Abstract
The BTB/POZ (BTB) domain is an approximately 120 residue sequence that is conserved at the N-terminus of many proteins in both vertebrates and invertebrates. We found that the protein encoded by a lethal allele of the Drosophila modifier of mdg4 [mod(mdg4)] gene has two mutated residues in its BTB domain. The identities of the residues at the positions of these mutations are highly conserved in the BTB domain family of proteins, and when the corresponding mutations were engineered into the BTB domain-containing GAGA protein, the activity of GAGA as a transcription activator in a transient transfection assay was severely reduced. The functional equivalence of the BTB domains was established by showing that the BTB domain of the mod(mdg4) protein can effectively substitute for that of GAGA.
INTRODUCTION
Many proteins are mosaic composites constructed from functionally discrete domains. One such domain that has been identified due to its conservation among several zinc finger proteins (e.g. Bric à brac, Tramtrack, Broad) is the BTB domain (1–3). The BTB domain has approximately 120 residues and has been identified in a variety of species, including nematode, chicken, rat and human. Several of the human BTB proteins have been implicated in malignant disease (e.g. PLZF, BCL-6 and HIC-1; 4–7).
The BTB domain most commonly occurs near the N-terminus of proteins that have either a demonstrated role in activating or repressing transcription, or in proteins that are believed to have such a role. It is also in the Kelch protein, an actin binding protein localized to the ring canals of Drosophila oocytes (8). The function of the BTB domain is therefore likely to be a general one that is not specific to DNA binding proteins or to other transcription factors. It is currently believed that the BTB domain facilitates homodimer (9–11) or heterodimer formation (3,7,12–22) as well as oligomerization (23,24).
Trithorax-like (trl) and mod(mdg4) are two Drosophila genes that encode BTB domain-containing proteins with roles in transcription regulation (25,26). trl encodes GAGA protein (26). GAGA was first identified as a factor that stimulates in vitro transcription of the Drosophila engrailed and Ultrabithorax promoters (27,28). Subsequent studies have shown that GAGA has a general and significant role in transcription regulation. Its single Cys2–His2 zinc finger can mediate high affinity specific binding to a consensus sequence containing the GAGAG/CTCTC purine pentamer (29). It binds cis-regulatory elements in many genes (30) as well as to heterochromatic sequences (31). Its capacity to function as an anti-repressor is thought to be related to its role in chromatin remodeling (32–35). These results have led to the proposal that GAGA functions to maintain a conformational state of chromatin that allows general access to DNA by transcription factors. This proposal is consistent with the finding that the GAGA protein remains associated with chromatin throughout the cell cycle. It is also consistent with the observed enhancement of Ultrabithorax and engrailed mutants and the apparent loss of homeotic gene function of trl mutants.
Twenty-four alleles of mod(mdg4) have been isolated. These alleles have a variety of different phenotypes, including neurons with abnormal synapse specificity and synapse morphology, abnormal apoptosis and developmental defects such as segmental transformations. Several enhance position effect variegation (PEV). PEV is a phenomenon that can result when a euchromatic gene has translocated to a heterochromatic region of the chromosome. When mutated, genes encoding structural components of heterochromatin, or genes that function to promote the formation of heterochromatin, would be expected to suppress a PEV phenotype. On the other hand, genes that normally promote transcriptional activation and thereby antagonize the formation of heterochromatin would be expected to enhance a variegated phenotype when mutant. Both mod(mdg4) and trl enhance PEV, and their similar phenotypes suggest both functional and mechanistic relatedness.
As part of an effort to understand the role of the BTB domain, particularly with respect to its role in chromatin organization, we investigated the BTB domains of the GAGA and Mod(mdg4) proteins. From among a collection of eight mutant alleles of mod(mdg4), we identified one in which two highly conserved amino acids in the BTB domain are mutated to biochemically dissimilar residues. Through the use of a transient co-transfection assay, we found that substitution of these same residues in GAGA protein abolished the ability of GAGA to activate transcription of a reporter gene. To provide additional direct evidence for the functional relatedness of BTB domains, the BTB domain of GAGA was replaced by that of the Mod(mdg4) protein. The resulting hybrid protein retained a high level of activity in the co-transfection assay.
