Abstract
KRAS is a commonly mutated oncogene in human gastric cancer and is often associated with drug resistance and poor prognosis. Co-clinical trial of combined MEK-CDK4/6 inhibition in KRAS mutated cancers demonstrated therapeutic efficacy in patient-derived xenografts and safety in patients. Here, present research focuses on targeting CDK4/6 and MEK synergistically block the proliferation of KRAS-mutated gastric cancer cells in vitro and in vivo and induced autophagy through the AMPK/mTOR pathway. Furthermore, autophagy inhibitor combined with targeting CDK4/6 and MEK therapy had significant antitumor effects on KRAS mutant gastric cancer cells. Clinical trials are needed to determine the mechanism behind this finding and its clinical utility. In conclusion, our results demonstrate autophagy inhibitor combined targeting MEK and CDK4/6 that concurrently block multiple metabolic processes may be an effective therapeutic approach for gastric cancer.
Keywords: Gastric cancer, KRAS, MEK, CDK4/6, AMPK/mTOR
1. Introduction
Gastric cancer is one of the leading causes of cancer morbidity and mortality worldwide, particularly in Asia, Eastern Europe, and Central America [1]. KRAS mutations often lead to drug resistance and a poor prognosis in human cancers. It is estimated that 3%–13 % of gastric cancers contain mutations in the oncogenic pathway KRAS [2]. However, the direct mutation of KRAS or the inhibition of pan-KRAS has a limited effect on gastric cancer proliferation and the downstream RAF-MEK-ERK pathway [3]. In a genetically engineered melanoma mouse model based on synthetic lethal extinction, MEK and CDK4/6 inhibition could inhibit KRAS mutation induction [4]. Treatment of KRAS amplified gastroesophageal cancer with combination inhibition of SHP2 and MEK could be effective [5]. Phase I clinical trials were initiated to examine the effect of an ERK inhibitor combined with a CDK4/6 inhibitor in patients with advanced pancreatic ductal adenocarcinoma [6], which showed that genes were enriched diverse signaling nodes, including CDK2 activation and autophagy-activating pathways [7]. A co-clinical trial of MEK-CDK4/6 inhibition demonstrated therapeutic efficacy in xenografts from patients with RAS-mutated colorectal cancer [8]. Autophagy is required for the maintenance of oxidative metabolism and tumorigenesis in activated RAS [7]. However, there is no prior knowledge of how CDK4/6, MEK, and autophagy are targeted in gastric cancers with KRAS mutations.
Here, CDK4/6 and MEK promote AMPK/mTOR-driven autophagy in KRAS-mutated gastric cancer cells in vitro and in vivo. Furthermore, combining autophagy inhibitors with CDK4/6 and MEK inhibition led to significant antitumor effects in KRAS-mutated gastric cancer cells. As a result, we conclude that autophagy inhibitors combined with MEK and CDK4/6 inhibitors may present a novel therapeutic approach for gastric cancer.
2. Materials and methods
2.1. Cell lines
Human gastric cancer cell lines AGS and SNU1(KRAS- mutated), MKN45 (non-KRAS-mutated) as well as their control normal gastric cell line (GES-1) were cultured in Dulbecco's modified Eagle's medium (Cat: SH30243, DMEM, HyClone) supplemented with 10 % fetal bovine serum (Cat: 10100147, FBS, Gibco). Cell lines were authenticated using Short Tandem Repeat (STR) analysis and tested for the potential mycoplasma contamination. All cell lines were cultured in a 5 % CO2 incubator at 37 °C.
2.2. Antibodies and chemical reagents
Chemical reagents were palbociclib (HY-50767), trametinib (HY-10999), CellCountingKit-8 (HY–K0301) and Chloroquine (HY-17589) from MedChemExpress. Antibodies were p62 (23214T), LC3 (12741T), Beclin-1 (3495T), ATG5 (Cat:12994T), AMPK (5831T), P-AMPK(2535T), mTOR (2983T), P-mTOR (2974T) and GAPDH (5174S) from Cell Signaling Technologies. Blots were probed with species-appropriate horseradish peroxidase–conjugated secondary antibodies (7076P2or7074P2, Cell Signaling Technologies) at a 1:2000 dilution.
2.3. Cell proliferation detection
The cellular proliferation of gastric cancer was detected using CCK-8 assay. The transfected gastric cancer cells and controls were cultured in a 96-well plate (3000 cells per well), respectively. The cell viability was collected every 24 h using cell counting kit-8 (CCK-8) reagent (CK04, Dojindo Laboratories, Kumamoto, Japan). 10 μl CCK-8 reagent was administrated to each well. Cellular activity of the absorbance was measured at 450 nm. Triplicate assays were conducted for each group.
2.4. Cell cloning experiment
In the process of cell culture, the growth state of the cells in the 6-well plate was observed frequently and the drug was changed every 3 days. Discard the old medium in the 6-well plate and wash it twice with PBS solution. In each well, 1 ml of 4 % paraformaldehyde solution was added, 1 ml of crystal violet solution was added, and the dyeing solution was discarded after 10 min. For counting and statistics, the excess dye was gently washed with PBS solution and dried at room temperature.
2.5. Cell wound scratch assay
Make three horizontal lines behind each hole on the 6-hole plate using a marker, under the condition of a ruler ratio. The cell density was adjusted to 1 × 106/ml using complete medium after collecting cancer cells in good growth state. After 24 h of cell growth, the scratch healing was observed with an inverted microscope and photographed.
