Abstract
Effects of d(CAG)n·d(CTG)n repeats on expression of a reporter gene in human cell culture were studied using transient transfection, RNase protection and coupled transcription/translation assays. Cloning these repeats into the reporter 3′-UTR did not affect gene functioning. In contrast, placing the repeats in the reporter 5′-UTR led to strong inhibition of expression. This inhibition depended on the repeat orientation, being prominent only when the (CTG)n tracts were in the sense strand for transcription. Further, the strength of inhibition increased exponentially with an increase in repeat length. Our data indicate that expanded (CTG)n repeats prevent efficient translation of the reporter mRNA both in vitro and in vivo. We suggest that formation of stable hairpins by (CUG)n runs of increasing length in the 5′-UTR of a mRNA progressively inhibits the scanning step of translation initiation. This points to a novel mechanism of regulating gene expression by expandable d(CTG)n repeats.
INTRODUCTION
Simple DNA repeats are enormously over-represented in eukaryotic genomes (1,2). The length polymorphism of these repeats might affect the pattern of gene expression, as was most vividly demostrated for trinucleotide repeats (reviewed in 3). Specific trinucleotide repeats, (CGG)n·(CCG)n, (CTG)n·(CAG)n and (GAA)n·(TCC)n, have attracted wide attention due to their role in several human hereditary neurological disorders (reviewed in 4). These repeats have been detected within at least a dozen human genes. The number of repeats at each of these loci is polymorphic but relatively small in the normal human population (n ≤ 30). If the number of a given repeated unit exceeds ∼30, the length of this repeat can be expanded during intergenerational transmission. Remarkably, both the scale and likelihood of expansion increase with length of the repeat. While the mechanisms of expansion are still unknown, most hypotheses involve mispairing during DNA replication or recombination (reviewed in 5–7).
Expanded trinucleotide repeats disturb the expression and/or function of the corresponding gene products. It appears that different repeats can affect gene expression at various levels. In the case of Fragile X syndrome, caused by expansion of a CGG repeat in the 5′-UTR of the FMR1 gene (8–10), expansion induces de novo methylation which spreads, leading to heterochromatinization of the FMR1 gene and adjacent DNA (11,12). Expansion of CAG repeats situated in the coding regions of various human genes is linked to Huntington disease (13), spinal and bulbar muscular atrophy (Kennedy disease) (14), spinocerebral ataxia (15,16) and dentatorubral pallidoluysian atrophy (17). This expansion does not seem to affect transcription or translation of the corresponding genes, but repeat-encoded polyglutamine stretches in the protein products lead to their aggregation (reviewed in 18,19). Friedreich’s ataxia is caused by expansion of a GAA repeat within the first intron of the frataxin gene (20). Amplified (GAA)n repeats block transcription of the host gene, presumably due to triplex formation (21,22). Myotonic dystrophy is caused by an expansion of the CTG stretch located in the 3′-UTR of the myotonic dystrophy protein kinase (DMPK) gene (23,24). This expansion alters the ability of the DMPK primary transcript to be processed into mature mRNA (25), decreases transcription of the adjacent SIX5/DMAHP gene (26,27) and blocks splicing of several non-related RNAs (28).
Trinucleotide repeats are common elements of the human genome (29). It is becoming increasingly clear that they are present in numerous genes that are not currently implicated in human diseases and in positions differing from those described above. To give just one example, 12 and 8 (CTG)n repeats, respectively, were found in the 5′-UTRs of the human SHMT gene, encoding cytosolic serine hydroxymethyltransferase (30), and the BPGM gene, encoding erythrocyte 2,3-biphosphoglycerate mutase (31). This warrants studies of the effects of different trinucleotide repeats located in various gene segments on gene functioning. Here we describe the results of such studies on the effects of (CTG)n repeats on reporter gene expression in human cells. Contrary to what one might expect based on the myotonic dystrophy case, we observed a drastic decrease in reporter expression when those repeats were situated in the 5′-UTR, rather than in the 3′-UTR, of our reporter. The extent of inhibition increased exponentially with increasing repeat length. Expanded (CTG)n repeats appear to interfere with translation, rather than transcription, of the reporter gene. We suggest that formation of a stable hairpin by the (CUG)n repeat in the 5′-UTR of the luciferase mRNA might be responsible for this effect.
MATERIALS AND METHODS
Plasmids
d(CAG)n·d(CTG)n repeats cloned into the pcDNA3 vector (Invitrogen) have been described by Richard et al. (32). To obtain constructs with these repeats in our reporter 5′-UTR, NotI–PstI fragments from the pcDNA3 derivatives were recloned into a modified pGL2-Promoter vector (Promega) containing a NotI–PstI linker in two orientations at the HindIII site. As a result, we obtained sets of plasmids containing either (CTG)n runs or (CAG)n runs in the sense strand of the luciferase 5′-UTR. Subsequent sequencing revealed that (CAG)n-containing plasmids also had an extra AUG codon upstream of the trinucleotide repeat and the luciferase translation start codon. To avoid potential artifacts, this extra AUG codon was removed by Bal31 exonuclease digestion.