MATERIALS AND METHODS
Sequence alignments
The sequence of GAGA was used in a BLAST search against the completed Drosophila genome to assemble a small family of three BTB domain-containing proteins, Broad, Kelch and Lola. These were then individually used in BLAST searches against the Drosophila genome. These searches were run with default parameters except that the expectation parameter was raised to 1000. Hits from these searches were processed using a custom program (M.J.Butte and R.P.Otillar, unpublished results) that identifies overlaps in the hits across multiple searches. The hits were then subjected to statistical filtering and a structurally biased, multiple sequence alignment against the starting four members of the set using CLUSTAL X with default parameters (36). The hits identified in the NCBI database were cross-referenced against the Berkeley Drosophila Genome Project database, which provided the CG numbers of all the proteins. We also cross-checked our BTB superfamily against the Pfam database (37) and the BDGP list of BTB-domain containing proteins. Neither of these latter two lists was complete, and neither had any listings that we did not find using our program. The alignment figure was made using Alscript (38).
Transactivation assays
Schneider 2 (S2) cells were grown in 75-cm2 T flasks, using Schneider’s Drosophila medium plus 10% heat-inactivated fetal calf serum. For transfections, 4 × 106 cells were plated into 6-cm-diameter tissue culture dishes in a volume of 5 ml of medium 24 h prior to the start of a transfection procedure. Calcium phosphate precipitation of the plasmid DNA and addition to the cells were carried out as described (30). Typically, the total amount of plasmid added per 6-cm-diameter plate was ~4 µg and the total amount of DNA was equalized with parental vector. This included pD33-CAT reporter plasmid (30) and a Bluescript-derived expression plasmid (actin 5C-GAGA). Chloramphenicol acetyltransferase (CAT) was assayed using a phase-partition procedure (39).
Homology model
The crystal structure of the PLZF BTB domain (PDB code 1BUO, 7) was used as a template to generate a homology model of GAGA using the program Modeller which uses spatial restraints to place homologous residues (40). The BTB domains from PLZF and GAGA share 76% identical amino acids. Figure 1 was made using Bobscript (41,42) and Raster3D (43). Hydrogen bonds were verified using the program hbplus (44).
Figure 1.
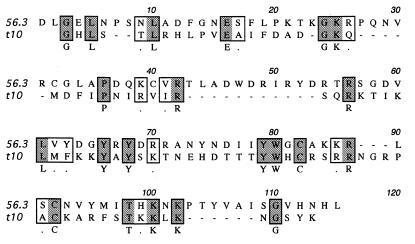
Multiple sequence alignment of the 56 putative BTB domain-containing proteins from Drosophila, ordered in decreasing similarity to GAGA. The 44 sequences most closely related to the BTB consensus are listed above the horizontal line and the 12 whose inclusion in the BTB family is less certain are listed below the line. The three mutated residues, D35, G93 and L112, show considerable homology across the superfamily and are indicated with bullets above the alignment. The secondary structure assignments, derived from the homology model and the PLZF crystal structure, are also indicated above the alignment (tubes are α-helices and arrows are β-strands). The annotation on the left is the Berkeley Drosophila Genome Project CG code and the common name is included where possible. Colors indicate amino acid conservation at each position: blue, hydrophobic residues; green, serine and threonine; magenta, negatively charged; orange, positively charged; olive, glycine; yellow, proline; purple, histidine.
Microscopy
Embryos were collected and prepared by standard methods of methanol/formaldehyde fixation. Fixed embryos were stained with DAPI and examined with epi-fluorescent optics. Cells were grown in 50-mm Petri dishes on sterile 22 × 40 mm glass microscope coverslips that were first treated with TESPA (3-aminopropyltriethoxysilane, Sigma) to improve adhesion. Cells were fixed in the dishes by addition of 3.7% formaldehyde in PBS for 5 min, post-fixed for 2 min by replacement of the buffered formaldehyde with 100% methanol, rehydrated in PBS and then processed for FISH and/or immunofluorescence as described by Dernburg and Sedat (45). For simultaneous detection of the transfected and native GAGA factor, the samples were incubated with a mouse monoclonal α-Glu antibody and a rabbit anti-GAGA antibody and stained with FITC-conjugated anti-mouse and Texas Red-conjugated anti-rabbit secondary antibodies (Jackson Immunoresearch). Three-dimensional data stacks were collected at 0.2-µm Z-spacing using a wide-field optical sectioning microscope. Images were deconvolved with an appropriate point-spread function and projected using a maximum-intensity algorithm.
