Abstract
Some regions of nucleic acid targets are not accessible to heteroduplex formation with complementary oligonucleotide probes because they are involved in secondary structure through intramolecular Watson–Crick pairing. The secondary conformation of the target may be destabilised to assist its interaction with oligonucleotide probes. To achieve this, we modified a DNA target, which has self-complementary sequence able to form a hairpin loop, by replacing dC with N4-ethyldeoxycytidine (d4EtC), which hybridises specifically with natural dG to give a G:4EtC base pair with reduced stability compared to the natural G:C base pair. Substitution by d4EtC greatly reduced formation of the target secondary structure. The lower level of secondary structure allowed hybridisation with complementary probes made with natural bases. We confirmed that hybridisation could be further enhanced by modifying the probes with intercalating groups which stabilise the duplex.
INTRODUCTION
Arrays of oligonucleotides are used for the analysis of gene expression (1–3), resequencing genes to find mutations and for polymorphism detection (4). However, a number of problems must be addressed before they can be used optimally. One limitation of the method is that stable secondary structures in the target nucleic acid can make the target sequence inaccessible to intermolecular Watson–Crick base pairing (5–7). Several techniques have been explored to alleviate this problem, but none appears to be totally successful. Fragmenting the nucleic acid sequence, preferably to a size close to that of the oligonucleotides on the array, by heating the DNA or RNA in the presence of Mg2+ (8) or by the use of uracil-N-glycosylase (9) can reduce the secondary structure effect. However, the extent of fragmentation is difficult to control. Oligonucleotide analogues such as peptide nucleic acid (PNA) exhibit PNA–DNA heteroduplexes with a high thermal stability at low salt concentration. The authors suggest that under these conditions the DNA strands are less likely to fold to form stable secondary structure and therefore would be accessible to the oligonucleotide probes (10). However, there is an increasing tendency for non-specific DNA binding. We here describe a new strategy for minimising the secondary structure in the target DNA by introduction of modified nucleosides into the target that form weak base pairs and show that targets modified in this way have a good affinity for oligonucleotide probes.
MATERIALS AND METHODS
Phosphoramidite monomers were from Cruachem. N4-ethyl-2′-deoxycytidine (d4EtC) and 6-chloro-2-methoxyacridine phosphoramidites were from Glen Research. T4 polynucleotide kinase was from BioLabs. [γ-32P]ATP was from Amersham. Acrylamide solution (40%, 19:1 acrylamide:bisacrylamide) was from Anachem (Luton, UK).
Oligonucleotides preparations
Chain assembly was carried out on an Applied Biosystems 394 synthesiser on a CPG (controlled pore glass) solid support functionalised with a nucleoside using phosphoramidite chemistry. Oligonucleotides 1–3 were deprotected by overnight treatment with concentrated ammonia solution at 55°C. Oligonucleotide 4 was deblocked using a deprotection procedure with 0.4 M methanolic sodium hydroxide (4:1 methanol/water) for 16 h at room temperature to avoid cleavage of the bond between the C9 atom of the acridine ring and the N atom of the linker (11). The oligonucleotides were then purified on a 15% denaturing polyacrylamide gel (TBE, 150 V, 5 h). The oligonucleotides (10 pmol) were 5′-terminal-labelled with T4 polynucleotide kinase (5 U) for 1 h at 37°C in the presence of [γ-32P]ATP.
Melting temperature experiment
Changes in absorbance with temperature of 2 µM oligonucleotides in 10–2 M sodium cacodylate buffer, pH 7, containing 1 M NaCl and 2 × 10–4 M EDTA were measured at λ = 260 nm on a Beckman DU 640 cell changer spectrophotometer. The samples and the reference were slowly heated at a rate of 0.5°C/min from 15 to 90°C. Melting temperature (Tm) was taken as the temperature corresponding to half-dissociation of the structure.
Electrophoresis
Aliquots of 0.5 pmol oligonucleotides 1 and 2 were mixed with the appropriate amount of probes 3 and 4 in 10–2 M sodium cacodylate buffer, pH 7, containing 20 mM NaCl, 30% glycerol and bromophenol blue. Samples were heated to 90°C and then cooled slowly before being kept at 4°C for 30 min prior to loading. PAGE experiments were carried out for 3 h on a 15% non-denaturing gel in TBE buffer, pH 7, containing 20 mM NaCl at 4°C. Gels were exposed to a storage phosphor plate and scanned using a phosphorimager (Molecular Dynamics Storm 860).
