Abstract
Motivation
Proteins, the molecular workhorses of biological systems, execute a multitude of critical functions dictated by their precise three-dimensional structures. In a complex and dynamic cellular environment, proteins can undergo misfolding, leading to the formation of aggregates that take up various forms, including amorphous and ordered aggregation in the shape of amyloid fibrils. This phenomenon is closely linked to a spectrum of widespread debilitating pathologies, such as Alzheimer’s disease, Parkinson’s disease, type-II diabetes, and several other proteinopathies, but also hampers the engineering of soluble agents, as in the case of antibody development. As such, the accurate prediction of aggregation propensity within protein sequences has become pivotal due to profound implications in understanding disease mechanisms, as well as in improving biotechnological and therapeutic applications.
Results
We previously developed Cordax, a structure-based predictor that utilizes logistic regression to detect aggregation motifs in protein sequences based on their structural complementarity to the amyloid cross-beta architecture. Here, we present a dedicated web server interface for Cordax. This online platform combines several features including detailed scoring of sequence aggregation propensity, as well as 3D visualization with several customization options for topology models of the structural cores formed by predicted aggregation motifs. In addition, information is provided on experimentally determined aggregation-prone regions that exhibit sequence similarity to predicted motifs, scores, and links to other predictor outputs, as well as simultaneous predictions of relevant sequence propensities, such as solubility, hydrophobicity, and secondary structure propensity.
Availability and implementation
The Cordax webserver is freely accessible at https://cordax.switchlab.org/.
1 Introduction
Proteins are the fundamental building blocks of life, playing pivotal roles in an array of biological processes. They are versatile molecules, executing functions ranging from catalyzing chemical reactions to providing structural support. However, the proper functioning of these biomolecules is inherently linked to their three-dimensional structure and stability (Dill and MacCallum 2012). In recent years, there has been a growing realization that misfolding and aggregation of proteins, including the formation of amyloid structures, are critical determinants of both debilitating diseases and valuable biotechnological applications (Chiti and Dobson 2017, Louros et al. 2023). Protein aggregation refers to the non-native, multimeric assembly of protein molecules, which often culminates in the formation of amyloid fibrils. These fibrils are characterized by their cross-β-sheet structure and have been implicated in a wide range of diseases including neurodegenerative disorders, such as Alzheimer’s, Parkinson’s, and Huntington’s disease, as well as localized or systemic amyloidosis, such as type-II diabetes or light-chain (AL) amyloidosis, respectively (Chiti and Dobson 2017, Buxbaum et al. 2022). The accumulation of misfolded protein aggregates in various tissues is a hallmark of these disorders and is associated with cellular dysfunction and organ failure. Conversely, in the field of biotechnology, protein aggregation and amyloid formation have emerged as both challenges and opportunities. Aggregation can reduce the yield and efficacy of recombinant protein production, affecting biopharmaceutical manufacturing processes and biotherapeutic product quality (Hamrang et al. 2013). Conversely, amyloid-like protein structures have found utility in the development of functional materials (Chakraborty et al. 2019, Jin et al. 2022), including nanotechnology, drug delivery or enzymatic catalysis (Ghosh et al. 2023, Yuan et al. 2023), and tissue engineering (Das et al. 2018), as well as a strategy for the targeted inactivation of hard-to-drug cellular factors related to diseases (Michiels et al. 2020, Janssen et al. 2023).
