. FEBS Open Bio. 4, 11–17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alarcon-Martinez L., Yilmaz-Ozcan S., Yemisci M., Schallek J., Kilic K., Can A., Di Polo A., Dalkara T., Capillary pericytes express α-smooth muscle actin, which requires prevention of filamentous-actin depolymerization for detection. eLife 7, e34861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muchir A., Shan J., Bonne G., Lehnart S. E., Worman H. J., Inhibition of extracellular signal-regulated kinase signaling to prevent cardiomyopathy caused by mutation in the gene encoding A-type lamins. Hum. Mol. Genet. 18, 241–247 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart C. L., Roux K. J., Burke B., Blurring the boundary: The nuclear envelope extends its reach. Science 318, 1408–1412 (2007). [DOI] [PubMed] [Google Scholar]
- 46.West G., Gullmets J., Virtanen L., Li S. P., Keinanen A., Shimi T., Mauermann M., Helio T., Kaartinen M., Ollila L., Kuusisto J., Eriksson J. E., Goldman R. D., Herrmann H., Taimen P., Deleterious assembly of the lamin A/C mutant p.S143P causes ER stress in familial dilated cardiomyopathy. J. Cell Sci. 129, 2732–2743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sano R., Reed J. C., ER stress-induced cell death mechanisms. Biochim. Biophys. Acta 1833, 3460–3470 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haze K., Yoshida H., Yanagi H., Yura T., Mori K., Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787–3799 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R. J., IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16, 452–466 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Misra J., Kim D. K., Choi W., Koo S. H., Lee C. H., Back S. H., Kaufman R. J., Choi H. S., Transcriptional cross talk between orphan nuclear receptor ERRγ and transmembrane transcription factor ATF6α coordinates endoplasmic reticulum stress response. Nucleic Acids Res. 41, 6960–6974 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K., XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Samali A., Fitzgerald U., Deegan S., Gupta S., Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int. J. Cell. Biol. 2010, 830307 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu X., Thai P. N., Lu S., Pu J., Bers D. M., Intrafibrillar and perinuclear mitochondrial heterogeneity in adult cardiac myocytes. J. Mol. Cell. Cardiol. 136, 72–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sbodio J. I., Snyder S. H., Paul B. D., Golgi stress response reprograms cysteine metabolism to confer cytoprotection in Huntington's disease. Proc. Natl. Acad. Sci. U.S.A. 115, 780–785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eisenberg-Lerner A., Benyair R., Hizkiahou N., Nudel N., Maor R., Kramer M. P., Shmueli M. D., Zigdon I., Cherniavsky Lev M., Ulman A., Sagiv J. Y., Dayan M., Dassa B., Rosenwald M., Shachar I., Li J., Wang Y., Dezorella N., Khan S., Porat Z., Shimoni E., Avinoam O., Merbl Y., Golgi organization is regulated by proteasomal degradation. Nat. Commun. 11, 409 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dinter A., Berger E. G., Golgi-disturbing agents. Histochem. Cell Biol. 109, 571–590 (1998). [DOI] [PubMed] [Google Scholar]
- 57.Reiling J. H., Olive A. J., Sanyal S., Carette J. E., Brummelkamp T. R., Ploegh H. L., Starnbach M. N., Sabatini D. M., A CREB3-ARF4 signalling pathway mediates the response to Golgi stress and susceptibility to pathogens. Nat. Cell Biol. 15, 1473–1485 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sasaki K., Yoshida H., Golgi stress response and organelle zones. FEBS Lett. 593, 2330–2340 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Farber-Katz S. E., Dippold H. C., Buschman M. D., Peterman M. C., Xing M., Noakes C. J., Tat J., Ng M. M., Rahajeng J., Cowan D. M., Fuchs G. J., Zhou H., Field S. J., DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3. Cell 156, 413–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larrieu D., Britton S., Demir M., Rodriguez R., Jackson S. P., Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science 344, 527–532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sela D., Conkright J. J., Chen L., Gilmore J., Washburn M. P., Florens L., Conaway R. C., Conaway J. W., Role for human mediator subunit MED25 in recruitment of mediator to promoters by endoplasmic reticulum stress-responsive transcription factor ATF6α. J. Biol. Chem. 288, 26179–26187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saunders J., Sikder K., Phillips E., Ishwar A., Mothy D., Margulies K. B., Choi J. C., Med25 limits master regulators that govern adipogenesis. Int. J. Mol. Sci. 24, 6155 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oh-Hashi K., Takahashi K., Hirata Y., Regulation of the ER-bound transcription factor Luman/CREB3 in HEK293 cells. FEBS Lett. 593, 2771–2778 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Lin H. J., Herman P., Lakowicz J. R., Fluorescence lifetime-resolved pH imaging of living cells. Cytometry A. 52, 77–89 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Velho R. V., De Pace R., Klunder S., Di Lorenzo G., Schweizer M., Braulke T., Pohl S., Site-1 protease and lysosomal homeostasis. Biochim. Biophys. Acta Mol. Cell. Res. 1864, 2162–2168 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Marschner K., Kollmann K., Schweizer M., Braulke T., Pohl S., A key enzyme in the biogenesis of lysosomes is a protease that regulates cholesterol metabolism. Science 333, 87–90 (2011). [DOI] [PubMed] [Google Scholar]