RESULTS AND DISCUSSION
Cloning novel members
In order to gain insight into the role of the BTB domain, we first sought to establish the size of the BTB domain-encoding gene family in Drosophila. We screened a Drosophila genomic library with hybridization probes derived from the BTB domains of two BTB domain-encoding genes, tramtrack (ttk) (46) and trl. Duplicate filters were probed separately under conditions of low stringency with labeled DNA fragments derived from cDNA clones, and following purification of positive clones, the isolates representing ttk, trl and broad (br) cDNAs were identified. The remaining clones were then assigned to groups based on restriction mapping and Southern analysis, and in the case of seven of them, by cytogenetic mapping to polytene chromosomes. In total, the ttk probe detected nine novel isolates per genome equivalent, and the trl probe detected two. To verify that these sequences represent members of the BTB sequence family, genomic fragments containing the hybridizing sequences from five were subcloned and sequenced. Predicted amino acid sequences from these five clones indicate that they represent mod(mdg4), lola, a gene required for proper axon growth in the embryonic central and peripheral nervous systems (47), abrupt, and the proteins BTB-II and BTB-VII predicted by annotation of the Drosophila genome sequence (48).
Subsequent to the completion of this search for novel BTB domain-containing proteins, the Drosophila genome sequence was released and its annotation by Interpro statistical measures identified 64 BTB domain-containing protein sequences (48). We independently queried the Drosophila genome sequence using a technique that combines information from BLAST queries, multiple sequence alignments and the structural comparisons. This approach identified 44 different sequences in the NCBI database that we judge to have the conserved hydrophobic patches, charged residues and predicted secondary structure motifs that can be considered as hallmarks of the BTB domain (Fig. 1). Twelve additional sequences were not as closely related to the consensus BTB domain and their inclusion in the family is less certain. Eight sequences were either variants produced from a single gene, were deleted for the N- or C-terminal portions of the BTB domain, or were truncated coding sequences that either represent pseudogenes or mistakes in open reading frame predictions. We suggest that the 44 identified sequences listed above the line in Figure 1 represent the best estimate for this gene family in the Drosophila genome. The significant difference between this estimate of 44 and the 64 sequences cited previously (48) suggests that the published estimates from the Interpro analysis should be interpreted with caution.
Examination of the Interpro analysis of the 44 proteins that we designate as bone fide BTB domain-containing proteins reveals that 10 are predicted to contain zinc fingers, two are predicted to have helix–turn–helix domains of the Fis1 type, and five are predicted to have Kelch domains. All of the BTB domains are located at or near the N-termini of the protein sequences. None is predicted to have a signal sequence or transmembrane segment. We conclude that the BTB domain is distributed among a diverse set of intracellular proteins that function in both the cytoplasmic and nuclear compartments. This distribution is consistent with the proposal that the BTB domain has a protein interaction function.
Mutants of mod(mdg4)
In the course of a genetic and molecular analysis of the tinman (tin) gene (49), eight members of an embryonic lethal complementation group were found that map to a location just proximal to tin. An open reading frame in this region was identified and a corresponding cDNA was isolated and sequenced. Over most of its length, this cDNA, t10, is identical to cDNAs derived from mod(mdg4) (50). mod(mdg4) produces at least 21 distinct cDNAs that have a common 5′ region that encodes 402 residues, and a variable 3′-end. A sequence motif present in some members of the mod(mdg4) family includes a Cys2–His2 of unknown function (50), but the 87 residue variable region at the C-terminus of t10 lacks this motif and is a novel variant (Fig. 2). This variable region is encoded by a single exon, identified in the Drosophila sequence as 144 462–144 833. t10 has no recognized DNA binding domain or sequence motif other than the BTB domain.
Figure 2.
