Abstract
Different chemical methods used to attach oligonucleotides by their 5′-end on a glass surface were tested in the framework of solid phase PCR where surface-bound instead of freely-diffusing primers are used to amplify DNA. Each method was first evaluated for its capacity to provide a high surface coverage of oligonucleotides essentially attached via a 5′-specific linkage that satisfyingly withstands PCR conditions and leaves the 3′-ends available for DNA polymerase activity. The best results were obtained with 5′-thiol-modified oligonucleotides attached to amino-silanised glass slides using a heterobifunctional cross-linker reagent. It was then demonstrated that the primers bound to the glass surface using the optimal chemistry can be involved in attaching and amplifying DNA molecules present in the reaction mix in the absence of freely-diffusing primers. Two distinct amplification processes called interfacial and surface amplification have been observed and characterised. The newly synthesised DNA can be detected and quantified by radioactive and fluorescent hybridisation assays. These new surface amplification processes are seen as an interesting approach for attachment of DNA molecules by their 5′-end on a solid support and can be used as an alternative route for producing DNA chips for genomic studies.
INTRODUCTION
DNA chip technology is becoming an important area of high-throughput research in basic biological and disease pathways. The technology used to generate DNA chips is evolving rapidly. Historically, gene analysis was performed by the hybridisation of labelled probes to DNA targets that were passively adsorbed to solid supports such as nitrocellulose, nylon membranes or lysine-coated glass slides (see 1 for review). Covalent linkage of DNA to the surface became the preferred approach as it allows a more stable attachment under conditions used for hybridisation. Direct 5′-end attachment of DNA is of interest but liquid PCR is usually a prerequisite for generating 5′-end functionalised DNA before attachment. Alternatively, short nucleic acid molecules can be directly synthesised onto the solid support by the 3′-end (2). In situ synthesis of 5′-end oligonucleotides has also been described (3,4) and Kwiatkowski et al. (5) have recently reported the possibility of inverting the attachment orientation, from the 3′- to the 5′-end, after synthesis on latex beads. An alternative approach for attaching target DNA for analysis on a DNA chip is to amplify a captured DNA target in situ [6–8; C.P.Adams and S.J.Kron (1997) US patent no. 5641658].
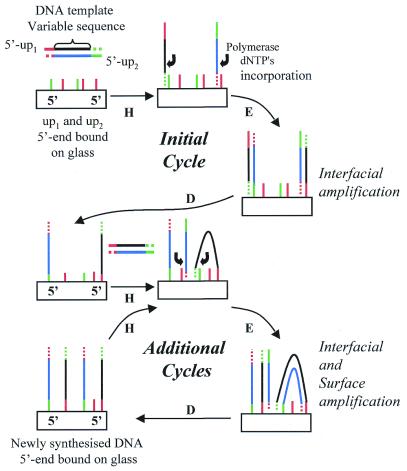
Here we propose a new method for generating DNA chips that utilises 5′-bound oligonucleotide primers directly attached on glass slides and solid phase PCR (Fig. 1). Primers are covalently bound specifically at their 5′-end, allowing the free 3′-end to prime DNA synthesis. DNA templates are able to hybridise to the surface bound primers. The primers are elongated with the DNA polymerase to produce a copy of the hybridised template. This copy is covalently attached to the surface to which the primer was originally attached. The amplification takes place via two distinct mechanisms. First, at each cycle a given proportion of the DNA molecules present in solution participate in the priming elongation process before being released back into solution after denaturation. We call this process ‘interfacial amplification’ as the DNA target present in solution will repeatedly hybridise to attached primers. Secondly, the surface bound copies of the DNA molecules can also hybridise to attached primers and form additional copies in the vicinity of the initial copies. We call this second process ‘surface amplification’.
Figure 1.
Amplification of DNA by solid phase PCR. DNA primers (up1 or up2) are 5′-end covalently attached to the solid support. Thermocycling the surface (H, hybridisation; E, elongation; D, denaturation) in the presence of a solution of DNA target template, nucleotides and a thermostable polymerase leads to the amplification of DNA molecules on the solid surface. During the initial cycle only interfacial amplification occurs, as DNA target template is present only in solution. During subsequent cycles, in addition to interfacial amplification, surface amplification also occurs between the copied DNA template (5′-end attached to the surface) and attached primers in the vicinity.
In order to have solid phase PCR, the attached primers must satisfy three main constraints. First, the surface density of attached oligonucleotides should be high enough for detecting immobilised DNA amplification products by hybridisation assay after solid phase PCR. Secondly, the covalent linkage between the primer and the surface should not be affected by the repeated heating/cooling cycles during the nucleic acid amplification procedure. Thirdly, each primer must be coupled via a 5′-specific linkage to ensure that it can participate in the solid phase PCR process.