Fabrication of arrays
The oligonucleotide array was prepared directly on the surface of aminated polypropylene (12) in an adapted Applied Biosystem’s DNA synthesiser (ABI) using a 49.5 mm diamond-shaped mask as described previously (13–14). Standard phosphoramidites were used in synthesis. The offset between each of the 23 coupling steps was 1.98 mm. The arrays were deprotected in 30% ammonia solution at 55°C for 16 h in a closed chamber.
Hybridisation conditions and analysis
Hybridisations were performed in 1 M NaCl in the presence of 10 mM Tris, pH 7, 1 mM EDTA and 0.01% SDS at 4°C for 3 h. Approximately 2 pmol of oligonucleotide was diluted in 4 ml hybridisation solution. The array was then briefly washed in the hybridisation solution and dried with tissue paper. Autoradiography and analysis were carried out as described (15). The array was stripped in 100 mM sodium carbonate/bicarbonate buffer (pH 10) containing 0.01% SDS for 2–3 min at 90–95°C and re-used.
RESULTS
Our strategy involves modification of the target, incorporating nucleic acid analogues to reduce the stability of intramolecular base pairs. For practical applications it will be necessary to incorporate the modified bases in the nucleic acid fragments to be analysed by an enzymatic reaction, constraining the choice of modified nucleosides to those whose 5′-triphosphates can be used as substrates for DNA polymerases. From the large number of modified nucleosides described in the literature we chose d4EtC to replace natural deoxycytidine (dC); d4EtC is known to hybridise specifically with natural deoxyguanosine (dG) to give a G:4EtC base pair having reduced stability as compared to the natural G:C base pair, but very close to that of A:T. Moreover, it has been shown that its triphosphate can be used as a substrate by DNA polymerases (16).
Choice of target sequence
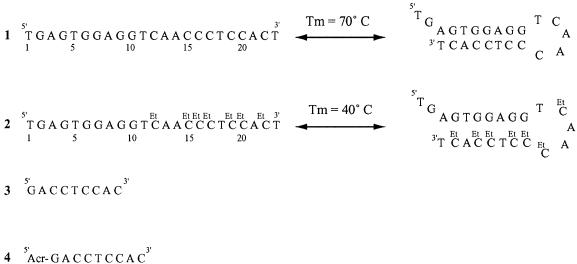
Most secondary structures include hairpin loops composed of duplex stems and a terminal loop of unpaired nucleotides (17–18). Because GC-rich sequences are more likely to form stable intramolecular structures, we chose to decrease the stability of the G:C base pair. The target sequence is 23 nt in length; an unmodified sequence containing only natural nucleosides (sequence 1) and a modified sequence including d4EtC instead of dC (sequence 2). Both sequences can be folded to form a hairpin structure containing a stem of 8 bp (five of which are G:C or G:4EtC base pairs) and a terminal loop of 5 nt (Fig. 1).
Figure 1.
Single-stranded structure in equilibrium with the hairpin structure of unmodified 23mer oligodeoxynucleotide 1 and modified 23mer oligodeoxynucleotide 2 involving d4EtC. Tm was determined at an oligomer strand concentration of 2 µM in 10–2 M sodium cacodylate buffer, pH 7, containing 1 M NaCl and 2 × 10–4 M EDTA. Sequences of the nonamer oligodeoxynucleotide probe 3 and probe 4 bearing at its 5′-end a 6-chloro-2-methoxyacridine molecule (Acr). The acridine molecule is covalently attached to the 5′-terminus of the oligonucleotide via a hexaethylene spacer. All oligodeoxynucleotide sequences are in the ‘deoxy’ series.
Effect of d4EtC on the stability of the DNA target secondary structure
The stability of the hairpin structure was determined by absorption spectroscopy. In 1 M NaCl the native sequence 1 exhibited a half-dissociation temperature (Tm) of 70°C, clearly higher than the Tm of the modified oligonucleotide 2, which was 40°C.
