Abstract
We have developed a protocol for rapid sequencing of short DNA stretches (15–20 nt) using MALDI-TOF-MS. The protocol is based on the Sanger concept with the modification that double-stranded template DNA is used and all four sequencing reactions are performed in one reaction vial. The sequencing products are separated and detected by MALDI-TOF-MS and the sequence is determined by comparing measured molecular mass differences to expected values. The protocol is optimized for low costs and broad applicability. One reaction typically includes 300 fmol template, 10 pmol primer and 200 pmol each nucleotide monomer. Neither the primer nor any of the nucleotide monomers are labeled. Solid phase purification, concentration and mass spectrometric sample preparation of the sequencing products are accomplished in a few minutes and parallel processing of 96 samples is possible. The mass spectrometric analyses and subsequent sequence read-out require only a few seconds per template.
INTRODUCTION
The use of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) for DNA sequencing has been extensively explored over the last decade (1–12). The main driving force for these studies was the enormous sample throughput capacity of a single mass spectrometer. The realization that this approach is limited to short DNA stretches (<100 nt), however, has changed the direction of development over the past 5 years. Instead of shaping a tool that accelerates genomic sequencing, the focus was on other, new applications of DNA sequencing. For instance, combination of a primer extension reaction or related methods with MALDI-TOF-MS has been shown to be an efficient means for the detection of single nucleotide polymorphisms (SNPs) (13–20). Several thousand samples can be analyzed per instrument per day and multiplexing promises even higher sample throughputs. Other potential, not yet well explored, applications are comparative sequencing of short DNA stretches, identification of gene transcription products and verification of recombinant cDNA expression libraries. Twenty nucleotides provide a genomic flag (landmark), 15 nt identify a gene at the cDNA level and a few nucleotides answer the question of whether a recombinant gene was cloned in the right reading frame. With respect to sequencing, these applications put stress on the number of templates analyzed per day per instrument. To determine 100 000 × 10 nucleobases instead of 1000 × 1000 nucleobases adds up to the same total number, but changes the analytical challenge. In contrast to genomic sequencing, this challenge appears well suited to be addressed by MALDI-TOF-MS concerning the necessary accessible mass range and sample throughput.
Among the many proposed concepts for sequencing DNA using mass spectrometry, the most successful has been to combine Sanger cycle sequencing with MALDI-TOF-MS (1,2,4–12). Four nucleobase-specific oligonucleotide ladders are generated in separate reaction vials, which are then separated and detected inside the mass spectrometer. The sequence is determined by comparing the recorded spectra (vertical sequencing). A disadvantage of this approach with respect to throughput and cost is that for each sample four reactions need to be performed and analyzed. The obvious alternative is to perform all four reactions in one vial and determine the sequence by comparing determined oligonucleotide mass differences with expected data (horizontal sequencing).
MALDI-TOF-MS horizontal sequencing has already been demonstrated in combination with limited exonuclease digestion (21–23). The target DNA is degraded by a 3′- or 5′-specific exonuclease and the reaction products mass analyzed. The main drawbacks of this approach are that it is limited to single-stranded DNA and that the kinetics of exonucleolytic digestion are sequence dependent. The latter makes multiple analyses necessary, either of aliquots removed after different reaction times or of a series of reactions performed in parallel, stopped at the same time, but catalyzed by different amounts of the exonuclease. Consequently, the method has been applied to chemically synthesized oligonucleotides and not to typical double-stranded templates such as PCR amplicons or plasmids.
We have explored the one vial Sanger cycle sequencing strategy and have developed a simple and cost-effective protocol. This includes the sequencing reaction, subsequent concentration and purification of the generated products and MALDI sample preparation. Applying this protocol to 300 fmol double-stranded DNA template using 10 pmol primer and 200 pmol each nucleotide monomer (5% ddNTP), 15–20 nt could be determined reliably in a short time. Compared to the use of four sequencing reactions, our protocol increases sample throughput, simplifies the experimental effort and reduces the total cost per template.