To obtain constructs with d(CAG)n·d(CTG)n repeats in the reporter 3′-UTR, NotI–PstI fragments excised from the pcDNA3 derivatives were blunt-ended and recloned into the PflMI site of the pGL2-Promoter vector.
Constructs for in vitro translation were made from the pGL2 derivatives containing (CAG)n·(CTG)n repeats in the luciferase 5′-UTR by replacing the SV40 promoter between XhoI and SfiI sites with a synthetic sequence corresponding to the T7 promoter (33). As a result, T7 polymerase generated 5′-UTRs of the same length and structure as in the original SV40-derived constructs. To generate RNA probes for RNase protection assays, the EcoRV–AvaI fragment of the firefly luciferase gene or the Bsp14071–BpiI fragment of the Renilla luciferase gene were cloned in the antisense orientation into the EcoRV site of the pBluescript vector downstream of the T7 promoter.
To generate plasmids without a luciferase start codon, NotI–NarI fragments from the pGL2 derivatives carrying (CTG)n repeats in their 5′-UTRs were replaced with synthetic DNA sequences lacking the AUG codon but otherwise identical to the original fragments. AUG– plasmids with out-of-frame (CUG)n repeats were then generated by NotI digestion, end-filling and religation of the AUG– plasmids. Plasmids with a translation stop codon upstream of the luciferase AUG codon were obtained by replacing NotI–NarI fragments from the corresponding (CTG)n-containing plasmids with a synthetic DNA sequence carrying a TGA triplet 30 bp downstream from the repeat.
Transient transfection assay
The SW480 colon carcinoma cell line was used for the transient transfection assay. Cells were grown on Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal calf serum (Invitrogen). A day before transfection, cells were split into 6-well (20 mm) plates to give a confluency of ∼40%. DMRCI transfection reagent (Gibco) was used for transfection according to the manufacturer’s recommendations. Specifically, we co-transfected 2 µg of each reporter construct (a control pGL2 plasmid encoding firefly luciferase and its repeat-containing derivatives) with 0.2 µg of a reference plasmid (pRL, encoding Renilla luciferase) per plate. Repeat-containing plasmids were sequenced prior to transfection in order to confirm repeat lengths and lack of interruptions. After 48 h incubation cells were lysed and enzymatic activities of both firefly and Renilla luciferase were measured by luminometer, using substrates and procedures from the Dual Luciferase Assay Kit (Promega). The values for firefly luciferase activity for every reporter construct were normalized to the corresponding values of Renilla luciferase activity to account for varying transfection efficiency. Relative expression values for different repeat-containing plasmids were obtained by comparing their normalized luciferase activities with those for the control pGL2 plasmid. For the majority of our constructs transfections were repeated at least three times with at least two independently isolated DNA samples.
For RNA isolation, cells were grown on 100 mm plates. Transfections were carried out as described above, with 10 µg firefly and 10 µg Renilla DNA (total of 20 µg DNA/plate).
RNase protection assay
Total RNA was isolated from ∼107 cells using Trizol reagent (Gibco). To eliminate plasmid DNA used for transfection, each RNA sample was treated with DNase I (1–2 U/µg DNA) (Ambion) followed by extraction with phenol/chloroform. Probes for RNase protection were generated using the Riboprobe Combination System–T3/T7 (Promega) in the presence of [32P]CTP (800 Ci/mmol; Amersham). pBluescript derivatives containing antisense fragments of the firefly and Renilla genes were linearized with EcoRI for in vitro transcription with T7 RNA polymerase. This gave 280 and 150 nt long riboprobes for the firefly and Renilla luciferases, respectively. In addition to luciferase sequences these riboprobes contained ∼60 nt from the pBluescript vector. Radiolabeled probes were separated in a 5% denaturing polyacrylamide gel followed by elution using Ambion elution buffer. RNase protection was performed using the RPII kit (Ambion) as recommended by the manufacturer. To normalize for transfection efficiency, we first quantitated Renilla mRNA and adjusted the amounts of RNA used for hybridization with the firefly probe accordingly. Between 20 and 30 µg total RNA and ∼300–400 pg (4 × 104 c.p.m.) riboprobes were typically used for hybridization. Upon hybridization and RNase digestion, samples were fractionated in 8% denaturing polyacrylamide gel. Gels were dried and the amounts of protected fragments were estimated using a 445 SI PhosphorImager (Molecular Dynamics). Hybrydization with yeast RNA was used to control for specificity of the probe.
In vitro translation
In vitro translation experiments were performed using the TnT Quick Coupled Transcription/Translation System (Promega). An aliquot of 2 µg plasmid template DNA was used for each reaction. Proteins were labeled using [35S]methionine (1000 Ci/mmol, 10 mCi/ml; Amersham). Products were separated by PAGE in Bio-Rad precast 4–20% Tris–glycine polyacrylamide gels. Upon fixation, gels were dried under vacuum in a conventional gel drier and analyzed with a phosphorimager. The reaction was repeated at least three times with each construct.