2.6. Immunofluorescence assay
Incubate gastric cancer cells at 4 °C overnight in the wet box. The cell crawling tablets were removed and placed on a shaker set at 80 g/min, washed with PBS solution for 5 min and cleaned 3 times. Add 15 μl of diluted secondary antibody to a flat bottom box with a pre-lined sealing film at the bottom, incubate in the wet box at room temperature for 1 h. Afterwards, the slipper was cleaned with PBS solution three times, placed on a shaker and removed again. Drop the anti-fluorescence quenched sealing sheet containing DAPI on the slide after it has been labeled, and then take photos in a dark room using a positive fluorescence microscope.
2.7. Western blotting
Cell lysates were collected in RIPA buffer [50 mmol/L TRIS-HCL pH 7.4, 50 mmol/L NaCl, 2 mmol/L EDTA, 0.1 % SDS supplemented with protease inhibitor (Complete EDTA-Free, Roche)] and phosphatase inhibitor (Phosphatase Inhibitor Cocktail Sets 1 and 2, Millipore). Equal amount of lysate (18 μg) was boiled in SDS protein loading buffer supplemented with 10 mmol/L DTT, separated by SDS-PAGE, transferred to Immobilon-FL PVDF membrane (Millipore, Billerica), and blotted for indicated protein.
2.8. ATP assay
Clean the cells twice with cold PBS solution, remove the old culture medium from the culture bottle, and add 500 ml of cell lysate. Blowing repeatedly with a sterile pipette will fully contact the lysate and lyse the cells. Sterilized cell scrapers were used to scrape off cracked cells, and the liquid was transferred to a cooled EP tube at 4 °C at 12,000 rpm/min. The supernatant was taken into a new cooled EP tube after centrifugation for 10 min. To prevent degradation, store samples on ice for a short time before testing. Follow the directions for preparing the standard for the concentration gradient. The reagent was melted on ice, then diluted 1:9 with reagent diluent, then stored on ice. Set three multiple holes for each sample on a black light-proof 96-well plate. Place 100 μl of ATP detection solution in the detection hole and leave at room temperature for 3 min. Using a multifunctional enzyme marker with luminometer function, mix 20 μl of sample or standard product into each detection hole. Based on the standard curve, ATP concentration in the sample is calculated.
2.9. Reverse Transcription qPCR
RNA is extracted according to the instructions provided with the reagent. Mix 80 μl chloroform thoroughly, then place it in a refrigerator at 4 °C for 10 min. Transfer the supernatant to a cold, enzyme-free EP tube with a pipette after centrifuging at 12,000 rpm/min at 4 °C for 15 min. The concentration of each group of RNA was adjusted to about 500 ng/μl, and then the system of 20 μl mixture was prepared in the enzyme-free EP8 tube. Among them, 2 μl template RNA + 4 μL 4×gDNA wiper Mix + 4 μL 5 × qRT SuperMixⅡ+ 10 μl RNase free ddH2O. Note that the reagent should be thoroughly mixed when adding. Prepare each cDNA template and the upper and lower primers of the target genes.Primer sequence of each target gene:Gene name Upstream primer Downstream primer:
GAPDH:GGAAGCTTGTCATCAATGGAAATC TGATGACCCTTTTGGCTCCC.
LC3: CCTCAGACCGGCCTTTCAA CAGCTGCTTCTCACCCTTGTAG.
Beclin1:GGCACAATCAATAACTTCAGGCCCGTAAGGAACAAGTCGGTATCTC.
ATG5: GCACACCACTGAAATGGCAT CGGGTAGCTCAGATGTTCACTC.
P62: GGAAGGTGAAACACGGACACTT CTCTTCTCCTCTGTGCTGGAACT.
AMPK:GGCAGAAGTATGTAGAGCAATCAAA GCAGTCCCTGATTTGGCTTCT
mTOR: AATGCTGTCCCTGGTCCTTATG GCGAACAAATTGGGTCAGAGA.
Each sample was set with 3 multiple holes, and the results were analyzed. ΔCt = [Ct (target gene) -Ct (GAPDH reference gene)]; ΔΔCt = [ΔCt (experimental cells) -ΔCt (control cells)], 2-ΔΔCt was calculated as the difference multiple of the target gene in the experimental group and the control group.
2.10. Xenograft nude mice model
The Xenograft mice experiment was approved by the Committee on Animal Northern Subei People's Hospital. All procedures were performed in accordance with the ethical standards following the licensing guidelines. Male BALB/c nude mice (4–6 weeks) were provided by Nanjing Medical University (Nangjing, China) and maintained in specific pathogen free condition. A sufficient amount of MKN1 or AGS gastric cancer cells were harvested and re-suspended in PBS solution, and 5 × 106 cells were injected subcutaneously into each nude mouse. When the tumor size of 100–200 mm3, according to the initial tumor volume of nude mice were randomly divided into 4 groups (control, trametinib, palbocicilb, combined group), four nude mice in each group. Depending on the group, vehicle, trametinib (3 mg/kg), and/or palbociclib (150 mg/kg) were administered orally 4 days a week, followed by discontinuation for 3 days. The nude mice were killed 21 days later, and the tumor tissue of the nude mice was harvested.
2.11. Statistical analysis
Statistical analyses were performed with SPSS 20.0 software (SPSS, Chicago, IL, USA). Data was analyzed with unpaired Student's t-test. P-value <0.05 was considered statistically significant.
3. Result
3.1. Targeting CDK4/6 and MEK synergistically inhibits the proliferation of KRAS mutant gastric cancer cells
In our experiments, a CDK4/6 targeting drug was employed in combination with trametinib, a MEK targeting drug, which is sensitive to KRAS-mutated cancer cells [[9], [10], [11], [12]], for the treatment of AGS and SNU1, two strains of gastric cancer cells with KRAS mutations. The IC50 concentration values are presented in Table 1.