Variable C-terminal domain of Mod(mdg4)-t10 and Mod(mdg4)-56.3. The N-terminal 402 residues of t10 and 56.3 (50) include the BTB domain and are identical. The C-terminal residues are aligned according to CLUSTAL X and the residues that conform to the consensus (50) are in bold. Mod(mdg4)-56.3 is also designated as Mod(mdg4)-Alt1 in GenBank.
Three of the lethal alleles of mod(mdg4) were analyzed thoroughly. All were embryonic lethal and had phenotypes that were essentially indistinguishable. None survived to a stage that secreted an embryonic cuticle, and all had abnormalities during pre-cellular blastoderm stages that affected the normal distribution of nuclei (not shown). These phenotypes indicate that mod(mdg4) has an essential embryonic function and are consistent with the proposed role of the Mod(mdg4) protein in regulating chromatin architecture (25). These phenotypes are also similar to those described for mutants of trl (51).
To ascertain whether the lethal phenotype of any of the mod(mdg4) alleles is associated with mutations in the BTB domain, cDNA was prepared from embryos of each mod(mdg4) mutant line, and the BTB domain-encoding regions were sequenced after amplification by PCR. Of the eight lines, seven had an unchanged BTB sequence, and one, mod(mdg4)351, contained two mutations: G→A transitions at nucleotide positions 252 and 429 resulting in D35→N and G93→S substitutions, respectively. Among the Drosophila family of BTB domain-containing proteins, D35 is conserved in 42/44, with one domain substituting E→D and two domains deleted for the N-terminal region. G93 is conserved in 32/44. This high degree of conservation suggests that D35 and G93 are essential for the domain’s normal function.
To assess the functional significance of the mutations in D35 and G93 of mod(mdg4)351, we made use of the ability of the GAGA factor to activate binding site-dependent transcription in transfected S2 cells. Site-directed mutagenesis was used to generate GAGA cDNAs with mutations corresponding to those in the BTB domain of mod(mdg4)351. A double mutant D35N+G93S was made, as well as the single mutant D35N. The mutant cDNAs were each placed under the control of the Drosophila actin 5C promoter, and the abilities of the expressed mutant proteins to activate transcription were compared with that of wild-type GAGA.
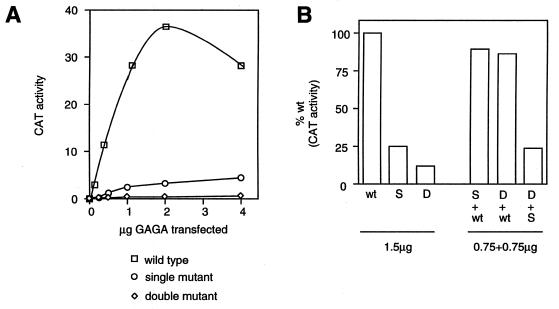
The results depicted in Figure 3 show that the GAGA protein containing the D35N substitution retains <25% of the wild-type transcription activation activity. The G93S mutation in the context of the double mutant results in a further reduction in GAGA activity, to a level of ~10% of wild-type. Thus, the D35 and G93 residues each contribute to the ability of GAGA to activate transcription. Cells were also transfected with a mixture of wild-type and mutant GAGA constructs. The results of these experiments (Fig. 3) indicate that levels of transcription activation were additive, arguing against any trans-dominant action of the mutant proteins.
Figure 3.
Transactivation is reduced by mutations in the GAGA BTB domain. S2 cells transfected with wild-type and mutant GAGA protein expressed under the actin 5C promoter were analyzed for the transcriptional activity of a CAT gene regulated by a promoter containing multiple GAGA binding sites. Proteins containing single (S, D35N) and double (D, D35N and G93S) substitutions were transfected alone in increasing amounts (A), or alone in various combinations at 1.5 µg per assay (B). Activity is expressed as 1000 c.p.m. in (A).
Functional equivalence of BTB domains
In order to determine whether the Mod(mdg4) BTB domain is functionally equivalent to the BTB domain of the GAGA protein, we performed a domain swap. A chimeric GAGA cDNA was constructed which replaces the first 121 residues of GAGA by the first 122 residues of the Mod(mdg4) protein, almost precisely retaining the position of the BTB domain with respect to the GAGA protein. In addition to this chimeric construct, we obtained a clone in which L112 was replaced by a glutamine residue, apparently as a result of mis-incorporation of A for T (sense strand) during PCR synthesis of the Mod(mdg4)-encoding substitution fragment. Allowing for conservative substitutions, this residue is conserved in all members of the Drosophila BTB protein family whose domains are not C-terminally deleted.