MATERIALS AND METHODS
Double-stranded DNA templates and primers
50 bp template: 5′-d(ACT GGA AGA CTT TAA CTG ATG TCA CGG TTA GCG AAG GTT GCG AAT GTC AA)-3′. Primers: 16mer (i), 5′-d(GCA ACC TTC GCT AAC C)-3′; 16mer (ii), 5′-d(CA TTC GCA ACC TTC GC)-3′; 20mer, 5′-d(TTG ACA TTC GCA ACC TTC GC)-3′; 25mer, 5′-d(TTG ACA TTC GCA ACC TTC GCT AAC C)-3′.
299 bp PCR product (bp 3130–3428) of the human cytochrome P450 gene, containing exon 3. Primer, 5′-d(GGC CCA AGT TGC GCA AGG TG)-3′.
PCR products of 2–5 kb inserts of a genomic pUC19 SmaI plasmid shotgun library of Bombus pratorum. Primer, 5′-d(GTG AAT TCG AGC TCG GTA CCC)-3′.
Plasmids containing the above inserts. The same primers as above were used.
The single strands of the 50 bp template and all primers were custom synthesized by Metabion GmbH (Martinsried, Germany). The purified and quantified 299 bp PCR product was provided by S. Vente and B. Timmermann (GenProfile AG, Berlin, Germany). The other PCR products and the plasmids were provided as purified and quantified stock solutions by M. Kube (Sequencing Service Department, Max Planck Institute for Molecular Genetics, Berlin, Germany).
Sequencing reactions
Stock solutions of single-stranded DNA were 10 pmol/µl in double distilled water. The stock solution of the 50 bp template was 1 pmol/µl in 25 mM Tris–HCl, pH 9.5. Stock solutions of purified PCR amplicons and plasmid DNA ranged from 200 to 500 fmol/µl. Cycle sequencing reactions were performed in 10 µl volumes containing 2 pmol 50 bp template or 300 fmol PCR amplicons or plasmid DNA, 10 pmol sequencing primer, 190 pmol each dNTP, 10 pmol each ddNTP, 2 U Thermosequenase (Amersham Life Science, UK) and 1 µl of reaction buffer stock solution. As replacement for the buffer stock solution supplied by the enzyme manufacturer the following buffer stock solution was used: 200 mM (NH4)2SO4, 20 mM MgCl2, 750 mM Tris–HCl, pH 9.5. The reactions were heated to 94°C for 2 min before they were subjected to 20 cycles of annealing temperature (Tanneal) for 20 s, 72°C for 30 s, 94°C for 20 s. Tanneal was 48, 54 and 60°C for the 50 bp template and the 16mer, 20mer and 25mer primers, respectively. For the 299 bp PCR product Tanneal was 60°C. For all PCR products of the shotgun clone library as well as the plasmids Tanneal was 58.5°C.
Sample preparation and mass spectrometry
All following steps were performed with individual samples or eight samples in parallel using a 0.5–10 µl single channel or a 0.5–10 µl eight channel Eppendorf Research Pro laboratory pipette. At ambient temperature, 2 µl of 1 M TEAA stock solution, pH 7.0 (HPLC grade; Merck, Darmstadt, Germany) were added to each reaction vial and the oligonucleotides were purified and concentrated on a micro reverse phase column integrated in the outlet of a P10 pipetting tip (ZipTip-C18; Millipore, USA) following the instructions supplied by the manufacturer. Bound molecules were washed twice with 10 µl of 200 mM TEAA, pH 7.0, and once with 10 µl of 20 mM TEAA, pH 7.0, and eluted with 1 µl of matrix solution directly onto a Bruker Scout 384 stainless steel MALDI sample support. The matrix solution was 400 mM 3-hydroxypicolinic acid (mass spectrometry grade; Fluka, Neu-Ulm, Germany) in 50% acetonitrile. All mass spectra were recorded on a Bruker Scout MTP Biflex III or Reflex III mass spectrometer (Bruker Daltonik, Bremen, Germany). Exclusively positively charged ions were analyzed and 50–150 single shot spectra were accumulated. All samples were analyzed in linear mode using delayed ion extraction and a total acceleration voltage of 20 (Biflex III) or 25 kV (Reflex III). Baseline correction of the recorded spectra was performed using the software package XMASS 5.0, provided by the manufacturer. For all analyses the same set of calibration constants was used (default calibration).