Consequently, it is essential to attain a comprehensive grasp of the factors that govern protein aggregation. The propensity of proteins to form amyloid structures is intrinsically encoded within their amino acid sequences (Tartaglia et al. 2008, Navarro and Ventura 2022). These sequences contain local motifs, historically referred to as “aggregation-prone regions” (APRs), “amyloid motifs” or “amyloidogenic determinants,” which have been demonstrated to actively facilitate the assembly of amyloid fibrils (Fernandez-Escamilla et al. 2004, Ventura et al. 2004, Teng and Eisenberg 2009). APRs are ubiquitously distributed throughout the vast spectrum of proteins (Sawaya et al. 2007, Teng and Eisenberg 2009, Goldschmidt et al. 2010, Louros et al. 2020, Sawaya et al. 2021). They are evolutionarily tied to the functional fold of soluble protein domains (Prabakaran et al. 2017, Langenberg et al. 2020), and are associated with the function of intrinsically disordered proteins (Santos et al. 2021), while also often acting as integral parts of transmembrane domains or protein-protein interaction interfaces (Castillo and Ventura 2009). In addition, short amyloid motifs have been shown to drive the formation of functional amyloid scaffolds, as for instance in the case of bacterial curli (Louros et al. 2016, Perov et al. 2019) or RHIMs, which form the necrosome complex or are employed by viruses attempting to hijack the same pathway (Mompeán et al. 2018, Baker et al. 2020). Numerous studies have elucidated the capacity of APRs to autonomously self-assemble into aggregates with characteristic amyloid-like morphologies when studied in isolation as peptide fragments (Sawaya et al. 2007, Guenther et al. 2018, Louros et al. 2020, Rawat et al. 2020). Their pivotal role in orchestrating the assembly of proteins is underscored by studies in which the introduction of APRs into proteins that typically do not aggregate induces their self-assembly (Ventura et al. 2002, Ivanova et al. 2004). Furthermore, mutational experiments have reinforced this link, demonstrating that altering specific residues within APRs with the intent of deactivating them results in the prevention of parental protein aggregation (Ventura et al. 2004, Teng and Eisenberg 2009, Guthertz et al. 2022). Recent research endeavours have also unveiled that APRs are capable of forming early intermediate species that are shared among various amyloid conformations of the same protein (Lövestam et al. 2024), known as polymorphs, form homotypic interfaces that act as protofilament contacts and establish common interactions that bolster the stability of fibril polymorphs extracted from the cerebral tissues of patients afflicted with various amyloid-related diseases (Sawaya et al. 2021, Louros et al. 2022, van der Kant et al. 2022, Mullapudi et al. 2023, Louros et al. 2024).
We recently developed a logistic regression model to predict amyloid propensity in protein sequences with high sensitivity and specificity (Louros et al. 2020). As a structure-based approach, this tool named Cordax was shown to uncouple protein aggregation propensity from traditional sequence propensities, such as hydrophobicity and solubility, thus, increasing its ability to detect less common APRs in protein sequences (Hughes et al. 2018, Santos et al. 2021) and to outperform current state-of-art software dedicated to detecting protein aggregation (Louros et al. 2020). Here, we report the development of a dedicated freely accessible webserver for Cordax that supports both the prediction and 3D visualization of predicted APRs in protein sequences.
2 Availability and implementation
The Cordax web server is accessible to users online at https://cordax.switchlab.org/. This platform was designed and implemented using Netlify and is compatible with all devices and web browsers. While email registration is optional for users, it provides registered users the ability to maintain a personalized dashboard, enabling them to monitor the status of submitted tasks and access the outcomes of previous executions. The new job submission page, as well as the personalized dashboard, are both accessible through dedicated buttons that are permanently displayed on the web server title bar (Fig. 1A, arrows). Briefly, in the operational framework of Cordax, an input protein sequence is dissected into hexapeptides via a sliding window technique. Cordax employs the FoldX energy force field (Schymkowitz et al. 2005) to execute all-atom modelling of sequences against its structural database, as described previously (Louros et al. 2020), and the resulting free energies are converted into scores for each peptide fragment, using a recursive feature elimination algorithm and a logistic regression model trained against experimentally determined amyloid motifs (Louros et al. 2020). This process generates an amyloidogenic profile by assigning the highest score obtained for each residue within the input sequence (Cordax Score). A structural model that best represents the predicted amyloid fibril core topology is also selected for windows exceeding its scoring threshold (0.61). This operation is notably computationally intensive. However, computed energies are systematically recorded within an expanding database, facilitating subsequent retrieval. This engenders an efficient interface that circumvents redundant computational tasks for recurring sequence segments in future submissions. More information on the above, as well as a detailed description of the features offered through the webserver interface is provided in an “About” and “Help” page available online.