Several chemical strategies have been described for the direct attachment of 5′-end oligonucleotides on glass surfaces. Beattie and colleagues have described the use of an epoxide opening reaction to generate a covalent linkage between 5′-amino-modified oligonucleotides and epoxy silane-derivatised glass (9–11). Other studies have used 5′-succinylated target oligonucleotides immobilised on amino-derivatised glass slides (12) or 5′-disulfide modified oligonucleotides bound via disulfide bonds onto thiol-derivatised glass slides (13). Another strategy for covalent DNA attachment via the 5′-termini utilises cross-linkers, such as phenyldiisothiocyanate (14) or maleic anhydride (7). The chemistry of 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC) was first described by Gilham (15) who attached DNA templates to cellulose membranes via the 5′-phosphate group. Carbodiimide chemistry has been employed with other supports such as amino controlled-pore glass (16), latex beads (17,18), dextran supports (19) and polystyrene microwells (20). Heterobifunctional cross-linkers are designed to form stable covalent links in aqueous solution and their use in binding oligonucleotides onto glass surfaces by their 3′- (21) or 5′-ends (22) has been reported. Synthesis of more flexible linkers of different lengths terminating with a hydroxyl group or a primary amino group have been described in the work of Maskos and Southern (23) and Beier and Hoheisel (24), respectively.
The first part of this paper focuses on the selection of an appropriate chemical method for primer attachment on glass slides for solid phase PCR. We evaluate the thermal stability of the oligonucleotide linkage under PCR conditions and the specificity of attachment toward the 5′-end of the oligonucleotides. In the second part of the paper, we characterise the solid phase amplification process using oligonucleotide glass supports prepared with the selected chemical method and hybridisation assays.
MATERIALS AND METHODS
Microscope glass slides (76 × 26 × 1 mm3, Knittel, Merck ABS, Postfach, Germany) or glass cover slips (4 mm diameter, 0.15 mm thick, PolyLabo, Strasbourg, France) were obtained from commercial sources. All solvents were purchased from Fluka Chemicals (Buchs, Switzerland) unless otherwise specified. Buffers were prepared with deionised water and micro-filtered (0.22 µ, Millipore GmbH, Eschborn, Germany). Oligonucleotides were obtained from Eurogentec S.A. (Brussels, Belgium). Water soluble heterobifunctional cross-linkers, sulfonated analogues (s-), s-MBS (m-maleimidobenzoyl-N-hydroxysulfo-succinimide ester), s-SIAB [sulfosuccinimidyl(4-iodoacetatyl)aminobenzoate], s-SMCC [sulfosuccinimidyl 4-(N-maleimidomethyl)-cyclohexane-1 carboxylate], s-GMBS [N-(γ-maleimidobutyryloxy)sulfo succinimide ester], s-SMPB [sulfosuccinimidyl 4-(p-maleimidophenyl)-butyrate] and the corresponding non-water soluble forms (MBS, SIAB, SMCC, GMBS) were purchased from Pierce Chemicals (Rockford, IL). Fluorescent measurements were performed using an inverted epi-fluorescence microscope equipped with an arc mercury lamp (Axiovert S100TV + HBO 100W/2, Carl Zeiss, Oberkochen, Germany) and coupled to a CCD camera (Princeton Instrument Inc., Trenton, NJ). Fluorescent signals were measured by integration of the signal in an image with in-house developed software. HPLC analyses were run on a HP1090 Liquid Chromatography (Hewlett-Packard, Agilent Technologies, Palo Alto, CA) using an Aquapore C18 column (RP300, 7 µm particle size, ABI, Foster City, CA). Phosphorimaging was performed on the Molecular Imager FX (BioRad, Richmond, CA) and radioactive signals analysed with the Quantity One Biorad software.
Nucleic acid preparation: (i) synthetic oligonucleotides
Oligonucleotides were designed as follows;
up1: 5′-TTTTTTTTTTCACCAACCCAAACCAACCCAAACC
up2: 5′-TTTTTTTTTTAGAAGGAGAAGGAAAGGGAAAGGG
Rup1: 5′-GGTTTGGGTTGGTTTGGGTTGGTG
Rup2: 5′-CCCTTTCCCTTTCCTTCTCCTTCT
The 10 Ts serve as a link to enhance hybridisation efficiency as previously shown (14). The 5′-ends of oligonucleotides up1 and up2 were modified during synthesis with one of the following groups: thiol (5′-SH), amino (5′-NH2), phosphate (5′-P), hydroxyl (5′-OH), dimethoxytrityl (5′-DMT) and fluorescein isothiocyanate (5′-FITC). Rup1 and Rup2 are the complementary sequences of up1 and up2, respectively. All the oligonucleotides were desalted and purified by HPLC by the manufacturer (Eurogentec). The purity level of all oligonucleotides was systematically checked by HPLC before use.
Nucleic acid preparation: (ii) 5′-end radiolabelled oligonucleotides
Primers Rup1 and Rup2 were enzymatically phosphorylated with [γ-32P]ATP (Amersham Pharmacia, Buckinghamshire, UK) using the bacteriophage T4 polynucleotide kinase (New England Biolabs, Beverly, MA). Excess [γ-32P]ATP was removed with a chroma-spin column TE-10 (Clontech, Palo Alto, CA).