Using the co-transfection assay described above, we found that the chimeric Mod(mdg4)/GAGA protein retained ~70% of wild-type GAGA activity. In contrast, the mutant chimera L112Q mutant had essentially no transcription activation activity. These results formally establish the functional relatedness of BTB domains, and, together with the results with the point mutants described above, support the hypothesis that the D35N and G93S mutations contribute to, if they do not entirely account for, the lethal phenotype of mod(mdg4)351.
Structural implications of the mod(mdg4) mutations
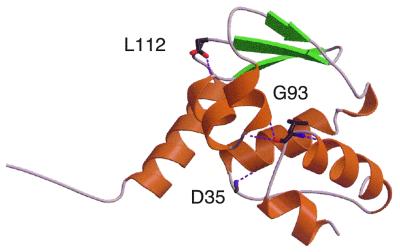
The high degree of sequence identity between the BTB domains of GAGA, Mod(mdg4) and PLZF (>76%) allowed us to model the GAGA and Mod(mdg4) BTB domains based on the structure of the PLZF BTB domain (7). Figure 4 depicts the homology model and illustrates the predicted positions of the three residues mutated in the defective proteins. We propose that each of these residues, D35, G93 and L112, form hydrogen bonds to residues in adjacent helices. Although studies on the stability of the mutant proteins have not been carried out, the hydrogen bonds putatively formed by these residues suggest a ‘helix capping’ role. Elimination of these hydrogen bonds in the mutant proteins might disrupt these helices. Based in part on our identification of D35N, G93S and L112Q as inactivating mutations, site directed mutagenesis engineered equivalent mutations into PLZF. These mutations severely reduced the stability and function of PLZF (6). In a separate study, the D35N mutation was shown to inhibit dimerization and to impair the repression function of PLZF constructs (13).
Figure 4.
Homology model of a GAGA protomer derived from the PLZF crystal structure. The camera is where the dimer-mate would be, showing the three mutated residues and their hydrogen bonds to nearby α-helices.
D35 is situated in a groove in the PLZF dimer and the proposal has been made that this groove is a binding site (7). We found that changing D35 to N in GAGA reduced transcriptional transactivation activity to <25% of wild-type. This suggests that the otherwise electroneutral nature of the groove is critical to function. The high degree of conservation of D35 among the family of Drosophila BTB proteins (42/44) suggests that this residue has a critical role that does not tolerate change.
G93 is both at the end of a helix and at a juncture at which the polypeptide chain makes a sharp turn, almost doubling back in direction. The severity of this turn is needed in order to make a two-stranded β-sheet with the dimer-mate. The presence of G at this point in the polypeptide chain is likely to be important, as G lacks a sidechain and has maximum conformational flexibility. The G93S mutation might not allow the necessary fold to form and may compromise dimer interactions.
L112 is at the base of two helices. It forms the C-terminal cap on the helix from 92 to 105, and it interacts with the N-terminus of adjacent helix from 51 to 60. More importantly, the sidechain of L112 lies buried in a hydrophobic patch consisting of L53, L69, L0, A102, M68 and F52. Since buried polar residues are very uncommon, the severity of the L112Q mutation is not surprising.
