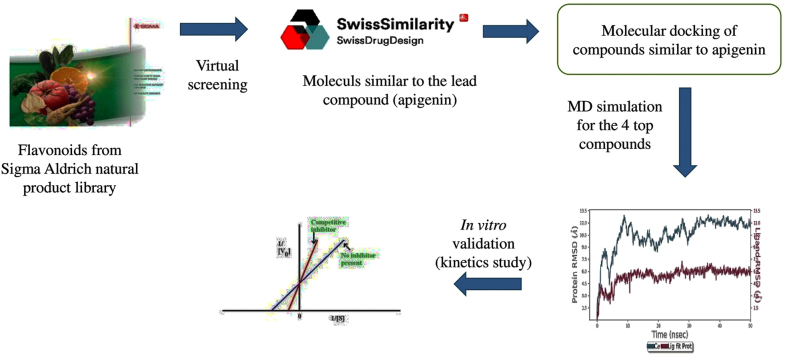
Abstract
Existing inhibitors of kynurenine-3-monooxygenase (KMO) have side effects and poorly cross the blood-brain barrier. Therefore, the discovery of new molecules targeting KMO isnecessary.This study aims to develop a novel therapeutic drug targeting KMO using computational methods and experimental validation of natural compounds.The results of our study show that the top four compounds, namely, 3′-Hydroxy-alpha-naphthoflavone exhibited the best docking scores with KMO (−10.0 kcal/mol), followed by 3′-Hydroxy-ss-naphthoflavone (−9.9 kcal/mol), genkwanin (−9.2 kcal/mol) and apigenin(-9.1 kcal/mol) respectively. Molecular dynamics was used to assess the stability of the primary target, KMO, and inhibitor complexes. We found stable interactions of 3′-Hydroxy-ss-naphthoflavone and apigenin with KMO up to 100 ns. Further, kinetic measurements showed that 3′-Hydroxy-alpha-naphthoflavone and 3′-Hydroxy-ss-naphthoflavone induce competitive inhibition with a good IC50 activity (15.85 ± 0.98 μM and 18.71 ± 0.78, respectively), while Genkwanin and Apigenin exhibit non-competitive inhibition mechanism (21.61 ± 0.97 μM and 24.14 ± 1.00 μM, respectively).Drug-likeness features and ADME analysis features also showed that the top four compounds could be used as potential candidates to replace the synthetic KMO inhibitor drugs with known side effects and poor brain-blood barrier penetration.
Keywords: Molecular docking, Molecular dynamics, Kinetic assays, Flavonoids, KMO inhibition
Graphical abstract
Highlights
-
•
The inhibitory effect of natural compounds with a similar structure to apigenin on KMO was investigated using molecular modeling.
-
•
The inhibitory effect of four lead compounds underwent a kinetic study to confirm the findings from docking and molecular dynamic simulations.
-
•
3′-Hydroxy-alpha-naphthoflavone, 3′-Hydroxy-ss-naphthoflavone, genkwanin and apigenin appeared to have the best potential to act as KMO inhibitors and display a suitable pharmacokinetic profile.
Abbreviations
- ADME
absorption distribution metabolism and excretion
- BBB
blood-brain-barrier
- MD simulations
molecular dynamic simulations
- L-Kyn
l-kynurenine
- H-bond
hydrogen bond
- ns
nanosecond
- LD50
Lethal Dose 50
- Km
Michaelis constant
- Vmax
maximum velocity
1. Introduction
By converting tryptophan to neuroinflammatory/neurotoxic metabolites, the kynurenine pathway (KP) is critically involved in several neurodegenerative diseases and behavioral disorders [[1], [2], [3], [4]]. As part of this pathway, the enzyme kynurenine-3-monooxygenase (KMO) converts kynurenine (KYN) to 3-hydroxykynurenine (3HK), a free radical generator that causes neuronal cell death and overstimulation of glutamate receptors [3,5]. Compounds that decrease the levels of 3HK, especially KMO inhibitors such as UPF-648 (IC50 = 20 nM), KNS366, and GSK 366 (IC50 = 2.3 nM), are being considered for treating several diseases [3,[5], [6], [7], [8], [9], [10], [11]].
Until now, no reports have suggested the inhibitory activity of flavonoids on KMO enzyme using computational tools to discover new inhibitors and drug candidates for neuropsychiatric and neurodegenerative diseases.
The present study determined if natural flavonoid compounds significantly inhibit KMO in molecular simulation and kinetic studies. Flavonoids are part of the polyphenol class of phytonutrients with well-documented biological properties. The polyphenol structure of flavonoids is responsible for their pharmacological activities, including antimicrobial, antiviral, antioxidant, and anti-inflammatory activities [12,13]. They are potent inhibitors of several brain enzymes, including indoleamine 2,3-dioxygenase [14] and monoamine oxidase [15].
We have measured the inhibitory activity of flavonoids on KMO using computational tools to discover new inhibitors and drug candidates for neurodegenerative diseases and behavioral disorders.
2. Materials and methods
2.1. Chemicals and reagents
3′-Hydroxy-alpha-naphthoflavone and 3′-Hydroxy-ss-naphthoflavone were bought from Indofine (Township, NJ, USA), Genkwanin, Apigenin,Ro 61–8048, and DSMOwere obtained from Sigma-Aldrich Co. LLC (Germany). KMO kit and buffers were bought from (Thermo Fisher Scientific (Waltham, MA, USA)).
2.2. Computational method
2.2.1. Receptors preparation
Three-dimensional structures of the protein-ligand complexes were downloaded from the Protein Data Bank (http://www.rcsb.org/). KMO has the PDB code 4J31.
AutoDock Vina was used to perform molecular docking studies. Each protein-ligand complex was prepared by using AutoDock Tools-1.5.6. The first step involves removing water molecules, cofactors, and ligands from the protein's crystal structure. The chain A of the protein was also removed. We added polar hydrogens, and Kollman united atom-type charges to the structure. BIOVIA Discovery Studio Visualizer (2020) was used to picture the docked complexes. Before further analysis, we selected the best position for the ligand to bind to the target protein.