Sequence determination
Adaptive background correction was applied to the raw data using the freeware program M/Z provided at www.proteometrics.com . Adaptive signal expansion, peak picking and all data computation necessary to extract sequence information was performed using in-house developed software routines.
RESULTS AND DISCUSSION
The performance of the developed protocol is documented in Figures 1–4. A 50 bp DNA template was chosen for optimization of the protocol. The following parameters were varied: amount of template, total nucleotide concentration, percentage of ddNTP, magnesium ion, primer and buffer concentrations, number of polymerase units, reaction volume, primer length, annealing temperature and number of reaction cycles. In addition, an in-house developed PCR buffer was tested as a replacement for the reaction buffer supplied by the polymerase manufacturer. The best results were obtained with the following reaction conditions: a reaction volume of 10 µl containing 1 pmol double-stranded template DNA and 75 mM Tris–HCl, pH 9.5, 20 mM (NH4)2SO4, 2 mM MgCl2, 10 pmol primer, 2 U Thermosequenase, 190 pmol each dNTP and 10 pmol each ddNTP. The reaction was incubated at 94°C for 2 min and subsequently 20 cycles of Tanneal for 20 s, 72°C for 30 s, 94°C for 20 s were performed. Tanneal was set 2–4° below the expected melting temperature of the primer provided by the manufacturer. The reaction volume could be reduced to 6 µl or increased to 20 µl with little effect on the sequencing results. The nucleotide monomer concentration proved not to be critical in the range 10–50 µM. Higher values favored incorrect nucleotide incorporation (see below). The magnesium ion concentration could be varied from 1.2 to 6 mM without loss of performance. The amounts of template, enzyme and primer were minimized because these parameters determine the total cost of the sequencing reaction.
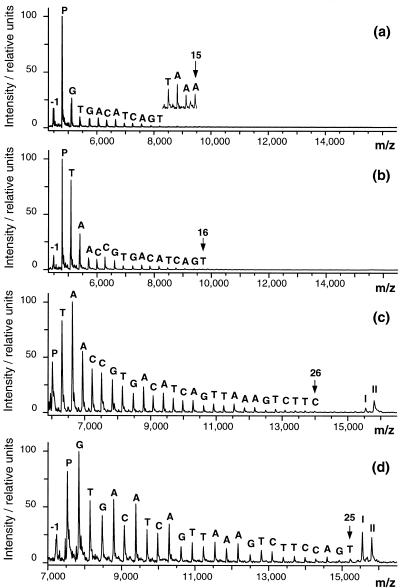
Figure 1.
Influence of primer length on sequencing performance. A 50 bp DNA template was sequenced using different, overlapping primers. (a–d) MALDI-TOF mass spectra of the sequence ladders obtained with the 16mer (i), 16mer (ii), 20mer and 25mer primers, respectively. P, the signal of the sequencing primer; I+C and II+C, the signals of the template strands each extended by one dCMP. To aid visual spectrum interpretation all spectra were baseline corrected.
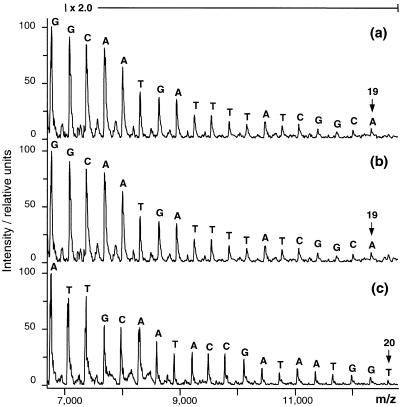
Figure 4.