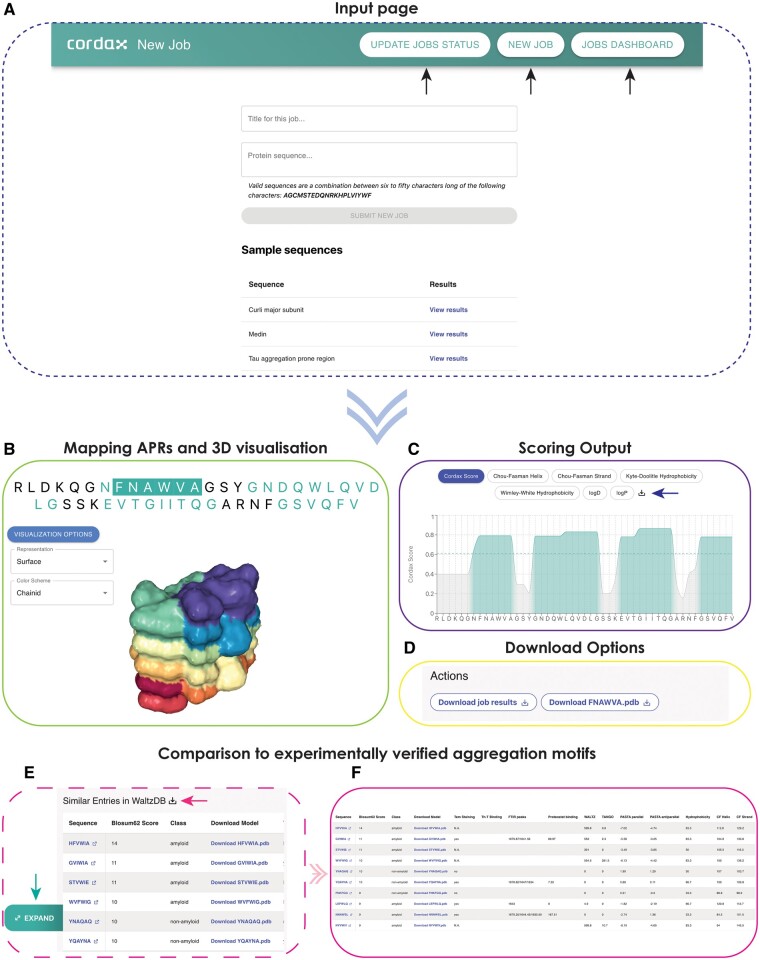
Figure 1.
The Cordax web server interface. (A) Users can submit new jobs or track current and previous jobs through dedicated buttons on the webserver title page (indicated by arrows). Job submission requires a protein sequence as input, with an optional title. (B–F) Representative example of the information provided as output by the Cordax webserver. (B) The main interaction panel of the output page shows the query sequence, with predicted aggregation-prone regions highlighted. By selecting identified hexapeptide sequences, users can activate the 3D visualization plugin indicating the predicted steric zipper topology of the segment. (C) The scoring plot indicates by default the Cordax score per residue but can also be used to plot additional relevant sequence propensities. Users can access per-residue information through a box that appears by browsing over the query sequence shown on the x-axis. Access to the raw data is also provided through a download option (arrow). (D) Download options for information shown in (B and C) are also provided in the “Actions” submenu. (E) For predicted hexapeptides selected from the query sequence shown in (A), an interactive table is generated with experimentally determined aggregation-prone regions derived from WALTZ-DB (Louros et al. 2020) that are sorted based on sequence similarity scores. Information contained can be access using the download option at the top of the table (left-directed arrow). Using the expand option (down arrow), (F) a pop-up window appears for improved visualization of the table contents.