The docking results of the four top-ranked ligands were confirmed by re-docking them into the same defined regions to legitimize the virtual molecular docking protocol for accuracy. The binding scores and protein-ligand interactions were then re-validated using the BINDSURF Achilles blind docking server http://bio-hpc.eu/software/blind-docking-server/. [16].
2.2.2. Ligands preparation
A library of flavonoids was constructed from Sigma-Aldrich's natural product portfolio (https://www.sigmaaldrich.com/DZ/en/products/chemistry-and-biochemicals/biochemicals/natural-products) and was screened using molecular docking.
Before molecular docking, we filtered all compounds using the Lipinski and Veber rules [17,18]; those with reactive functional groups were removed.
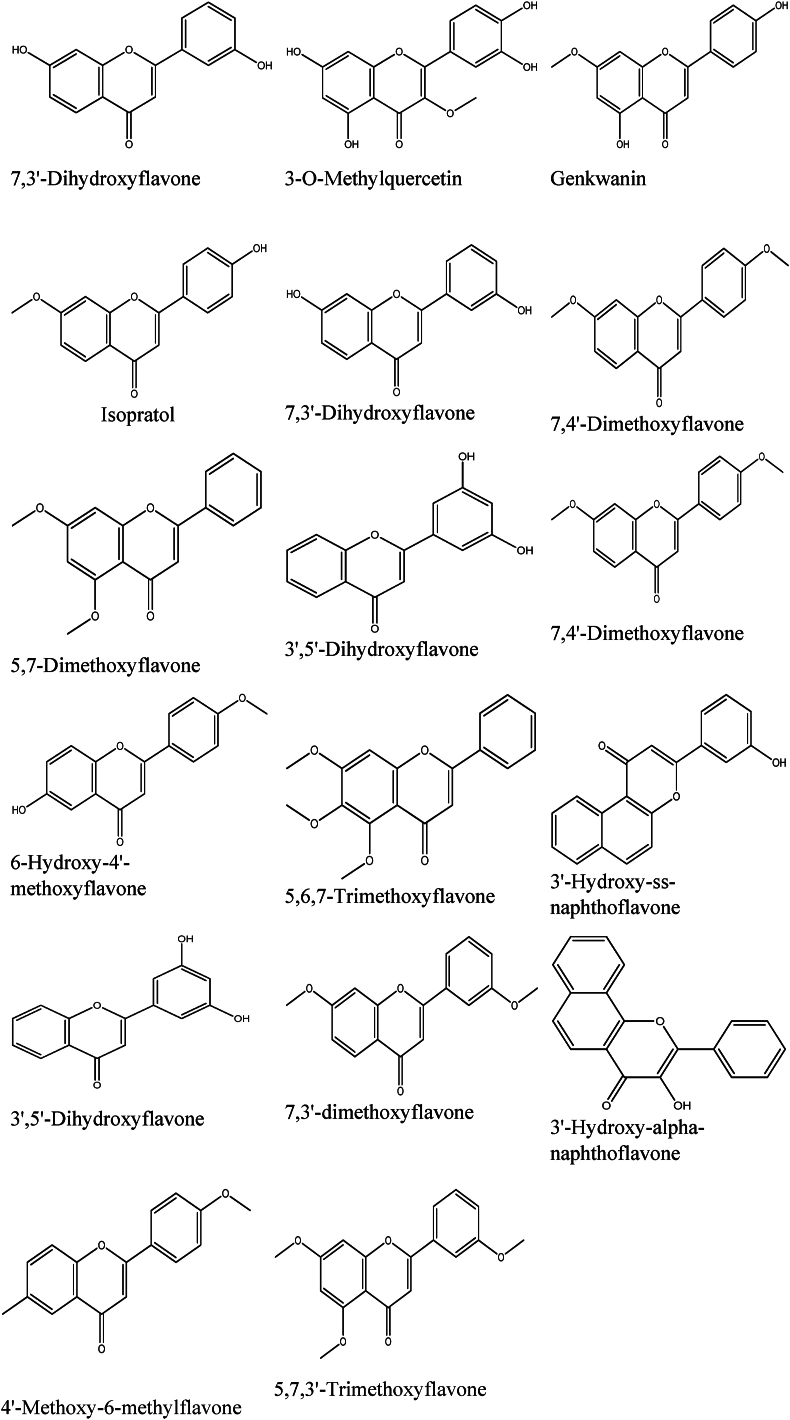
Virtual screening results show that apigenin possesses a high binding affinity. Then, compounds structurally similar to apigenin were searched using the Swiss similarity web tool (http://www.swisssimilarity.ch/) [19]. Seventeen flavonoid compounds (Fig. 1), with a score similarity higher than 0.5, were selected and docked to the KMO inhibition site after being drawn by ChemDraw Ultra (version 12.0) optimized using the Avogadro software. We minimized the energies by using the MMFF94 force field.
Fig. 1.
2D structures of 17 selected compounds similar to apigenin.
2.2.3. Drug-likeness and ADMET analysis
We used the SwissADME(http://www.swissadme.ch/) online server to evaluate the pharmacokinetic features such as lipophilicity, gastrointestinal absorption, BBB permeation, and drug-likeness of the best-scored flavonoid compounds [20]. We predicted the physicochemical properties: topological polar surface area, number of hydrogen bond donors, number of hydrogen bond acceptors, and number of rotatable bonds. Additionally, we assessed the toxicity parameters of the selected flavonoids with the ProTox-II web tool(https://tox-new.charite.de/protox_II/).