Table 1.
The combined index value (CI) of palbociclib and trametinib targeting KRAS-mutated gastric cancer cells.
| Cancer cells | palbociclib (nM) | trametinib(nM) | CI Value |
|---|---|---|---|
| AGS | 5000 | 2 | 0.2885 |
| SNU1 | 100 | 20 | 0.4559 |
CI Value: The combined index value (CI) of two drugs on cells calculated by CompuSyn software. If CI < 1, it indicates that the combined use of the two drugs has a synergistic inhibitory effect on cells, and the smaller of the CI value, the more significant of the combined inhibitory effect.
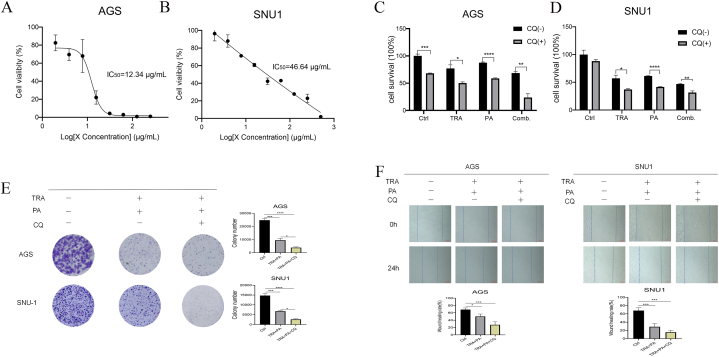
In this study, we investigated the possibility of inducing autophagy in gastric cancer cells with KRAS mutations by targeting CDK4/6 (palbociclib) and MEK (trametinib), thereby offering a new perspective for the treatment of KRAS-mutated gastric cancers. Our findings demonstrate that the combined administration of palbociclib and trametinib effectively suppressed the colony formation of KRAS-mutated gastric cancer cells, surpassing the efficacy of individual drugs or control groups (Fig. 1A and B). Moreover, the combination of palbociclib and trametinib demonstrated a significant inhibitory effect on the proliferation and wound healing rate of gastric cancer cells with KRAS mutations (Fig. 1C and D).
Fig. 1.
Combination of palbociclib and trametinib significantly inhibited the proliferation and healing rate of KRAS-mutated gastric cancer cells.
(A) AGS and SNU1 cells were divided into control group (Ctrl), trametinib group (TRA), palbociclib group (PA) and trametinib + palbociclib group Fig. 4E (Comb), respectively. After 14 days of drug treatment, clone formation was observed. (B) Combination of palbociclib and trametinib treatment significantly inhibited the proliferation and healing rate of KRAS-mutated AGS and SNU1 gastric cancer cells analyzed in the histogram. (C) The scratch healing rates of AGS and SNU1 cells treated with trametinib, palbociclib and trametinib + palbociclib were compared with controls before treatment. (D) The scratch healing rates revealed that combination of palbociclib and trametinib treatment significantly inhibited the proliferation in the KRAS-mutated AGS and SNU1 gastric cancer cells analyzed in the histogram. Scale bar: 100 μm; Data are expressed as the mean ± SD of three independent experiments. *, P < 0.05; ****, P < 0.0001.
3.2. The concurrent inhibition of CDK4/6 and MEK effectively induces autophagy in gastric cancer cells harboring KRAS mutations
Tumorigenesis and oxidative metabolism are maintained by autophagy in activated RAS [7]. However, the relationship between targeting CDK4/6 and MEK in KRAS-mutated gastric cancer and autophagy metabolic processes has not previously been examined.
In order to clarify the relationship between dual targeting of KRAS mutant gastric cancer cells and autophagy, we conducted immunofluorescence experiments and found that the green fluorescence of LC3, a marker of autophagy, was significantly higher in the dual drug group than in the single drug group or no drug group after 72 h of KRAS mutant gastric cancer cells (Fig. 2A). Western Blot (Fig. 2B and C) and RT-qPCR (Fig. 2E) showed that the expression levels of autophagy related proteins LC3II/LC3I, Beclin-1 and ATG5 and their mRNA were up-regulated in KRAS mutant gastric cancer cells after 72 h in the double-drug group compared with the single-drug group or no-drug group, while the expression levels of P62 and its mRNA (Fig. 2D and E) were down-regulated. By adding chloroquine to AGS cells for 24 h and 48 h, we found that P62 protein levels gradually increased, and we could conclude that chloroquine inhibited autophagy (Fig. 2F).
Fig. 2.
Palbociclib and trametinib synergistically induced autophagy in KRAS-mutated gastric cancer cells.
