Abstract
Poly(ADP-ribose) polymerase 1 (PARP-1) is a nuclear enzyme that is activated by binding to DNA breaks induced by ionizing radiation or through repair of altered bases in DNA by base excision repair. Mice lacking PARP-1 and, in certain cases, the cells derived from these mice exhibit hypersensitivity to ionizing radiation and alkylating agents. In this study we investigated base excision repair in cells lacking PARP-1 in order to elucidate whether their augmented sensitivity to DNA damaging agents is due to an impairment of the base excision repair pathway. Extracts prepared from wild-type cells or cells lacking PARP-1 were similar in their ability to repair plasmid DNA damaged by either X-rays (single-strand DNA breaks) or by N-methyl-N′-nitro-N-nitrosoguanidine (methylated bases). In addition, we demonstrated in vivo that PARP-1-deficient cells treated with N-methyl-N′-nitro-N-nitrosoguanidine repaired their genomic DNA as efficiently as wild-type cells. Therefore, we conclude that cells lacking PARP-1 have a normal capacity to repair single-strand DNA breaks inflicted by X-irradiation or breaks formed during the repair of modified bases. We propose that the hypersensitivity of PARP-1 null mutant cells to γ-irradiation and alkylating agents is not directly due to a defect in DNA repair itself, but rather results from greatly reduced poly(ADP-ribose) formation during base excision repair in these cells.
INTRODUCTION
The genome is constantly exposed to damaging agents of both endogenous and environmental sources that lead to the formation of modified bases, abasic sites or single-strand breaks in DNA. Without an efficient repair mechanism, these damages would occur with a frequency too high to maintain cell viability (1,2). In mammalian cells one of the multi-enzymatic processes that protects the genome from the potentially mutagenic and cytotoxic effects of such DNA lesions is called base excision repair (BER) (2–4). Release of altered bases by BER is initiated by a DNA N-glycosylase that cleaves the base–deoxyribose bond of the modified base, thereby creating an apurinic/apyrimidinic (AP) site. The phosphate group 5′ to the AP site is subsequently incised by an AP endonuclease, followed by excision of deoxyribose phosphate. DNA polymerases fill the resulting gap and, finally, the nick left in the DNA strand is sealed by DNA ligases (2–4). The BER pathway has been previously shown to depend strongly on the presence of nicotinamide adenine dinucleotide (NAD+) and this dependency was proposed to be mediated through the catalytic activation of the nuclear enzyme poly(ADP-ribose) polymerase 1 (EC 2.4.2.30; PARP-1) following its transient binding to BER-induced DNA strand breaks (5,6).
PARP-1 is an abundant nuclear enzyme found in many eukaryotes, with the exception of yeast. This enzyme has a high affinity for single- and double-strand DNA breaks (for a review see 7). Upon binding to DNA strand breaks, PARP-1 catalyzes extensive synthesis of poly(ADP-ribose) from NAD+ and covalently modifies many nuclear proteins, including itself. The massive automodification of PARP-1 effects its dissociation from DNA strand breaks and inhibition of its catalytic activity. A modulatory role for poly(ADP-ribose) formation and PARP-1 automodification in BER has been proposed by Satoh and Lindahl (5). According to this model, the unmodified enzyme binds tightly to DNA strand interruptions formed either by ionizing radiation or through incision of AP sites during BER and interferes with the BER process since the bound PARP-1 molecule hinders access of the repair machinery to the lesion. Automodification and release of PARP-1 from the breaks allow the repair process to proceed. In the absence of NAD+ or upon inhibition of poly(ADP-ribose) synthesis, PARP-1 persists on DNA breaks and DNA repair is abrogated.
PARP-1 null mice have recently been established by three independent laboratories (8–10). PARP-1-deficient animals are highly susceptible to whole body irradiation (9,11) and, in some cases, immortalized cells derived from these mice exhibit extreme sensitivity to γ-irradiation and alkylating agents and arrest in G2/M (9,12).
In the present study we examined the possible involvement of PARP-1 in BER and whether the lack of PARP-1 may contribute to the increased sensitivity of PARP-1(–/–) cells to genotoxic agents via an impairment of this repair pathway. We compared DNA repair efficiencies of wild-type and PARP-1 null mouse embryonic fibroblasts (i) by an in vitro repair assay using whole cell extracts and damaged plasmid DNAs and (ii) in living cells by measuring the rate of strand-break rejoining in the genomic DNA by post-labeling. Our results clearly demonstrate that PARP-1(–/–) and (+/+) cells have a similar capacity to repair DNA damage inflicted by X-irradiation or the alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), indicating that BER is not affected in the absence of PARP-1. These findings imply that the hypersensitivity of PARP-1(–/–) cells to DNA damage may be related to the dramatically reduced repair-associated poly(ADP-ribose) synthesis in these cells.
MATERIALS AND METHODS
Cell culture and reagents
The mouse embryonic fibroblast cell line A1 [PARP-1(–/–)] and the parent cell line F20 [PARP-1(+/+)] were kindly provided by Dr Z.-Q. Wang (IARC, Lyon, France) (8,11) and were grown at 37°C in a humidified 5% CO2 atmosphere in DMEM low glucose medium (Gibco BRL) supplemented with 1% l-glutamine, 0.2% bicarbonate, antibiotics and 10% fetal bovine serum. For PARP-1(–/–) cells 600 µg/ml neomycin (Gibco BRL) was also added to the medium (8). PARP-1 was purified from calf thymus as described by Zahradka and Ebisuzaki (13). MNNG was purchased from Sigma-Aldrich. All reagents were of analytical grade.
Preparation of plasmid DNA substrates
The 3 kb Bluescript II KS+ plasmid (pBS) (Stratagene) was propagated in Escherichia coli strain DH5α and purified by a sodium dodecyl sulfate (SDS)/alkali lysis procedure followed by two ethidium bromide/cesium chloride gradient ultra-centrifugations (14). For X-irradiation, pBS at 3.4 mg/ml was resuspended in 10 mM Tris–HCl, pH 8.0, and 1 mM EDTA (TE buffer) and exposed to 50 Gy X-rays (dose rate 16 Gy/min) from a linear accelerator at 0°C. An open circular plasmid containing an average of one single-stranded DNA break per plasmid molecule (5) was purified by two successive ethidium bromide/cesium chloride gradient ultracentrifugations. For MNNG treatment, pBS at 0.2 mg/ml was incubated with 0.4 mM MNNG at 37°C for 30 min in TE buffer. The MNNG-treated pBS was precipitated with ethanol and dissolved in TE buffer. The X-irradiated pBS was stored at –30°C whereas the methylated pBS was freshly prepared before use.