2.2.4. Molecular dynamic (MD)
We investigated the binding stability of protein-ligand docking complexes using MD, which also offers information on intermolecular interactions within a reference period. We used the Maestro tool System Builder to generate the protein-ligand complex structure of KMO and the top four potential molecules for MD simulations. We immersed the complexes created in a box filled with water molecules via the simple point charge (SPC) scheme. We performed the MD simulations using Maestro-Desmond (Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2020). We set the box size to 10 x 10 × 10 Å. We added sodium and chlorine ions to obtain a final NaCl concentration of 0.15 M. Energy minimization was conducted by 2000 steps using the steepest descent method with a cutoff of 9 Å for van der Waals interactions. We used the Particle Mesh Ewald (PME) method with a tolerance of 10 -9 in the electrostatic part. We did NPT simulations at 300K using the Nosé-Hoover algorithm [21], keeping the pressure at 1 bar using the Martyna-Tobias-Kelinbarostat. The force field OPLS3e was used to do all the runs. The simulation time of each run was 100 ns.
2.3. Enzymatic inhibition assay
We performed the inhibition assay of KMOusingthe method described by Puopolo et al. [22] with a slight modification, using Ro 61–8048 as the positive control. The test molecules were dissolved in 100 mM DMSO and diluted in assay buffer to 1 mM. We incubated in a 96-well plate a reaction made from a mixture of assay buffer (50 μL), human KMO (20 g/mL; 50 μL), test compounds (1 mM; 10 μL), and substrate (40 μL NADPH (10 mM) plus L-kyn (L-Kyn, 20 mM) for 1.5 h at room temperature. All assays were performed in triplicate. The optical densities of each sample were measured on a microplate reader at a wavelength of 340 nm. We determined the enzymatic activity by using the formula below:
| % Activity = [(Abssample − Absblank)/(Abscontrol − Absblank)] × | 100 |
The results are reported as the inhibition rate (%) of KMO. A graphical analysis was done to find the IC50 value of each compound.
2.4. Kinetics study
We determined the inhibitory strength of the most potent compounds using the same procedure as the enzyme inhibition assay. We measured the kinetic parameters by creating Lineweaver-Burk plots (double reciprocal plots) using various concentrations of the tested compounds (0 μM–40 μM) with a concentration range of 0–20 μM for the substrate (L-Kyn).
3. Results and discussion
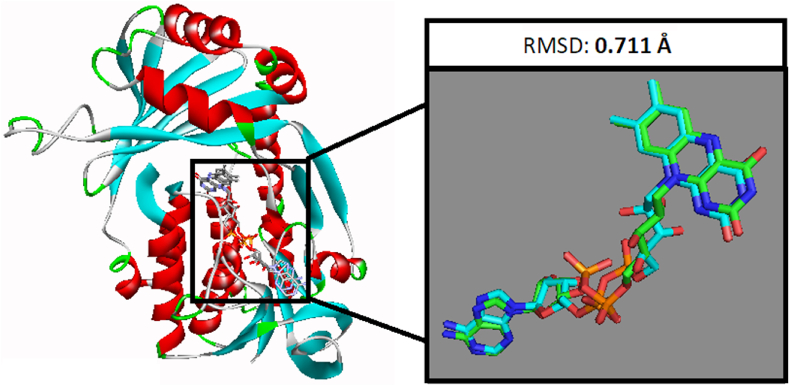
Flavonoids are a broad class of polyphenolic compounds that have neuroprotective properties. They are widely used to prevent cancer, Alzheimer's disease, viral disease, and atherosclerosis. These beneficial effects are attributed to their action on enzymes involved in inflammation [[23], [24], [25]]. In this study, we assessed the inhibitory effect of phyto-ligand compounds belonging to the flavonoids on KMO. After validating the docking procedure, we did a structure-based virtual screening of the co-crystal ligand by evaluating the analogy between the lowest energy state predicted by AutoDock Vina and the binding mode of the native ligand (Fig. 2). The root mean square deviation (RMSD) value after the docking procedure was 0.711 Å, which is within the desired range (i.e., less than 2.0 Å).
Fig. 2.
Validation of docking study; the docked co-crystal ligand and the native ligand with RMSD of 0.711 Å
3.1. Computational method
3.1.1. Molecular docking
In the present study, 17 flavonoids, similar to apigenin, were identified from the Swiss Similarity server. These phytochemicals were docked in the binding site of the KMO enzyme.
Table 1 lists the docking scores of the studied compounds. The top four compounds in terms of affinity (Fig. 3)are 3′-Hydroxy-alpha-naphthoflavone, which exhibited the lowest docking score (−10.0 kcal/mol), followed by 3′-Hydroxy-ss-naphthoflavone (−9.9 kcal/mol), genkwanin (−9.2 kcal/mol) and apigenin (−9.1 kcal/mol), respectively. Interestingly, these flavonoid ligands showedhigher docking scores for KMO thanthe reference inhibitor, Ro 61–8048, which scored −9.1 kcal/mol.
Table 1.
Molecular docking score of 17 flavonoid phyto-ligands for KMO'sactive site.
| Rank | Compound | PubChem CID | Binding Energy (kcal/mol) |
|---|---|---|---|
| 1 | 3′-Hydroxy-alpha-naphthoflavone | 13707046 | −10.0 |
| 2 | 3′-Hydroxy-ss-naphthoflavone | 688843 | −9.9 |
| 3 | Genkwanin | 5281617 | −9.2 |
| 4 | Apigenin | 5280443 | −9.1 |
| 5 |
Ro 61–8048 (reference inhibitor) 3′,5′-Dihydroxyflavone |
5282337 45933941 |
−9.1 −9.0 |
| 6 | Isopratol | 676307 | −8.9 |
| 7 | 7,3′-Dihydroxyflavone | 5391140 | −8.8 |
| 8 | 4′-Methoxy-6-methylflavone | 688682 | −8.8 |
| 9 | 2,6,7-Trihydroxy-9-methylxanthen-3-one | 72721 | −8.6 |
| 10 | 3-O-Methylquercetin | 5280681 | −8.6 |
| 11 | 5,7,3′-Trimethoxyflavone | 12150586 | −8.5 |
| 12 | 7,4′-Dimethoxyflavone | 5322058 | −8.4 |
| 13 | 7,3′-dimethoxyflavone | 688672 | −8.4 |
| 14 | 6-Hydroxy-4′-methoxyflavone | 688679 | −8.4 |
| 15 | 5,7-Dimethoxyflavone | 88881 | −8.4 |
| 16 | 5,6,7-Trimethoxyflavone | 442583 | −8.4 |
| 17 | 4′,5-Dimethoxyflavone | 688669 | −8.2 |
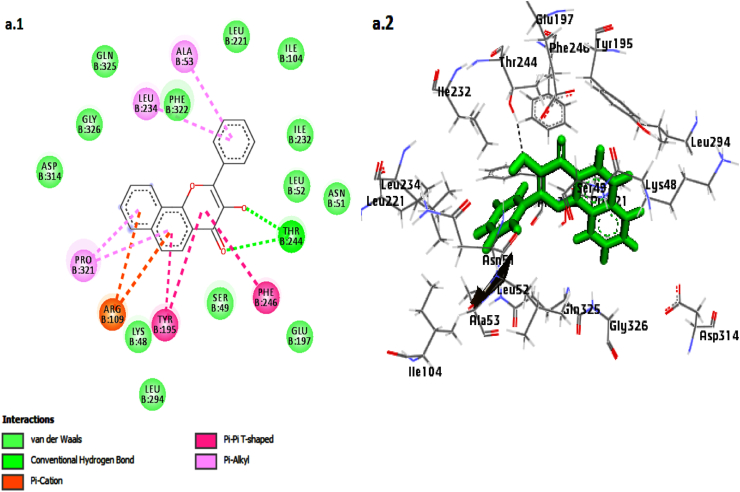
Fig. 3.