Nuclear localization of GAGA
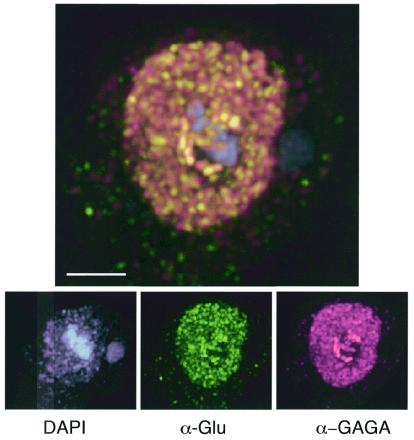
The distribution of the GAGA protein was examined in embryos, S2 cells and larval salivary glands. In older embryos and S2 cells, a diffuse pattern of staining throughout the nucleus was seen to overlay a speckled distribution and areas of intense staining (Fig. 5). We assume that the punctate staining represents the association of GAGA protein with chromosomal sites where the protein binds and that the blocks of intense staining represent its binding to centromeric heterochromatin. GAGA has been shown to associate with centromeric heterochromatin in early embryos (31), and a speckled distribution of the BTB domain-containing PLZF protein has been previously noted (52). We were interested to know whether the inability of the mutant GAGA protein to activate transcription of a co-transfected reporter gene might be reflected in an altered subnuclear distribution. In order to be able to visualize the protein expressed from the transfecting plasmid over the background of endogenous GAGA protein, we inserted an ‘epitope tag’ downstream of the GAGA zinc finger region. The presence of the epitope tag had no significant effect on the ability of either the wild-type or mutant protein to activate transcription in our assays (not shown). When transfected into cells, the distribution of the mutant form of the exogenous GAGA factor was indistinguishable from that of the endogenous protein (Fig. 5). Apparently, the mutations that inactivate GAGA did not de-stabilize it enough to prevent it from localizing in the nucleus or from binding to chromosomes in an apparently normal manner. This suggests that the inactivation phenotype of these mutant proteins cannot be attributed solely to reduced stability.
Figure 5.
Nuclear distribution of wild-type and mutant GAGA protein. An S2 cell transfected with mutant GAGA protein substituted with two replacements (D35N and G93S) and carrying the ‘Glu’ epitope tag was fixed and stained with DAPI, anti-GAGA and anti-Glu antibodies. Both the mutant and wild-type GAGA proteins localize to the nucleus and concentrate in a similar distribution of discrete accumulations.
CONCLUSION
This study was undertaken to explore the diversity of the BTB domains in Drosophila proteins and to examine the functional relationship between them. We used low stringency hybridization to identify members of the BTB domain-containing family and identified several that are among the ones most closely related in sequence to the hybridization probes. The complete set was compiled using a routine that combines sequence alignment and structure comparisons. Our studies suggest that both the structure and function of the BTB domain are conserved between the different members of the set.
Several observations support this conclusion. First, a mutant allele of one of the members of the family of Drosophila BTB domain-containing proteins, Mod(mdg4)351, has two altered residues, both of which are highly conserved among members of the BTB family. This mutant has a lethal phenotype in Drosophila embryos, underscoring the importance of these conserved residues to the function of the BTB domain. Second, GAGA protein mutants engineered to have the homologous mutations have a severely diminished capacity to activate transcription of a target promoter. Third, substitution of the Mod(mdg4) BTB domain for that of GAGA did not alter the transcriptional activation activity of GAGA, highlighting the conservation of BTB domain function, at least in these two proteins. The family of BTB domain proteins includes transcriptional activators, such as GAGA, and transcriptional repressors, such as Tramtrack. It will be interesting to determine whether the BTB domains are specific to each of these two classes or have functional equivalence in both.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by grant GM30637 from the NIH to T.B.K.