RESULTS
(CAG)n·(CTG)n repeats cause length- and orientation-dependent inhibition of reporter gene expression in vivo
To study the effects of (CTG)n·(CAG)n repeats on reporter gene expression in vivo, the repeats were inserted into different positions of the test plasmid pGL2-Promoter (Promega) (Fig. 1A) containing the firefly luciferase gene under control of the SV40 early promoter. Cloning into the HindIII site positioned these repeats within the 5′-UTR, 53 bp downstream of a transcription start site and 29 bp upstream of the initiator AUG codon. Cloning into the PflM1 site placed repeats within the 3′-UTR, 65 bp downstream of the translation termination codon and ∼500 bp upstream of a polyadenylation signal. The repeats of varying lengths (35 ≤ n ≤ 160) were cloned into these positions in both orientations, leading to the appearance of either (CTG)n or (CAG)n runs in the sense strand (the plasmids were named accordingly).
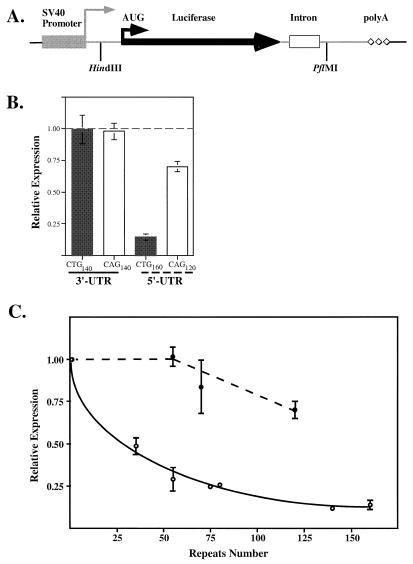
Figure 1.
Effects of (CTG)n·(CAG)n repeats on reporter gene expression in human SW480 cells. (A) Schematic representation of the luciferase reporter gene with its regulatory regions. (B) Effects of the longest (CTG)n·(CAG)n repeats placed in the 3′- or 5′-UTRs of the reporter. Expression levels are relative to the control pGL2 plasmid. The dashed line represents the control level of expression. (C) (CTG)n·(CAG)n repeats in the reporter 5′-UTR inhibit its expression in a length- and orientation-dependent manner. All expression values are relative to the control pGL2 plasmid. Closed circles, (CAG)n repeats in the sense strand; open circles, (CTG)n repeats in the sense strand.
Each of the repeat-containing plasmids as well as the control pGL2 plasmid were co-transfected into SW480 human colon carcinoma cells together with the reference plasmid pRL. The latter contains the Renilla luciferase gene under control of the same SV40 promoter and serves as a sensor for transfection efficiency so that the data on firefly luciferase activity for the control and repeat-containing plasmids were normalized for Renilla luciferase activity in every transfection. Effects of the longest (CTG)n·(CAG)n repeats in different positions and orientations on reporter expression relative to the control pGL2 plasmid are shown in Figure 1B. A significant (∼10-fold) inhibition of reporter expression was observed only when these repeats were situated in one position (the 5′-UTR) and one orientation [the (CTG)n run in the sense strand].
In all trinucleotide repeat diseases there is a clear correlation between the size of an expanded repeat and the severity of the disease phenotype. Therefore, we investigated the dependence of (CTG)n-caused inhibition on repeat length. Since the only effects we saw were caused by repeats in the 5′-UTR, we cloned d(CTG)n·d(CAG)n stretches with n ranging from 35 to 160 into this location. Figure 1C shows the results of these transient transfection experiments. A smooth exponential decrease in the level of reporter gene expression compared to the control pGL2 plasmid was observed with an increase in the length of the CTG repeats in the sense strand. The maximum (∼10-fold) inhibition was achieved when the number of repeats exceeded 100. CAG repeats in the sense strand for transcription, in contrast, did not significantly affect luciferase expression: only a 1.4-fold inhibition was observed for the longest (n = 120) repeat tested. We conclude, therefore, that the effects of (CTG)n·(CAG)n repeats on gene activity depend both upon repeat length and orientation.
The mechanisms of repeat-caused repression of the reporter gene
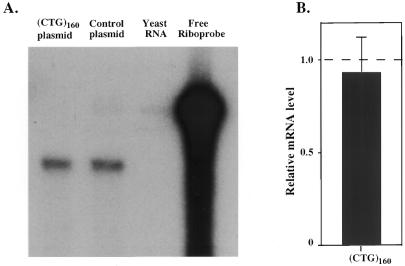
We then studied whether (CTG)n repeats in the 5′-UTR interfered with transcription or translation of our reporter gene. To test the effects of CTG repeats on transcription of the luciferase gene in a transient expression system, we used an RNase protection assay to compare the levels of luciferase mRNA in cells transfected with either the control plasmid or the construct containing a (CTG)160 repeat. Figure 2A shows typical levels of reporter mRNA for the control (lane 2) and (CTG)160-containing plasmid (lane 1) that were normalized for differences in transfection efficiency as described in Materials and Methods. One can see that there is only a slight difference in mRNA levels in the two cases. Figure 2B shows a statistical analysis of five independent experiments comparing the control with the (CTG)160-containing plasmid. Clearly, the presence of even the longest CTG repeat did not affect reporter transcription.