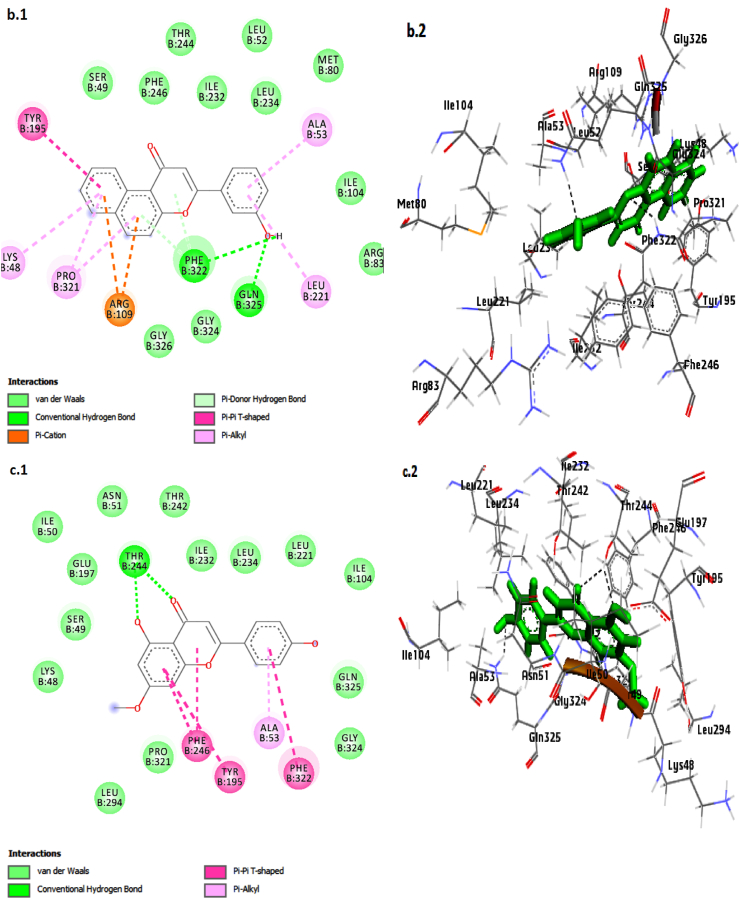
Interaction of the compounds, 3′-Hydroxy-alpha-naphthoflavone (a1, a2), 3′-Hydroxy-ss-naphthoflavone (b1, b2), genkwanin (c1, c2),apigenin (d1, d2) and Ro 61-8048in the binding site of KMO shown in 3D and 2D representation.
Molecular docking analysis revealed several molecular interactions, notably H-bonds and hydrophobic interactions between the ligand molecules and the target enzyme active site residues.Furthermore, the binding poses of the lead KMO inhibitors and standard compounds are depicted in Fig. 3, Fig. 4.
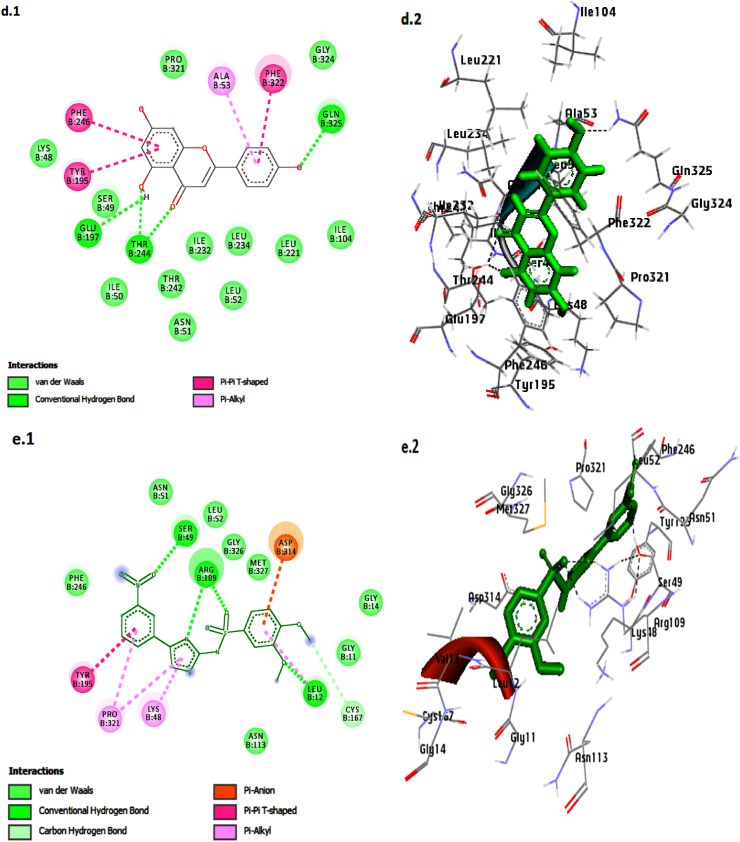
Fig. 4.