Preparation of cell extracts
Cell-free extracts were prepared using a slightly modified version of the procedure originally described by Tanaka et al. (15) and recently adapted for in vitro DNA repair studies by Biade et al. (16). Exponentially growing A1 PARP-1(–/–) and F20 PARP-1(+/+) mouse fibroblasts were washed three times with ice-cold phosphate-buffered saline (PBS) and then resuspended at 106 cells/10 µl in buffer I (10 mM Tris–HCl, pH 7.8, 200 mM KCl). An equal volume of buffer II [10 mM Tris–HCl, pH 7.8, 600 mM KCl, 2 mM EDTA, 40% (v/v) glycerol, 0.2% (v/v) Nonidet P-40, 2 mM dithiotreitol (DTT), 0.5 mM phenylmethylsulphonyl fluoride (PMSF) and 2× antiprotease cocktail (Boehringer)] was added to the cell suspension and the mixture was shaken at 4°C for 1.5 h to allow cell lysis. The lysed cells were spun at 16 000 g for 10 min to remove cellular debris and DNA (P1). The supernatant (S1) was recovered and dialyzed overnight at 4°C against buffer III (25 mM HEPES–KOH, pH 8.0, 100 mM KCl, 12 mM MgCl2, 1 mM EDTA, 17% (v/v) glycerol, 1 mM DTT). After centrifugation at 16 000 g for 10 min to remove insoluble material (P2), the supernatant (S2) was aliquoted and stored at –80°C.
Cell-free DNA repair assay
Repair of single-strand DNA breaks induced by X-rays and of methylated DNA bases induced by MNNG was analyzed as previously described (5). Briefly, the damaged pBS (150 ng) was incubated in 50 µl of reaction mixture containing 45 mM HEPES–KOH, pH 7.8, 70 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.4 mM EDTA, 2 mM ATP, 20 µM each dCTP, dTTP and dGTP, 8 µM dATP, 40 mM phosphocreatine, 2.5 µg creatine phosphokinase, 3% glycerol, 20 µg/ml bovine serum albumin (BSA) and 30 µg cell extract in the presence or absence of 2 mM NAD+ at 30°C for various times. Plasmid DNA was then purified with phenol/chloroform and ethanol precipitated. Open circular and closed circular forms of the plasmid were resolved by electrophoresis through a 1% agarose gel containing ethidium bromide to analyze DNA repair activity. DNA was visualized with UV light using an AlphaImager system and AlphaEase software (Alpha Innotech Inc.) was used to quantify the DNA. Signals for covalently closed circular DNA were corrected by an experimentally determined factor of 1.6 to compensate for the reduced binding of ethidium bromide.
In vitro DNA repair synthesis
To monitor repair synthesis, 2 µCi [α-32P]dATP (3000 Ci/mmol; DuPont NEN) was added to the reaction mixture in the presence of 0.25 mM NAD+. The repaired pBS DNA was purified as described above, then linearized with EcoRI and applied to a 1% agarose gel containing ethidium bromide. The linearized plasmid DNA was visualized and quantified as described above. After gel drying, the amount of [32P]dAMP incorporated into the pBS plasmid was visualized by autoradiography and quantified with an Instant Imager (Canberra-Packard).
Poly(ADP-ribose) formation
Synthesis of poly(ADP-ribose) was carried out in the same reaction mixture as described above, supplemented with 2 µCi [32P]NAD+. After various incubation times the reactions were terminated by addition of 500 µl of ice-cold 20% trichloroacetic acid and the precipitated material was collected by filtration on glass filters. After washing and drying the filters, the radioactivity was quantified by liquid scintillation counting (LKB-Wallac RackBeta liquid scintillation counter).
Immunoblotting of PARP-1
Aliquots of the pellet and supernatant fractions following the first (P1 and S1) and second (P2 and S2) centrifugation steps of the cell extract preparation procedure were taken, the protein concentration in each sample was determined and the proteins were dissolved by adding an equal volume of PAGE loading buffer containing 62.5 mM Tris–HCl, pH 6.8, 6 M urea, 10% (v/v) glycerol, 2% (v/v) SDS, 5% (v/v) β-mercaptoethanol, 1 mM DTT and 0.00125% bromophenol blue. After sonication for 30 s and incubation at 65°C for 10 min the proteins were resolved through an 8% polyacrylamide gel and electrotransferred onto Hybond C nitrocellulose membranes (Amersham). The membranes were stained with Ponceau S (0.5%) to confirm equal loading and transfer of proteins. The filters were blocked in PBS-TM (140 mM NaCl, 3.7 mM KCl, 2.9 mM KH2PO4, 7.7 mM Na2HPO4, 5% skimmed milk, 0.1% Tween 20, pH 7.4) for 1 h at room temperature and then incubated overnight in PBS-TM containing CII-10 monoclonal antibody diluted 1:15 000 and 1 mM sodium azide. After three washes of 15 min in PBS-TM the membranes were incubated for 35 min with a secondary antibody coupled to peroxidase diluted 1/3000 in PBS-TM. Finally, the filters were washed with PBS for 1 h and the immune complexes were detected by enhanced chemilumeniscence (Renaissance ECL-Plus kit; DuPont).
For quantification of PARP-1 in cell extracts, 30 µg protein extracted from PARP-1(+/+) cells and 10, 20, 40, 60 and 80 ng purified PARP-1, loaded as standards, were separated by SDS–PAGE, transferred to nitrocellulose membrane and blotted as described above. An AlphaImager system controlled by AlphaEase software (Alpha Innotech Inc.) was used to determine the amount of PARP-1 present in the cell extract.
Rate of DNA break rejoining in PARP-1(–/–) and PARP-1(+/+) cells
Cells grown to 80% confluence in 30 mm dishes were treated with 100 µM MNNG in serum-free DMEM at 37°C for 20 min. After treatment, the medium containing MNNG was replaced with a serum-completed medium and cells incubated at 37°C for different time intervals as indicated. The genomic DNA was extracted at each time point essentially as described by Legault et al. (17). Briefly, cells were lyzed by the addition of 400 µl of 50 mM Tris–HCl, pH 7.4, 1% SDS, 20 mM EDTA, 10 µM deferoxamine mesylate (Sigma) and 0.5 mg/ml proteinase K (Gibco BRL) and incubated overnight at 37°C. Proteins were removed by the protein salting out method of Miller et al. (18) and the genomic DNA was ethanol precipitated and dissolved in TE buffer. After complete resuspension, the DNA was quantified and stored at 4°C up to a maximum of 2 weeks. The combined activities of E.coli endonuclease IV and T4 DNA polymerase were employed to cleave the abasic sites in genomic DNA and to label 3′-ends of DNA strand breaks in a nucleotide exchange reaction as described previously (17). Briefly, 200 ng genomic DNA was incubated at 37°C for 60 min in a reaction mixture containing 33 mM Tris–acetate, 66 mM potassium acetate, 10 mM magnesium acetate, pH 8.0, 0.5 mM DTT, 0.1 mg/ml BSA, 100 µM each dATP, dTTP and dGTP, 0.165 µM [α-32P]dCTP (300 Ci/mmol; DuPont NEN), 0.1 U E.coli endonuclease IV and 0.5 U T4 DNA polymerase (Pharmacia). The reaction was stopped by heating at 65°C for 5 min. DNA was then applied to a Hybond N+ membrane (Amersham) under vacuum by using a dot-blot manifold and the amount of [32P]dCMP incorporated was quantified by electronic autoradiography on an Instant Imager (Canberra-Packard).