Partial sequencing of a 2300 and a 1600 bp insert of a genomic pUC19 SmaI plasmid shotgun library of B.pratorum. All sequencing reactions started at the 3′-end of the inserted reverse strand using the same 21mer primer. (a and b) Results obtained with 300 fmol PCR amplicons of the 2300 bp insert versus 300 fmol of the 5000 bp plasmid DNA containing the insert, respectively. (c) Results obtained with 300 fmol of the plasmid DNA harboring the 1600 bp insert. To aid visual interpretation, all spectra were baseline corrected and the intensity was doubled throughout the marked m/z range.
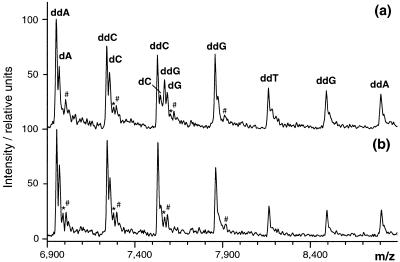
A frequently observed problem with the 50 bp template was abundant incorporation of G instead of the correct nucleotide C at position 25 of the template strand as documented in Figure 2a. This error concerns incorporation of ddNTP as well as dNTP (premature chain termination). In the latter case, however, the synthesized strand was not further extended. The observation that close to the primer specific sites favor incorporation of G instead of C is in agreement with a previous report (10). The above problem was overcome by replacing the reaction buffer provided by the enzyme manufacturer (10× stock solution: 260 mM Tris–HCl, pH 9.5, 64 mM MgCl2) by an in-house optimized PCR buffer [10× stock solution: 750 mM Tris–HCl, pH 9.5, 20 mM MgCl2, 200 mM (NH4)2SO4]. The improvement is documented in Figure 2b. The false incorporation of G was detected 5 nt following the primer, a region that is usually not analyzed in polyacrylamide or capillary electrophoresis based DNA sequencers. The proposed sequencing method, however, focuses on this region. The modified buffer is inexpensive and can be used for both PCR amplification and sequencing. Another important aspect is that the sequencing buffer does not contain a high molecular weight detergent such as Tween 20 or NP 40, because these additives negatively affect the reverse phase purification of oligonucleotides (10).
Figure 2.
Suppression of incorrect nucleotide incorporation. (a) Incorrect incorporation of ddG and dG in addition to the correct nucleotides ddC and dC (non-specific termination event) at position 25 of the 50 bp DNA template (strand II) was frequently observed using the sequencing buffer provided by the polymerase manufacturer. (b) Incorrect incorporation of ddG and dG was suppressed when, instead of the supplied buffer, the sequencing reactions contained 20 mM (NH4)2SO4, 2 mM MgCl2, 75 mM Tris–HCl, pH 9.5. In both cases the 20mer primer was used. *, #, satellite peaks typically observed in MALDI-TOF mass spectra of oligonucleotides due to replacement of acidic protons by a potassium or an iron cation, respectively.
The length of the sequencing primer is an important parameter for the developed sequencing method because it determines the minimum size of the sequencing products. In MALDI-TOF-MS of DNA the lifetime of the generated molecular ions limits the accessible mass range (1). If the analyte ions decompose before they enter the mass-over-charge analyzer the signal resolution drops dramatically or, in the worst case, the signal is lost. It has been shown that the lifetime of DNA molecular ions generated by MALDI strongly declines with increasing number of nucleotides. In addition, it depends on the nature of the nucleotides. G and A are most prone to fragmentation reactions, followed by C and T (1). As a consequence, purine-rich molecular ions are likely to have shorter lifetimes than pyrimidine-rich molecular ions. It follows that shorter and pyrimidine-rich primers favor analysis of sequencing products by MALDI-TOF-MS.
The primer length, however, also directly affects Tanneal of the cycle sequencing reaction and a minimum length is required to start the reaction at a specific site. Tanneal increases if a longer primer is used. As a direct consequence, following heat denaturation the primer competes with the template counterstrand for annealing to the template strand at higher temperature. Consequently, the two strands of the template have less time to reanneal before primer annealing. After the first reaction cycle, already formed sequencing products also compete for annealing to the template strand. In addition, the more Tanneal falls below the reaction temperature optimum of the polymerase (72°C), the more annealed primer will escape from the sequencing reaction by heat denaturation prior to extension.