Interaction and binding pose of the four lead compounds and reference inhibitor (Ro 61–8048)in the active site; 3′-Hydroxy-alpha-naphthoflavone (Grey), 3′-Hydroxy-ss-naphthoflavone (Black), Genkwanin(Red), Apigenin (Yalow) and Ro 61–8048 (Blue).
Arg 109, Ser 49, and Leu 12 residues were involved in hydrogen bond formation with Ro 61–8048. Besides, the amino acids Lys 48, Asn 51, Leu 52, Asn 113, Cyt 167, Tyr 195 Phe 246, Pro 321, Gly 326, and Met 327 were involved inVan der Waals andhydrophobic interactions.
The lead compound, 3′-Hydroxy-alpha-naphthoflavone,exhibited a combination of hydrogen bonding, van der Waals, hydrophobic interactions, π–π T-shaped, and π-Alkyl interaction in the docked complex. It formed two hydrogen bonds with Thr 244 amino acid residue, a π-cation bond with Arg 109, π-Alkyl interaction with residues Ala 53, Leu 234, and Pro 231, and two π–π T-shaped bonds with Tyr 195 andPhe 246 residues (Fig. 3; a1, a2).
In addition, 3′-Hydroxy-ss-naphthoflavone formed two conventional hydrogen bonds with Gln, 325 andPhe 322 residues, two π-donor hydrogen bonds with the Phe 322 residue, two π-Cation with Arg 109, four π-Alkyl interactions with Lys 48, Ala 53, Leu 221, and Pro 321 residues, and made one π–π T-shaped bond with Tyr 195 amino acid residue (Fig. 3; b.1, b2).
Furthermore, the bonding of genkwanin with the active site of KMO involves various intriguing interactions: two hydrogen bonds with Thr 244 residue, π-Sigma bound with Ile 232 residue, three Pi-Pi-T-shaped bonds with Tyr 195, Phe 246, and Phe 322 residues, and three Pi-Alkyl interactions with Lys 48, Ala 53, and Pro 321 amino acids residues.
We found almost identical interactions between apigenin and the KMO receptor (Fig. 3; d1, d2). Apigenin formed four hydrogen bonds, two each with Thr 244, Glu 197, Gln 325, one with Pi-Sigma (Ile 232), one π-Alkyl with Ala 53 residue, and three π–π T-shaped bonds with Tyr 195, Phe 246, and Phe 322 amino acid residues.
3.1.2. Drug-likeness proprieties and ADMET analysis
The pharmacokinetics and properties of the selected compounds are summarized in Table 2. We determined the drug-likeness using the in silico SwissADME server to test for Lipinski's rule of five parameters, including lipophilicity, the number of H-bond donors and acceptors, and the molecular weight.
Table 2.
Drug-likeness and ADME proprieties of the lead compounds.
| Compound | MW g/mol | AccptHB | DonorHB | Nrot | TPSA (Å2) | LogP< 5 | GI absorption | BBB permeability | Bioavailability score |
|---|---|---|---|---|---|---|---|---|---|
| 3′-Hydroxy-alpha-naphthoflavone | 288.30 | 3 | 1 | 1 | 50.44 | 4.32 | High | Yes | 0.55 |
| 3′-Hydroxy-ss-naphthoflavone | 288.30 | 3 | 1 | 1 | 50.44 | 4.32 | High | Yes | 0.55 |
| Genkwanin | 284.26 | 5 | 2 | 2 | 79.90 | 2.88 | High | No | 0.55 |
| Apigenin | 270.24 | 5 | 3 | 1 | 90.90 | 2.58 | High | No | 0.55 |
| Ro 61-8048 | 421.45 | 7 | 1 | 7 | 159.96 | 4.43 | Low | No | 0.55 |
MW: molecular weight; AccptHB: acceptor hydrogen bond; DonorHB: donor H-bond; Nrot:Rotatable Bond; TPSA: Topological Polar Surface Area; Log Po/w: lipophilicity octanol/water; GI: Gastrointestinal absorption.
The results show that the four top flavonoids similar to apigenin (listed in Table 1) do not violate Lipinski and Veber rules (Table 2); they will be good candidates as orally active drugs.
Blood-brain barrier permeability (BBB) for 3′-Hydroxy-alpha-naphthoflavone and 3′-Hydroxy-ss-naphthoflavone was higher, as well as the gastrointestinal absorption for the four compounds (Table 2).The lead compounds with the highest affinity for KMO exhibited enough lipophilicity level (logP) to suggest that they diffuse through the cell membrane.
Furthermore, the Ld50 value, toxicity class, hepatotoxicity, carcinogenicity, immunotoxicity, and mutagenicity of the four top compounds have been analyzed on the ProTox-II web server results (Table 3).3′-Hydroxy-ss-naphthoflavone and genkwanin exhibited the highest LD50 value of 4000 mg/kg and 3919 mg/kg, respectively, followed by 3′-Hydroxy-alpha-naphthoflavone and apigenin with LD50 of 2500 mg/kg, respectively. These compounds belong to the same class (Class V) and are found to be inactive for hepatotoxicity and immunotoxicity, except for 3′-Hydroxy-ss-naphthoflavone and genkwanin, which are active for carcinogenicity.
Table 3.