Figure 2.
Comparison of luciferase mRNA levels for the pGL2-Promoter and (CTG)160-containing plasmids. (A) Typical experimental data. (B) Statistical analysis of five independent experiments. RNA amounts were normalized to that for the control pGL2 plasmid. The dashed line shows the expression level for the control plasmid.
It was reasonable to assume, therefore, that (CTG)n repeats in the 5′-UTR might decrease luciferase gene expression at the level of translation. Thus, we examined the influence of these repeats on the efficiency of luciferase mRNA translation. We used an in vitro system of coupled transcription–translation where transcription of the luciferase gene was carried out by T7 RNA polymerase followed by translation in a rabbit reticulocyte lysate (34). For use in a cell-free system, the SV40 promoter in our control and CTG-containing plasmids was replaced with the T7 promoter without changing the 5′-UTR region. In each reaction the same amounts of DNA template were added to the reaction mixture, newly synthesized protein was labeled with [35S]methionine and amounts of this protein were estimated after separation by PAGE.
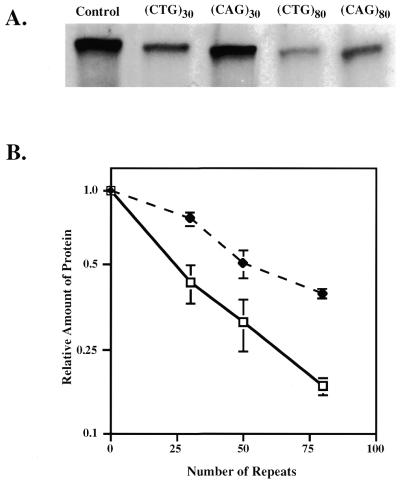
The results of a typical experiment are shown in Figure 3A. One can see that the (CUG)30 repeat leads to a substantial inhibition of protein synthesis, while the (CUG)80 repeat blocks it severely. (CAG)n repeats, while also inhibitory, were less detrimental: 30 repeats showed little effect, while 80 repeats behaved similarly to (CUG)30. Note that for all the above templates the amounts of luciferase RNA transcripts produced by the T7 RNA polymerase were very similar (data not shown). Thus, inhibition of protein synthesis in our system occurred at the translation stage.
Figure 3.
Effects of (CTG)n and (CAG)n repeats on reporter translation in vitro. (A) Typical experimental data. (B) Statistical analysis of four to five independent experiments. Protein amounts were normalized to that of the control plasmid without (CUG)n repeats. Closed circles, (CAG)n repeats in the sense strand; open squares, (CTG)n repeats in the sense strand.
Figure 3B shows a statistical analysis of several in vitro experiments with (CAG)n·(CAG)n repeats of varying length. Relative translation efficiency compared to the control template declines rapidly with lengthening of (CUG)n repeats in the 5′-UTR, but substantially more slowly with lengthening of (CAG)n repeats. Thus, these experiments show that the length- and orientation-dependent effects of d(CTG)n·d(CAG)n repeats on translation in vitro are qualitatively similar to those for luciferase expression in vivo. It is plausible to speculate, therefore, that inhibition of translation by these repeats might be responsible for reporter repression in vivo.
The configuration of the CUG-containing 5′-UTR of the reporter mRNA is shown in Figure 4A. There is a 65 nt long sequence between the 5′-end and the (CUG)n run and another 59 nt long sequence following the (CUG)n run prior to the luciferase AUG start codon. Note that most of the CUG triplets, including those in the repeated stretch and one immediately downstream, are upstream of the luciferase ORF but in-frame with it. Additionally, there are two out-of-frame CUG codons. CUG codons are known to occasionally serve as minor translation intiation sites in eukaryotes (reviewed in 35). One may ask, therefore, whether multiply repeated CUG codons in our 5′-UTRs can interfere with translation initiation from the luciferase start codon, thus obstructing gene expression.
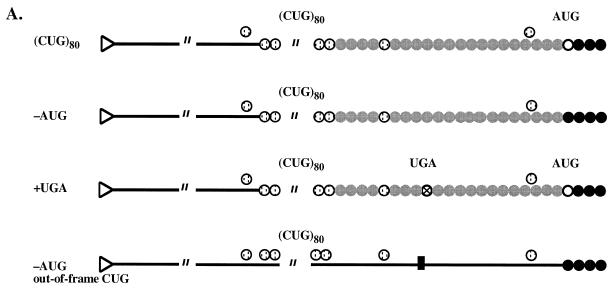
Figure 4.