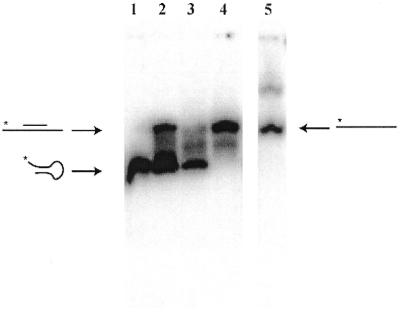
The structures of the native and d4EtC-modified oligonucleotides 1 and 2 were analysed by mobility shift on a non-denaturing polyacrylamide gel (Fig. 2). The unmodified oligonucleotide 1 (lane 1) migrated as a single band faster than the linear oligonucleotide reference 23mer oligodeoxythymidylate (T23) (lane 5), suggesting that it contained a hairpin. The d4EtC-modified oligonucleotide 2 also migrated as a major band at the same rate as unmodified oligonucleotide 1 but also showed a smear between the positions of the hairpin and linear molecule, suggesting an equilibrium between the two forms (lane 3).
Figure 2.
Electrophoretic analysis of 32P-labelled sequences 1 and 2 (0.5 pmol) in the absence and presence of nonadeoxynucleotide probes 3 and 4 complementary to the region G4–C13 (4EtC13). PAGE experiments were carried out for 3 h on a 15% non-denaturing gel in TBE buffer, pH 7, and containing 20 mM NaCl at 4°C. Lane 1, sequence 1; lane 2, sequence 1 + 600 equiv. probe 4; lane 3, sequence 2; lane 4, sequence 2 + 600 equiv. probe 4; lane 5, T23.
To see if a short probe can displace the target secondary structure to form a heteroduplex, mixtures of the targets with a large excess of nonadeoxynucleotide probe bearing a 6-chloro-2-methoxyacridine molecule at its 5′-end (oligonucleotide 4) were analysed by polyacrylamide gel retardation (Fig. 2). We chose to add the acridine moiety because a short oligonucleotide might not have a strong enough affinity for its target sequence and it has been shown that affinity can be greatly increased by covalently linking an intercalating molecule, like acridine, at the end of the oligonucleotide (19–20). In the presence of probe 4 the unmodified sequence 1 presented two well-resolved bands (Fig. 2, lane 2). The fast moving band corresponding to the hairpin structure was much more intense than the slow moving band attributed to the heteroduplex formed with the acridine-modified probe 4. In contrast, the d4EtC-modified target (sequence 2) in the presence of the acridine probe 4 migrated as a unique retarded band corresponding to the heteroduplex (lane 4). Under these experimental conditions the unmodified target 1 exists primarily as a hairpin structure and a large excess of probe 4 (600 equiv.) did not completely displace the intramolecular structure to heteroduplex formation. We estimate that only 30% of unmodified sequence 1 was involved in heteroduplex, in contrast to the d4EtC-modified sequence 2 containing d4EtC; for this target only the linear duplex was formed.
The quantity of probe necessary to induce complete heteroduplex formation was estimated by gel retardation analysis of d4EtC-modified oligonucleotide 2 in the presence of varying concentrations of the acridine probe 4. It was found that 1 equiv. of probe 4 is sufficient to open most of the intramolecular structure, allowing the formation of heteroduplex. The experiment was also carried out with a nonadeoxynucleotide probe lacking the acridine moiety (sequence 3). In this case ∼10 equiv. of probe 3 were necessary to form the linear duplex, confirming the stabilising influence of the acridine. We noted that this mixture produced a smear, suggesting that the heteroduplex might not be very stable under the experimental conditions (data not shown).