3 Features of the Cordax webserver interface
3.1 Main scoring display
The tool accepts simple protein sequences as input, with a minimum length of six residues and a maximum of 50 residues (Fig. 1A). All-atom modelling is a computationally intensive operation; hence, this length limitation has been set to expedite the webserver queue processing and to reduce output waiting times. The structural context of protein sequences is retained, as Cordax uses local sequence information to profile aggregation propensity. However, considering that it employs a hexapeptide sliding window, scoring of residues at the end of queries derived from longer sequences will derive only from the subset of hexapeptide windows included in the sequence query. To adjust for this, users can run sequence queries with overlapping ends, or alternatively use the standalone version of the tool that can be applied locally with no length constraints. Users are prompted to provide a title for each submitted job request, while completed processes can be accessed through the job dashboard.
Once accessed, each results page displays the query sequence on the top, with residues scoring higher than the Cordax threshold (0.61) (Louros et al. 2020) colored green (Fig. 1B). A graphical representation of the results shown at the bottom of the output page better illustrates this. Specifically, this interactive plot contains the amino acid query sequence on the x-axis, while alternative options are available to the user for display on the y-axis (Fig. 1C). Starting with the Cordax scoring as the default representation, by hovering over the query sequence a box appears labeling both individual residues, their corresponding Cordax aggregation scores, and the defined threshold of prediction. The latter is also shown with a dashed green light. The same interactive features are available for additional sequence properties that can be selected by the user and displayed on the interactive plot (Fig. 1C). For secondary structure propensity, we used the Chou–Fasman empirical technique (Chou and Fasman 1974). Sequence hydrophobicity is calculated based on two different scales, namely the Kyte–Doolittle (Kyte and Doolittle 1982) and the Wimley–White scale which holds considerable importance as it considers the combined contributions of both the peptide bonds and the sidechains in absolute values, providing a direct and empirical foundation based on experimentally determined values for the transfer free energies of polypeptides (Wimley and White 1996). Finally, considering the ability of Cordax to predict with high accuracy aggregation-prone sequence segments of higher solubility, we have included per residue calculations of partition coefficients calculated using PlogP, a method that calculates peptide coefficients by a residue-addition method and also considers blocked termini, as well as partition as a function of the pH (ionizable and non-ionizable) (Tao et al. 1999). A download option is also available for obtaining and analyzing the data presented in the interactive plot locally.
3.2 Modelling the structural topology of predicted aggregation-prone regions
The sequence presented at the top of the output page is interactive, whereby individual predicted residues can be engaged by a user. This interaction serves to illuminate the protein sequence segments that score above the threshold. Clicking on predicted residues highlights the hexapeptide window of prediction starting with this residue in position 1. If this window scores above the threshold of prediction, this selection concurrently activates a graphical plugin interface situated beneath the query sequence (Fig. 1B). Within this graphical interface, various modes for representing the structural topology of selected hexapeptides that surpass the Cordax aggregation propensity threshold are supported. These modes encompass options such as cartoon, ball and stick, ribbon, space-fill models, and surface representations, among others. Furthermore, a range of distinctive color themes are provided predicated on diverse properties, including chain ID, atom and residue types, and hydrophobicity (Fig. 1B).
3.3 Comparison to peptides with experimentally determined amyloid-forming properties