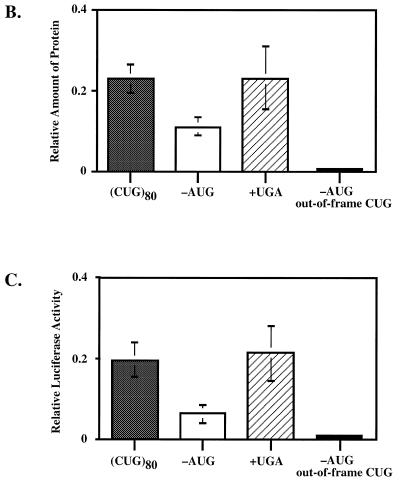
Effects of modifications in the 5′-UTR of the (CTG)80-containing plasmid on reporter expression in vivo and translation in vitro. (A) Schematic representation of the 5′-UTRs of modified plasmids. Open circles, initiator AUG codon; striped circles, CUG codons (on line, in-frame; above line, out-of-frame); solid circles, luciferase amino acids; gray circles, hypothetical amino acids resulting from initiation at CUG codons; crossed circle, translation stop codon; solid bar, small duplication placing all CUG codons out-of-frame; triangle, capped 5′-end of mRNA. (B) Effects of 5′-UTR modifications on translation in vitro. Protein levels for all experimental plasmids were normalized to that of the control plasmid without (CUG)n repeats. (C) Effects of 5′-UTR modifications on luciferase expression in vivo. Luciferase levels for all experimental plasmids were normalized to that of the control plasmid without (CUG)n repeats.
In order to address this problem, we have constructed three derivatives of our plasmid with the d(CTG)80 repeat. First, we removed the luciferase AUG codon. As a result, luciferase translation could either start from any of the in-frame CUG codons or from the next available AUG codon located 84 nt downstream of the original one. Second, we inserted a UGA stop codon between the CUG run and the luciferase AUG codon. In this case, translation from an in-frame CUG codon should not result in luciferase synthesis. In a final construct the luciferase AUG start codon was removed and CUG triplets were placed out-of-frame. Consequently, no luciferase expression in vitro or in vivo is expected.
The experimental data on translation in vitro and luciferase expression in vivo obtained for those constructs are shown in Figure 4B and C, respectively. One can see a clear correlation between in vitro and in vivo data. Incorporation of a stop codon between the CUG run and the first AUG codon has little, if any, effect on luciferase synthesis or expression. Elimination of the first AUG codon, in contrast, leads to an ∼5-fold drop in luciferase expression in vivo and an ∼2-fold decrease in translation in vitro. The residual translational activity seems to be due to initiation from some CUG codon(s), rather than from the downstream AUG codons, since the plasmid with the out-of-frame CUG codons and without the first AUG codon gave no detectable luciferase synthesis. In fact, our preliminary data indicate that the individual CUG codon situated downstream of the (CUG)n run is primarily responsible for translation initiation in the absense the AUG codon (data not shown).
Altogether, these results demonstrate that initiation of translation from repeated CUG codons in our 5′-UTR is possible. However, its efficiency is not high and never exceeds 10% of the AUG-dependent translation in the control pGL2-Promoter plasmid. It is unlikely, therefore, that this inefficient process would cause major inhibition of luciferase synthesis in vitro and in vivo. It is clear, at the same time, that some peculiarities of the (CUG)n-containing 5′-UTR prevent efficient translation from the AUG start codon.
DISCUSSION
Current ideas on the mechanisms of gene repression by expanded (CTG)n repeats came from studies of two hereditary disorders, myotonic dystrophy (23,24) and SCA 8 (36). In the first case, long (CTG)n repeats situated in the 3′-UTR of the DMPK gene block its RNA processing (25). They also lead to repression of the downstream SIX5/DMAHP gene promoter (26,27) and prevent maturation of several CUG-containing RNAs in trans (28). In the second case, expanded (CTG)n repeats are found in a transcribed (but untranslated) RNA produced from the antisense strand of a distinct gene (36). This points to inhibition of gene expression by virtue of RNA interference.
Here we placed (CTG)n repeats in different locations relative to the coding part of a reporter luciferase gene, in either the 5′- or 3′-UTR. Positioning repeats in the 3′-UTR of our reporter gene mimicked the situation with the DMPK gene. It was previously suggested that specific CUG-binding proteins cause nuclear retention and/or aberrant splicing of the DMPK hnRNA (28,37,38). Surprisingly, in our experiments there was no inhibition of gene expression from placing (CTG)n repeats in this position. Note that the DMPK studies were performed in muscle biopsies or cultured differentiated myoblasts from patients with myotonic dystrophy. Thus, the differences with our data could result from two reasons. First, the number of (CTG)n repeats affecting DMPK gene expression in clinical studies usually exceeds several hundred and only modest effects were observed with ∼200 repeats. Our longest (CTG)n stretch contained 160 repeats, which is below the expansion length in clinical samples. Second, most of the CUG-binding proteins described so far are expressed predominantly in differentiated cells of skeletal and cardiac muscle. Thus, the de-differentiated colon carcinoma cell line that we used in our transfection experiments may not contain such proteins.