Nucleic acid preparation: (iii) DNA templates
Fragments of human genomic DNA corresponding to the receptor for advanced glycation end-products (RAGE; GenBank accession no. D28769) were generated by PCR using PCR primers located at 6550–7403 (exon 1–2 template: E1-2) and 8400–9266 (intron 8 template: I8). The sequences of the PCR primers contain the primer sequences of up1 or up2 without the 10 Ts as follows: for E1-2, AGAAGGAGAAGGAAAGGGAAAGGGGCGGCCGCTCGCCTGGTTCTGGAAGACA and CACCAACCCAAACCAACCCAAACCGCGGCCGCTGAGGCCAGTGGAAGTCAGA; and for I8, AGAAGGAGAAGGAAAGGGAAAGGGAGCTGAGGAGGAAGAGAGG and CACCAACCCAAACCAACCCAAACCGAGCTCAAGGCAGGCTAGGAGCTGAG.
Nucleic acid preparation: (iv) labelled nucleic acid probes
RAGE DNA probes were labelled radioactively or with digoxygenin by PCR using PCR primers located at 6550–7091 for probe E1-2 and primers located at position 8400–9266 for the I8 template. Primers for probe E1-2, CCTGTGACAAGACGACTGAA and AGTGAGACGGAGTGTCAGGA; primers for probe I8, AAGGCAGGCTAGGAGCTGAG and AGGGAACTTGACAAGACCGGAG.
Probes were labelled with digoxygenin using a commercial labelling mix (Boehringer-Mannheim, Mannheim, Germany). Probes were radiolabelled by PCR with [α-32P]dCTP (Amersham Pharmacia).
Attachment of oligonucleotides to the glass surface: (i) glass slide pre-treatment
Glass slides were soaked in basic Helmanex solution (HelmanexIIR, 0.25%, 1 N NaOH) for 1 h (HelmanexIIR, Hellma, Müllhein/Baden, Germany), rinsed with water, immersed overnight in 1 N HCl, rinsed again in water and treated for 1 h in sulfuric acid solution (H2SO4/H2O, 1/1, v/v) (H2SO4 95–97%) with a small amount of fresh ammonium persulfate added (BioRad). Acid-treated slides were submitted to extensive washes in water followed by rinsing in ethanol, dried and stored under vacuum for further use.
Attachment of oligonucleotides to the glass surface: (ii) silanisation of glass slide surface
Pre-treated glass slides were immersed into a 5% solution of amino-silane reagents, aminopropyltriethoxysilane (ATS, Aldrich Chemicals, Steinheim, Germany), bis(trimethoxysilylpropyl)amine (BTS, ABCR, Karlsruhe, Germany) or 3-mercaptopropyltrietoxysilane (SHS) in dry acetone. Silanisation was carried out at room temperature for 1 h. After three washes in acetone (5 min/wash), the slides were rinsed once in ethanol, dried and stored under vacuum. We refer to amino-silanised glass slides as ‘derivatised’ glass slides.
Attachment of oligonucleotides to the glass surface: (iii) EDC attachment
Oligonucleotides were prepared as a 1 µM solution in 10 mM 1-methyl-imidazole (pH 7.0) (Sigma Chemicals, Deisenhofen, Germany) containing 40 mM EDC (Pierce, Rockford, IL). Drops of 1–5 µl were spotted on the amino-silanised glass slides. During the 2 h of attachment, the glass slides were kept under a humid atmosphere to avoid evaporation of the drops. The glass slides were washed twice (5 min each) in 5× SSC buffer (0.75 M NaCl, 0.075 M Na citrate pH 7) with 0.1% Tween 20 (Fluka Chemicals) and rinsed in 5× SSC. Slides were finally washed (10 min) in 1.5 M NaCl, 10 mM NaH2PO4 (pH 7) to remove non-covalently attached oligonucleotides (13), rinsed and stored in 5× SSC at 4°C for further use.
Attachment of oligonucleotides to the glass surface: (iv) cross-linkers chemistry
Functionalisation of silanised glass slides with heterobifunctional cross-linkers. Cross-linker refers to a heterobifunctional cross-linking reagent. Functionalised glass slides refer to a derivatised glass surface on which the cross-linker has been covalently attached by one end. Water soluble cross-linkers were prepared at 20 mM in PBS (0.1 M NaH2PO4, 0.15 M NaCl, pH 7.2). The corresponding non-water-soluble reagents were prepared at 20 mM in DMSO:DMF (9:1 v/v). Derivatised glass slides were covered with the cross-linker solution, protected with a clean micro cover glass (Esco, Erie Scientific, Portsmouth, NH) and reacted for 5 h at 20°C. Glass slides functionalised with water soluble cross-linkers were rinsed in PBS, briefly immersed in water and rinsed in ethanol. Glass slides functionalised with non-water soluble cross-linkers were rinsed in ethanol. Functionalised slides were dried and stored under vacuum for further use. As SIAB and s-SIAB are light sensitive, preparation of SIAB functionalised glass slides, storage and attachment of oligonucleotides were done in the dark.