- 67.De Tito S., Hervas J. H., van Vliet A. R., Tooze S. A., The Golgi as an Assembly Line to the Autophagosome. Trends Biochem. Sci. 45, 484–496 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Bekier M. E. II, Wang L., Li J., Huang H., Tang D., Zhang X., Wang Y., Knockout of the Golgi stacking proteins GRASP55 and GRASP65 impairs Golgi structure and function. Mol. Biol. Cell 28, 2833–2842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Engelsberg A., Hermosilla R., Karsten U., Schulein R., Dorken B., Rehm A., The Golgi protein RCAS1 controls cell surface expression of tumor-associated O-linked glycan antigens. J. Biol. Chem. 278, 22998–23007 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Drake K. R., Kang M., Kenworthy A. K., Nucleocytoplasmic distribution and dynamics of the autophagosome marker EGFP-LC3. PLOS ONE 5, e9806 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang R., Xu Y., Wan W., Shou X., Qian J., You Z., Liu B., Chang C., Zhou T., Lippincott-Schwartz J., Liu W., Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 57, 456–466 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., Shiosaka S., Hammarback J. A., Urano F., Imaizumi K., Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 26, 9220–9231 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fu S., Yalcin A., Lee G. Y., Li P., Fan J., Arruda A. P., Pers B. M., Yilmaz M., Eguchi K., Hotamisligil G. S., Phenotypic assays identify azoramide as a small-molecule modulator of the unfolded protein response with antidiabetic activity. Sci. Transl. Med. 7, 292ra298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chai R. J., Werner H., Li P. Y., Lee Y. L., Nyein K. T., Solovei I., Luu T. D. A., Sharma B., Navasankari R., Maric M., Sim L. Y. E., Loh Y. J., Aliwarga E., Cheong J. W. L., Chojnowski A., Autio M. I., Haiyang Y., Boon Tan K. K., Keng C. T., Ng S. L., Chew W. L., Ferenczi M., Burke B., Foo R. S. Y., Stewart C. L., Disrupting the LINC complex by AAV mediated gene transduction prevents progression of Lamin induced cardiomyopathy. Nat. Commun. 12, 4722 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leong E. L., Khaing N. T., Cadot B., Hong W. L., Kozlov S., Werner H., Wong E. S. M., Stewart C. L., Burke B., Lee Y. L., Nesprin-1 LINC complexes recruit microtubule cytoskeleton proteins and drive pathology in Lmna-mutant striated muscle. Hum. Mol. Genet. 32, 177–191 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ostlund C., Bonne G., Schwartz K., Worman H. J., Properties of lamin A mutants found in Emery-Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J. Cell Sci. 114, 4435–4445 (2001). [DOI] [PubMed] [Google Scholar]
- 77.Raharjo W. H., Enarson P., Sullivan T., Stewart C. L., Burke B., Nuclear envelope defects associated with LMNA mutations cause dilated cardiomyopathy and Emery-Dreifuss muscular dystrophy. J. Cell Sci. 114, 4447–4457 (2001). [DOI] [PubMed] [Google Scholar]
- 78.Vigouroux C., Auclair M., Dubosclard E., Pouchelet M., Capeau J., Courvalin J. C., Buendia B., Nuclear envelope disorganization in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene. J. Cell Sci. 114, 4459–4468 (2001). [DOI] [PubMed] [Google Scholar]
- 79.Cho S., Vashisth M., Abbas A., Majkut S., Vogel K., Xia Y., Ivanovska I. L., Irianto J., Tewari M., Zhu K., Tichy E. D., Mourkioti F., Tang H. Y., Greenberg R. A., Prosser B. L., Discher D. E., Mechanosensing by the lamina protects against nuclear rupture, DNA damage, and cell-cycle arrest. Dev. Cell 49, 920–935.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Earle A. J., Kirby T. J., Fedorchak G. R., Isermann P., Patel J., Iruvanti S., Moore S. A., Bonne G., Wallrath L. L., Lammerding J., Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. Nat. Mater. 19, 464–473 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murakami T., Saito A., Hino S., Kondo S., Kanemoto S., Chihara K., Sekiya H., Tsumagari K., Ochiai K., Yoshinaga K., Saitoh M., Nishimura R., Yoneda T., Kou I., Furuichi T., Ikegawa S., Ikawa M., Okabe M., Wanaka A., Imaizumi K., Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat. Cell Biol. 11, 1205–1211 (2009). [DOI] [PubMed] [Google Scholar]