Equally unexpectedly, we found that moderately expanded (CTG)n repeats placed in the reporter 5′-UTR led to a drastic inhibition of its expression. To the best of our knowledge this is the first time that (CTG)n repeats within a 5′-UTR have been shown to impede gene functioning. The profundity of the inhibitory effect (an order of magnitude at the modest expansion number n ≥ 100) encouraged us to study its mechanisms. We analyzed the level of luciferase mRNA in vivo using a RNase protection assay and the translation efficiency of this RNA in vitro. It appeared that even the longest repeated stretch studied, (CTG)160, does not significantly affect the amount of luciferase mRNA in the cell. At the same time, translation of the luciferase mRNA was strongly inhibited by even relatively short repeats.
During translation in eukaryotes the small ribosomal subunit is first attached to the mRNA capped 5′-end and then translocates to the first suitable AUG codon (reviewed in 35,39). The large ribosomal subunit joins the preinitiation complex there and translation initiation occurs. Since our (CTG)n tracts were situated between the mRNA 5′-end and the first AUG codon, their inhibitory effect on translation can be explained by several different mechanisms.
First, while initiation of translation in eukaryotes primarily occurs at the first AUG codon from the 5′-end of mRNA, there are notable exceptions. Specifically, CUG codons are known to serve as translation initiation sites in several cases, including the human c-myc (40,41), WT1 (42) and FGF-2 (43) genes. It is possible, therefore, that placing multiply repeated CUG codons upstream of the luciferase start codon might promote translation initiation from the CUG stretch, thus preventing proper initiation. Since the CUG run is in-frame with the luciferase gene, the resulting protein would contain extra amino acids at its N-terminus (potentially including repeated leucines), which could be detrimental to luciferase activity. Based on our data, however, this explanation is unlikely. We do not see any effect of placing the stop codon between the (CUG)n run and luciferase initiation codon on translation in vitro or luciferase expression in vivo. Further, when the original luciferase initiation codon is deleted, all upstream CUG codons together provide only 50% of residual translation in vitro and 20% of residual luciferase activity in vivo. Finally, the level of translation from combined CUG codons does not exceed 10% that for a control plasmid without CUG repeats. Altogether, these data indicate that the initiator AUG codon of the luciferase gene is responsible for the majority of, if not all, translation initiation events even in the presence of expanded (CUG)n runs. Only when the AUG codon is deleted do CUG codons become translation initiators.
Second, (CUG)n repeats might affect the process of translocation of the small ribosomal subunit from a RNA 5′-end to the start codon, called scanning (reviewed in 35). It is well documented that eukaryotic mRNAs with highly structured 5′-UTRs are relatively inefficient translationally (44,45). This is likely due to the inability of the 40S ribosomal subunit and/or associated RNA helicases to unwind stable secondary structures in the 5′-UTR during scanning. Supporting this, it was found that formation of a strong RNA hairpin (ΔG = –61 kcal/mol) within the 5′-UTR abolished translation almost completely (46). Also, hairpin formation by expanded (CGG)n repeats in the 5′-UTR of the human FMR1 gene is believed to impede translation (47). (CUG)n repeats have been shown to form RNA hairpins in vitro (48,49). The stability of these hairpins increases with the length of the (CUG)n run and becomes very substantial for long repeats. For example, the free energy of a hairpin formed by a (CUG)49 run is –55 kcal/mol (48). Thus, formation of stable hairpins by (CUG)n repeats of increasing length in our reporter RNA could progressively inhibit the scanning step of translation initiation. This prediction is in excellent agreement with our experimental data. If true, the strikingly good correlation between our results in vitro and in vivo would indicate that stable CUG hairpins are formed in intracellular RNA as well.
Can the above hypothesis explain the much more modest inhibitory effects of (CAG)n runs? In single-stranded DNA CAG repeats form hairpins (50), though these are less stable than their CTG counterparts (51,52). While no data are available for RNA, one might expect the same to be true. We show that (CAG)n stretches inhibit translation in a reticulocyte lysate, though more weakly than (CUG)n runs. This is consistent with the formation of a CAG hairpin that is thermodynamically or kinetically less favorable than a CUG hairpin under the given conditions. Yet in vivo (CAG)n repeats practically do not inhibit luciferase expression. This might be due to an inability of (CAG)n repeats to form hairpins in the intracellular environment for thermodynamic or kinetic reasons or due to the existence of CAG-binding proteins preventing hairpin formation (53,54).
