Abstract
Acute respiratory infections remain a leading cause of death among young children in low- and middle-income countries. The etiological diagnosis of these infections is challenging due to the similarity in clinical presentations and overlapping symptoms caused by various pathogens. This database provides comprehensive epidemiological, clinical, paraclinical, and biological data on 801 Moroccan children admitted to the Children's Hospital of Rabat for the management of Clinical Severe Pneumonia. Identification of the pathogens responsible of respiratory infections was carried out using blood samples for hemoculture, standard bacterial culture and multiplex RT-PCR using the TrueScience RespiFinder Pathogen Identification Panel (Applied Biosystems).
Keywords: Bacteria, Respiratory viruses, Nasopharyngeal aspirate, Hemoculture, RespiFinder, RT-PCR
Specifications Table
| Subject | Pulmonary and Respiratory Medicine. |
| Specific subject area | Pediatrics, Respiratory diseases, Microbiology |
| Type of data | Excel file |
| Data collection | All patient data were obtained from both the medical records and from the interviews conducted with the children' parents.Identification of the pathogens responsible of respiratory infection was carried out using blood samples for hemoculture, standard bacterial culture and multiplex RT-PCR using the TrueScience RespiFinder Pathogen Identification Panel (Applied Biosystems).Patient data were recorded in a data file containing the following information:
|
| Data source location |
Institutions: Medical Research Laboratory, Children's Hospital, Ibn Sina University Hospital Center, Rabat, Morocco. City: Rabat Country: Morocco |
| Data accessibility | Repository name: Mendeley Data Data identification number: 10.17632/2d9dvnycjw.1 Direct URL to data: https://data.mendeley.com/datasets/2d9dvnycjw/1 |
1. Value of the Data
-
•
Acute respiratory infections represent a major cause of morbidity and mortality in children aged under five years old, particularly in developing countries, where they account for almost two-thirds of childhood illnesses.
-
•
The current study was conducted in Morocco, generating a dataset that encompasses comprehensive epidemiological, clinical, paraclinical, biological, and etiological data on 801 children admitted to the Children's Hospital of Rabat for the management of clinical severe pneumonia.
-
•
This database can be used by the World Health Organization's (WHO) Department of Immunization, Vaccines, and Biologicals to support a project aimed at developing a database detailing the age distribution of respiratory syncytial viruses (RSV) in children under 5 years of age in low- and middle-income countries. This database can also be used by policy makers and healthcare professionals.
-
•
These data will provide a valuable resource for other researchers to conduct further studies, design experiments to investigate specific mechanisms of disease development, assess the effectiveness of interventions, and develop predictive models for better management of acute respiratory infections.
2. Background
In 2014, the Ministry of Health in Morocco estimated that 32,251 children aged between 0 and 59 months presented acute respiratory infections across the country. Pneumonia stands as the primary cause of death from infectious diseases in children under 5 years old, contributing significantly to morbidity and mortality among newborns. Annually, between 152,000 and 490,000 infants aged under one die of pneumonia [1,2].
Clinical diagnosis of acute respiratory infections has focused on detecting and identifying influenza, RSV and parainfluenza viruses (PIV), which are important pathogens in community-acquired pneumonia. However, emerging evidence suggests that other viruses, such as coronaviruses and human metapneumovirus (hMPV), are frequently associated with lower respiratory tract infections. Surprisingly, rhinoviruses have also shown a high prevalence in lower respiratory tract infections in both children and adults [3]. Given this evolving understanding, there is an increasing importance in detecting a wide range of respiratory viruses, highlighting the utility of multiplex RT-PCR.
This data aims to describe the epidemiological and clinical features of respiratory infections, with a specific focus on clinical severe pneumonia, and to identify the profile of respiratory pathogens responsible for the disease in Moroccan children.
3. Data Description
The current dataset represents the initial comprehensive compilation of epidemiological, clinical and laboratory data describing cases of Moroccan children admitted with severe clinical pneumonia. Patient information was meticulously documented in a file containing the following details:
-
-
Epidemiological data;
-
-
Patient environment;
-
-
Clinical examinations and assessment;
-
-
Biological data;
-
-
Radiological data;
-
-
Patient progress;
-
-
Etiological data.
The results of multiplex RT-PCR revealed the presence of mixed infections in 39 % of the hospitalized patients. Rhinovirus was the most frequently detected virus, followed by RSV and adenovirus.
4. Experimental Design, Materials and Methods
4.1. Patients
This prospective study was conducted at the Children's Hospital of Rabat, Ibn Sina University Hospital (CHUIS). A total of 801 children admitted to the children's hospital from November 2010 to December 2011, presenting respiratory symptoms, were recruited for this study. The enrolled children met the WHO definition of severe clinical pneumonia, characterized by history of cough or reported breathing difficulties, elevated respiratory rate relative to age, and chest induration.
4.2. Chest X-ray interpretation
The utilization of chest X-rays is an indispensable tool for evaluating paediatric pulmonary conditions. In our study, chest X-rays were interpreted by pediatricians according to the WHO protocol [4]. The identification of consolidation and/or pleural effusion on a chest X-ray is defined as a "pneumonia assessment criterion". This indicates that the presence of these features strongly indicates the presence of pneumonia in the child.