REFERENCES
- 1.Zollman S., Godt,D., Prive,G.G., Couderc,J.L. and Laski,F.A. (1994) Proc. Natl Acad. Sci. USA, 91, 10717–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albagli O., Dhordain,P., Deweindt,C., Lecocq,G. and Leprince,D. (1995) Cell Growth Differ., 6, 1193–1198. [PubMed] [Google Scholar]
- 3.Bardwell V.J. and Treisman,R. (1994) Genes Dev., 8, 1664–1677. [DOI] [PubMed] [Google Scholar]
- 4.Ye B.H., Lista,F., Lo Coco,F., Knowles,D.M., Offit,K., Chaganti,R.S. and Dalla-Favera,R. (1993) Science, 262, 747–750. [DOI] [PubMed] [Google Scholar]
- 5.Wales M.M., Biel,M.A., el Deiry,W., Nelkin,B.D., Issa,J.P., Cavenee,W.K., Kuerbitz,S.J. and Baylin,S.B. (1995) Nature Med., 1, 570–577. [DOI] [PubMed] [Google Scholar]
- 6.Melnick A. and Licht,J.D. (1999) Blood, 93, 3167–3215. [PubMed] [Google Scholar]
- 7.Ahmad K.F., Engel,C.K. and Prive,G.G. (1998) Proc. Natl Acad. Sci. USA, 95, 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue F. and Cooley,L. (1993) Cell, 72, 681–693. [DOI] [PubMed] [Google Scholar]
- 9.Dhordain P., Albagli,O., Ansieau,S., Koken,M.H., Deweindt,C., Quief,S., Lantoine,D., Leutz,A., Kerckaert,J.P. and Leprince,D. (1995) Oncogene, 11, 2689–2697. [PubMed] [Google Scholar]
- 10.Davies J.M., Hawe,N., Kabarowski,J., Huang,Q.H., Zhu,J., Brand,N.J., Leprince,D., Dhordain,P., Cook,M., Morriss-Kay,G. and Zelent,A. (1999) Oncogene, 18, 365–375. [DOI] [PubMed] [Google Scholar]
- 11.Morrison D.J., Pendergrast,P.S., Stavropoulos,P., Colmenares,S.U., Kobayashi,R. and Hernandez,N. (1999) Nucleic Acids Res., 27, 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson D.N. and Cooley,L. (1997) J. Cell Biol., 138, 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., Peng,H., Schultz,D.C., Lopez-Guisa,J.M., Rauscher,F.J.,III and Marmorstein,R. (1999) Cancer Res., 59, 5275–5282. [PubMed] [Google Scholar]
- 14.Kobayashi A., Yamagiwa,H., Hoshino,H., Muto,A., Sato,K., Morita,M., Hayashi,N., Yamamoto,M. and Igarashi,K. (2000) Mol. Cell. Biol., 20, 1733–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muto A., Hoshino,H., Madisen,L., Yanai,N., Obinata,M., Karasuyama,H., Hayashi,N., Nakauchi,H., Yamamoto,M., Groudine,M. and Igarashi,K. (1998) EMBO J., 17, 5734–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinzel T., Lavinsky,R.M., Mullen,T.M., Soderstrom,M., Laherty,C.D., Torchia,J., Yang,W.M., Brard,G., Ngo,S.D., Davie,J.R., Seto,E., Eisenman,R.N., Rose,D.W., Glass,C.K. and Rosenfeld,M.G. (1997) Nature, 387, 43–48. [DOI] [PubMed] [Google Scholar]
- 17.Chen J.D. and Evans,R.M. (1995) Nature, 377, 454–457. [DOI] [PubMed] [Google Scholar]
- 18.Dhordain P., Albagli,O., Lin,R.J., Ansieau,S., Quief,S., Leutz,A., Kerckaert,J.P., Evans,R.M. and Leprince,D. (1997) Proc. Natl Acad. Sci. USA, 94, 10762–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huynh K.D. and Bardwell,V.J. (1998) Oncogene, 17, 2473–2484. [DOI] [PubMed] [Google Scholar]
- 20.Hong S.H., David,G., Wong,C.W., Dejean,A. and Privalsky,M.L. (1997) Proc. Natl Acad. Sci. USA, 94, 9028–9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grignani F., De Matteis,S., Nervi,C., Tomassoni,L., Gelmetti,V., Cioce,M., Fanelli,M., Ruthardt,M., Ferrara,F.F., Zamir,I., Seiser,C., Lazar,M.A., Minucci,S. and Pelicci,P.G. (1998) Nature, 391, 815–818. [DOI] [PubMed] [Google Scholar]
- 22.He L.Z., Guidez,F., Tribioli,C., Peruzzi,D., Ruthardt,M., Zelent,A. and Pandolfi,P.P. (1998) Nature Genet., 18, 126–135. [DOI] [PubMed] [Google Scholar]
- 23.Espinas M.L., Jimenez-Garcia,E., Vaquero,A., Canudas,S., Bernues,J. and Azorin,F. (1999) J. Biol. Chem., 274, 16461–16469. [DOI] [PubMed] [Google Scholar]