(A) Immunofluorescence staining was performed after drug treatment for 72 h in AGS gastric cancer cells. The autophagy protein LC3 in each group was observed under the fluorescence microscope with green fluorescence, and the nucleus was stained with DAPI. Scale bar: 20 μm; (B) Western Blot and RT-qPCR were used to analyze the protein and mRNA expression levels of autophagy-related proteins LC3II/LC3I, Beclin1 and ATG5 in each group after 72 h of drug treatment in AGS gastric cancer cells, respectively. (C) Western Blot and RT-qPCR were used to analyze the protein and mRNA expression levels of autophagy-related proteins LC3II/LC3I, Beclin1 and ATG5 in each group after 72 h of drug treatment in SUN1 gastric cancer cells, respectively., respectively. (D) Western Blot were used to analyze P62 protein in each group after 72 h of drug treatment in SUN1 gastric cancer cells, respectively, respectively. (E) mRNA expression levels of autophagy related proteins LC3, Beclin1, ATG5 and P62 were analyzed by RT-qPCR after 72 h of drug treatment. (F) The P62 protein level was measured by adding 6.5 μM chloroquine to AGS cells for 24 h and 48 h, respectively. Data are expressed as the mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Targeting CDK4/6 and MEK induces autophagy in KRAS- mutated gastric cancer cells through AMPK/mTOR pathway
We examined the canonical AMPK/mTOR pathway in order to characterize how the combination of drugs induces autophagy. Autophagy is triggered by insufficient cell energy production by the AMPK/mTOR pathway [13,14]. An increase in the AMP to ATP ratio in cells in turn activates the AMPK pathway [15].
Therefore, we investigated the effects of dual drug, single drug and controls on ATP levels in KRAS-mutated gastric cancer cells drug therapy for 72 h. Targeting CDK4/6 and MEK synergistically induced autophagy in KRAS-mutated gastric cancer cells through down-regulating cellular ATP levels (Fig. 3A), activating AMPK, and inhibiting the mTOR pathway (Fig. 3B–F). Our finding supported that drug inducing protective autophagy can inhibit the growth of gastric cancer with the decreased the level of p-mTOR protein through the AMPK/mTOR pathway [16].
Fig. 3.
The combination of palbociclib and trametinib induces autophagy in KRAS mutant gastric cancer cells by down-regulating cellular ATP levels, activating AMPK, and inhibiting the mTOR pathway.
(A) The level of ATP in AGS and SNU1 gastric cancer cells treated with palbociclib and trametinib for 72 h. (B) The protein expression levels of AMPK, P-AMPK, mTOR and P-mTOR in AGS gastric cancer cells of each group were detected by Western Blot analysis after 72h treatment. (C) Protein quantization map of AMPK, P-AMPK, mTOR and P-mTOR in AGS cells after 72h of drug action (D) The protein expression levels of AMPK, P-AMPK, mTOR and P-mTOR in SNU1 gastric cancer cells of each group were detected by Western Blot analysis after 72h treatment. (E) Protein quantization map of AMPK, P-AMPK, mTOR and P-mTOR in SNU1 cells after 72h of drug action. The mRNA expression levels of AMPK and mTOR were analyzed by RT-qPCR in AGS. (E)The mRNA expression levels of AMPK and mTOR were analyzed by RT-qPCR in AGS and SNU1. Data are expressed as the mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
3.4. Autophagy inhibitor (CQ) enhances the inhibitory effect of targeting CDK4/6 and MEK on the proliferation of KRAS mutant gastric cancer cells
CQ (chloroquine) was first discovered and used to treat malaria, followed by inflammatory diseases [17]. CQ inhibits autophagy flux by reducing autophagosome lysosome fusion and its derivative hydroxychloroquine are the only autophagy inhibitors approved by the US Food and Drug Administration (FDA) [18]. Therefore, we detected and calculated the IC50 (semi-inhibitory concentration) of CQ on KRAS-mutated gastric cancer cells through CCK8 assay (Fig. 4A and B). Furthermore, CQ enhanced the inhibitory effect of trametinib combined with palbociclib on the survival (Fig. 4C and D), colony formation (Fig. 4E), and wound healing rate (Fig. 4F) of KRAS-mutated gastric cancer cells. It implies that autophagy may be a protective phenomenon in KRAS-mutated gastric cancer cells exposed to trametinib combined with palbociclib treatment. Combinations of pharmacologic inhibitors that concurrently block both ERK-MAPK and autophagic processes that are upregulated in response to ERK inhibition may be effective treatments for pancreatic ductal adenocarcinoma [19].
Fig. 4.
CQ enhanced the inhibitory effect of trametinib combined with palbociclib on the proliferation of KRAS-mutated gastric cancer cells.