Oligonucleotide attachment on cross-linker-functionalised glass slides. Oligonucleotides were prepared as 50 µM solutions in saline buffer (NaPi: 0.1 M NaH2PO4 pH 6.5, 0.15 M NaCl) and drops (1–5 µl) were spotted on functionalised glass slides. A pH 6.5 solution was necessary to limit disulfide bond formation between 5′-SH modified oligonucleotides. During the 5 h of attachment at room temperature, the glass slides were kept under a humid atmosphere to avoid evaporation. The glass slides were washed twice (5 min each) on a shaker in NaPi, then a solution of mercaptoethanol (Sigma Chemicals), 10 mM in NaPi was used to cap residual maleimide active moieties (1 h treatment). Glass slides were washed (5 min) in NaPi buffer and submitted to 10 min exposure to a 1.5 M NaCl solution (10 mM NaH2PO4, pH 7) to eliminate non-specific attachment. They were washed in 5× SSC buffer with 0.1% Tween (5 min), rinsed in 5× SSC and stored in 5× SSC at 4°C for further use.
Cover slips were used for quantitative analyses relative to loading density determination (using radioactive hybridisation assay) and attachment was performed on both sides.
Solid phase DNA amplification
Glass slides on which oligonucleotides were covalently bonded were subjected to a 1 h blocking step using 5× SSC buffer containing 0.1% Tween 20, 0.1% BSA (New England Biolabs), washed in 5× SSC buffer and finally washed in water to remove salts. DNA amplification was initiated on glass slides with a PCR mix containing 1× PCR buffer, the four dNTPs at 0.1 mM each, 0.1% BSA, 0.1% Tween supplemented with 1 nM DNA and ampliTaq Gold polymerase (0.05 U/ml, PE Biosystems, Foster City, CA). Cover slip glass slides (4 mm diameter) were immersed into 100 µl of the PCR mix placed into PCR tubes (200 µl, MicroAmpTM, PE Biosystems). For the microscope glass slides, the PCR mix was placed in a frame seal chamber (MJ Research, Waterton, MA). The tubes and the microscope glass slides were put into the 16×16 twin tower block thermocycler (PTC 200, MJ Research). Thermocycling was carried out as follows: 94°C for 9 min and 50 cycles (94°C for 45 s, 65°C for 3 min and 72°C for 4 min). Glass slides were washed in 5× SSC, 0.1% Tween 20 (2 × 10 min) and rinsed and stored in 5× SSC until used.
Hybridisation
Radioactively or fluorescently labelled oligonucleotide probes (0.5 µM in 5× SSC, 0.1% Tween 20) were hybridised onto glass slides for 2 h at room temperature. Following hybridisation, the glass slides were rinsed on a shaker in 5× SSC for 5 min and twice in 2× SSC, 0.1% SDS for 5 min at 37°C. Long probes, radioactively-labelled or labelled with digoxygenin, (1 nM in Easyhyb, Boehringer-Mannheim GmbH) were applied to glass slides and hybridised on the MJR PCR machine (95°C for 5 min, 0.1°C/min to 50°C, 50°C for ever). Glass slides were rinsed at 37°C twice in 2× SSC, 0.1% SDS for 15 min, once in 0.2× SSC, 0.1% SDS and twice in 2× SCC. Fluorescent signal of the probe labelled with digoxygenin is obtained by a sandwich assay using an anti-digoxygenin mouse antibody (Boehringer-Mannheim) and a FITC labelled sheep anti mouse (Boehringer-Mannheim).
The amount of hybridisation of the radiolabelled probe was assayed by scintillation counting (Beckman Instruments, Fullerton, CA). The amount of fluorescently labelled probe was detected using the epi-fluorescence microscope setup. To protect the fluorescent labelling dye against photobleaching during irradiation, the glass surface is overlaid with the ProLongTM antifade mix solution (Molecular Probes, Eugene, OR).
RESULTS
Evaluation of the stability of the oligonucleotide attachment under PCR conditions
We have tested several chemical approaches to covalently attach the 5′-end of oligonucleotides to a modified glass support. The chemical procedures described in the literature (10,12,13,20,21) among others (23,24) have been selected because they were previously shown to give reasonably high yields of covalently bound oligonucleotides using only two to three chemical steps. The attached oligonucleotides were subjected to a thermocycling protocol identical to solid phase PCR conditions (see Materials and Methods). The percentage of attached oligonucleotides remaining after such treatment was measured by hybridisation using a fluorescent complementary probe (Table 1).
Table 1. Thermal stability study and 5′-end selectivity.
| Chemistry | 5′-end | Derivatised glass surface | |||||
|---|---|---|---|---|---|---|---|
| ATS | BTS | ||||||
| Loading densitya fmol/mm2 | 5′-specifica fmol/mm2 | Thermal stabilityb | Loading densitya fmol/mm2 | 5′-specifica fmol/mm2 | Thermal stabilityb | ||
| EDC | P | 9 4 | 2 2 | 40 6% | 14 1 | 4 4 | 60 5% |
| OH | 7 1 | 9 1 | |||||
| DMT | 8 2 | 10 4 | |||||
| s-MBS | SH | 25 5 | 22 4 | 56 5% | 28 5 | 24 4 | 52 3% |
| OH | 3 1 | 5 0 | |||||
| DMT | 3 1 | 2 1 | |||||
| s-SIAB | SH | 47 21 | 32 21 | 51 6% | 43 14 | 26 4 | 53 8% |
| OH | 10 4 | 15 4 | |||||
| DMT | 20 15 | 18 10 | |||||
| s-SMCC | SH | nd | nd | 45 8% | nd | nd | 40 3% |
| s-GMBS | SH | nd | nd | 45 6% | nd | nd | 45 4% |
| s-MPB | SH | nd | nd | 50 8% | nd | nd | 48 6% |
Oligonucleotide, up1. with 5′-SH, 5′-OH, 5′-P, 5′-NH2 or 5′-DMT were coupled to glass which was derivatised with ATS or BTS according to the type of chemistry used.