As expected, the maximum reading length was found to be critically dependent on the length of the primer. This is demonstrated in Figure 1. Using two different 16mer primers that share a sequence of 11 nt, 15 (Fig. 1a) and 16 nt (Fig. 1b) could be determined, respectively. For both primers the expected melting temperature was 51.8°C and Tanneal was set to 48°C. A 20mer primer (expected Tm 57.3°C, Tanneal 54°C) that contained the sequence of the second 16mer primer yielded a maximum of 26 nt (Fig. 1c). In most cases, however, not more than 20 nt could be determined. The abundance ratio of longer versus shorter ladder oligonucleotides was further improved if the primer was extended by an additional 5 nt at the 3′-end (expected Tm 63.0°C, Tanneal 60°C). In that case, the template restricted the maximum possible reading length to 25 nt (Fig. 1d). Although the signal-to-noise ratio of the sequencing products suggests that a longer sequence could have been determined, this example demonstrates the current limit of the proposed method. The reason for this is lack of a signal-to-noise ratio and signal resolution for the longest sequencing product. In some cases the mass difference determined for incorporation of T50 exceeded the correct value by up to 4 Da. Considering that a deviation of 4.5 Da makes the distinction A (+313.2 Da) versus T (+304.2 Da) impossible, it is clear that horizontal sequence determination above 50–55 nt becomes uncertain. This observation is in good agreement with previous reports (10,22,23). The template was specifically designed such that T is the last nucleotide incorporated because the mass difference of 9 Da between A and T is smallest, followed by C/T (15 Da), A/G (16 Da), C/A (24 Da), T/G (25 Da) and C/G (40 Da).
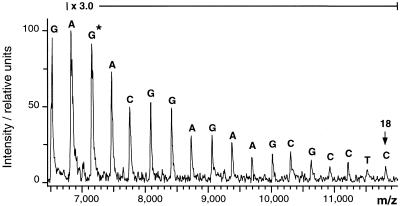
Following optimization, performance of the protocol was evaluated with different DNA templates. These included a 299 bp PCR product of the human cytochrome P450 gene (bp 3130–3428) containing exon 3 from an individual known, from DNA sequencing, to have C3280→G on both alleles. Figure 3 shows the recorded spectrum demonstrating correct determination of 18 nt, including the substitution at position 3 following the primer. Since only one signal is recorded at this position, it is clear that the same mutation is present in both alleles. The presence of G in one of the two alleles would have led to a stop event with ddC in addition to ddG at that position. Because C and G differ in mass by 40 Da, a signal doublet would be the consequence at that position as well as the following positions.
Figure 3.
Partial sequencing of the forward strand of a 299 bp PCR product containing exon 3 of the human cytochrome P450 gene (bp 3130–3428) of an individual harboring two alleles with a C3290→G transition. The sequencing reaction started at G3292 using a 20 nt primer (bp 3293–3302). *, the signal detecting the point mutation. To aid visual interpretation, the baseline was corrected and the intensity was tripled throughout the marked m/z range.
Furthermore, the protocol was applied to the inserts of 20 different clones of a genomic pUC19 SmaI plasmid shotgun library of Bombus pratorum which had been previously sequenced in the service department of our institute. All sequencing reactions started at the 3′-end of the reverse strand. Both PCR amplicons of the inserts as well as intact plasmids were used as template material. In no case did the protocol fail and in all cases the determined sequence was correct and exceeded 15 nt. Typically, 17–20 nt were determined. Figure 4 shows three examples, including a comparison of the results obtained for one clone using PCR amplicons versus plasmid DNA as sequencing template. As expected, the results obtained with the plasmid DNA were very similar to or better than those obtained with the PCR amplicons. An important observation was that the sensitivity of the method, with respect to number of template molecules required, increased considerably when PCR products or plasmids replaced the 50 bp template. In all cases 300 fmol or less yielded good results compared to 1 pmol required for the 50 bp template. This corresponds to ∼50 ng of the 299 bp amplicon, <0.5 µg of PCR-amplified insert and <1 µg of plasmid DNA.