(A) The IC50 (half-inhibitory concentration) of CQ on cells was detected in AGS. (B) The IC50 of CQ on cells was detected in SUN1. (C) CCK8 assay was used to detect the cell survival activity of each group after 72 h of drug treatment in AGS. (D) CCK8 assay was used to detect the cell survival activity of each group after 72 h of drug treatment in SNU1. (E) AGS and SNU1 (F) were divided into blank control group, trametinib + palbociclib group and CQ + trametinib + palbociclib group, respectively, and the clones were formed after 14 days of drug treatment. Scale bar: 100 μm; Data are expressed as the mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
3.5. Targeting CDK4/6 and MEK on gastric cancer in vivo
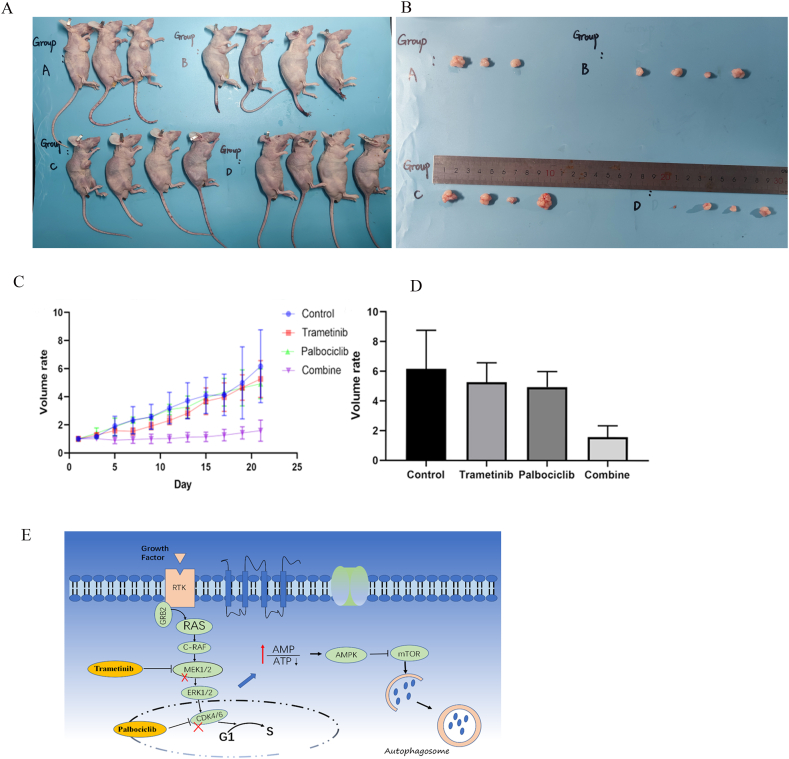
In order to further investigate the effect of dual targets on gastric cancer in vivo, we selected male BALB/c nude mice aged 4–6 weeks to inject gastric cancer cells subcutaneously to construct a gastric cancer mouse model. The nude mice were randomly divided into 4 groups (control, trametinib, palbocicilb, combined group). Control (solvent-promoting), trametinib (3 mg/kg), palbociclib (150 mg/kg) and combine (trametinib 3 mg/kg and palbociclib 150 mg/kg) were administered orally 4 days a week. The drug was then discontinued for 3 days for 3 weeks, and the weight and tumor volume of the nude mice were recorded the next day. In vivo experiment results, compared with the control group and the single administration group, the growth state of nude mice in the combined administration group was significant, and the tumor growth was slower as compared with single drug therapy and controls (P < 0.05) (Fig. 5A–D). In conclusion, the combination of MEK inhibitors and CDK4/6 inhibitors can inhibit the growth of gastric cancer in vivo.
Fig. 5.
Targeting CDK4/6 and MEK significantly inhibits the growth of gastric cancer in nude mouse in vivo. (A) Four groups of nude mice. (B) Tumors of nude mice. (C) Curve of tumor volume initial volume ratio. (D) Histogram of tumor final volume/initial volume ratio, compared with control group. *, P<0.05; **, P<0.01 (n ≥ 3). (E) Mechanistic diagram of the signaling pathway targeting CDK4/6 and MEK to induce autophagy in KRAS-mutant gastric cancer cells.
4. Discussion
Patients diagnosed with gastric cancer and harboring KRAS mutations exhibit a reduced overall survival rate [20]. According to reports, the inhibition of autophagy in cancer cells has been found to facilitate tumor regression in KRAS-driven lung cancer [21]. The proliferation of tumors is linked to metabolic abnormalities in tumor cells that rely on nutrients from the host, and these abnormalities can be altered by inhibiting autophagy in the tumor microenvironment [21]. The inclination of autophagy to facilitate the metabolic requirements of aggressive tumors, whether inherent or involuntary, has been replicated across multiple models and has unveiled discernible patterns wherein autophagy promotes the advancement of cancer [22,23]. Autophagy serves as a stress response pathway that facilitates cellular survival and the restoration of cellular homeostasis in the majority of instances [22]. The growth of solid tumors from precancerous to malignant stages is accompanied by a restriction in the availability of oxygen, glucose, and amino acids, as the nutrient supply through normal blood vessels becomes insufficient to meet the increased demands [24]. The capacity of nutrients confers a competitive advantage to tumor cells, enhancing autophagy in response to conditions of limited nutrient availability [24]. In order to counteract the growth-inhibitory consequences of cancer-related stresses, the upregulation and activation of autophagy frequently occur in cancer. Additionally, autophagy assumes a pivotal function in facilitating tumor growth and proliferation [[21], [22], [23]].
The simultaneous administration of CDK4/6 inhibitors and ERK inhibitors exhibits a synergistic effect in inhibiting the proliferation of pancreatic cancer cell lines and organoids. These cell lines and organoids are characterized by a high concentration of various signaling nodes, such as cell-cycle regulatory proteins primarily associated with CDK2 activation and pathways that induce autophagy [6]. To enhance the efficacy of drug regimens for gastric cancer treatment, our study aimed to investigate the mechanisms underlying the induction of autophagy in KRAS-mutated gastric cancer through the targeting of CDK4/6 and MEK. Previous research has demonstrated that cancer cells can up-regulate autophagy as a survival mechanism in response to chemotherapy [25].
The inhibition of autophagy as a means to enhance the susceptibility of cancer cells to targeted therapies has been substantiated, while the induction of autophagy is frequently observed in pancreatic cancer due to the prevalent activation of KRAS signaling [14]. Autophagy is activated through pharmacological or genetic inhibition of RAS-ERK signaling in cancers driven by KRAS [14,26,27]. The activation of autophagosome formation and the translocation of ubiquitin proteins to autophagy vesicles are dependent on the crucial involvement of autophagy-associated proteins LC3II/LCI, Beclin-1, ATG5, and P62 [21,23,26]. P62, a versatile adaptor protein, serves as a discerning substrate for autophagy, undergoing self-degradation upon the delivery of ubiquitinated proteins to autophagosomes [14]. The findings from our research provide unequivocal evidence that targeting CDK4/6 and MEK effectively triggers autophagy in KRAS-mutated gastric cancer. Furthermore, our in vivo mouse model demonstrates a significant reduction in tumor growth upon targeting CDK4/6 and MEK.