aThe loading density of bound oligonucleotides (fmol/mm2) was determined by hybridisation with a radioactively labelled complementary oligonucleotide, Rup1. As a control, a non-complementary radioactively labelled oligonucleotide (Rup2) was used. The loading density of up1 specifically attached via the 5′-end is calculated as the difference between the density obtained with 5′-SH or 5′-P oligonucleotides and the average of densities obtained with the control oligonucleotides (5′-OH and 5′-DMT).
bThe thermal stability parameter was calculated as a percentage of attached oligonucleotides remaining after the PCR experiment. Oligonucleotides were hybridised using a complementary fluorescently labelled oligonucleotide (5′-FITC Rup1) before and after PCR treatment and the fluorescence signal measured using the epi-fluorescence micrsocope. A non-complementary labelled oligonucleotide was used to check the specificity of the hybridisation (5′-FITC Rup2).
nd, not determined.
The chemical attachment obtained with di-sulfide bound oligonucleotides (13) or with 5′-succinylated oligonucleotides (12), on thiol- (SHS) or amino- (ATS or BTS) derivatised glass slides, respectively, did not withstand the repeated heat treatments (0% measured thermal stability). Although the attachment of 5′-amino-modified oligonucleotides on epoxy-derivatised slides (10) is relatively thermostable (50%), the sensitivity of the epoxy ring to moisture was a major drawback and led to reproducibility problems.
Hydroxylated glass slides were derivatised by silanisation using the primary amino-silane compound, ATS (Fig. 2, top), or the secondary amino-silane compound, BTS, to give reactive amino groups on the surface. Attachment using EDC chemistry (Fig. 2a) leads to phosphoramidate bond formation between the amino group on the derivatised glass surface and the 5′-phosphoramidate oligonucleotide (20). More than 40% of the molecules coupled to the surface using ATS-derivatised glass slides were still attached after PCR treatment. Using BTS-derivatised glass slides, the stability improved and reached 60% (Table 1).
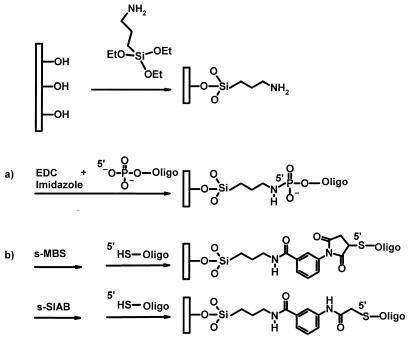
Figure 2.
Process to covalently attach nucleic acid molecules to a glass support. Hydroxylated glass surfaces were silanised using amino-silane reagents exemplified here by ATS. EDC and imidazole reagents lead to the formation of a phophoramidate linkage between amino-derivatised glass surfaces and 5′-phosphate modified oligonucleotides in a one-step reaction (a). For cross-linker chemistry (b), an amide bond was formed between amino groups of the surface and the succinimidyl ester moiety of the cross-linker compounds exemplified with s-MBS or s-SIAB reagents. Oligonucleotides, 5′-thiol modified, reacted with either the maleimide portion of s-MBS-like compounds or the iodoacetamide portion of s-SIAB.
We have tested five different heterobifunctional cross-linkers that have been previously described (21,22) as well as their corresponding water-soluble forms (which contain a sulfonyl modification) (see Materials and Methods). These cross-linkers are capable of forming a covalent bond between the amino and thiol groups (Fig. 2b). We explored two ways of attaching oligonucleotides to solid surfaces using these cross-linkers. The first way is to cross-link the 5′-thiol modified oligonucleotides on amino-derivatised glass slides that are activated by the cross-linkers (Fig. 2b). The second way is to activate the 5′-amino oligonucleotide with the cross-linker and then attach the activated oligonucleotide to mercapto-silanised glass slides. The first procedure gave highest coupling yields and best reproducibility, in coherence with previous reports (21,24). Interestingly, the water-soluble reagents gave consistently higher coupling yields than their insoluble water counterparts, up to ∼10% when assayed by hybridisation with fluorescently-labelled probes (data not shown). This may reflect a better compatibility of the amino-derivatised glass surface with reactions in aqueous buffers such as phosphate buffer (pH 6.5–7.5). The water-soluble cross-linkers s-MBS and s-SIAB gave linkages with the highest thermal stability, 56 and 51% respectively (Table 1). The robustness of the linkage under PCR conditions was equivalent with both silanes (ATS and BTS), indicating that in this case the amide bond was not the most sensitive part of the linkage.