RESULTS
Repair of single-strand DNA breaks induced by X-rays
In a preliminary experiment we compared the repair activity of cell extracts prepared by two different methods. The procedure described by Manley et al. (19) is most widely used for in vitro DNA repair studies but is time consuming and requires large amounts of cells, which renders it incovenient when adherent cells are to be used for cell extract preparation. Recently, Biade et al. (16) adapted a technique, originally reported by Tanaka et al. (15), for cell extract preparation from detergent lyzed cells to be utilized in an in vitro repair assay. The method of Tanaka et al. is much more rapid and can be accomplished starting with as few as several million cells. Since PARP-1 is tightly associated with chromatin (20), we first tested different salt conditions for cell extraction by the modified method of Tanaka et al. (Fig. 1A). An optimal concentration of 400 mM KCl in the cell lysis step was found to allow complete extraction of PARP-1. During the final dialysis step (see Materials and Methods) insoluble aggregates usually formed and we observed 15–20% loss of PARP-1 at this step (Fig. 1B). A similar dialysis step is present in Manley’s method as well but, in our experience, the total loss of PARP-1 in Manley cell extract preparations is 2-fold higher, presumably because of the much longer, multistep procedure (data not shown). Proteolysis or PARP-1 degradation products, which are known to strongly inhibit DNA repair (21,22), were not detected in our cell extract preparations (Fig. 1).
Figure 1.
Optimization of the procedure for cell extract preparation for studies implicating PARP-1. (A) Western blot analysis of the pellet (P1) and supernatant (S1) fractions after the first centrifugation step of the modified method of Tanaka et al. (15) (see Materials and Methods). The detergent-lyzed PARP-1(+/+) cells were extracted with different salt concentrations and the samples were analyzed for the presence of PARP-1 by CII-10 monoclonal anti-PARP-1 antibody to determine the optimal salt conditions for complete extraction of PARP-1. Purified PARP-1 was loaded in the first lane as a control. (B) Immunoblotting with CII-10 antibody of pellet (P2) and supernatant (S2) fractions following the second centrifugation step of the modified Tanaka procedure. In PARP-1(+/+) cell extracts ∼15% loss of PARP-1 was noted at this step; no immunodetectable PARP-1 was observed by CII-10 antibody in the P1, P2 and S2 fractions from PARP(–/–) cells. Purified PARP-1 was run in the first lane as a control.
To compare the cell extracts prepared by these two alternative methods, we conducted an in vitro repair assay with 150 ng X-irradiated plasmid DNA and equivalent amounts of the two cell extracts. As shown in Table 1, both Manley’s cell extracts and the extracts prepared according to the modified procedure of Tanaka et al. (15) provided similar repair activity. We therefore found the Tanaka procedure much more advantageous for work with fibroblasts and we carried out subsequent experiments in our study using cell extracts prepared by this procedure.
Table 1. Comparison of two alternative methods for cell extract preparation.
| Method | PARP-1 status in cell extract | Repair of X-irradiated DNA(%) |
|---|---|---|
| I | (+/+) | 38 |
| (–/–) | 37 | |
| II | (+/+) | 36 |
| (–/–) | 39 |
Repair activity of cell extracts prepared from PARP-1(+/+) and PARP-1(–/–) mouse fibroblasts by the modified method of Tanaka et al. (15) (I) or by the classical method of Manley et al. (19) (II) was examined in a cell free repair assay. Repair reactions were carried out with X-irradiated plasmid DNA for 60 min at 30°C in the presence of 2 mM NAD+ as described in Materials and Methods.
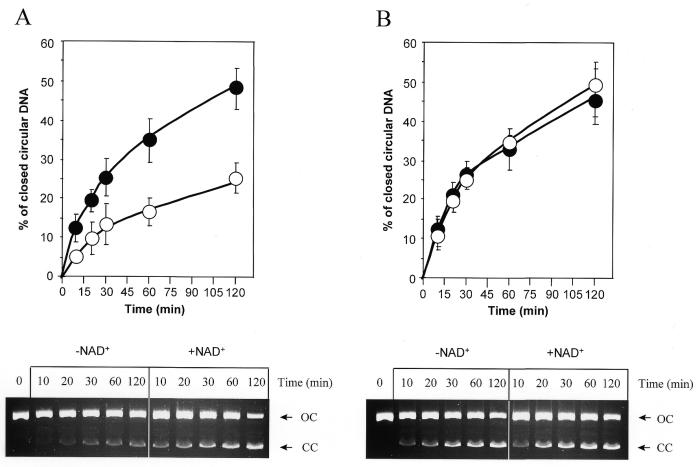
We next investigated the efficiency of PARP-1(–/–) cells to repair radiation-induced DNA strand breaks in a cell-free assay using X-irradiated plasmid DNA bearing an average of one single-strand break per molecule. The rejoining of DNA strand breaks by cell extracts results in conversion of open circular (damaged pBS) to covalently closed circular DNA (repaired pBS) and can be quantified by agarose gel electrophoresis in the presence of ethidium bromide (5). Reaction mixtures containing 150 ng X-irradiated pBS were incubated with 30 µg PARP-1(+/+) (Fig. 2A) or PARP-1(–/–) (Fig. 2B) cell extract for various times at 30°C, in the presence or absence of 2 mM NAD+. In the presence of NAD+ ∼50% of the open circular pBS molecules were converted to a closed circular form by both PARP-1(–/–) and PARP-1(+/+) cell extracts within 120 min of incubation (Fig. 2). About 40–50% rejoining of single-strand DNA breaks by 50 µg of extracts from HeLa and lymphoblastoid cells has been reported previously (5,6), which is consistent with our findings. These results thus indicate that extracts from PARP-1(–/–) cells have a normal capacity to repair single-strand DNA breaks induced by X-rays.
Figure 2.