Toxicity parameters prediction of the lead compounds.
| Parameters |
3′-Hydroxy-alpha-naphthoflavone |
3′-Hydroxy-ss-naphthoflavone |
Genkwanin |
Apigenin |
Ro 61-8048 |
|
|---|---|---|---|---|---|---|
| LD50(mg/kg) |
2500 |
4000 |
3919 |
2500 |
4500 |
|
| Prediction class | Class V | Class V | Class V | Class V | Class V | |
| Hepatotoxicity | Prediction | Inactive | Inactive | Inactive | Inactive | Active |
| Probability | 0.65 | 0.63 | 0.72 | 0.68 | 0.61 | |
| Carcinogenicity | Prediction | Inactive | Active | Active | Inactive | Active |
| Probability | 0.50 | 0.56 | 0.51 | 0.62 | 0.56 | |
| Immunotoxicity | Prediction | Inactive | Inactive | Inactive | Inactive | Inactive |
| Probability | 0.94 | 0.85 | 0.95 | 0.99 | 0.98 | |
| Mutagenicity | Prediction | Inactive | Inactive | Active | Inactive | Active |
| Probability | 0.60 | 0.62 | 0.57 | 0.57 | 0.52 | |
| Prediction accuracy | 69.26 % | 70.97 % | 70.97 % | 70.97 % | 67.38 % | |
3.1.3. Molecular dynamic (MD)
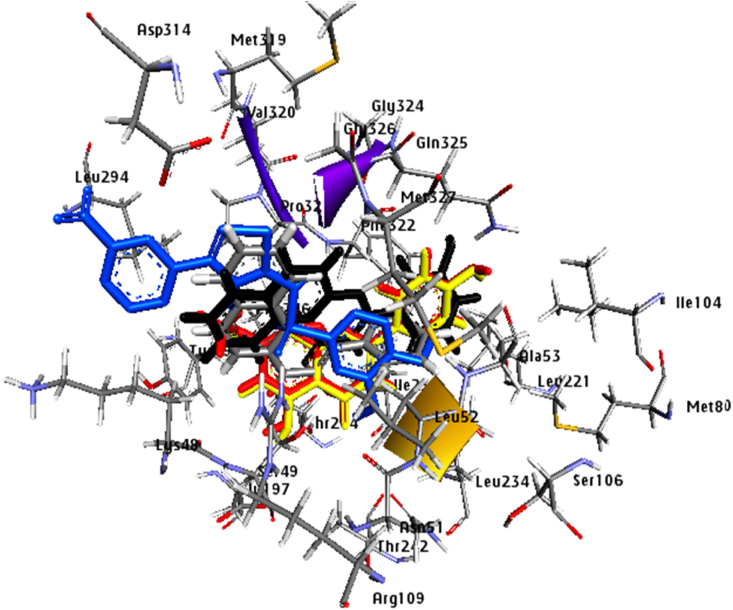
An MD study of the top four docked complexes at 100 ns was conducted to assessthe stability of ligand-KMO complexes. We evaluated the MD simulations using the root mean square deviation (RMSD), root mean square fluctuation (RMSF), and protein-ligand interactions. The RMSD is used to assess the conformational stability of a structure during simulation by evaluating the average change in atom displacement relative to a reference.
The RMSD graph (Fig. 5, A1) of the KMO-3′-Hydroxy-alpha-naphthoflavone complex shows a slight fluctuation, less than 3 Å during the first 60 ns of MD simulation, then the complex ligand-protein shows significant stability. However, it was slightly divergent after 60 ns of simulation, within the acceptable range of 0–3 Å. The RMSD plot of the KMO-3′-Hydroxy-ss-naphthoflavone complex in Fig. 5, B1 shows that the complex is stable throughout the simulation period except for a slight deviation between 40 and 60 ns. The RMSD plot of (Fig. 5, C1) shows that the KMO- genkwanin complex was unstable at the beginning of the simulation; there was high fluctuation from 0 to 10 ns of simulation, and after 10 ns, it became stable up to 80 ns. Although a slight deviation was seen between 80 and 90 ns, the complexes stabilized at the end of the simulation. The KMO-apigenin complex plot (Fig. 5, D1) shows good stability from the simulation's start to end. We observed only a slight fluctuation between 90 and 95 ns, indicating that the complex has not undergone significant conformational changes.
Fig. 5.
MD for the 4 top KMO-complexes, (A1): RMSD plot of the KMO-3′-Hydroxy-alpha-naphthoflavone, (A2): RMSF plot of KMO-3′-Hydroxy-alpha-naphthoflavone complex, (B1): RMSD plot of KMO-3′-Hydroxy-ss-naphthoflavone, (B2): RMSF plot of KMO-3′-Hydroxy-ss-naphthoflavone complex. (C1): RMSD plot of KMO-genkwanin complex, (C2) RMSF plot of KMO-genkwanin complex. (D1): RMSD plot of KMO-apigenin complex, (D2): RMSF plot of KMO-apigenin complex.
We performed RMSF analyses to assess the flexibility per residue using the docked complex structures. The N and C terminals of A2, B2, C2, and D2 fluctuate more than other regions of the protein (Fig. 5). Peaks indicate protein fluctuations; the lower the associated fluctuations, the more stable the complexes during simulation, showing that the system is in equilibrium [26].The RMSF did not vary much over the 100 ns simulation period, and the average RMSF values were kept constant for all complexes except for the higher RMSF seen in the 3′-Hydroxy-alpha-naphthoflavone-KMO and 3′-Hydroxy-ss-naphthoflavone-KMO complexes, at the backbone residue positions between Gly81 and Tyr96.
3.2. KMO inhibitory activity
All flavonoid compounds displayed a good inhibitory potential of KMO, with IC50 values varying between 15.85 ± 0.98 μM and 24.14 ± 1.00 μM at concentrations ranging from 2.5 to 40 μM. Among the flavonoid compounds, 3′-Hydroxy-alpha-naphthoflavone was identified as the lead compound with the highest IC50 activity (15.85 ± 0.98 μM), while apigenin (IC50 value 24.14 ± 1.00 μM) showed the least inhibitory effect. However, Ro 61–8048 (IC50 value 4.91 ± 0.31 μM) was the most effective KMO inhibitor (Table 4).
Table 4.
