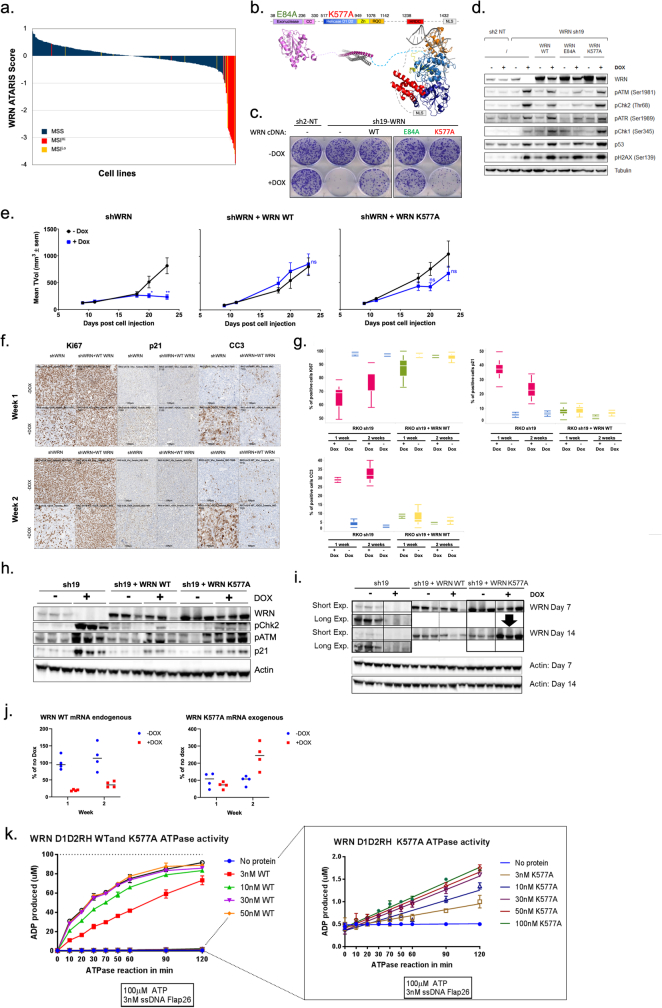
Extended Data Fig. 1. Genetic validation of WRN helicase dependence.
a. WRN ATARIS dependency scores from DRIVE coloured by MSI (high-red and Low-orange) and MSS (blue) status. b. Schematic representation of WRN helicase domains. Clonogenic assay (c) or immunoblot (d) of RKO cells doxycycline induction of a non-targeting negative control (sh2-NT), and one shRNA against WRN (sh19) without (-) or with a wild type (WT), exonuclease mutant (E84A) or helicase mutant (K577A) WRN cDNA. Clonogenic assay shown 15 days and immunoblot 4 days after dox induction. e. RKO xenograft growth ± doxycycline induction of WRN sh19 with WT or helicase dead (K577A) WRN cDNA. Mice were randomized (n = 6). Differences between the means of TVol were assessed on the endpoint ∆TVol using a two-tailed t-test (* P < 0.05, ** P < 0.01 and ns is non-significant). f. Ki67, p21 and cleaved caspase 3 (CC3) expression were evaluated on RKO xenograft sections by immunohistochemistry 7 and 14 days after treatment ± doxycycline and ± WT WRN cDNA. g. Quantification of Ki67, p21 and CC3 positive cells from FFPE sections. The white centre line denotes the median value, while the filled boxes contain the first quartile (25th) and third quartile (75th) percentiles of the dataset. The coloured whiskers mark the 5th and 95th percentiles. h. Immunoblot for WRN, pCHK2, ATM, p21 and actin of RKO tumour ± doxycycline and ± WT WRN cDNA. i. Immunoblot for WRN from samples from panel h treated for 7 and 14 days at different exposures. j. RTqPCR for WRN WT (endogenous) and WRN K577A mutant from tumours treated with doxycycline for 1 and 2 weeks. Data are mean of expression compared to no Dox treatment ± SD, n = 4 tumours. k. Quantification of ADP production over time in an ATPase assay at indicated conditions for both WRN WT and WRN K577A D1D2RH helicase domain protein. Data represent mean ± SD, n = 3.