Kinetics of rejoining of X-ray-induced single-strand breaks in plasmid DNA by PARP-1(+/+) and PARP-1(–/–) cell extracts. The cell free assay was performed in the presence (filled symbols) or absence (empty symbols) of 2 mM NAD+ with 30 µg protein extract from (A) PARP-1(+/+) or (B) PARP-1(–/–) cells. The primary data, obtained after electrophoretic separation of nicked plasmid DNA (OC) to closed circular repaired plasmid DNA (CC) on a 1% agarose gel in the presence of ethidium bromide, are shown below the corresponding graphics.
NAD+-independent repair in PARP-1(–/–) cells
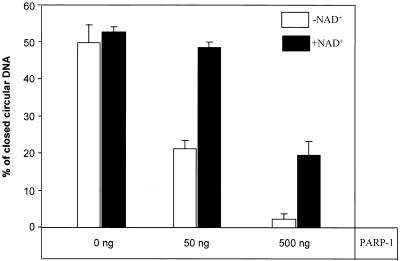
In our experiments with X-irradiated plasmids the major difference between the DNA repair kinetics in PARP-1(+/+) and PARP-1(–/–) cell extracts was the dependence of the repair process on NAD+. As shown in Figure 2, rejoining of DNA strand breaks in PARP-1(–/–) cell extracts was independent of NAD+ added to the reaction, whereas in PARP-1(+/+) cell extracts DNA repair was suppressed in the absence of NAD+ and the addition of NAD+ strongly promoted DNA strand break rejoining up to a level comparable to that in PARP-1(–/–) extracts. These observations suggest that PARP-1 is responsible for the NAD+ dependence of BER and are in agreement with the previously proposed model where, in the absence of NAD+, PARP-1 persistently binds to DNA breaks and interferes with BER (5,6). To test this model in a PARP-1 null mutant background, we studied the effect of purified PARP-1 addition to extracts from PARP-1(–/–) cells. We estimated, by western blotting, that 30 µg protein from PARP-1(+/+) cell extract contains roughly 50 ng PARP-1. Therefore reactions carried out with 30 µg protein extract from PARP-1(–/–) cells were supplemented with 50 and 500 ng purified PARP-1. The reactions were performed for 120 min at 30°C in the presence or absence of 2 mM NAD+. As shown in Figure 3, the addition of 50 ng purified PARP-1 to the reaction mixture results in restoration of NAD+-dependent DNA repair in PARP-1(–/–) cell extracts and ∼50% inhibition of DNA break rejoining in the absence of NAD+, which is similar to the suppression observed in PARP-1(+/+) cell extracts (Fig. 2). The addition of a 10-fold excess of purified PARP-1 exacerbated DNA repair inhibition in reactions both with and without NAD+ (Fig. 3). These results indicate that PARP-1 is responsible for the NAD+ dependence of BER and that this enzyme may compete with the BER process under conditions where its substrate is limited or not available.
Figure 3.
Restoration of the NAD+ dependence of DNA break rejoining in PARP-1(–/–) cell extract after the addition of purified PARP-1. Reaction mixtures contained 150 ng plasmid DNA with a single-strand break generated by X-irradiation, 30 µg protein extract from PARP-1(–/–) cells, either 2 mM or no NAD+ and different amounts of purified PARP-1 as indicated.
Repair of alkylated DNA bases in the absence of PARP-1
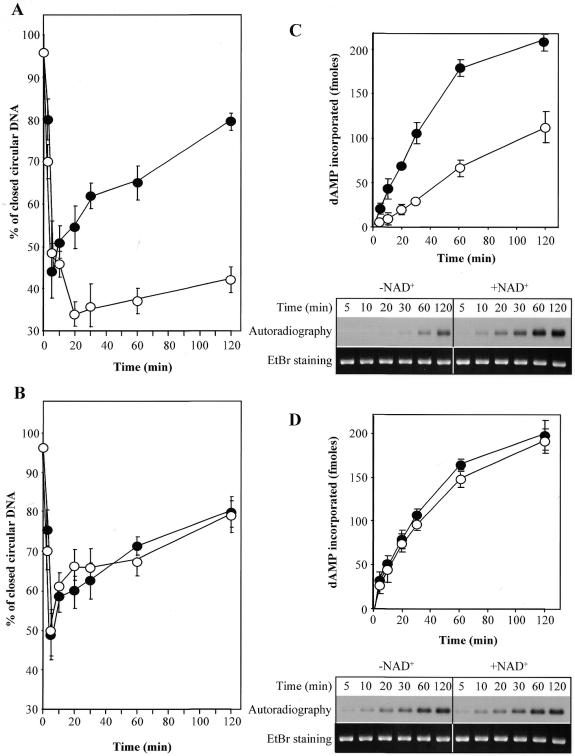
The monofunctional alkylating agent MNNG generates a variety of methylated bases in DNA. We next analyzed the capacity of PARP-1(–/–) cell extracts to repair these types of damage, which are normally corrected through the BER pathway. MNNG-treated plasmid, which is introduced in our DNA repair assay in the form of covalently closed circular DNA, is first converted to an open circular DNA molecule by removal of modified bases and incision of the AP sites, mediated by DNA N-glycosylases and AP endonuclease, respectively. Following DNA repair synthesis by DNA polymerases, the breaks are rejoined by a DNA ligase, resulting in conversion of open circular to repaired closed circular plasmid. The incision of AP sites and subsequent DNA break rejoining can be monitored by measurement of the relative proportions of covalently closed to open circular plasmid molecules at different time points (6). Figure 4A and B shows the repair kinetics of MNNG-treated plasmid in PARP-1(+/+) and PARP-1(–/–) cell extracts, as determined by agarose gel electrophoresis in the presence of ethidium bromide. Similar rates of conversion of alkylated DNA to the open circular form were observed for PARP-1(+/+) and PARP-1(–/–) cell extracts, indicating that the extracts from PARP-1(–/–) cells contain normal levels of the activities required to remove damaged bases and to incise AP sites. Subsequently, the generated DNA breaks were rejoined at similar rates in both cell extracts (Fig. 4A and B) when the reaction mixture was supplemented with NAD+. Consistent with our observations on the repair of X-ray induced single-strand DNA breaks, these results demonstrate that the rejoining step for DNA breaks induced by BER is normal in the absence of PARP-1.
Figure 4.
Repair of alkylated plasmid DNA by cell extracts from PARP-1(+/+) and PARP-1(–/–) cells. The cell free assay was performed under standard conditions with MNNG-treated plasmid DNA in the presence (filled symbols) or absence (empty symbols) of 2 mM NAD+. (A and B) The relative proportions of closed versus open circular plasmid DNA determined after agarose gel electrophoresis in the presence of ethidium bromide at each time point of a time course with PARP-1(+/+) and PARP-1(–/–) cell extracts, respectively. (C and D) DNA repair replication in PARP-1(+/+) and PARP-1(–/–) extracts was monitored at various times by incorporation of [α-32P]dAMP. The plasmid DNA was linearized at the end of the repair reaction, run on a 1% agarose gel and the incorporated radioactivity quantified. The primary data shown below the graphs are representative autoradiographs from three independent experiments.