Subsequently, our investigation aimed to elucidate the impact of CDK4/6 and MEK targeting on autophagy in KRAS-mutated gastric cancer cells by exploring the associated pathways. Prior studies have established the responsiveness of the AMPK/mTOR pathway to inadequate cellular energy production and autophagy [17,25]. AMPK is a cellular energy state sensor that is highly conserved and becomes activated in instances of low intracellular ATP levels. Activation of AMPK induces energy stress by impeding cell growth and biosynthesis processes, partially through the inhibition of the rapamycin sensitive mTOR pathway [17,25]. Intriguingly, our study revealed a noteworthy decrease in ATP levels within KRAS-mutated gastric cancer cells following a 72-h exposure to the combined administration of the two drugs, in contrast to the single drug or control groups. This reduction in ATP levels led to an augmented AMP to ATP ratio within the cells, consequently activating the AMPK pathway and inhibiting mTOR through the phosphorylation of two distinct substrate proteins. Ultimately, this cascade of events induced autophagy [21,26]. The study revealed a significant increase in the expression levels of P-AMPK and mTOR, alongside a decrease in the expression levels of P-mTOR and AMPK. These findings indicate that the activation of autophagy can be achieved by targeting CDK4/6 and MEK, which subsequently reduces cellular energy metabolism. Consequently, this activation is facilitated through AMPK phosphorylation and the inhibition of mTOR phosphorylation. In this study, we opted for gastric cancer cell lines and animal models to replicate the effects of anti-autophagy, MEK, and CDK4/6 treatment on KRAS mutant gastric cancer, both in vitro and in vivo. This approach successfully altered the metabolic profile of the gastric cancer microenvironment, suppressed cellular proliferation, and resulted in a reduction in tumor size. These findings hold promising implications for potential clinical therapeutic interventions.
Despite the limited efficacy of KRAS inhibition as a standalone approach in the treatment of pancreatic cancer, the concurrent inhibition of autophagy binding to MAPK or ERK1/2, which are downstream targets of RAS, has been found to synergistically diminish tumor formation [11,[27], [28], [29]]. Additionally, RAS activation also plays a role in the activation of BRAF, a gene commonly mutated in melanoma, central nervous system tumors, and colorectal cancer [30,31]. Cancers subjected to treatment with the BRAF inhibitor vemurafenib frequently exhibit resistance towards chemotherapy, concomitant with elevated levels of autophagy [30]. The inhibition of autophagy by CQ has been reported to restore the sensitivity of tumors to vemurafenib or other inhibitors that specifically target the downstream kinases of BRAF [32]. It is hypothesized that the inhibition of autophagy by CQ may render KRAS-mutated gastric cancer cells susceptible to the regulation of the cell cycle and the MAPK signaling pathway. Consequently, a CCK8 assay was employed to determine the IC50 of CQ against KRAS-mutated gastric cancer cells. The experimental findings indicate that the targeting of CDK4/6 and MEK can induce autophagy in KRAS-mutated gastric cancer cells, and the inhibition of autophagy can further augment the inhibitory efficacy of the combined drug treatment on KRAS-mutated gastric cancer cells.
To the best of our knowledge, we present novel findings indicating that the simultaneous inhibition of autophagy, MEK, and CDK4/6 can effectively impede various metabolic processes and suppress the proliferation of gastric cancer. These results collectively imply that the integration of autophagy inhibitors with targeted chemotherapy holds promise for the treatment of KRAS-mutated gastric cancer exhibiting an "autophagy addicted" phenotype. Nevertheless, it is imperative to validate these outcomes through real-world clinical trials involving both KRAS-mutated and KRAS-non-mutated gastric cancer patients.
Ethics approval
The research was approved by the Clinical Research Ethics Committee in Northern Jiangsu People's Hospital (Yangzhou, China, approval no. 2021ky075).
Funding
This work was supported by grants from the National Natural Science Foundation of China (82,172,345, 81,573,220), Natural Science Foundation of Jiangsu Province (BK20221281), Foundation of Yangzhou Science and Technology Planning(YZ2020076). Crosscooperation project of Subei Peoples' Hospital of Jiangsu Province (SBJC22003).
Data availability statement
Data will be made available on request.
CRediT authorship contribution statement
Hong Zhou: Investigation, Formal analysis, Data curation, Conceptualization. Guiling Li: Writing – review & editing, Visualization, Validation, Data curation, Conceptualization. Liuyue Kan: Writing – review & editing, Methodology, Investigation, Data curation. Mingyu Yang: Writing – review & editing, Visualization, Validation, Data curation, Conceptualization. Yu Liu: Validation, Resources, Methodology, Investigation, Formal analysis, Data curation. Xiaye Miu: Validation, Resources. Lei Shi: Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Zhanjun Yang: Writing – review & editing, Validation, Resources, Data curation, Conceptualization. Xucai Zheng: Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Hui Chen: Project administration, Methodology, Investigation, Data curation. Chuanli Ren: Writing – review & editing, Writing – original draft, Visualization, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Zhanjun Yang, Email: zjyang@yzu.edu.cn.
Xucai Zheng, Email: ahszlyyzxc@163.com.
Hui Chen, Email: Chenhuimd@foxmail.com.