KMO inhibition (IC50) by the lead compounds.
| Compound | IC50 (μM) | Inhibition type |
|---|---|---|
| 3′-Hydroxy-alpha-naphthoflavone | 15.85 ± 0.98 | Competitive |
| 3′-Hydroxy-ss-naphthoflavone | 18.71 ± 0.78 | Competitive |
| Genkwanin | 21.61 ± 0.97 | Non-competitive |
| Apigenin | 24.14 ± 1.00 | Non-competitive |
| Ro 61–8048 (positive control) | 4.96 ± 0.31 | NT (not tested) |
3.3. Inhibition mechanisms of KMO
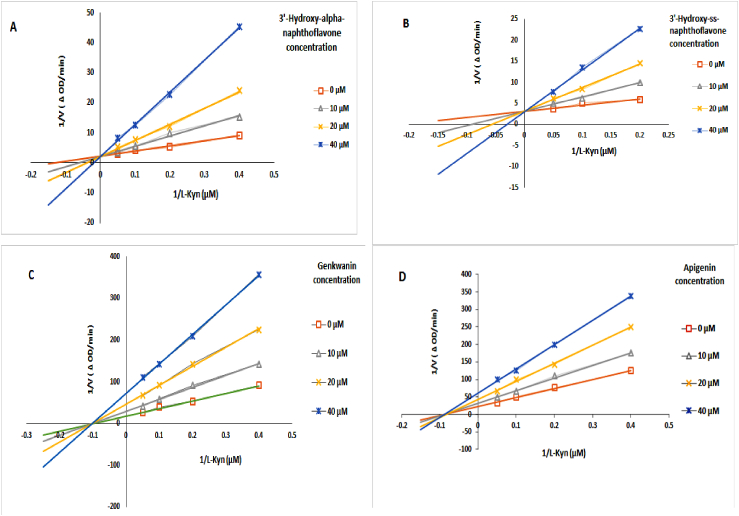
The kinetics of the tested flavonoids were quantitatively analyzed using Lineweaver-Burk plots to investigate the type of inhibition. As noted in (Fig. 6, A, and Fig. 6, B), the value of Km increased without changing Vmax in the presence of increasing concentrations of the inhibitors. In addition, the y-intercept of the curves was unaffected as the concentration of the compounds increased, while the addition of inhibitors changed the x-intercept. This behavior shows that 3′-hydroxy-alpha-naphthoflavone and 3′-hydroxy-ss-naphthoflavone are competitive inhibitors.
Fig. 6.
Inhibitory kinetics of flavonoids (3′-Hydroxy-alpha-naphthoflavone, 3′-Hydroxy-ss-naphthoflavone, genkwanin, and apigenin). KMO enzyme was treated with 0–40 μM of each compound using 0–20 μM of L-Kyn as the substrate. Experiments were performed in triplicate and repeated three times with similar results.
In Fig. 6C and D, the y-intercept changed by adding inhibitors, but there was no effect on the x-intercept, showing that the Vmax values decreased, and the Km values remained constant. We conclude that genkwanin and apigenin are non-competitive inhibitors.
The studied compounds exhibit a strong affinity and effectively engage with the critical residues located within the inhibitor binding site of the protein, demonstrating a remarkable stability within the binding pocket without unnecessary fluctuations. These findings suggest that the selected compounds have the ability to trigger biological reactions within the KMO enzyme, demonstrating their promising inhibitory effects to enhance specific activities within the biological system.
4. Conclusion
Four potent KMO inhibitors from Sigma-Aldrich chemical library of natural compounds were identified based on docking score, predicted binding energies, and in vitro validation by establishing IC 50s. The four compounds exhibited favorable pharmacokinetic proprieties and acceptable RMSD and RMSF stability. The results presented here could help select a lead molecule for future in vivo research, drug discovery, and development targeting KMO.
Funding
This work was supported by the University of Mohamed Khider, Biskra, Algeria (RedouaneRebai and Abdennacer Boudah). Funding was also obtained from the Painless Research Foundation (Redouane Rebai, Luc Jasmin, Abdennacer Boudah). Funding sources were not involved in conducting the research and preparing the article.
Institutional review Board statement
Not applicable.
Informed consent statement
Not applicable.
Data availability
Data will be made available on request.
CRediT authorship contribution statement
Redouane Rebai: Writing – review & editing, Writing – original draft, Visualization, Software, Investigation, Formal analysis, Conceptualization. Miguel Carmena-Bargueño: Writing – original draft, Validation, Software, Methodology. Mohammed Esseddik Toumi: Validation, Software, Methodology. Imene Derardja: Visualization, Validation, Software, Methodology. Luc Jasmin: Writing – review & editing, Validation, Supervision, Conceptualization. Horacio Pérez-Sánchez: Writing – review & editing, Writing – original draft, Validation, Supervision. Abdennacer Boudah: Writing – review & editing, Writing – original draft, Validation, Supervision, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Hahn B., Reneski C.H., Pocivavsek A., Schwarcz R. Prenatal kynurenine treatment in rats causes schizophrenia-like broad monitoring deficits in adulthood. Psychopharmacol. 2018;235(3):651–661. doi: 10.1007/s00213-017-4780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kindler J., Lim C.K., Weickert C.S., Boerrigter D., Galletly C., Liu D., Jacobs K.R., Balzan R., Bruggemann J., O'Donnell M., Lenroot R., Guillemin G.J., Weickert T.W. Dysregulation of kynurenine metabolism is related to proinflammatory cytokines, attention, and prefrontal cortex volume in schizophrenia. Mol. Psychiatry. 2020;25(11):2860–2872. doi: 10.1038/s41380-019-0401-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savitz J. The kynurenine pathway: a finger in every pie. Mol. Psychiatry. 2020;25(1):131–147. doi: 10.1038/s41380-019-0414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith J.R., Jamie J.F., Guillemin G.J. G. J. Kynurenine-3-monooxygenase: a review of structure, mechanism, and inhibitors. Drug Discov. Today. 2016;21(2):315–324. doi: 10.1016/j.drudis.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S., Sakuma M., Deora G.S., Levy C.W., Klausing A., Breda C., Read K.D., Edlin C.D., Ross B.P., Muelas M.W., Day P.J., O'Hagan S., Kell D.B., Scharcz R., Leys D., Heyes D.J., Giorgini F., Scrutton N.S. A brain-permeable inhibitor of the neurodegenerative disease target kynurenine 3-monooxygenase prevents accumulation of neurotoxic metabolites. Commun. Biol. 2019;2(1):271. doi: 10.1038/s42003-019-0520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai M.Y., Lovejoy D.B., Guillemin G.J., Kozak R., Stone T.W., Koola M.M., Galantamine-Memantine M.M. Combination and kynurenine pathway enzyme inhibitors in the treatment of neuropsychiatric disorders. Complex Psychiatry. 