We further investigated DNA repair synthesis in PARP-1(–/–) cell extracts by the addition of an [α-32P]-labeled deoxynucleoside triphosphate to the repair reaction mixtures, which allowed us to monitor this process by incorporation of radioactive material. As shown in Figure 4C and D, DNA repair synthesis during BER in extracts prepared from PARP-1(–/–) cells was as efficient as in PARP-1(+/+) cell extracts. In agreement with the data shown in Figure 3, proficient DNA repair synthesis in PARP-1(+/+) extracts required NAD+, whereas DNA repair replication was independent of NAD+ in PARP-1(–/–) cell extracts. Taken together, our results demonstrate that the PARP-1(–/–) cell extracts are fully competent to repair methylated bases in DNA.
Repair of DNA breaks in cultured PARP-1(–/–) cells
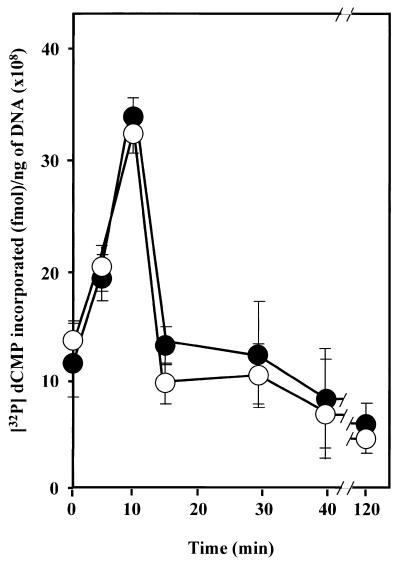
We next asked whether the BER pathway in living PARP-1 null cells functions as well as was found for cell extracts prepared from these cells. We studied the repair of MNNG-induced methylation damage in cultured PARP-1(–/–) and PARP-1(+/+) cells. The exponentially growing cells were treated for 20 min with 100 µM MNNG and the relative number of DNA strand breaks and abasic sites in their genomic DNA was estimated at different time intervals following the damage. The 3′-end of transient DNA strand breaks, formed either by the combined action of cellular DNA N-glycosylase and AP endonuclease at the site of methylated bases or in vitro by the activity of E.coli endonuclease IV on the abasic sites in the genomic DNA, was labeled by an [α-32P]dCTP exchange reaction catalyzed by T4 DNA polymerase (17; Fig. 5). The rate of [32P]dCMP incorporation, which mainly occured within the first 10 min of post-treatment culture, was similar for the PARP-1(–/–) and PARP-1(+/+) cell lines, confirming that PARP-1(–/–) cells contain a normal activity to remove damaged DNA bases and incise the resulting AP sites. The amount of incorporated [32P]dCMP per ng genomic DNA decreased dramatically for both cell lines after 15 min of cell recovery and reached the basal level of DNA strand breaks within 120 min. The kinetics suggest that the majority of the genomic DNA is repaired in both PARP-1(–/–) and PARP-1(+/+) cell lines as early as 15 min after MNNG removal. Thus, consistent with our results from in vitro assays, no apparent abnormality in BER was found in living PARP-1(–/–) cells.
Figure 5.
DNA repair kinetics in living PARP-1(–/–) and PARP-1(+/+) cells following MNNG treatment. PARP-1(–/–) (filled symbols) and PARP-1(+/+) (empty symbols) cells were treated for 20 min with 100 µM MNNG in DMEM. Genomic DNA was extracted at various times following this treatment and incubated with E.coli endonuclease IV and T4 DNA polymerase in the presence of [32P]dCTP (see Materials and Methods). Results are expressed as the amount of [32P]dCMP incorporated per ng genomic DNA as a function of time following removal of MNNG. The basal level of incorporation due to genomic DNA strand breaks was similar for both cell lines and was subtracted.
DNA repair-associated poly(ADP-ribose) synthesis in PARP-1(–/–) cells
Since we found that BER is not impaired in the absence of PARP-1, we concluded that the increased sensitivity of PARP-1(–/–) cells to DNA damaging agents is not related to a BER defect in these cells. Previous studies have shown that extensive poly(ADP-ribose) synthesis accompanying DNA repair is triggered in cells following genotoxic treatments (7,23). A putative role for poly(ADP-ribose) has been proposed as a stress-induced signal and/or an anti-recombinogenic factor (24,25). PARP-1 null cells were initially reported to be devoid of poly(ADP)ribosylation activity (8) but, more recently, residual poly(ADP-ribose) synthesis and poly(ADP)ribosylating enzymes were found in these cells (26–30). We therefore sought to compare BER-associated poly(ADP-ribose) formation in PARP-1(–/–) and PARP-1(+/+) cells as a potential factor which may contribute to the augmented sensitivity of PARP-1(–/–) cells to DNA damage. The analysis of poly(ADP-ribose) synthesis using our cell-free DNA repair assay with X-irradiated plasmid DNA revealed 10–15 times less poly(ADP-ribose) formed during BER in PARP-1(–/–) cell extracts in comparison to PARP-1(+/+) extracts (Fig. 6). Even the undamaged plasmid introduced in PARP-1(+/+) extracts stimulated 3–4 times more poly(ADP-ribose) synthesis than the damaged plasmid in PARP-1(–/–) extracts, reflecting the presence of intrinsic nicking activities in the cell extracts that cause immediate PARP-1 activation. Similar kinetics of poly(ADP-ribose) synthesis stimulation were also observed with MNNG-treated plasmid (data not shown). The dramatic reduction in poly(ADP-ribose) in PARP-1(–/–) cells after DNA damage may abolish important functions proposed for poly(ADP-ribose) (see Discussion for details) and thereby render these cells hypersensitive to DNA damage.
Figure 6.
Reduction in DNA repair-associated poly(ADP-ribose) formation in PARP-1(–/–) cells. The stimulation of repair-related poly(ADP-ribose) synthesis was followed in standard in vitro repair reactions containing 150 ng of either X-irradiated plasmid DNA (filled symbols) or undamaged plasmid (empty symbols), 30 µg protein extract from PARP-1(+/+) (circles) or PARP-1(–/–) (diamonds) cells in the presence of 0.2 mM NAD+ and 2 µCi [32P]NAD+. The reactions were performed for various times at 30°C. The reaction was stopped by adding trichloroacetic acid and acid-insoluble material was collected on a glass filter for counting.