- 82.Raggo C., Rapin N., Stirling J., Gobeil P., Smith-Windsor E., O'Hare P., Misra V., Luman, the cellular counterpart of herpes simplex virus VP16, is processed by regulated intramembrane proteolysis. Mol. Cell. Biol. 22, 5639–5649 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stirling J., O'Hare P., CREB4, a transmembrane bZip transcription factor and potential new substrate for regulation and cleavage by S1P. Mol. Biol. Cell 17, 413–426 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brown M. S., Goldstein J. L., A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. U.S.A. 96, 11041–11048 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen C. Y., Chi Y. H., Mutalif R. A., Starost M. F., Myers T. G., Anderson S. A., Stewart C. L., Jeang K. T., Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell 149, 565–577 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Santos J. M., Martinez-Zaguilan R., Facanha A. R., Hussain F., Sennoune S. R., Vacuolar H+-ATPase in the nuclear membranes regulates nucleo-cytosolic proton gradients. Am. J. Physiol. Cell Physiol. 311, C547–C558 (2016). [DOI] [PubMed] [Google Scholar]
- 87.Zhang X., Wang L., Ireland S. C., Ahat E., Li J., Bekier M. E. II, Zhang Z., Wang Y., GORASP2/GRASP55 collaborates with the PtdIns3K UVRAG complex to facilitate autophagosome-lysosome fusion. Autophagy 15, 1787–1800 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maltese W. A., Overmeyer J. H., Methuosis: Nonapoptotic cell death associated with vacuolization of macropinosome and endosome compartments. Am. J. Pathol. 184, 1630–1642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Overmeyer J. H., Kaul A., Johnson E. E., Maltese W. A., Active ras triggers death in glioblastoma cells through hyperstimulation of macropinocytosis. Mol. Cancer Res. 6, 965–977 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu Y., Shoji-Kawata S., Sumpter R. M. Jr., Wei Y., Ginet V., Zhang L., Posner B., Tran K. A., Green D. R., Xavier R. J., Shaw S. Y., Clarke P. G., Puyal J., Levine B., Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc. Natl. Acad. Sci. U.S.A. 110, 20364–20371 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mbah N. E., Overmeyer J. H., Maltese W. A., Disruption of endolysosomal trafficking pathways in glioma cells by methuosis-inducing indole-based chalcones. Cell Biol. Toxicol. 33, 263–282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nah J., Fernandez A. F., Kitsis R. N., Levine B., Sadoshima J., Does autophagy mediate cardiac myocyte death during stress? Circ. Res. 119, 893–895 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guenantin A. C., Jebeniani I., Leschik J., Watrin E., Bonne G., Vignier N., Puceat M., Targeting the histone demethylase LSD1 prevents cardiomyopathy in a mouse model of laminopathy. J. Clin. Invest. 131, e136488 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arimura T., Onoue K., Takahashi-Tanaka Y., Ishikawa T., Kuwahara M., Setou M., Shigenobu S., Yamaguchi K., Bertrand A. T., Machida N., Takayama K., Fukusato M., Tanaka R., Somekawa S., Nakano T., Yamane Y., Kuba K., Imai Y., Saito Y., Bonne G., Kimura A., Nuclear accumulation of androgen receptor in gender difference of dilated cardiomyopathy due to lamin A/C mutations. Cardiovasc. Res. 99, 382–394 (2013). [DOI] [PubMed] [Google Scholar]
- 95.Bharath L. P., Agrawal M., McCambridge G., Nicholas D. A., Hasturk H., Liu J., Jiang K., Liu R., Guo Z., Deeney J., Apovian C. M., Snyder-Cappione J., Hawk G. S., Fleeman R. M., Pihl R. M. F., Thompson K., Belkina A. C., Cui L., Proctor E. A., Kern P. A., Nikolajczyk B. S., Metformin Enhances Autophagy and Normalizes Mitochondrial Function to Alleviate Aging-Associated Inflammation. Cell Metab. 32, 44–55.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.LaMoia T. E., Shulman G. I., Cellular and Molecular Mechanisms of Metformin Action. Endocr. Rev. 42, 77–96 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matsuda T., Cepko C. L., Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. U.S.A. 104, 1027–1032 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanbe A., Gulick J., Hanks M. C., Liang Q., Osinska H., Robbins J., Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ. Res. 92, 609–616 (2003). [DOI] [PubMed] [Google Scholar]
- 99.Louch W. E., Sheehan K. A., Wolska B. M., Methods in cardiomyocyte isolation, culture, and gene transfer. J. Mol. Cell. Cardiol. 51, 288–298 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Durkin M. E., Qian X., Popescu N. C., Lowy D. R., Isolation of Mouse Embryo Fibroblasts. Bio. Protoc. 3, e908 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 102.Abramoff M. D., Magalhaes P. J., Ram S. J., Image Processing with ImageJ. Biophotonics Int. 11, 36–42 (2004). [Google Scholar]
- 103.Choi J. C., Wu W., Phillips E., Plevin R., Sera F., Homma S., Worman H. J., Elevated dual specificity protein phosphatase 4 in cardiomyopathy caused by lamin A/C gene mutation is primarily ERK1/2-dependent and its depletion improves cardiac function and survival. Hum. Mol. Genet. 27, 2290–2305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bolte S., Cordelieres F. P., A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Filippi-Chiela E. C., Oliveira M. M., Jurkovski B., Callegari-Jacques S. M., da Silva V. D., Lenz G., Nuclear morphometric analysis (NMA): Screening of senescence, apoptosis and nuclear irregularities. PLOS ONE 7, e42522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Tables S1 to S4