Hybridisation of the unmodified and modified targets to a scanning array
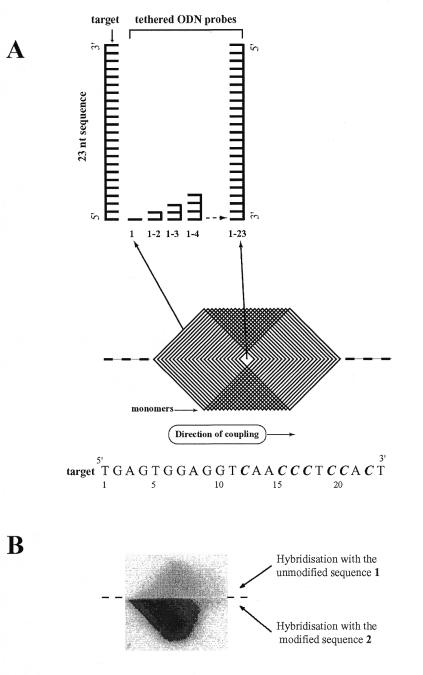
To further characterise the effect of d4EtC on the stability of secondary structure and the effect on its ability to form heteroduplex, unmodified oligonucleotide 1 and d4EtC-modified oligonucleotide 2 were hybridised to a scanning array of complementary oligonucleotides made with natural nucleotides (13–15). The scanning array of oligonucleotides, ranging in size from monomers to 23mers, represented complements of both 23 nt targets (sequences 1 and 2). The organisation and structure of the scanning array are described in Figure 3A. The series of oligonucleotides was made by coupling the bases through a mask to a polypropylene surface in the order in which they occur in the complement of the target sequence, starting from the first base at the 5′-ends. The 3′-ends of the tethered oligonucleotides are attached to the surface. The longest oligonucleotides were in the centre of the array and the tips of the large diamond shapes were occupied by mononucleotides. In between were cells containing all lengths between 1 and 23. The array as synthesised was symmetrical above and below the centre line of the template and each oligonucleotide was represented twice. The diamond-shaped template creates a series of chevron-shaped patches flanking the two ends of the array of diamond-shaped cells. Successive chevrons are occupied by oligonucleotides differing in length by 1 nt. The symmetrical duplication of the array above and below the centre line allowed us to compare and quantify the hybridisation yields of unmodified oligonucleotide 1 and d4EtC-modified oligonucleotide 2 in a single hybridisation experiment. This was achieved by dividing the array along the center line with a thin rubber seal: one half was hybridised with oligonucleotide 1 and the other half with oligonucleotide 2 containing d4EtC (Fig. 3B). The hybridisation patterns for sequences 1 and 2 are similar but differ markedly in intensity. The signal from unmodified sequence 1 was barely detectable. Hybridisation signals of heteroduplexes formed between oligonucleotides 1 and 2 with the set of 23 immobilised probes of varying length were quantified and are shown in Figure 4.
Figure 3.
Organisation of a scanning array. (A) A template of overlapping diamonds representing the footprints of a diamond-shaped mask used to make a scanning array. The tiling path on the top shows the positions of some of the oligonucleotides in the grid. Successive chevrons are occupied by oligonucleotides differing in length by 1 nt. The series of oligonucleotides is made by coupling the bases through a mask to a polypropylene surface in the order in which they occur in the complement of the target sequence, starting from the first base at the 5′-end. Synthesis is carried out on an automated DNA synthesiser, such as an ABI 394. Using standard nucleotide phosphoramidites in the synthesis, the oligonucleotides are attached to the solid support at their 3′-ends. After each coupling step the mask is moved along the substrate by a 1.98 mm size step. The process is continued until an array of 23mer length is obtained. (B) Image obtained after hybridisation of 32P-labelled unmodified sequence 1 (top half) and d4EtC-modified sequence 2 (bottom half) to the complementary oligonucleotide scanning array. Hybridisations were performed in 1 M NaCl in the presence of 10 mM Tris, pH 7, 1 mM EDTA and 0.01% SDS at 4°C for 3 h. Approximately 2 pmol of oligonucleotide was diluted in 4 ml of hybridisation solution. The array was then washed briefly in the hybridisation solution, dried with tissue paper and exposed to a phosphor storage screen and scanned with a phosphorimager. C = C or 4EtC.
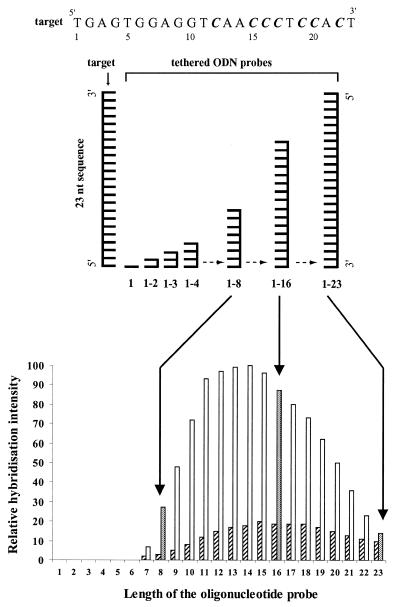
Figure 4.
Relative hybridisation intensity of unmodified target sequence 1 (striped rectangle) and d4EtC-modified target sequence 2 (open rectangle) as a function of the length of the tethered oligonucleotide probes varying in length from 1 to 23 nt. C = C or 4EtC.