4.3. Laboratory methods
Samples were analyzed in the medical research laboratory of Children's Hospital of Rabat.
-
-
Hemoculture
To detect the presence of bacteria in the bloodstream, a minimum of 2 ml of venous blood was collected. Blood samples were incubated using an automated blood culture system (BD Bactec®). Subsequently, bacterial isolates were identified either by the BD Phoenix Automated Microbiology System (PHX system, Becton Dickinson) or by colony morphology and biochemical tests.
-
-
Bacterial culture
A standard bacterial culture was performed on Nasopharyngeal aspirate samples (NPAs). The samples were cultured on blood and chocolate agar plates for 72 h at 37°C in an atmosphere of 5 % CO2. Bacterial isolates were identified using conventional biochemical methods (Api 20E, Biomerieux laboratories).
-
-
Qualitative multiplex PCR
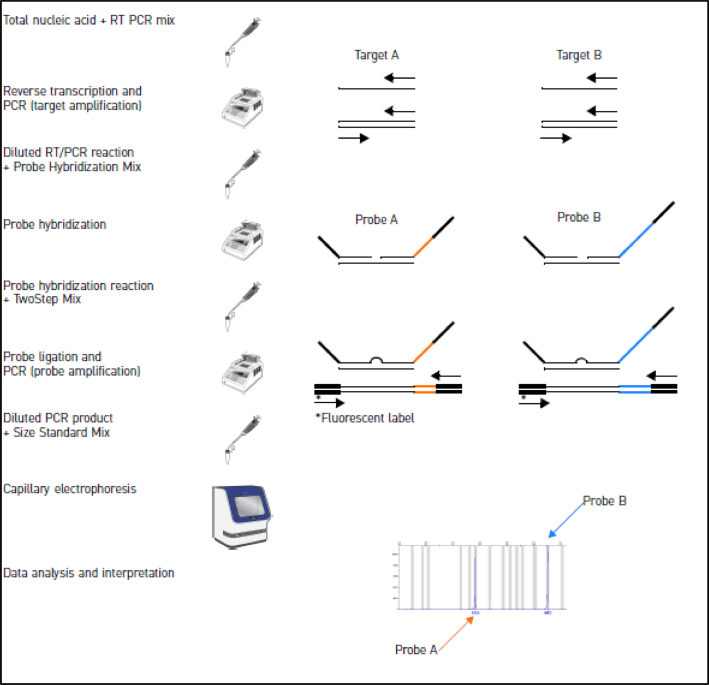
The NPAs were analyzed according to the manufacturer's instructions using the TrueScience™ RespiFinderR 19 Pathogen Identification Panel (CATALOGUE N° 4460382) (Fig. 1), a qualitative multiplex PCR assay used to detect and differentiate pathogens responsible for respiratory tract infection. This assay can detect and differentiate simultaneously 19 respiratory pathogens: 15 viruses, and 4 bacteria (Table 1; Table2).
-
1.
Pre-analysis phase
Fig. 1.
Overview of TrueScience™ RespiFinderR assay [4].
Table 1.
Pathogens detected by RespiFinder® 19.
| Virus |
Bacteria | |
|---|---|---|
| RNA viruses | DNA viruses | |
| Rhinovirus Influenza A Influenza B Influenza A H5N1 Parainfluenza 1 Parainfluenza 2 Parainfluenza 3 Parainfluenza 4 RSV- A RSV-B Coronavirus NL63 Coronavirus OC43 Coronavirus 229E hMPV |
Adenovirus |
Bordetella pertussis Chlamydophila pneumoniae Legionella pneumophila Mycoplasma pneumoniae |
RSV: respiratory syncytial virus; hMPV: human metapneumovirus.
Table 2.
Target genes used for probe and primer design.
| Target | Gene |
|---|---|
| Internal Amplification Control | Polyprotein gene (PP) of encephalomyocarditis virus |
| Rhinovirus | 5’untranslated region Polyprotein gene (PP) |
| Influenza A | Matrix protein gene (M1) |
| Influenza B | Matrix protein gene (M1) |
| Influenza A H5N1 | Matrix protein gene (M1) |
| Adenovirus | Hexon (H) gene |
| Parainfluenza 1 | Haemagglutinin-neuraminidase gene (HN) |
| Parainfluenza 2 | Haemagglutinin-neuraminidase gene (HN) |
| Parainfluenza 3 | Haemagglutinin-neuraminidase gene (HN) |
| Parainfluenza 4 | Haemagglutinin-neuraminidase gene (HN) |
| RSV-A | Major nucleocapsid protein gene (N) |
| RSV-B | Major nucleocapsid protein gene (N) |
| Coronavirus NL63 | Nucleocapsid protein gene (NP) |
| Coronavirus OC43 | Nucleocapsid protein gene (NP) |
| Coronavirus 229E | Nucleocapsid protein gene (NP) |
| hMPV | Nucleocapsidp rotein gene (NP) |
| Bordetella pertussis | Pertussis Toxin promoter (PT) gene |
| Chlamydophila pneumoniae | Major outer membrane protein (OmpA) gene |
| Legionella pneumophila | Macrophage inhibitor potentiator (Mip) gene |
| Mycoplasma pneumoniae | Cytadhesin protein gene (P1) |
RSV: respiratory syncytial virus; hMPV: human metapneumovirus.