In summary, we show that moderately expanded (CUG)n repeats within a reporter 5′-UTR inhibit its expression at the translation level. While the explicit mechanism of this translational block remains to be understood, we believe that formation of RNA hairpins by these repeats is likely to be responsible. Whatever the mechanism, however, one might expect that expansion of (CTG)n repeats within 5′-UTRs of conceptual human genes could lead to their inactivation. Our preliminary search has already revealed two cognate human genes: the SHMT gene, encoding cytosolic serine hydroxymethyltransferase, with 12 CUG-repeats (30); the BPGM gene, encoding erythrocyte 2,3-biphosphoglycerate mutase, with 8 repeats (31). Expanding this search may lead to revealing new human genetic disorders.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Nahum Sonenberg, Alexander Mankin and Louis Deiss for helpful discussions, Kotla Kumar for his help with the in vitro translation assay, Michael Kharas for technical assistance, Alexander Mankin for critical reading of the manuscript and Gerald Buldak for editorial help. This work was supported by a grant from the Council for Tobacco Research (4468) to S.M.M. G.R. is a recipient of the Dean’s scholarship from the University of Illinois at Chicago.
REFERENCES
- 1.Schroth G.P. and Ho,P.S. (1995) Nucleic Acids Res., 23, 1977–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox R. and Mirkin,S.M. (1997) Proc. Natl Acad. Sci. USA, 94, 5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Timchenko L.T. and Caskey,C.T. (1999) Cell. Mol. Life Sci., 55, 1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wells R.D. and Warren,S.T. (eds) (1998) Genetic Instabilities and Hereditary Neurological Disorders. Academic Press, San Diego, CA.
- 5.McMurray C.T. (1995) Chromosoma, 104, 2–13. [DOI] [PubMed] [Google Scholar]
- 6.Wells R.D. (1996) J. Biol. Chem., 271, 2875–2878. [DOI] [PubMed] [Google Scholar]
- 7.McMurray C.T. (1999) Proc. Natl Acad. Sci. USA, 96, 1823–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kremer E.J., Pritchard,M., Lynch,M., Yu,S., Holman,K., Baker,E., Warren,S.T., Schlessinger,D., Sutherland,G.R. and Richards,R.I. (1991) Science, 252, 1711–1714. [DOI] [PubMed] [Google Scholar]
- 9.Oberle I., Rousseau,F., Heitz,D., Kretz,C., Devys,D., Hanauer,A., Boue,J., Bertheas,M.F. and Mandel,J.L. (1991) Science, 252, 1097–1102. [DOI] [PubMed] [Google Scholar]
- 10.Verkerk A.J.M.H., Pieretti,M., Sutcliffe,J.S. et al. (1991) Cell, 65, 905–914.1710175 [Google Scholar]
- 11.Hornstra I.K., Nelson,D.L., Warren,S.T. and Yang,T.P. (1993) Hum. Mol. Genet., 2, 1659–1665. [DOI] [PubMed] [Google Scholar]
- 12.Subramanian P.S., Nelson,D.L. and Chinault,A.C. (1996) Am. J. Hum. Genet., 59, 407–416. [PMC free article] [PubMed] [Google Scholar]
- 13.The Huntington’s Disease Collaborative Research Group (1993) Cell, 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 14.LaSpada A.R., Wilson,E.M., Lubahn,D.B., Harding,A.E. and Fischbeck,K.H. (1994) Nature, 352, 77–79. [DOI] [PubMed] [Google Scholar]
- 15.Chung M.Y., Ranum,L.P., Duvick,L.A., Servadio,A., Zoghbi,H.Y. and Orr,H.T. (1993) Nature Genet., 5, 254–258. [DOI] [PubMed] [Google Scholar]
- 16.Orr H.T., Chung,M.Y., Banfi,S. et al. (1993) Nature Genet., 4, 221–226. [DOI] [PubMed] [Google Scholar]
- 17.Koide R., Ikeuchi,T., Onodera,O. et al. (1994) Nature Genet., 6, 9–13. [DOI] [PubMed] [Google Scholar]
- 18.Bates G. (1996) Bioessays, 18, 175–178. [DOI] [PubMed] [Google Scholar]
- 19.Lunkes A., Trottier,Y. and Mandel,J.L. (1998) Essays Biochem., 33, 149–163. [DOI] [PubMed] [Google Scholar]
- 20.Campuzano V., Montermini,L., Molto,M.D. et al. (1996) Science, 271, 1423–1427. [DOI] [PubMed] [Google Scholar]
- 21.Bidichandani S.I., Ashizawa,T. and Patel,P.I. (1998) Am. J. Hum. Genet., 62, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oshima K., Montermini,L., Wells,R.D. and Pandolfo,M. (1998) J. Biol. Chem., 273, 14588–14595. [DOI] [PubMed] [Google Scholar]
- 23.Brook J.D., McCurrach,M.E., Harley,H.G. et al. (1992) Cell, 68, 799–808.1310900 [Google Scholar]