The proposed protocol is optimized for economical determination of short DNA sequences. Sample purification, concentration and mass spectrometric sample preparation are performed in a few minutes using exclusively commercially available supplies. Following the protocol supplied by the manufacturer, ZipTip columns could be used more than five times without loss of performance. Information on the market availability of tested automated pipettors for ZipTip purification is available at www.millipore.com/ziptip . This includes single, eight and 96 channel pipetting stations for low, medium and high throughput applications.
Sequence determination
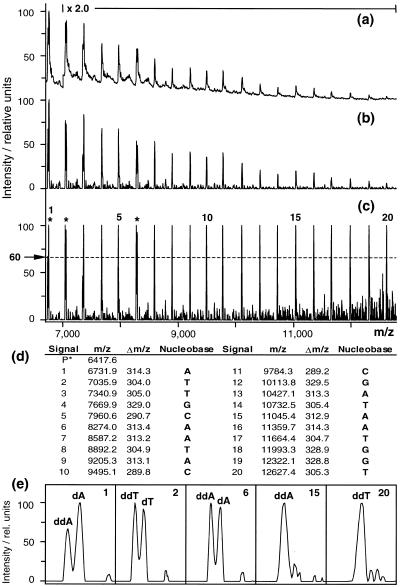
It has been shown by Carroll and Beavis that matrix convolution filters, commonly used in image enhancement to sharpen blurry images and remove noise, provide efficient and very fast algorithms that aid in the extraction of molecular mass information from MALDI-TOF mass spectra (24). One class of filters, designated by the authors adaptive background correction, efficiently removes slowly varying background from the spectra, leaving rapidly varying components, such as peaks. We applied these filters to our sequencing spectra using the corresponding function of the excellent freeware program M/Z and observed a significant improvement in the subsequent peak picking routine. Best results were obtained when the two parameters that control the correction, filter width and filter factor, were set to 2 and 10, respectively. Figure 5a and b compares the unprocessed and background corrected mass spectra obtained from the 1600 bp insert of the shotgun library. It is clearly apparent that the correction filter removed slowly varying components.
Figure 5.
Sequence determination. (a) The unprocessed raw data of the spectrum obtained with the 1600 bp insert of the shotgun library (Fig. 4c). (b) After adaptive background correction. To aid visual interpretation, in both spectra the intensity was doubled throughout the marked m/z range. (c) For each additionally incorporated nucleotide the relative intensity of the highest signal was set to 100% and the central m/z value of all peaks exceeding 60% was determined. (d) Determined m/z values, derived Δm/z values and assigned nucleotides according to: 285.1 Da < Δm/z (C) < 293.3 Da; 300.1 Da < Δm/z (T) < 308.3 Da; 309.1 Da < Δm/z (A) < 317.3 Da; 325.1 Da < Δm/z (G) < 333.3 Da. P*, the m/z value 6433.6 Da determined for the primer was reduced by 16 Da because, in contrast to the sequencing products, the primer terminates with dNTP. (e) For incorporation of 1, 2 and 6 nt, abundant erroneous dNTP termination events accompanied correct ddNTP termination. In these cases both peaks were picked, but only the lower values were used for sequence determination. The two windows to the right show that for incorporation of 15 and 20 nt the signal resolution is sufficient to unambiguously detect abundant dNTP termination events.
To read sequence information from the raw mass spectrometric data the following strategy was added to our protocol. The raw data are first subjected to the above adaptive background correction filter followed by adaptive signal intensity expansion and peak picking using an in-house developed software routine (Fig. 5c). From the established m/z peak list relevant m/z differences (Δm/z) are computed and compared to the expected values according to: 285.1 Da < Δm/z (C) < 293.3 Da; 300.1 Da < Δm/z (T) < 308.3 Da; 309.1 Da < Δm/z (A) < 317.3 Da; 325.1 Da < Δm/z (G) < 333.3 Da. The m/z values of molecular ions differ from the molecular mass only by the addition of one proton, with the consequence that Δm/z is equal to Δm. Figure 5 exemplifies the different steps of the developed strategy. All necessary routines, including adaptive background correction, are currently integrated in a small program that recognizes new incoming data in the raw data directory of the mass spectrometer and automatically extracts the contained DNA sequence information.