The hybridisation results are in agreement with those obtained in solution. There are sharp transitions between regions that give high yield and those that give a yield close to the background. The 8, 9, 21, 22 and 23 nt oligonucleotides showed weaker hybridisation while the 10mers to 19mers produced much stronger hybridisation. The shortest oligonucleotide showing detectable hybridisation was an 8mer. Hybridisation with oligonucleotides shorter than 8mer was not detectable. The oligonucleotides showing maximum hybridisation were the 11mers to 15mers. For oligonucleotides longer than 15 nt the intensity of the hybridisation signals decreased progressively and merged with the background for the 23mer.
DISCUSSION
Hairpin structures are a common feature of single-stranded DNA and RNA molecules. These stable structures probably explain why some regions of targets are not accessible for heteroduplex formation with complementary oligonucleotides. Heteroduplex formation between a hairpin structure and a complementary oligonucleotide probe implies two main steps: opening of the structure to allow formation of one or a few base pairs in a transient nucleation complex followed by further base pairing between the two complementary strands to form a heteroduplex which is more stable than the starting structures. The secondary structure of nucleic acid targets can be minimised by using a hybridisation temperature above the melting temperature of the intramolecular structure or low salt concentration solutions. However, these conditions are not favourable for hybridisation of nucleic acid molecules with short oligodeoxynucleotides. We modified the target sequence to destabilise its secondary conformation to assist interactions with oligonucleotides probes.
We chose in this work to reduce the stability of the G:C base pair, which is the mainstay of hairpin structure. We chose to modify C because the preparation of analogues of C is simpler and easier than those of G. Among the modified nucleosides which fulfilled the criteria mentioned above, we chose d4EtC, rather than N4-methyl-2′-deoxycytidine (d4MeC), which has been used to suppress band compression in sequencing gels (21), because it has been shown previously that the destabilisation imparted to the G:C base pair by d4EtC appears to be much greater than the destabilisation due to d4MeC (22). As expected, replacement of dC with d4EtC in a 23mer oligodeoxynucleotide sequence which can form hairpin structure composed of a double-stranded stem of 8 nt and a 5 nt loop led to a decrease in the hairpin stability. The difference in stability between unmodified sequence 1 (Tm 70°C) and d4EtC-modified sequence 2 made with d4EtC (Tm 40°C) is 30°C. Therefore, replacement of a G:C base pair in the sequence by a G:4EtC base pair led to an average Tm decrease of ∼6°C per substitution. This Tm decrease of 6°C is higher than the value of 4–5°C described in a previous study carried out with a set of nonamer intermolecular duplexes (16). The difference could be due to the particular hairpin structure of the sequence. The Tm decrease of the hairpin structure increased availability of the target for hybridisation with complementary probes. A gel shift experiment carried out with nonadeoxynucleotide probe 3 showed that ∼10 equiv. were able to open most of the hairpin structure of modified sequence 2 involving d4EtC and form heteroduplex. When the affinity of the probe was increased by adding the intercalating molecule 6-chloro-2-methoxyacridine, which improves binding due to the stacking interaction between the intercalator and the base pairs of the probe–target duplex, heteroduplex was formed with only equimolar quantities of probe.
In contrast to this result, only 30% of unmodified sequence 1 were involved in heteroduplex in the presence of 600 equiv. of probe 4.