Several critical factors are involved in the accurate diagnosis of respiratory pathogens. Firstly, it requires the collection of high-quality samples. Once collected, samples must be transported rapidly to the laboratory. Timely delivery is essential to preserve sample integrity and prevent any degradation that could compromise the accuracy of the testing process. In addition, appropriate storage conditions are essential prior to laboratory testing.
-
2.
Extraction of total nucleic acids (RNA and DNA)
-
3.Preparation of RT-PCR mix
-
•RT-PCR mix component: Dilution Buffer; 5✕ RT-PCR Buffer; Pre-Amplification Primer mix; dNTP Mix; RT-PCR Enzyme Mix.
-
•Probe hybridization mix component: Dilution Buffer; Hybridization Buffer; Probe Mix.
-
•
-
4.
Pre-amplification phase: reverse transcription/PCR to convert viral RNA into complementary DNA and amplify cDNA
In this phase, 10 µL of total nucleic acid template is added to each PCR tube that contains 15 µL of RT/PCR mix, then the PCR tubes are kept on ice until the thermal cycler is pre-heated. When the When the temperature of thermal cycler reaches 50°C, the PCR tubes are placed in the thermal cycler.
-
5.
Probe hybridization phase
In this phase 100 µL of sterile water is added to each reverse transcription/PCR reaction, then 2 µL of the diluted reverse transcription/PCR reaction is added to each tube containing 6 µL of probe hybridization mix. PCR tubes are placed in the thermal cycler, before the probe hybridization program is run.
-
6.
Probe ligation and amplification phase (PCR)
Immediately after the probe hybridization, the prob hybridization program should be stopped and the ligation and PCR program started.
When the temperature of the thermocycler reaches 54°C, pause it to maintain the temperature of the thermal cycler block at 54°C, then prepare the probe hybridization reactions for ligation and PCR and restart the paused ligation and PCR program.
After the program completes, you can store the PCR products at 4°C for up to 1 week or at −20°C for longer periods.
-
7.
Probe detection by capillary electrophoresis
Targets are detected using capillary electrophoresis.
An internal amplification control is included in the assay to differentiate true negative samples from false negatives generated by nucleic acid degradation, PCR inhibition or handling errors.
Limitations
Although this study involved a relatively large number of patients, it is essential to conduct additional studies on a broader scale at various sites to gain a clearer understanding of the epidemiology of respiratory infections in our country. This will be critical for implementing effective case notification plans, as well as for timely patient management. A more comprehensive approach will facilitate a better grasp of the various factors involved, such as pathogen prevalence, associated risk factors, and transmission patterns. Such expanded data will be crucial for informing public health policies and guiding medical interventions, ultimately helping to reduce the incidence and severity of respiratory infections in our population.
Ethics Statement
The patients' parents gave their informed consent to the anonymous use of their data, those of their children, and to the collection of NPAs to be used in the molecular study. All parents were clearly informed of the aim of the study and received a brief summary of the protocol. They participated in the study only after signing an informed consent form.
The protocol was approved by the ethics committee of the Hospital Clinic (Barcelona Spain) and by the ethics committee of Faculty of Medicine and Pharmacy in Rabat and (N° 1252-16Dec2009).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Houssain Tligui: Conceptualization, Methodology, Validation, Supervision, Writing – review & editing. Kenza Hattoufi: Methodology, Writing – review & editing. Imane Jroundi: Methodology, Validation, Investigation, Writing – original draft. Chafiq Mahraoui: Investigation, Validation. Quique Bassat: Conceptualization, Methodology, Supervision, Formal analysis, Writing – review & editing.
Data Availability
References
- 1.Wang H., Naghavi M., Allen C., Barber R.M., Bhutta Z.A., Carter A., et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the global burden of disease study 2015. Lancet. 2016;388(10053):1459‑544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hattoufi K., Tligui H., Obtel M., El Ftouh S., Kharbach A., Barkat A. Molecular diagnosis of pneumonia using multiplex real-time PCR Assay RespiFinder® SMART 22 FAST in a group of moroccan infants. Adv. Virol. 2020;2020 doi: 10.1155/2020/6212643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reijans M., Dingemans G., Klaassen C.H., Meis J.F., Keijdener J., Mulders B., et al. RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses. J. Clin. Microbiol. 2008;46(4):1232‑40. doi: 10.1128/JCM.02294-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherian T., Mulholland E.K., Carlin J.B., Ostensen H., Amin R., de Campo M., et al. Standardized interpretation of paediatric chest radiographs for the diagnosis of pneumonia in epidemiological studies. Bull. World Health Organ. 2005;83(5):353‑9. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.