Following these preliminary results, we retained two approaches to attach DNA oligonucleotides on glass slides for further studies. The first approach, ‘EDC chemistry’, is based on the use of EDC-mediated coupling of 5′-phosphate oligonucleotides to amino-derivatised glass slides. The second approach, ‘cross-linker chemistry’, utilises the cross-linkers s-MBS and s-SIAB, to couple 5′-thiol modified oligonucleotides to amino-derivatised glass slides.
5′-End specificity of oligonucleotide attachment
Solid phase PCR requires attachment of oligonucleotide primers to the solid support specifically via the 5′-end. This will ensure that the free 3′-end is accessible for DNA polymerase activity and that base pairing is not hindered by internal attachment. The level of 5′-end selectivity of the cross-linker chemistry was determined using s-MBS and s-SIAB, and the results were compared with those obtained with EDC chemistry. We coupled 5′-SH or 5′-P to amino glass surfaces, using the cross-linker or EDC chemistry, respectively. As a control for 5′-specificity, oligonucleotides with a non-reactive 5′-end group (5′-OH) or a 5′-blocked group (5′-DMT) were used. The amount of hybridised probe to molecules attached on the glass slides (called loading density) was determined using a hybridisation assay with radioactively labelled complementary probes.
The most 5′-end-specific attachment was observed using 5′-SH modified oligonucleotides with the s-MBS cross-linker (Table 1). Using this cross-linker with ATS-derivatised glass slides, only 12% of the control oligonucleotides (5′-DMT and 5′-OH) can be bound to the surface, corresponding to 88% of 5′-end specific attachment. The EDC and s-SIAB chemistries showed lower 5′-end selectivity with as little as 28 and 68% of 5′-end specific attachment, respectively (Table 1). From these results, it appears that attachment using s-SIAB and EDC occurs not only via the 5′-end of the molecules, but also occurs via internal reactive moieties of the nucleic acids. Derivatisation of the glass surface with BTS does not significantly improve the 5′-end specificity of the EDC approach in contrast to previous work using polystyrene surface (20).
The loading density of oligonucleotides obtained with s-MBS reaches 25 fmol/mm2 and is in the range of the loading densities obtained on glass surfaces with other attachment protocols (9,10,13,21). The s-MBS chemistry, with both silanes, combines good 5′-end specificity, thermal stability and reproducibility. Therefore we selected s-MBS chemistry to attach oligonucleotides to glass slides used in the following steps for solid phase PCR. As similar results were obtained with both silanes we preferred to use the less expensive ATS in further studies.
Optimisation of the density of bound oligonucleotide primers in solid phase PCR
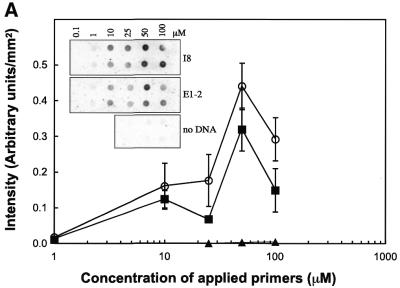
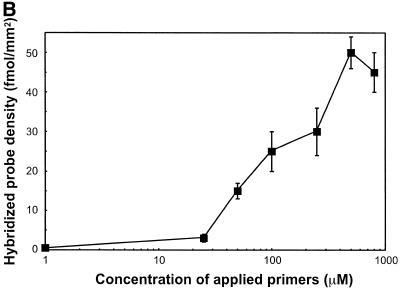
Glass slides for solid phase PCR were prepared using the s-MBS cross-linker on ATS-derivatised glass. Drops (5 µl) containing different concentrations of the oligonucleotide primers, up1 and up2, were spotted onto the glass slides. The slides were then subjected to solid phase PCR with or without a DNA template in solution and the incorporation of radioactively labelled nucleotides was monitored (Fig. 3A). Radioactive nucleotide incorporation in the absence of added template was not detectable indicating that self-priming between the oligonucleotide primers is negligible. The incorporation of radiolabelled nucleotides when the specific DNA template was present showed that the thermostable DNA polymerase activity is maintained when the oligonucleotides are attached to the chemically modified glass substrate. There was an optimum of radiolabelled nucleotide incorporation when a 50 µM solution of primers was spotted onto the glass surface. The corresponding loading densities of bound oligonucleotide primers were 15 and 25 fmol/mm2 when 50 and 100 µM oligonucleotide, respectively, were spotted onto the slide (Fig. 3B). This result suggests that there is an optimal surface primer density for solid phase PCR, and that higher densities of bound primers will decrease the yield of amplification.
Figure 3.
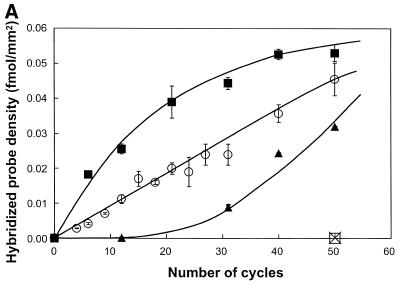
Effect of the concentration of oligonucleotides. A mix of two primers, 5′-SH up1 and 5′-SH up2, was applied to a glass surface derivatised with ATS and functionalised with cross-linker s-MBS. Concentration of the mixed primer solution ranged from 1 to 100 µM. Glass slides were submitted to solid phase PCR with 1 nM template E1-2 (square) or I8 (circle) in solution or with no template in solution (triangle). Radioactively-labelled nucleotides were incorporated during PCR to the solid phase amplified DNA. Glass slides were exposed for phosphor imaging (A, top left) and density of the radioactive signal analysed (A). The loading density of up1, after PCR, at different concentrations was analysed by hybridisation using radioactively-labelled Rup1 complementary oligonucleotide (B).