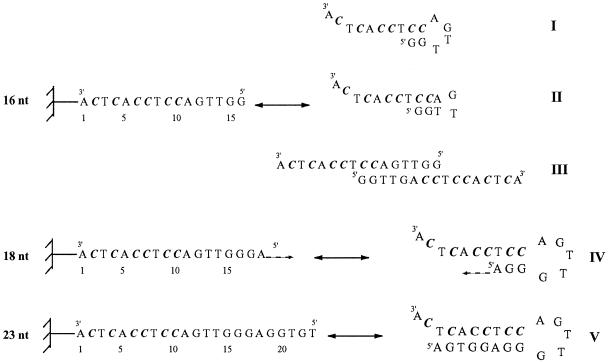
These results were verified by hybridisation to an oligonucleotide array. Under the same experimental conditions only d4EtC-modified oligonucleotide 2 gave heteroduplexes with complementary oligonucleotide probes from 8mers to 23mers. The oligonucleotide probes giving maximum hybridisation are those ranging from 11 to 15 nt. Oligonucleotide probes longer than 16mer have the potential to form self-associated complexes, which may not be available for heteroduplex formation with the target. For example, 16mer oligonucleotide could adopt two hairpin structures: the first with a stem of two G:C base pairs and a loop of 4 nt (Fig. 5, structure I), the second containing a stem of 3 nt and a loop of only 2 nt (Fig. 5, structure II). However, these hairpin structures are not very stable and it seems more likely that the tethered 16 nt oligonucleotides are close enough to form intermolecular structure due to the high surface density. In this way they can form a double-stranded structure involving four G:C base pairs, two A:T pairs and two G:T mismatches (Fig. 5, structure III). The G:T mismatch is well known as among the more stable mismatch base pairs. Similarly, the 17 nt oligonucleotide could adopt an intermolecular structure involving one more G:T mismatch. Oligonucleotide probes from 18 to 23 nt can form hairpin structures involving a loop of 5 nt and a stem with a length of from 3 to 8 nt, respectively (Fig. 5, structures IV and V). This explains the decrease in hybridisation yield when the length of the probe increases.
Figure 5.
Possible structures of the oligonucleotide probes as a function of their length. Hairpin structures (I and II) and double-stranded structure (III) of the 16 nt oligomer probe. Possible hairpin structures of oligomer probes vary in length from 18 to 23 nt (IV and V). C = C or 4EtC. All sequences are in the ‘deoxy’ series.
There are other possibilities for target modification to produce or to augment the effects illustrated above. In the case of the G:C base pair we can increase the destabilisation by using bulkier groups (22). We can also decrease the stability by substitution of deoxyguanosine with 7-deaza-2′-deoxyguanosine (23). The stability of the A:T base pair could be reduced by substituting deoxythymidine with deoxyuridine and deoxyadenosine with 7-deaza-2′-deoxyadenosine (24,25), for example. Modifying the target in this way leads at the same time to a decrease in heteroduplex stability with oligonucleotide probes.
Other chemical modifications can be introduced in oligonucleotide probes to increase their affinity for the target. We have shown that tethering an intercalating agent, such as a 6-chloro-2-methoxyacridine molecule, at the end of the oligonucleotide probe enhanced target sequence hybridisation. Nucleoside analogues, such as C5-methyl- and C5-propynyl-pyrimidine nucleosides (26) or 2,6-diaminopurine 2′-nucleoside (27), known to have stabilising properties, can be also incorporated into oligonucleotide probes. In a preliminary experiment we showed that an oligonucleotide array including d5MeC instead of dC, complementary to the tRNAPhe sequence, substantially enhanced signal intensity with all sequences relative to the unmodified array.
CONCLUSIONS
Secondary structure in target sequences can prevent duplex formation with short oligonucleotides, leading to difficulties in the design and interpretation of experiments using oligonucleotide arrays. For the analysis of gene expression levels by hybridisation to gene-specific oligonucleotides, it is necessary to use a set of up to 20 different oligonucleotides to ensure that one or a few will give strong and specific interaction with each mRNA or cDNA target (2,8). For resequencing or genotyping there is less choice of probe; probes must embrace actual or potential variant sites and if there is no probe that gives good duplex yield because of folding in the target, the method cannot be used without modification. Base substitutions in the target and/or probe would be a simple, potentially general method to overcome the problem. The tests described in this paper show that a target with a very stable stem, which almost completely prevents hybridisation to complementary oligonucleotides, can be induced to hybridise by base substitutions which weaken secondary structure in the target. The modified base used in these studies (4EtC) can be readily incorporated into targets by DNA polymerases, which should allow the method to be applied to all current applications of oligonucleotide arrays.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank T. Fell for construction of the apparatus used in the array synthesis. We are grateful to J.K. Elder for data analysis, software and help with the computer and to F.-X. Barre, K. Mir, V. Regnier and M. Sohail for helpful discussions. H.-K.N. was supported by a TMR Marie Curie research grant.