- 24.Mahadevan M., Tsilfidis,C., Sabourin,L. et al. (1992) Science, 255, 1253–1255. [DOI] [PubMed] [Google Scholar]
- 25.Davis B.M., McCurrach,M.E., Taneja,K.L., Singer,R.H. and Housman,D.E. (1997) Proc. Natl Acad. Sci. USA, 94, 7388–7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klesert T.R., Otten,A.D., Bird,T.D. and Tapscott,S.J. (1997) Nature Genet., 16, 402–406. [DOI] [PubMed] [Google Scholar]
- 27.Thornton C.A., Wymer,J.P., Simmons,Z., McClain,C. and Moxley,R. (1997) Nature Genet., 16, 407–409. [DOI] [PubMed] [Google Scholar]
- 28.Philips A.V., Timchenko,L.T. and Cooper,T.A. (1998) Science, 280, 737–741. [DOI] [PubMed] [Google Scholar]
- 29.Han J., Hsu,C., Zhu,Z., Longshore,J.W. and Finley,W.H. (1994) Nucleic Acids Res., 22, 1735–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chave K.J., Snell,K. and Sanders,P.G. (1997) Biochem. Soc. Trans, 25, 53S. [DOI] [PubMed] [Google Scholar]
- 31.Joulin V., Peduzzi,J., Romeo,P.H. et al. (1986) EMBO J., 5, 2275–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richard G.-M., Goellner,G.M., McMurray,C.T. and Haber,J.E. (2000) EMBO J., 19, 2831–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier F.W., Rosenberg,A.H., Dunn,J.J. and Dubendorff,J.W. (1990) Methods Enzymol., 185, 60–89. [DOI] [PubMed] [Google Scholar]
- 34.Hurst R., Maffitt,M., Murray,E., Kappelman,J., Xu,Q., Mendez,J., Butler,B., Ryan,A. and Beckler,G.S. (1996) Promega Notes, 58, 8–11. [Google Scholar]
- 35.Kozak M. (1999) Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 36.Koob M.D., Moseley,M.L., Schut,L.J., Benzow,K.A., Bird,T.D., Day,J.W. and Ranum,L.P.W. (1999) Nature Genet., 21, 379–384. [DOI] [PubMed] [Google Scholar]
- 37.Timchenko L.T. and Caskey,C.T. (1996) FASEB J., 10, 1589–1597. [DOI] [PubMed] [Google Scholar]
- 38.Timchenko L.T., Miller,J.W., Timchenko,N.A., DeVore,D.R., Datar,K.V., Lin,L., Roberts,R., Caskey,C.T. and Swanson,M.S. (1996) Nucleic Acids Res., 24, 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gingras A.-C., Raught,B. and Sonenberg,N. (1999) Annu. Rev. Biochem., 68, 913–963. [DOI] [PubMed] [Google Scholar]
- 40.Blackwood E.M., Lugo,T.G., Kretzner,L., King,M.W., Street,A.J., Witte,O.N. and Eisenman,R.N. (1994) Mol. Biol. Cell., 5, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nanbru C., Lafon,I., Audigier,S., Gensac,M.C., Vagner,S., Huez,G. and Prats,A.C. (1997) J. Biol. Chem., 272, 32061–32066. [DOI] [PubMed] [Google Scholar]
- 42.Bruening W. and Pelletier,J. (1996) J. Biol. Chem., 271, 8646–8654. [DOI] [PubMed] [Google Scholar]
- 43.Kevil C., Carter,P., Hu,B. and DeBenedetti,A. (1995) Oncogene, 11, 2339–2348. [PubMed] [Google Scholar]
- 44.Kozak M. (1980) Cell, 19, 79–90. [DOI] [PubMed] [Google Scholar]
- 45.Kozak M. (1986) Proc. Natl Acad. Sci. USA, 83, 2850–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozak M. (1989) Mol. Cell. Biol., 9, 5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Y., Zhang,F., Lokey,L.K., Chastain,J.L., Lakkis,L., Eberhart,D. and Warren,S.T. (1995) Science, 268, 731–734. [DOI] [PubMed] [Google Scholar]
- 48.Napieraa M. and Krzyosiak,W.J. (1997) J. Biol. Chem., 272, 31079–31085. [DOI] [PubMed] [Google Scholar]
- 49.Michalowski S., Miller,J.W., Urbinati,C.R., Paliouras,M., Swanson,M.S. and Griffith,J. (1999) Nucleic Acids Res., 27, 3534–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gacy A.M., Goellner,G., Juranic,N., Macura,S. and McMurray,C.T. (1995) Cell, 81, 533–540. [DOI] [PubMed] [Google Scholar]
- 51.Petruska J., Arnheim,N. and Goodman,M.F. (1996) Nucleic Acids Res., 24, 1992–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gacy A.M. and McMurray,C.T. (1998) Biochemistry, 37, 9426–9434. [DOI] [PubMed] [Google Scholar]
- 53.McLaughlin B.A., Spencer,C. and Eberwine,J. (1996) Am. J. Hum. Genet., 59, 561–569. [PMC free article] [PubMed] [Google Scholar]
- 54.Yano H., Wang,B.E., Ahmad,I., Zhang,J., Abo,T., Nakayama,J., Krempen,K. and Kohwi,Y. (1999) Exp. Cell Res., 251, 388–400. [DOI] [PubMed] [Google Scholar]