- 24.Katsani K.R., Hajibagheri,M.A. and Verrijzer,C.P. (1999) EMBO J., 18, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorn R., Krauss,V., Reuter,G. and Saumweber,H. (1993) Proc. Natl Acad. Sci. USA, 90, 11376–11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farkas G., Gausz,J., Galloni,M., Reuter,G., Gyurkovics,H. and Karch,F. (1994) Nature, 371, 806–808. [DOI] [PubMed] [Google Scholar]
- 27.Soeller W.C., Poole,S.J. and Kornberg,T. (1988) Genes Dev., 2, 68–81. [DOI] [PubMed] [Google Scholar]
- 28.Biggin M.D. and Tjian,R. (1988) Cell, 53, 699–711. [DOI] [PubMed] [Google Scholar]
- 29.Pedone P.V., Ghirlando,R., Clore,G.M., Gronenborn,A.M., Felsenfeld,G. and Omichinski,J.G. (1996) Proc. Natl Acad. Sci. USA, 93, 2822–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soeller W.C., Oh,C.E. and Kornberg,T.B. (1993) Mol. Cell. Biol., 13, 7961–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raff J.W., Kellum,R. and Alberts,B. (1994) EMBO J., 13, 5977–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerrigan L.A., Croston,G.E., Lira,L.M. and Kadonaga,J.T. (1991) J. Biol. Chem., 266, 574–582. [PubMed] [Google Scholar]
- 33.Croston G.E., Kerrigan,L.A., Lira,L.M., Marshak,D.R. and Kadonaga,J.T. (1991) Science, 251, 643–649. [DOI] [PubMed] [Google Scholar]
- 34.Ohtsuki S. and Levine,M. (1998) Genes Dev., 12, 3325–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsukiyama T. and Wu,C. (1995) Cell, 83, 1011–1020. [DOI] [PubMed] [Google Scholar]
- 36.Jeanmougin F., Thompson,J.D., Gouy,M., Higgins,D.G. and Gibson,T.J. (1998) Trends Biochem. Sci., 23, 403–405. [DOI] [PubMed] [Google Scholar]
- 37.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barton G.J. (1993) Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- 39.Rushlow C.A., Han,K., Manley,J.L. and Levine,M. (1989) Cell, 59, 1165–1177. [DOI] [PubMed] [Google Scholar]
- 40.Sali A. and Blundell,T.L. (1993) J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 41.Esnouf R.M. (1997) J. Mol. Graph. Model., 15, 112–113. [DOI] [PubMed] [Google Scholar]
- 42.Esnouf R.M. (1997) J. Mol. Graph. Model., 15, 132–134. [DOI] [PubMed] [Google Scholar]
- 43.Merritt E.A. and Bacon,D.J. (1997) Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- 44.McDonald I.K. and Thornton,J.M. (1994) J. Mol. Biol., 238, 777–793. [DOI] [PubMed] [Google Scholar]
- 45.Dernburg A.F. and Sedat,J.W. (eds) (1998) Mapping Three-Dimensional Chromosome Architecture in Situ, Vol. 53. Methods in Cell Biology. Academic Press, San Diego, CA. [DOI] [PubMed]
- 46.Read D. and Manley,J.L. (1992) EMBO J., 11, 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giniger E., Tietje,K., Jan,L.Y. and Jan,Y.N. (1994) Development, 120, 1385–1398. [DOI] [PubMed] [Google Scholar]
- 48.Rubin G.M., Yandell,M.D., Wortman,J.R., Gabor Miklos,G.L., Nelson,C.R., Hariharan,I.K., Fortini,M.E., Li,P.W., Apweiler,R., Fleischmann,W. et al. (2000) Science, 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azpiazu N. and Frasch,M. (1993) Genes Dev., 7, 1325–1340. [DOI] [PubMed] [Google Scholar]
- 50.Büchner K., Roth,P., Schotta,G., Krauss,V., Saumweber,H., Reuter,G. and Dorn,R. (2000) Genetics, 155, 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhat K.M., Farkas,G., Karch,F., Gyurkovics,H., Gausz,J. and Schedl,P. (1996) Development, 122, 1113–1124. [DOI] [PubMed] [Google Scholar]
- 52.Dong S., Zhu,J., Reid,A., Strutt,P., Guidez,F., Zhong,H.J., Wang,Z.Y., Licht,J., Waxman,S., Chomienne,C. et al. (1996) Proc. Natl Acad. Sci. USA, 93, 3624–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]