Solid phase amplification of DNA templates
The amount of amplified target DNA molecules following solid phase PCR with increasing concentrations of DNA template added in solution was quantified using hybridisation assays with radioactive and fluorescent probes. The amplification was specific for templates containing tails complementary to the primers bound to the surface (Fig. 4). The amount of solid phase amplification product varied with the concentration of DNA in solution and rose with increasing numbers of PCR cycles. This indicates that solid phase amplification is indeed dependent on the initial target DNA concentration and on the number of amplification cycles as suggested by the model in Figure 1. The amplification levels observed with the 1 and 10 nM DNA concentrated solutions (Fig. 4A) are compatible with a linear amplification behaviour, as predicted by the interfacial amplification model (Fig. 1). Increasing the number of PCR cycles allowed the detection of target DNA templates in the picomolar concentration range (Fig. 4B). A plateau in amplification product was observed with DNA concentration above 10 nM. In these conditions the loading density of solid phase amplified DNA was ∼0.05 fmol/mm2 (Fig 4A). Since the primer loading density is 15 fmol/mm2 (at 50 µM primer), we conclude that 1 in 300 primers are converted into target DNA. This absence of primer saturation could be explained by a screening effect of the previously attached DNA molecules that could prevent additional DNA molecules reaching the primers on the surface.
Figure 4.
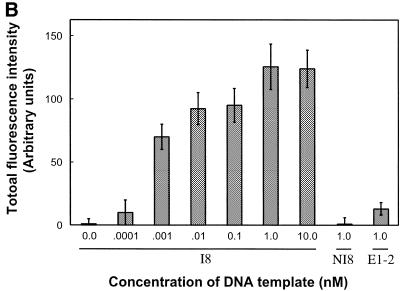
Effect of the concentration of DNA template in solution. A 50 µM primer solution (5′-SH up1 and 5′-SH up2) was applied on glass slides derivatised with ATS and functionalised with the bi-functional cross-linker s-MBS. (A) Glass slides were submitted to solid phase PCR with increasing concentrations of template I8: 0.1 nM (triangle), 1 nM (circle), 10 nM (square). The loading density of DNA molecules (fmol/mm2) amplified on glass surface was determined by hybridisation with radioactively-labelled probe. The negative control, 1 nM of NI8 template (open square), has the same sequence as I8 but does not contain up1 or up2 primer sequences. To check the specificity of the hybridisation assay a non-specific radioactive probe was used (cross). (B) Fifty PCR cycles were run with I8 template (0, 0.0001, 0.001, 0.01, 0.1, 1 and 10 nM) or non-specific templates (1 nM of NI8 or E1-2). Glass slides were submitted to hybridisation using specific I8 digoxygenin-labelled long probe and the fluorescence signal was measured using the epi-fluorescence microscope.
Contribution of interfacial and surface amplification during solid phase PCR
Solid phase amplification can occur by two processes, interfacial and surface amplification, depending on the presence or absence of the template in solution (Fig. 1). The contribution of surface amplification during solid phase amplification is characterised as follows. Two PCR cycles were performed with DNA templates in solution to covalently attach the templates to the glass slide by interfacial amplification. The initial PCR solution was replaced with a fresh PCR mix lacking the DNA template and amplification was continued for a further 28 cycles. The amount of surface amplification product was then determined by radioactive hybridisation (Table 2). The initial loading density of covalently attached DNA after the first two cycles was 0.0011 fmol/mm2. After 28 more cycles with no DNA in solution, the DNA coverage reached ~0.0029 fmol/mm2, indicating that the initially attached DNA was amplified 3-fold, on average, by surface amplification. We have shown previously that, during PCR, half of the attached nucleic acid molecules were released from the surface into the PCR mix. Therefore, amplification may take place in solution and the solution amplification product may be involved in interfacial amplification. However replacing the PCR solution after each cycle during the 28 cycles did not change the final surface coverage, ruling out the latter mechanism. When DNA template was maintained in PCR solution during PCR (30 cycles), the solid phase amplification result was 10-fold higher (0.019 fmol/mm2). We therefore conclude that interfacial amplification is predominant over surface amplification during solid phase PCR when both processes occur concomitantly.