2021;7(1–2):19–33. doi: 10.1159/000515066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutchinson J.P., Rowland P., Taylor M.R., Christodoulou E.M., Haslam C., Hobbs C.I., Holmes D.S., Homes P., Lidlle J., Mole D.J., Uings L., Walker A.L., Webster S.P., Mowat C.G., Chung C.W. Structural and mechanistic basis of differentiated inhibitors of the acute pancreatitis target kynurenine-3-monooxygenase. Nat. Commun. 2017;8(1):1–12. doi: 10.1038/ncomms15827. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noorbakhsh A., Hosseininezhadian K.E., Farshadfar C., Ardalan N. Designing a natural inhibitor against human kynurenine aminotransferase type II and a comparison with PF-04859989: a computational effort against schizophrenia. J. Biomol. Struct. Dyn. 2021:1–14. doi: 10.1080/07391102.2021.1893817. [DOI] [PubMed] [Google Scholar]
- 9.Pocivavsek A., Elmer G.I., Schwarcz R. Inhibition of kynurenine aminotransferase II attenuates hippocampus‐dependent memory deficit in adult rats treated prenatally with kynurenine. Hippocampus. 2019;29(2):73–77. doi: 10.1002/hipo.23040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sapko M.T., Guidetti P., Yu P., Tagle D.A., Pellicciari R., Schwarcz R. Endogenous kynurenate controls the vulnerability of striatal neurons to quinolinate: implications for Huntington's disease. Exp. Neurol. 2006;197(1):31–40. doi: 10.1016/j.expneurol.2005.07.004. 2006. [DOI] [PubMed] [Google Scholar]
- 11.Zwilling D., Huang S.Y., Sathyasaikumar K.V., Notarangelo F.M., Guidetti P., Wu H.Q., Muchowski P.J. Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell. 2011;145(6):863–874. doi: 10.1016/j.cell.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherrak S.A., Merzouk H., Mokhtari-Soulimane N. Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: a molecular docking and simulation studies. PLoS One. 2020;15(10) doi: 10.1371/journal.pone.024065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dias M.C., Pinto D.C., Silva A. Plant flavonoids: chemical characteristics and biological activity. Molecules. 2021;26(17):5377. doi: 10.3390/molecules26175377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon M., Ko S.K., Jang M., Kim G.H., Ryoo I.J., Son S., Ahn J.S. Inhibitory effects of flavonoids isolated from Sophora flavescens on indoleamine 2, 3-dioxygenase 1 activity. J Enzyme Inhib Med Chem. 2019;34(1):1481–1488. doi: 10.1080/14756366.2019.1640218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esfahani A.N., Mirzaei M. Flavonoid derivatives for monoamine oxidase–A inhibition. Adv. J. Chem. B. 2019;1(1):17–22. doi: 10.33945/SAMI/AJCB.2019.1.4. 2019. [DOI] [Google Scholar]
- 16.Sánchez-Linares I., Pérez-Sánchez H., Cecilia J.M., García J.M. High-throughput parallel blind virtual screening using BINDSURF. BMC Bioinf. 2012;13(14):1–14. doi: 10.1186/1471-2105-13-S14-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipinski C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods. 2000;44(1):235–249. doi: 10.1016/S1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 18.Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 19.Zoete V., Daina A., Bovigny C., Michielin O. SwissSimilarity: a web tool for low to ultra high throughput ligand-based virtual screening. J. Chem. Inf. Model. 2016;56(8):1399–1404. doi: 10.1021/acs.jcim.6b00174. [DOI] [PubMed] [Google Scholar]
- 20.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7 doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81(1):511–519. doi: 10.1063/1.447334. [DOI] [Google Scholar]
- 22.Puopolo T., Chang T., Liu C., Li H., Liu X., Wu X., Ma H., Seeram N.P. Gram-scale preparation of cannflavin A from hemp (Cannabis sativa L.) and its inhibitory effect on tryptophan catabolism enzyme kynurenine-3-monooxygenase. Biology. 2022;11(10):1416. doi: 10.3390/biology11101416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernardo J., Malheiro I., Videira R.A., Valentão P., Santos A.C., Veiga F., Andrade P.B. Trichiliacatigua and Turneradiffusa extracts: in vitro inhibition of tyrosinase, antiglycation activity and effects on enzymes and pathways engaged in the neuroinflammatory process. J. Ethnopharmacol. 2021;271 doi: 10.1016/j.jep.2021.113865. [DOI] [PubMed] [Google Scholar]
- 24.Hamsalakshmi A.M.A., ArehallyMarappa M., Joghee S., Chidambaram S.B. Therapeutic benefits of flavonoids against neuroinflammation: a systematic review. Inflammopharmacology. 2022:1–26. doi: 10.1007/s10787-021-00895-8. [DOI] [PubMed] [Google Scholar]
- 25.Mossine V.V., Waters J.K., Gu Z., Sun G.Y., Mawhinney T.P. Bidirectional responses of eight neuroinflammation-related transcriptional factors to 64 flavonoids in astrocytes with transposable insulated signaling pathway reporters. ACS Chem. Neurosci. 2022;13(5):613–623. doi: 10.1021/acschemneuro.1c00750. [DOI] [PubMed] [Google Scholar]
- 26.Akbar S., Das S., Iqubal A., Ahmed B. Synthesis, biological evaluation and molecular dynamics studies of oxadiazine derivatives as potential anti-hepatotoxic agents. J. Biomol. Struct. Dyn. 2022;40(20):9974–9991. doi: 10.1080/07391102.2021.1938233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.