For each hexapeptide region selected from the displayed query sequence, an adjacent right panel becomes active, offering several supplementary features. Primarily, users are provided with the option to download specific content at the top of this panel (Fig. 1D). This includes the Cordax scoring files in the .csv file format and the predicted structural topology in Protein Data Bank (wwPDB Consortium 2019) file format (.pdb files) for windows scoring above the threshold. Simultaneously, upon the selection of a hexapeptide, an interactive table is displayed on the right panel (Fig. 1E and F). This table, which can be expanded for improved visualization by moving the cursor over the table and selecting an expansion button option appearing on the left, enumerates peptide sequences that correspond to entries within WALTZ-DB 2.0, currently the largest openly accessible repository of peptides with experimentally ascertained amyloidogenic properties (Louros et al. 2020). The sequences are organized based on their sequence similarity to the selected hexapeptide, calculated using the Blosum62 matrix. This table further provides valuable data concerning the employed experimental techniques used to determine the aggregation properties of each peptide entry. This includes experimental validation obtained from diverse methodologies like Transmission electron microscopy (TEM), Fourier-Transform infrared spectroscopy (FTIR), and the binding of various fluorescence aggregation reporter dyes (such as Thioflavin-T and Proteostat binding). In addition, aggregation propensity prediction scores are listed, generated by other specialized high-specificity tools, such as WALTZ (Maurer-Stroh et al. 2010), TANGO (Fernandez-Escamilla et al. 2004), and PASTA 2.0 (Walsh et al. 2014) (for both parallel and anti-parallel orientation predictions, as described). Notably, the data presented in the interactive table can be downloaded locally using an option at the top of the table, and predicted topologies of the sequences can be downloaded in a .pdb format through a dedicated column containing links. Finally, each sequence presented in the table is hyperlinked, enabling direct access to the corresponding peptide entry within WALTZ-DB (Fig. 1E and F). This facilitates users in acquiring supplementary and pertinent information. Such information encompasses details regarding the source proteins from which the peptide matches originate and are initially analyzed within WALTZ-DB, denoted by their Uniprot identifiers (The UniProt Consortium 2023), along with their respective positions in the identified protein sequence. Additionally, users can access a comprehensive breakdown of individual energy components and the topological models predicted by Cordax for the specific peptide sequence entry. Moreover, these links provide access to additional aggregation prediction algorithms, such as Zipper-DB 3D-profiling method (Thompson et al. 2006) and Aggrescan (Conchillo-Solé et al. 2007). Lastly, they can access a visual representation of the experimental evidence confirming the aggregation propensity listed in the initial table.
Contributor Information
Nikolaos Louros, Switch Laboratory, VIB Center for Brain and Disease Research, VIB, 3000 Leuven, Belgium; Department of Cellular and Molecular Medicine, Switch Laboratory, KU Leuven, 3000 Leuven, Belgium; Switch Laboratory, VIB Center for AI & Computational Biology, VIB, 3000 Leuven, Belgium.
Frederic Rousseau, Switch Laboratory, VIB Center for Brain and Disease Research, VIB, 3000 Leuven, Belgium; Department of Cellular and Molecular Medicine, Switch Laboratory, KU Leuven, 3000 Leuven, Belgium; Switch Laboratory, VIB Center for AI & Computational Biology, VIB, 3000 Leuven, Belgium.
Joost Schymkowitz, Switch Laboratory, VIB Center for Brain and Disease Research, VIB, 3000 Leuven, Belgium; Department of Cellular and Molecular Medicine, Switch Laboratory, KU Leuven, 3000 Leuven, Belgium; Switch Laboratory, VIB Center for AI & Computational Biology, VIB, 3000 Leuven, Belgium.
Conflict of interest
None declared.
Funding
This work was supported by the Flanders Institute for Biotechnology (VIB); KU Leuven; the Fund for Scientific Research Flanders (FWO) [project grant numbers G0C2818N, G0C0320N, G053420N, and G0C3522N]; the Stichting Alzheimer Onderzoek [project numbers SAO‐FRA 2019/0015, SAO‐FRA 2020/0009, and SAO-FRA 2020/0013]; and the National Institute On Aging of the National Institutes of Health under Award Number R01AG079234. N.L. was furthermore supported by postdoctoral fellowships from the FWO [fellowship numbers 12P0919N and 12P0922N].