Chuanli Ren, Email: renchl@163.com.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer. J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Moore A.R., Rosenberg S.C., McCormick F., Malek S. RAS-targeted therapies: is the undruggable drugged? Nat. Rev. Drug Discov. 2020;19(8):533–552. doi: 10.1038/s41573-020-0068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samatar A.A., Poulikakos P.I. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat. Rev. Drug Discov. 2014;13(12):928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 4.Kwong L.N., Costello J.C., Liu H., Jiang S., Helms T.L., Langsdorf A.E., Jakubosky D., Genovese G., Muller F.L., Jeong J.H., Bender R.P., Chu G.C., Flaherty K.T., Wargo J.A., Collins J.J., Chin L. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat. Med. 2012;18(10):1503–1510. doi: 10.1038/nm.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong G.S., Zhou J., Liu J.B., Wu Z., Xu X., Li T., Xu D., Schumacher S.E., Puschhof J., McFarland J., Zou C., Dulak A., Henderson L., Xu P., O'Day E., Rendak R., Liao W.L., Cecchi F., Hembrough T., Schwartz S., Szeto C., Rustgi A.K., Wong K.K., Diehl J.A., Jensen K., Graziano F., Ruzzo A., Fereshetian S., Mertins P., Carr S.A., Beroukhim R., Nakamura K., Oki E., Watanabe M., Baba H., Imamura Y., Catenacci D., Bass A.J. Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat. Med. 2018;24(7):968–977. doi: 10.1038/s41591-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodwin C.M., Waters A.M., Klomp J.E., Javaid S., Bryant K.L., Stalnecker C.A., Drizyte-Miller K., Papke B., Yang R., Amparo A.M., Ozkan-Dagliyan I., Baldelli E., Calvert V., Pierobon M., Sorrentino J.A., Beelen A.P., Bublitz N., Lüthen M., Wood K.C., Petricoin E.F., Sers C., McRee A.J., Cox A.D., Der C.J. Combination therapies with CDK4/6 inhibitors to treat KRAS-mutant pancreatic cancer. Cancer Res. 2023;83(1):141–157. doi: 10.1158/0008-5472.CAN-22-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo J.Y., Chen H.Y., Mathew R., Fan J., Strohecker A.M., Karsli-Uzunbas G., Kamphorst J.J., Chen G., Lemons J.M., Karantza V., Coller H.A., Dipaola R.S., Gelinas C., Rabinowitz J.D., White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis, Genes. Dev. 2011;25(5):460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorokin A.V., Kanikarla Marie P., Bitner L., Syed M., Woods M., Manyam G., Kwong L.N., Johnson B., Morris V.K., Jones P., Menter D.G., Lee M.S., Kopetz S. Targeting RAS mutant colorectal cancer with dual inhibition of MEK and CDK4/6. Cancer Res. 2022;82(18):3335–3344. doi: 10.1158/0008-5472.CAN-22-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmann M.H., Gmachl M., Ramharter J., Savarese F., Gerlach D., Marszalek J.R., Sanderson M.P., Kessler D., Trapani F., Arnhof H., Rumpel K., Botesteanu D.A., Ettmayer P., Gerstberger T., Kofink C., Wunberg T., Zoephel A., Fu S.C., Teh J.L., Böttcher J., Pototschnig N., Schachinger F., Schipany K., Lieb S., Vellano C.P., O'Connell J.C., Mendes R.L., Moll J., Petronczki M., Heffernan T.P., Pearson M., McConnell D.B., Kraut N. BI-3406, a potent and selective SOS1-KRAS Interaction inhibitor, is effective in KRAS-driven cancers through combined MEK inhibition. Cancer Discov. 2021;11(1):142–157. doi: 10.1158/2159-8290.CD-20-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koga T., Suda K., Fujino T., Ohara S., Hamada A., Nishino M., Chiba M., Shimoji M., Takemoto T., Arita T., Gmachl M., Hofmann M.H., Soh J., Mitsudomi T. KRAS secondary mutations that confer Acquired resistance to KRAS G12C inhibitors, Sotorasib and Adagrasib, and overcoming strategies: insights from in vitro experiments. J. Thorac. Oncol. 2021;16(8):1321–1332. doi: 10.1016/j.jtho.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Kinsey C.G., Camolotto S.A., Boespflug A.M., Guillen K.P., Foth M., Truong A., Schuman S.S., Shea J.E., Seipp M.T., Yap J.T., Burrell L.D., Lum D.H., Whisenant J.R., Gilcrease G.W., 3rd, Cavalieri C.C., Rehbein K.M., Cutler S.L., Affolter K.E., Welm A.L., Welm B.E., Scaife C.L., Snyder E.L., McMahon M. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 2019;25(4):620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falcomatà C., Bärthel S., Widholz S.A., Schneeweis C., Montero J.J., Toska A., Mir J., Kaltenbacher T., Heetmeyer J., Swietlik J.J., Cheng J.Y., Teodorescu B., Reichert O., Schmitt C., Grabichler K., Coluccio A., Boniolo F., Veltkamp C., Zukowska M., Vargas A.A., Paik W.H., Jesinghaus M., Steiger K., Maresch R., Öllinger R., Ammon T., Baranov O., Robles M.S., Rechenberger J., Kuster B., Meissner F., Reichert M., Flossdorf M., Rad R., Schmidt-Supprian M., Schneider G., Saur D. Selective multi-kinase inhibition sensitizes mesenchymal pancreatic cancer to immune checkpoint blockade by remodeling the tumor microenvironment. Nat. Cancer. 2022;3(3):318–336. doi: 10.1038/s43018-021-00326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alers S., Löffler A.S., Wesselborg S., Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell Biol. 2012;32(1):2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y.C., Guan K.L. mTOR: a pharmacologic target for autophagy regulation. J. Clin. Invest. 2015;125(1):25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mihaylova M.M., Shaw R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011;13(9):1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rong L., Li Z., Leng X., Li H., Ma Y., Chen Y., Song F. Salidroside induces apoptosis and protective autophagy in human gastric cancer AGS cells through the PI3K/Akt/mTOR pathway. Biomed. Pharmacother. 2020;122 doi: 10.1016/j.biopha.2019.109726. [DOI] [PubMed] [Google Scholar]