DISCUSSION
The data in this paper demonstrate that PARP-1 is dispensable for BER and is not essential for the correction of DNA damage inflicted by X-irradiation and alkylating agents. We show, both in vitro and in vivo, that single-strand breaks and methylated bases in DNA are repaired with the same efficiency in PARP-1-deficient and wild-type cells. The BER enzymatic activities which act to remove the altered base, to incise the AP site and in the DNA synthesis and ligation steps appear to be unaffected in the absence of PARP-1. Consistent with this, Wang et al. (8) have reported the occurrence of unscheduled DNA synthesis in PARP-1 knock-out cells after exposure to the alkylating agent methylmethansulfonate (MMS). In the same study recovery of transcriptional activity from an in vitro damaged reporter plasmid was shown to occur following its transfection in PARP-1(–/–) cells, indicating normal repair ability in the absence of PARP-1. An alternative experimental approach has been used by Trucco et al. (12) to measure the number of DNA breaks in PARP-1(–/–) cells following MMS treatment. This technique, called the Comet assay, employs single cell alkaline gel electrophoresis, which results in the migration of fragmented DNA out of the cells towards the anode, giving rise to a DNA tail resembling a comet. The length and the amount of DNA in the tail are taken as a measure of the density of DNA breakage in the cells. The authors described a prolonged delay in DNA break resealing in PARP-1(–/–) cells following MMS administration and concluded that a severe defect in DNA repair due to the lack of PARP-1 is the primary cause for the observed increased sensitivity of these cells in response to genotoxic stress. The experimental approach used in this study, however, cannot distinguish DNA breaks caused by impaired rejoining of DNA ends from breaks induced during cell death, which has been reported to take place much more rapidly in PARP-1-deficient cells than in their counterparts following genotoxic treatment (31).
The major route for AP site processing in mammalian cells, which is thought to account for the repair of >80% of AP sites, is the DNA polymerase β-dependent single nucleotide patch (SNP) pathway (32). A small proportion of oxidized or reduced AP sites which are refractory to repair by the SNP pathway are corrected by an alternative, PCNA-dependent long patch (LP) pathway (33,34). A line of evidence implicating PARP-1 in BER comes from recent studies showing that PARP-1 may interact with the X-ray cross complementing-1 (XRCC-1) protein and DNA polymerase β through a specific domain homologous to the BRCA1 C-terminus motif (35,36). XRCC-1 is known to bind ligase III and DNA polymerase β and has been proposed to act as an adaptor protein in the SNP pathway.
A role for PARP-1 in this putative repair complex as a nick sensor, which in turn recruits XRCC-1 and the other BER proteins to the site of damage to facilitate the repair process, or as a factor acting directly on the activities of these repair enzymes through poly(ADP)ribosylation, has been proposed (35,36). In their very recent study de Murcia and co-workers investigated the repair patches generated during the repair of a single abasic site derived from uracil or 8-oxoguanine in plasmid DNA using PARP-1-deficient cell extracts (36). They found a considerable impairment at the polymerization step of the LP pathway but only partial inefficiency of SNP repair synthesis. These data must be interpreted with care since the incorporation of a labeled nucleotide, which was taken by the authors as a measure of repair ability, may actually reflect the length of the repair patch rather than the real repair efficiency. Indeed, if one assumed that long repair patches were preferentially produced in PARP-1(+/+) cell extracts, whereas in PARP-1(–/–) extracts the repair was completed by single nucleotide replacement, this could explain the dramatic difference between the two cell extracts in the amount of radioactivity incorporated during repair synthesis.
These observations raise the question of why cells would make more use of the LP pathway in the presence of PARP-1 (or, alternatively, why they would mostly employ the SNP pathway in the absence of PARP-1) given that: (i) the LP pathway is utilized by cells particularly to correct a minority of AP sites that are not amenable to cleavage by β-elimination; (ii) the potentially error-prone LP gap filling, especially when performed by low fidelity DNA polymerase β, may be dangerous and harmful to cells. Our results, obtained by methods that permit accurate estimation of overall BER efficiency, show similar rates of DNA strand break rejoining in PARP-1(–/–) and wild-type cell extracts (Fig. 2) and living cells (Fig. 5), as well as no apparent defect in DNA repair synthesis in PARP-1(–/–) cells (Fig. 4). These conflicting results could arise from the use of alternative pathways for processing of the distinct types of DNA damage (for example, DNA strand breaks and methylated bases in our experimental system could be predominantly repaired through the SNP pathway which is not significantly impaired in the absence of PARP-1), although the specific features of the two different PARP-1 knock-out model systems used, with respect to the site of PARP-1 gene disruption, may also be an important factor. The gene interruption within exon 4 in de Murcia’s model system leaves the possibility for expression of an N-terminal fragment of PARP-1 encompassing an intact zinc finger 1 and part of zinc finger 2 in those cells. This may be crucial for the outcome of studies on DNA repair using this knock-out model since zinc finger 1 is known to be sufficient for binding of single-strand breaks (37) and PARP-1 fragments bearing zinc fingers could have a significant impact on DNA repair by binding to DNA strand breaks and competing with DNA repair proteins (38,39). Concerning PARP-1 interactions with BER proteins, it is noteworthy that both the short (40,41) and long patch (42,43) BER pathways have recently been reconstituted with purified human proteins in the absence of PARP-1, clearly indicating that, at least in vitro, PARP-1 has no auxiliary role in BER. Furthermore, PARP-1-deficient mice do not exhibit an unusual incidence of tumor growth (9,10), which is likely to appear in the absence of efficient BER since the accumulation of DNA damage caused by a variety of reactive normal metabolites is an important contributory factor in spontaneous carcinogenesis (44,45).