References
- Baker MODG, Shanmugam N, Pham CLL. et al. RHIM-based protein:protein interactions in microbial defence against programmed cell death by necroptosis. Semin Cell Dev Biol 2020;99:86–95. [DOI] [PubMed] [Google Scholar]
- Buxbaum JN, Dispenzieri A, Eisenberg DS. et al. Amyloid nomenclature 2022: update, novel proteins, and recommendations by the International Society of Amyloidosis (ISA) Nomenclature Committee. Amyloid 2022;29:213–9. [DOI] [PubMed] [Google Scholar]
- Castillo V, Ventura S.. Amyloidogenic regions and interaction surfaces overlap in globular proteins related to conformational diseases. PLoS Comput Biol 2009;5:e1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P, Guterman T, Adadi N. et al. A self-healing, all-organic, conducting, composite peptide hydrogel as pressure sensor and electrogenic cell soft substrate. ACS Nano 2019;13:163–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM.. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem 2017;86:27–68. [DOI] [PubMed] [Google Scholar]
- Chou PY, Fasman GD.. Prediction of protein conformation. Biochemistry 1974;13:222–45. [DOI] [PubMed] [Google Scholar]
- Conchillo-Solé O, de Groot NS, Avilés FX. et al. AGGRESCAN: a server for the prediction and evaluation of "hot spots" of aggregation in polypeptides. BMC Bioinformatics 2007;8:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Jacob RS, Patel K. et al. Amyloid fibrils: versatile biomaterials for cell adhesion and tissue engineering applications. Biomacromolecules 2018;19:1826–39. [DOI] [PubMed] [Google Scholar]
- Dill KA, MacCallum JL.. The protein-folding problem, 50 years on. Science 2012;338:1042–6. [DOI] [PubMed] [Google Scholar]
- Fernandez-Escamilla A-M, Rousseau F, Schymkowitz J. et al. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol 2004;22:1302–6. [DOI] [PubMed] [Google Scholar]
- Ghosh C, Ghosh S, Chatterjee A. et al. Dual enzyme-powered chemotactic cross β amyloid based functional nanomotors. Nat Commun 2023;14:5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt L, Teng PK, Riek R. et al. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci USA 2010;107:3487–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther EL, Cao Q, Trinh H. et al. Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat Struct Mol Biol 2018;26:988–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthertz N, van der Kant R, Martinez RM. et al. The effect of mutation on an aggregation-prone protein: an in vivo, in vitro, and in silico analysis. Proc Natl Acad Sci USA 2022;119:e2200468119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamrang Z, Rattray NJW, Pluen A.. Proteins behaving badly: emerging technologies in profiling biopharmaceutical aggregation. Trends Biotechnol 2013;31:448–58. [DOI] [PubMed] [Google Scholar]
- Hughes MP, Sawaya MR, Boyer DR. et al. Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science 2018;359:698–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova MI, Sawaya MR, Gingery M. et al. An amyloid-forming segment of beta2-microglobulin suggests a molecular model for the fibril. Proc Natl Acad Sci USA 2004;101:10584–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen K, Claes F, Van de Velde D. et al. Exploiting the intrinsic misfolding propensity of the KRAS oncoprotein. Proc Natl Acad Sci USA 2023;120:e2214921120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Peydayesh M, Li M. et al. Functional coating from amyloid superwetting films. Adv Mater 2022;34:2205072. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF.. A simple method for displaying the hydropathic character of a protein. J Mol Biol 1982;157:105–32. [DOI] [PubMed] [Google Scholar]
- Langenberg T, Gallardo R, van der Kant R. et al. Thermodynamic and evolutionary coupling between the native and amyloid state of globular proteins. Cell Rep 2020;31:107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louros N, Konstantoulea K, De Vleeschouwer M. et al. WALTZ-DB 2.0: an updated database containing structural information of experimentally determined amyloid-forming peptides. Nucleic Acids Res 2020;48:D389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louros N, Orlando G, De Vleeschouwer M. et al. Structure-based machine-guided mapping of amyloid sequence space reveals uncharted sequence clusters with higher solubilities. Nat Commun 2020;11:3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louros N, Schymkowitz J, Rousseau F.. Mechanisms and pathology of protein misfolding and aggregation. Nat Rev Mol Cell Biol 2023;24:912–33. [DOI] [PubMed] [Google Scholar]
- Louros N, van der Kant R, Schymkowitz J. et al. StAmP-DB: a platform for structures of polymorphic amyloid fibril cores. Bioinformatics 2022;38:2636–8. [DOI] [PubMed] [Google Scholar]