An important advantage of the strategy to use m/z differences instead of absolute values for sequence determination is its robustness towards changes in the mass spectrometric calibration constants. Throughout the study the same calibration constants (default constants) were used, with the consequence that the measured molecular masses of the primers used and sequencing products generated in extreme cases deviated by up to 70 Da from the correct value. The mass differences between adjacent ladder oligonucleotides, however, deviated by a maximum of 2.7 Da from the true value. This observation is not surprising because changes in the calibration constants in MALDI-TOF-MS are linear over time as well as across the sample support, as has previously been shown (25). In a very extreme example the determined molecular mass of the longest sequencing product might exceed the true value by 40 Da, whereas the mass determined for the primer is correct (no data offset). In this case the deviations for the other sequencing products will stray closely around a linear graph from 0 to +40 Da, in the same way as they would stray around 0 if the above two oligonucleotides had been used for internal spectrum calibration. If the above scenario covers the incorporation of 20 nt, the maximum deviation of 40 Da means that the 20 Δm/z values will on average exceed the correct value by 2 Da. This deviation falls well within the maximum deviation of 4 Da allowed for by our protocol for nucleotide assignment.
A more realistic, extreme example is shown in Figure 5. The measured molecular mass of the primer (6432 Da) deviates by 70 Da from the correct value (6502 Da) due to a substantial data offset typical of measurements performed with calibration constants that have not been updated for some time. The determined mass differences listed in Figure 5c, in contrast, deviate by a maximum of 1.5 Da (290.7 versus 289.2 Da for C5).
Conclusions
This study demonstrates that one vial Sanger cycle sequencing followed by MALDI-TOF-MS provides a rapid means to determine 15–20 nucleobases. Compared to gel or capillary electrophoresis, the sequencing product analysis times are drastically shortened, which paves the way for high throughput identification of cloned DNA fragments in genomic and cDNA libraries, verification of in-frame cloning in expression libraries and preselection of shotgun libraries. These approaches are currently being evaluated in our laboratory.
Acknowledgments
ACKNOWLEDGEMENTS
We thank S. Vente and B. Timmermann (GenProfile AG, Berlin, Germany) for provision of the 299 bp PCR product and M. Kube (Sequencing Service Department, Max Planck Institute for Molecular Genetics) for provision of PCR amplicons and plasmids of the B.pratorum genomic shotgun library. We thank K.D. Kloeppel for assistance with the mass spectrometric instrumentation and valuable discussions and H. Rauth and R. Reinhardt (Sequencing Service Department, Max Planck Institute for Molecular Genetics) for general support of our studies. This work was funded by the German Ministry for Education and Research and the Max Planck Society.
REFERENCES
- 1.Nordhoff E., Kirpekar,F. and Roepstorff,P. (1996) Mass Spectrom. Rev., 15, 67–138. [DOI] [PubMed] [Google Scholar]
- 2.Limbach P.A. (1996) Mass Spectrom. Rev., 15, 297–336. [DOI] [PubMed] [Google Scholar]
- 3.Nordhoff E., Kirpekar,F., Roepstorff,P., Hahner,S. and Hillenkamp,F. (1996) Trends Anal. Chem., 15, 240–250. [Google Scholar]
- 4.Koster H., Tang,K., Fu,D.J., Braun,A., van den Boom,D., Smith,C.L., Cotter,R.J. and Cantor,C.R. (1996) Nature Biotechnol., 14, 1123–1128. [DOI] [PubMed] [Google Scholar]
- 5.Nordhoff E., Kirpekar,F., Hahner,S., Kristiansen,K., Roepstorff,P. and Hillenkamp,F. (1996) Proceedings of the 44th ASMS Conference on Mass Spectrometry and Allied Topics, Portland, OR, May 12–16, p. 402.