Why would PARP-1-deficient cells and mice be hypersensitive to γ-irradiation and alkylating agents when they clearly contain normal activities to repair their DNA following these injuries? Previous data have shown that extensive poly(ADP-ribose) synthesis takes place during the early stages of DNA repair following treatment with genotoxic agents (7,23). We hypothesize that the poly(ADP-ribose) level in cells after moderate DNA damage may play a crucial role in cell recovery and survival following exposure to genotoxic agents. Our results in Figure 6 demonstrate that the damaged plasmid DNA triggers ∼15 times less repair-associated poly(ADP-ribose) formation in PARP-1(–/–) cell extracts than in PARP-1(+/+) cell extracts. Thus, the numerous PARP-1 homologs reported recently (26–30) cannot compensate for PARP-1 to achieve the poly(ADP-ribose) levels that normally occur during repair in wild-type cells. In addition, unlike PARP-1, these new poly(ADP)ribosylating enzymes do not seem to interfere with BER, because the BER process is independent of NAD+ in PARP-1(–/–) cell extracts (Figs 2 and 4). The significant reduction in poly(ADP-ribose) level and/or the failure to timely poly(ADP)ribosylate proteins implicated in the cellular response to DNA damage could obliterate important DNA damage-initiated processes in PARP-1(–/–) cells and, thereby, result in extreme sensitivity of these cells to genotoxic treatment. In this regard, the covalent modification or non-covalent interaction of free poly(ADP-ribose) with other DNA damage-regulated proteins, like p53 and DNA-PK, are of special interest. The tumor suppressor phosphoprotein p53 reduces the emergence of cancer by mediating cell cycle arrest in G1 or G2/M to allow time for DNA repair before entering S phase, by inducing apoptosis in cells that have accumulated substantial DNA damage and by regulating genome stability. Both free and PARP-1-bound poly(ADP-ribose) has been shown to target three specific binding sites in the p53 protein for strong non-covalent binding (46). This binding of poly(ADP-ribose) prevents p53 specific binding to its consensus sequence and also interferes with its non-specific binding to single-stranded DNA ends. In addition, p53 has been shown to undergo extensive poly(ADP)ribosylation early during apoptosis and PARP-1 cleavage and degradation of covalently bound polymer by poly(ADP-ribose) glycohydrolase coincide with marked induction of expression of p53-responsive genes (47). The insufficient amount of poly(ADP-ribose) in PARP-1(–/–) cells may enhance p53-dependent transcription and the p53 response pathway and therefore sensitize these cells to DNA damage. The lack of poly(ADP)ribosylation of p53 could perturb other important functions of this protein, such as the maintenance of diploidy (48) and the regulation of chromosome duplication (49). Consistent with this, PARP-1(–/–) cells exhibit increased genomic instability and chromosomal aberrations (11,50). Despite some discrepancies in the literature on PARP-1–p53 interactions, it appears that the function of p53 as a multiple stress signaler controlling the cell cycle and apoptosis in DNA-damaged cells is regulated by poly(ADP)ribosylation.
Additional insights into the role of PARP-1 and poly(ADP)ribosylation in preserving genome integrity and preventing illegitimate recombination come from recently uncovered genetic and physical interactions between PARP-1 and DNA-PK (51–53). These studies demonstrate that the catalytic subunit of DNA-PK can be poly(ADP)ribosylated and that PARP-1 can be phosphorylated and infer that these two DNA break-activated molecules can mutually regulate each other. The increased general recombination activity in SCID mice after disruption of the PARP-1 gene has been shown to rescue the defective V(D)J recombination in these cells, indicating that PARP-1 and DNA-PK cooperate to minimize genomic damage caused by DNA strand breaks (51).
In summary, these data suggest that the dramatic reduction in poly(ADP-ribose) synthesis in PARP-1(–/–) cells is likely to affect many other events following DNA damage in these cells and to render them hypersensitive to genotoxic treatments. The poly(ADP-ribose) level in normal cells may directly reflect the extent of DNA damage and, in this way, influence the fate of a cell following exposure to DNA-damaging agents. Hence, early transient synthesis of poly(ADP-ribose) during BER, a phenomenon that is conserved in many cell types throughout evolution, apparently serves a useful and specific role.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Z.-Q. Wang for providing the PARP-1(–/–) and(+/+) cell lines. We thank Dr Alain Verreault for critical reading of the manuscript and Rashmi G. Shah and Claire Sevenhuysen for editorial work. We also thank Dr Marc-Edouard Mirault for discussions. This work was supported by grant MT-6128 from the Medical Research Council of Canada.
REFERENCES
- 1.Lindahl T. (1993) Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 2.Seeberg E., Eide,L. and Bjoras,M. (1995) Trends Biochem. Sci., 20, 391–397. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg E., Walker,G. and Siede,W. (1995) In DNA Repair and Mutagenesis. ASM Press, New York, NY.
- 4.Chatterjee S. and Berger,N. (1998) In Nickoloff,J.A. and Hoekstra,M.F. (eds), DNA Damage and Repair. Humana Press, Totowa, NJ, Vol. 2, pp. 487–515.
- 5.Satoh M.S. and Lindahl,T. (1992) Nature, 356, 356–358. [DOI] [PubMed] [Google Scholar]
- 6.Satoh M.S., Poirier,G.G. and Lindahl,T. (1993) J. Biol. Chem., 268, 5480–5487. [PubMed] [Google Scholar]
- 7.de Murcia G. and Menissier de Murcia,J. (1994) Trends Biochem. Sci., 19, 172–176. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z.Q., Auer,B., Stingl,L., Berghammer,H., Haidacher,D., Schweiger,M. and Wagner,E.F. (1995) Genes Dev., 9, 509–520. [DOI] [PubMed] [Google Scholar]
- 9.Ménissier-de Murcia J.M., Niedergang,C., Trucco,C., Ricoul,M., Dutrillaux,B., Mark,M., Oliver,F.J., Masson,M., Dierich,A., LeMeur,M., Walztinger,C., Chambon,P. and de Murcia,G. (1997) Proc. Natl Acad. Sci. USA, 94, 7303–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masutani M., Nozaki,T., Nishiyama,E., Shimokawa,T., Tachi,Y., Suzuki,H., Nakagama,H., Wakabayashi,K. and Sugimura,T. (1999) Mol. Cell. Biochem., 193, 149–152. [PubMed] [Google Scholar]