- Louros N, Wilkinson M, Tsaka G. et al. Local structural preferences in shaping tau amyloid polymorphism. Nat Commun 2024;15:1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louros NN, Bolas GMP, Tsiolaki PL. et al. Intrinsic aggregation propensity of the CsgB nucleator protein is crucial for curli fiber formation. J Struct Biol 2016;195:179–89. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Debulpaep M, Kuemmerer N. et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods 2010;7:237–42. [DOI] [PubMed] [Google Scholar]
- Michiels E, Roose K, Gallardo R. et al. Reverse engineering synthetic antiviral amyloids. Nat Commun 2020;11:2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mompeán M, Li W, Li J. et al. The structure of the necrosome RIPK1-RIPK3 core, a human hetero-amyloid signaling complex. Cell 2018;173:1244–53.e1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullapudi V, Vaquer-Alicea J, Bommareddy V. et al. Network of hotspot interactions cluster tau amyloid folds. Nat Commun 2023;14:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro S, Ventura S.. Computational methods to predict protein aggregation. Curr Opin Struct Biol 2022;73:102343. [DOI] [PubMed] [Google Scholar]
- Perov S, Lidor O, Salinas N. et al. Structural insights into curli CsgA cross-β fibril architecture inspire repurposing of anti-amyloid compounds as anti-biofilm agents. PLoS Pathog 2019;15:e1007978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran R, Goel D, Kumar S. et al. Aggregation prone regions in human proteome: insights from large-scale data analyses. Proteins: Struct Funct Bioinf 2017;85:1099–118. [DOI] [PubMed] [Google Scholar]
- Rawat P, Prabakaran R, Sakthivel R. et al. CPAD 2.0: a repository of curated experimental data on aggregating proteins and peptides. Amyloid 2020;27:128–33. [DOI] [PubMed] [Google Scholar]
- Santos J, Pallarès I, Iglesias V. et al. Cryptic amyloidogenic regions in intrinsically disordered proteins: function and disease association. Comput Struct Biotechnol J 2021;19:4192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Hughes MP, Rodriguez JA. et al. The expanding amyloid family: structure, stability, function, and pathogenesis. Cell 2021;184:4857–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R. et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 2007;447:453–7. [DOI] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F. et al. The FoldX web server: an online force field. Nucleic Acids Res 2005;33:W382–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lövestam S, Li D, Wagstaff JL. et al. Disease-specific tau filaments assemble via polymorphic intermediates. Nature 2024;625:119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao P, Wang R, Lai L.. Calculating partition coefficients of peptides by the addition method. Mol Model Annu 1999;5:189–95. [Google Scholar]
- Tartaglia GG, Pawar AP, Campioni S. et al. Prediction of aggregation-prone regions in structured proteins. J Mol Biol 2008;380:425–36. [DOI] [PubMed] [Google Scholar]
- Teng PK, Eisenberg D.. Short protein segments can drive a non-fibrillizing protein into the amyloid state. Protein Eng Des Sel 2009;22:531–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res 2023;51:D523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MJ, Sievers SA, Karanicolas J. et al. The 3D profile method for identifying fibril-forming segments of proteins. Proc Natl Acad Sci USA 2006;103:4074–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kant R, Louros N, Schymkowitz J. et al. Thermodynamic analysis of amyloid fibril structures reveals a common framework for stability in amyloid polymorphs. Structure 2022;30:1178–89.e3. [DOI] [PubMed] [Google Scholar]
- Ventura S, Lacroix E, Serrano L.. Insights into the origin of the tendency of the PI3-SH3 domain to form amyloid fibrils. J Mol Biol 2002;322:1147–58. [DOI] [PubMed] [Google Scholar]
- Ventura S, Zurdo J, Narayanan S. et al. Short amino acid stretches can mediate amyloid formation in globular proteins: the Src homology 3 (SH3) case. Proc Natl Acad Sci USA 2004;101:7258–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh I, Seno F, Tosatto SCE. et al. PASTA 2.0: an improved server for protein aggregation prediction. Nucleic Acids Res 2014;42:W301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley WC, White SH.. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol 1996;3:842–8. [DOI] [PubMed] [Google Scholar]
- wwPDB Consortium. Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res 2019;47:D520–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Chen L, Kong L. et al. Histidine modulates amyloid-like assembly of peptide nanomaterials and confers enzyme-like activity. Nat Commun 2023;14:5808. [DOI] [PMC free article] [PubMed] [Google Scholar]