REFERENCES
- 1.Duggan D.J., Bittner,M., Chen,Y., Meltzer,P. and Trent,J.M. (1999) Nature Genet., 21, 10–14. [DOI] [PubMed] [Google Scholar]
- 2.Lipshutz R.J., Fodor,S.P.A., Gingeras,T.R. and Lockart,D.J. (1999) Nature Genet., 21, 20–44. [DOI] [PubMed] [Google Scholar]
- 3.Bowtell D.D.L. (1999) Nature Genet., 21, 25–32. [DOI] [PubMed] [Google Scholar]
- 4.Hacia J.G. (1999) Nature Genet., 21, 42–47. [DOI] [PubMed] [Google Scholar]
- 5.Williams J.C., Case-Green,S.C., Mir,K.U. and Southern,E.M. (1994) Nucleic Acids Res., 22, 1365–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milner N., Mir,K.U. and Southern,E.M. (1997) Nature Biotechnol., 15, 537–541. [DOI] [PubMed] [Google Scholar]
- 7.Southern E.M., Mir,K. and Shchepinov,M. (1999) Nature Genet., 21, 5–9. [DOI] [PubMed] [Google Scholar]
- 8.Lockhart et al. (1996) Nature Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 9.Cronin M.T., Fucini,V., Kim,S.M., Masino,R.S., Wespi,R.M. and Miyada,C.G. (1996) Hum. Mutat., 7, 244–255. [DOI] [PubMed] [Google Scholar]
- 10.Weiler J., Gausepohl,H., Hauser,N., Jensen,O.N. and Hoheisel,J.D. (1997) Nucleic Acids Res., 25, 2792–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thuong N.T. and Asseline,U. (1991) In Eckstein,F. (ed.), Oligonucleotides and Analogues: A Practical Approach. IRL Press, Oxford, UK, pp. 283–308.
- 12.Matson R.S., Rampal,J.B. and Coassin,P.J. (1994) Anal. Biochem., 217, 306–310. [DOI] [PubMed] [Google Scholar]
- 13.Southern E.M., Case-Green,S.C., Elder,J.K., Johnson,M., Mir,K.U., Wang,L. and Williams,J.C. (1994) Nucleic Acids Res., 22, 1368–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sohail M. and Southern,E.M. (2000) Mol. Cell Biol. Res. Commun., in press. [DOI] [PubMed] [Google Scholar]
- 15.Elder K.J., Johnson,M., Milner,N., Mir,K.U., Sohail,M. and Southern,E.M. (1999) In Schena,M. (ed.), DNA Microarrays: A Practical Approach. IRL Press, Oxford, UK, pp. 61–76.
- 16.Nguyen H.-K., Bonfils,E., Auffray,P., Costaglioli,P., Schmitt,P., Asseline,U., Durand,M., Maurizot,J.-C., Dupret,D. and Thuong,N.T. (1998) Nucleic Acids Res., 26, 4249–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniatis T., Ptashne,M., Beckman,K., Kleid,D., Flashman,S., Jeffrey,A. and Maurer,R. (1975) Cell, 5, 109–113. [DOI] [PubMed] [Google Scholar]
- 18.Rosenberg M. and Court,D. (1979) Annu. Rev. Genet., 13, 319–351. [DOI] [PubMed] [Google Scholar]
- 19.Asseline U., Delarue,M., Lancelot,G., Toulmé,J.-.J., Thuong,N.T., Montenay-Garestier,T. and Hélène,C. (1984) Proc. Natl Acad. Sci. USA, 81, 3297–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toulmé J.-J., Krisch,H.M., Loreau,N., Thuong,N.T. and Hélène,C. (1986) Proc. Natl Acad. Sci. USA, 83, 1227–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S., Haces,A., Stupar,L., Gebeyehu,G. and Pless,R. (1993) Nucleic Acids Res., 21, 2709–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen H.-K., Auffray,P., Asseline,U., Dupret,D. and Thuong,N.T. (1997) Nucleic Acids Res., 25, 3059–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seela F., Tran-Thi,Q.-H. and Franzen,D. (1982) Nucleic Acids Res., 21, 4338–4343. [Google Scholar]
- 24.Ono A., Ohdoi,C., Matsuda,A. and Udea,T. (1992) Nucleosides Nucleotides, 11, 227–235. [Google Scholar]
- 25.Seela F. and Grein,T. (1992) Nucleic Acids Res., 20, 2297–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sagi J., Czuppon,A., Katjar,M., Szabolcs,A., Szembo,A. and Otwos,L. (1982) Nucleic Acids Res., 10, 6051–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chollet A., Chollet-Damerius,A. and Khawashima,E.H. (1986) Chem. Scr., 26, 37–40. [Google Scholar]