- 6.Cantor C.R., Tang,K., Graber,J.H., Maloney,M., Fu,D.J., Broude,N.E., Siddiqi,F., Koester,H. and Smith,C.L. (1997) Nucl. Nucl., 16, 591–598. [Google Scholar]
- 7.Taranenko N.I., Chung,C.N., Zhu,Y.F., Allman,S.L., Golovlev,V.V., Isola,N.R., Martin,S.A., Haff,L.A. and Chen,C.H. (1997) Rapid Commun. Mass Spectrom., 11, 386–392. [DOI] [PubMed] [Google Scholar]
- 8.Van den Boom D., Ruppert,A., Jurinke,C. and Koster,H. (1997) J. Biochem. Biophys. Methods, 35, 69–79. [DOI] [PubMed] [Google Scholar]
- 9.Koster H., van den Boom,D., Braun,A., Jacob,A., Jurinke,C., Little,D.P. and Tang,K. (1997) Nucl. Nucl., 16, 563–571. [Google Scholar]
- 10.Kirpekar F., Nordhoff,E., Larsen,L.K., Kristiansen,K., Roepstorff,P. and Hillenkamp,F. (1998) Nucleic Acids Res., 26, 2554–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van den Boom D., Jurinke,C., Ruppert,A. and Koster,H. (1998) Anal. Biochem., 256, 127–129. [DOI] [PubMed] [Google Scholar]
- 12.Fu D.J., Tang,K., Braun,A., Reuter,D., Darnhofer-Demar,B., Little,D.P., O’Donnell,M.J., Cantor,C.R. and Koster,H. (1998) Nat. Biotechnol., 16, 381–384. [DOI] [PubMed] [Google Scholar]
- 13.Harksen A., Ueland,P.M., Refsum,H. and Meyer,K. (1999) Clin. Chem., 45, 1157–1161. [PubMed] [Google Scholar]
- 14.Griffin T.J., Hall,J.G., Prudent,J.R. and Smith,L.M. (1999) Proc. Natl Acad. Sci. USA, 96, 6301–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Little D.P., Cornish,T.J., O’Donnell,M.J., Braun,A., Cotter,R.J. and Koster,H. (1997) Anal. Chem., 69, 4540–4546. [Google Scholar]
- 16.Haff L.A. and Smirnov,I.P. (1997) Genome Res., 7, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little D.P., Braun,A., O’Donnell,M.J. and Koster,H. (1997) Nature Med., 3, 1413–1416. [DOI] [PubMed] [Google Scholar]
- 18.Little D.P., Braun,A., Darnhofer-Demar,B. and Koster,H. (1997) Eur. J. Clin. Chem. Clin. Biochem., 35, 545–548. [PubMed] [Google Scholar]
- 19.Braun A., Little,D.P. and Koster,H. (1997) Clin. Chem., 43, 1151–1158. [PubMed] [Google Scholar]
- 20.Ross P., Hall,L., Smirnov,I. and Haff,L. (1998) Nat. Biotechnol., 16, 1347–1351. [DOI] [PubMed] [Google Scholar]
- 21.Pieles U., Zürcher,W., Schär,M. and Moser,H.E. (1993) Nucleic Acids Res., 21, 3191–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juhasz P., Roskey,M.T., Smirnov,I.P., Haff,L.A., Vestal,M.L. and Martin,S.A. (1996) Anal. Chem., 68, 941–946. [DOI] [PubMed] [Google Scholar]
- 23.Smirnov I.P., Roskey,M.T., Juhasz,P., Takach,E.J., Martin,S.A. and Haff,L.A. (1996) Biochemistry, 238, 9–25. [DOI] [PubMed] [Google Scholar]
- 24.Carroll J.A. and Beavis,R.C. (1996) Rapid Commun. Mass Spectrom., 13, 1683–1687. [Google Scholar]
- 25.Egelhofer V., Buessow,K., Luebbert,C., Lehrach,H. and Nordhoff,E. (2000) Anal. Chem., 72, 2741–2750. [DOI] [PubMed] [Google Scholar]