- 17.Al-Bari M.A. Chloroquine analogues in drug discovery: new directions of uses, mechanisms of actions and toxic manifestations from malaria to multifarious diseases. J. Antimicrob. Chemother. 2015;70(6):1608–1621. doi: 10.1093/jac/dkv018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manic G., Obrist F., Kroemer G., Vitale I., Galluzzi L. Chloroquine and hydroxychloroquine for cancer therapy. Mol. Cell. Oncol. 2014;1(1) doi: 10.4161/mco.29911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryant K.L., Stalnecker C.A., Zeitouni D., Klomp J.E., Peng S., Tikunov A.P., Gunda V., Pierobon M., Waters A.M., George S.D., Tomar G., Papke B., Hobbs G.A., Yan L., Hayes T.K., Diehl J.N., Goode G.D., Chaika N.V., Wang Y., Zhang G.F., Witkiewicz A.K., Knudsen E.S., Petricoin E.F., 3rd, Singh P.K., Macdonald J.M., Tran N.L., Lyssiotis C.A., Ying H., Kimmelman A.C., Cox A.D., Der C.J. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 2019;25(4):628–640. doi: 10.1038/s41591-019-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu X.H., Chen Z.T., Wang W.H., Fan X.J., Huang Y., Wu X.B., Huang J.L., Wang J.X., Lin H.J., Tan X.L., Wang L., Wang J.P. KRAS G12V mutation is an adverse prognostic factor of Chinese gastric cancer patients. J. Cancer. 2019;10(4):821–828. doi: 10.7150/jca.27899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karsli-Uzunbas G., Guo J.Y., Price S., Teng X., Laddha S.V., Khor S., Kalaany N.Y., Jacks T., Chan C.S., Rabinowitz J.D., White E. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4(8):914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amaravadi R.K., Kimmelman A.C., Debnath J. Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. 2019;9(9):1167–1181. doi: 10.1158/2159-8290.CD-19-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sui X., Chen R., Wang Z., Huang Z., Kong N., Zhang M., Han W., Lou F., Yang J., Zhang Q., Wang X., He C., Pan H. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4(10):e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker T.M., Henriques V., Beltran A., Nakshatri H., Gogna R. Cell competition and tumor heterogeneity. Semin. Cancer Biol. 2020;63:1–10. doi: 10.1016/j.semcancer.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Gwinn D.M., Shackelford D.B., Egan D.F., Mihaylova M.M., Mery A., Vasquez D.S., Turk B.E., Shaw R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., He S., Ma B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer. 2020;19(1):12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Infante J.R., Somer B.G., Park J.O., Li C.P., Scheulen M.E., Kasubhai S.M., Oh D.Y., Liu Y., Redhu S., Steplewski K., Le N. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur. J. Cancer. 2014;50(12):2072–2081. doi: 10.1016/j.ejca.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 28.Lee C.S., Lee L.C., Yuan T.L., Chakka S., Fellmann C., Lowe S.W., Caplen N.J., McCormick F., Luo J. MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival. Proc. Natl. Acad. Sci. U S A. 2019;116(10):4508–4517. doi: 10.1073/pnas.1817494116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma X.H., Piao S.F., Dey S., McAfee Q., Karakousis G., Villanueva J., Hart L.S., Levi S., Hu J., Zhang G., Lazova R., Klump V., Pawelek J.M., Xu X., Xu W., Schuchter L.M., Davies M.A., Herlyn M., Winkler J., Koumenis C., Amaravadi R.K. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J. Clin. Invest. 2014;124(3):1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulcahy Levy J.M., Zahedi S., Griesinger A.M., Morin A., Davies K.D., Aisner D.L., Kleinschmidt-DeMasters B.K., Fitzwalter B.E., Goodall M.L., Thorburn J., Amani V., Donson A.M., Birks D.K., Mirsky D.M., Hankinson T.C., Handler M.H., Green A.L., Vibhakar R., Foreman N.K., Thorburn A. Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. Elife. 2017;6 doi: 10.7554/eLife.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Awad M.M., Liu S., Rybkin I.I., Arbour K.C., Dilly J., Zhu V.W., Johnson M.L., Heist R.S., Patil T., Riely G.J., Jacobson J.O., Yang X., Persky N.S., Root D.E., Lowder K.E., Feng H., Zhang S.S., Haigis K.M., Hung Y.P., Sholl L.M., Wolpin B.M., Wiese J., Christiansen J., Lee J., Schrock A.B., Lim L.P., Garg K., Li M., Engstrom L.D., Waters L., Lawson J.D., Olson P., Lito P., Ou S.I., Christensen J.G., Jänne P.A., Aguirre A.J. Acquired resistance to KRAS(G12C) inhibition in cancer. N. Engl. J. Med. 2021;384(4):2382–2393. doi: 10.1056/NEJMoa2105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodall M.L., Wang T., Martin K.R., Kortus M.G., Kauffman A.L., Trent J.M., Gately S., MacKeigan J.P. Development of potent autophagy inhibitors that sensitize oncogenic BRAF V600E mutant melanoma tumor cells to vemurafenib. Autophagy. 2014;10(6):1120–1136. doi: 10.4161/auto.28594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.