- 11.Wang Z.Q., Stingl,L., Morrison,C., Jantsch,M., Los,M., Schulze-Osthoff,K. and Wagner,E.F. (1997) Genes Dev., 11, 2347–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trucco C., Oliver,F.J., de Murcia,G. and Menissier-de Murcia,J. (1998) Nucleic Acids Res., 26, 2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahradka P. and Ebisuzaki,K. (1982) Eur. J. Biochem., 127, 579–585. [PubMed] [Google Scholar]
- 14.Sambrook J., Fritsch,E. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Tanaka M., Lai,L.S. and Herr,W. (1992) Cell, 68, 755–767. [DOI] [PubMed] [Google Scholar]
- 16.Biade S., Sobol,R.W., Wilson,S.H. and Matsumoto,Y. (1998) J. Biol. Chem., 273, 898–902. [DOI] [PubMed] [Google Scholar]
- 17.Legault J., Tremblay,A., Ramotar,D. and Mirault,M.E. (1997) Mol. Cell. Biol. 17, 5437–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller S.A., Dykes,D.D. and Polesky,H.F. (1988) Nucleic Acids Res., 16, 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manley J.L., Fire,A., Samuels,M. and Sharp,P.A. (1983) Methods Enzymol., 101, 568–582. [DOI] [PubMed] [Google Scholar]
- 20.Althaus F.R. and Richter,C. (1987) Mol. Biol. Biochem. Biophys., 37, 1–237. [PubMed] [Google Scholar]
- 21.Molinete M., Vermeulen,W., Burkle,A., Menissier-de Murcia,J., Kupper,J.H., Hoeijmaker,J.H.J. and de Murcia,G. (1993) EMBO J., 12, 2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kupper J.H., Muller,M., Jacobson,M.K., Tatsumi-Miyajima,J., Coyle,D.L., Jacobson,E.L. and Burkle,A. (1995) Mol. Cell. Biol., 15, 3154–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cleaver J.E. and Morgan,W.F. (1991) Mutat. Res., 257, 1–18. [DOI] [PubMed] [Google Scholar]
- 24.Lindhal T., Satoh,M.S., Poirier,G.G. and Klungland,A. (1995) Trends Biochem. Sci. Sci., 20, 405–411. [DOI] [PubMed] [Google Scholar]
- 25.Satoh M.S., Poirier,G.G. and Lindahl,T. (1994) Biochemistry, 33, 7099–7106. [DOI] [PubMed] [Google Scholar]
- 26.Shieh W.M., Amé,J.-C., Wilson,M.V., Wang,Z.Q., Koh,D.W., Jacobson,M.K. and Jacobson,E.L. (1998) J. Biol. Chem., 273, 30069–30072. [DOI] [PubMed] [Google Scholar]
- 27.Smith S., Giriat,I., Schmitt,A. and de Lange,T. (1998) Science, 282, 1484–1487. [DOI] [PubMed] [Google Scholar]
- 28.Ame J.C., Rolli,V., Schreiber,V., Niedergang,C., Apiou,F., Decker,P., Muller,S., Hoger,T., Menissier-de Murcia,J. and de Murcia,G. (1999) J. Biol. Chem., 274, 17860–17868. [DOI] [PubMed] [Google Scholar]
- 29.Kirckhoefer V.A., Siva,A.C., Kedersha,N.L., Inman,E.M., Ruland,C., Streuli,M. and Rome,L.H. (1999) J. Cell Biol., 146, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sallmann F.R., Vodenicharov,M.D., Wang,Z.Q. and Poirier,G.G. (2000) J. Biol. Chem., 275, 15504–15511. [DOI] [PubMed] [Google Scholar]
- 31.Oliver F.J., de la Rubia,G., Rolli,V., Ruiz-Ruiz,C., de Murcia,G. and Menissier-de Murcia,J. (1998) J. Biol. Chem., 273, 33533–33539. [DOI] [PubMed] [Google Scholar]
- 32.Dianov G., Price,A. and Lindhal,T. (1992) Mol. Cell. Biol., 12, 1605–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto Y., Kim,K. and Bogenhagen,D.F. (1994) Mol. Cell. Biol., 14, 6187–6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frosina G., Fortini,P., Rossi,O., Carrozzino,F., Raspaglio,G., Cox,L.S., Lane,D.P., Abbondandolo,A. and Dogliotti,E. (1996) J. Biol. Chem., 271, 9573–9578. [DOI] [PubMed] [Google Scholar]
- 35.Masson M., Niedergang,C., Schreiber,V., Ménissier-de Murcia,J. and de Murcia,G. (1998) Mol. Cell. Biol., 18, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dantzer F., de la Rubia,G., Menissier-de Murcia,J., Hostomsky,Z., de Murcia,G. and Schreiber,V. (2000) Biochemistry, 39, 7559–7569. [DOI] [PubMed] [Google Scholar]
- 37.Ikejima M., Noguchi,S., Yamashita,R., Ogura,T., Sugimura,T., Gill,D.M. and Miuwa,M. (1990) J. Biol. Chem., 265, 21907–21913. [PubMed] [Google Scholar]
- 38.Kupper J.H., de Murcia,G. and Burkle,A. (1990) J. Biol. Chem., 265, 18721–18724. [PubMed] [Google Scholar]
- 39.Schreiber V., Hunting,D., Trucco,C., Gowans,B., Grunwald,D., de Murcia,G. amd Menissier-de Murcia,J. (1995) Proc. Natl Acad. Sci. USA, 92, 4753–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kubota Y., Nash,R.A., Klungland,A., Schar,P., Barnes,D.E. and Lindhal,T. (1996) EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholl I.D., Nealon,K. and Kenny,M.K. (1997) Biochemistry, 36, 7557–7566. [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto Y., Kim,K., Hurwitz,J., Gary,R., Levin,D.S., Tomkinson,A.E. and Park,M.S. (1999) J. Biol. Chem., 274, 33703–33708. [DOI] [PubMed] [Google Scholar]
- 43.Pascucci B., Stucki,M., Jonsson,Z.O., Dogliotti,E. and Hubscher,U. (1999) J. Biol. Chem., 274, 33696–33702. [DOI] [PubMed] [Google Scholar]
- 44.Lutz W.K. (1990) Mutat. Res., 238, 287–295. [DOI] [PubMed] [Google Scholar]
- 45.Loeb L.A. (1989) Cancer Res., 49, 5489–5496. [PubMed] [Google Scholar]
- 46.Malanga M., Pleschke,J.M., Kleczkowska,H.E. and Althaus,F.R. (1998) J. Biol. Chem., 273, 11839–11843. [DOI] [PubMed] [Google Scholar]
- 47.Simbulan-Rosenthal C.M., Rosenthal,D.S., Luo,R. and Smulson,M.E. (1999) Cancer Res., 59, 2190–2194. [PubMed] [Google Scholar]
- 48.Cross S., Sanchez,C., Morgan,C., Schimke,M., Ramel,S., Idzerda,R., Raskind,W. and Ried,B. (1995) Science, 267, 1353–1356. [DOI] [PubMed] [Google Scholar]
- 49.Fukasawa K., Choi,T., Kuriyama,R., Rulong,S. and Vande Woude,G. (1996) Science, 271, 1744–1747. [DOI] [PubMed] [Google Scholar]
- 50.Simbulan-Rosenthal C.M., Haddad,B.R., Rosenthal,D.S., Weaver,Z., Coleman,A., Luo,R., Young,H.M., Wang,Z.Q., Ried,T. and Smulson,M.E. (1999) Proc. Natl Acad. Sci. USA, 96, 13191–13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morrison C., Smith,G.C.M., Stingl,L., Jackson,S.P., Wagner,E.F. and Wang,Z.Q. (1997) Nature Genet., 17, 479–482. [DOI] [PubMed] [Google Scholar]
- 52.Ariumi Y., Matsutani,M., Copeland,T.D., Mimori,T., Sugimura,T., Shimotohno,K., Ueda,K., Hatanaka,M. and Noda,M. (1999) Oncogene, 18, 6416–6425. [DOI] [PubMed] [Google Scholar]
- 53.Galande S. and Kohwi-Shigematsu,T. (1999) J. Biol. Chem., 274, 20521–20528. [DOI] [PubMed] [Google Scholar]