Table 2. Contribution of surface amplification.
| A | B | C | D | E | ||||
|---|---|---|---|---|---|---|---|---|
| Cycle number | 1, 2 | 3–30 | 1, 2 | 3–30 | 1, 2 | 3–30 | 1–30 | 1–30 |
| DNA template I8 | + | – | + | – | + | – | + | |
| DNA template E1-2 | + | |||||||
| DNA polymerase | + | – | + | + | + | + | + | + |
| Amplified DNA coverage (fmol/mm2) | 0.0011 0.0002 | 0.0029 0.0002 | 0.0028 0.0003 | 0.0190 0.0002 | 0.0000 0.0001 |
Glass slides were prepared as described in Figure 4. Glass slides were submitted to two first solid phase PCR cycles (1, 2) with a 1 nM solution of template (I8) and Taq polymerase. Then 28 additional cycles (3–30) were run using the following procedures: A, without DNA template or DNA polymerase in solution; B, without DNA template but with DNA polymerase in solution; C, as B but with fresh PCR mix after each cycle; D, with DNA template (I8) and DNA polymerase in solution; E, as D but using control template (E1-2). The loading density of DNA molecules (fmol/mm2) amplified on the glass surface was determined by hybridisation with specific radioactively-labelled probe.
DISCUSSION AND CONCLUSION
We have determined that the most suitable solid phase chemistry for attachment of primers to amino-derivatised glass slides for solid phase PCR is via the s-MBS heterobifunctional cross-linker and 5′-thiol modified oligonucleotides. The s-MBS cross-linker provides a robust chemistry and can be easily scaled-up. With this method, we found that the oligonucleotides become densely attached to the glass surface. The s-MBS linkage is resistant to PCR conditions and the coupling to the solid support is highly specific for 5′-thiol modified oligonucleotides. The specificity of the attachment towards the 5′-end is chemical-dependent and, as previously shown with di-sulfide linked oligonucleotides (13), is vastly improved when the 5′-functional group involved in the bond, such as the thiol group, is different to the functional groups present on the DNA. Using the s-MBS linkage, the functionalised glass surface did not inhibit PCR and the subsequent amplification of DNA strands occurred at the surface of the glass slide.
The thermal stability of oligonucleotides bound to a functionalised glass surface is dependent on the chemistry of attachment and large losses are observed for several chemistries. It is important to note that, even with the most thermally stable chemistries such as EDC or the s-MBS and s-SIAB cross-linker chemistries, the retention of bound oligonucleotides following PCR thermocycling seems to reach a maximum of 50–60%. This compares well with the few reports where stability to heat treatment has been studied. For example, Chrisey et al. (21) have reported that 60% of oligonucleotides attached to glass chips using the SMPB cross-linker are released from the glass surface during a 10 min treatment at 80°C. We believe that the release of oligonucleotide is not necessarily due solely to the stability of the covalent bond between oligonucleotide and the amino-silane. We suspect that the cleavage of the bond between the amino-silane and the glass surface, as well as a potential instability of the glass surface itself contributes significantly to the observed release of bound oligonucleotides. Exploring alternative materials such as silicon (22) could be a route for improved support of DNA chips and are under development.
We have shown that when the DNA target is in solution, the DNA template is mainly amplified on the solid surface by an interfacial amplification process. As interfacial amplification takes place at each cycle, using the same amount of DNA molecules present in solution, the number of attached DNA copies due to this process increases linearly until surface saturation. Amplification is linear using concentrated DNA template solution up to 1 nM. This is another indication of the predominance of interfacial amplification during solid phase PCR with these template concentrations. However, when DNA target is used at lower concentrations, and even removed completely from the PCR solution, the DNA molecules that are already 5′-end covalently attached are amplified by the surface amplification process.
To our knowledge, this study presents the first evidence of DNA amplification occurring on the surface of a solid. In other words, we present the first study of solid phase PCR on a glass surface with no primers freely diffusing in solution. In previous studies, freely-diffusing primers were also present in the PCR mix to participate in DNA target amplification in solution in order to increase the amount of DNA target attached to the surface (7,8). In our case, DNA template, which is present in solution in the picomolar concentration range, can be highly amplified exclusively on the glass surface and detected by hybridisation assays. In contrast with previously reported in situ PCR in which amplification was in the bulk of porous polyacrylamide gel (8) with one primer attached to the polymer, the surface amplification on glass slides reported here occurs strictly at the solid–liquid interface. Micro-structuring effects similar to those reported in gels (8) have also been observed during surface amplification and preliminary results can be found in patent literature [E.H.Kawashima, L.Farinelli and P.Mayer (1998), WO patent no. 9844152]. Demonstration of the micro-structuring effects as well as practical applications of this technology are presently under development and will be reported at a later stage.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Gary Buell, Dr Serge Halazy and Professor Gerard Adessi for careful reading of the manuscript; Denis Fahmi, Sandrine Da Cruz and Geneviève Guedu for their technical support of this project; and Christopher Herbert for help in the preparation of Figures.
REFERENCES
- 1.Ramsay G. (1998) Nature Biotechnol., 16, 40–44. [DOI] [PubMed] [Google Scholar]
- 2.Pease A.C., Solas,D., Sullivan,E.J., Cronin,M.T., Holmes,C.P. and Fodor,S.P.A. (1994) Proc. Natl Acad. Sci. USA, 91, 5022–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shchepinov M.S., Case-Green,S.C. and Southern,E.M. (1997) Nucleic Acids Res., 25, 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mir K.U. and Southern,E.M. (1999) Nature Biotechnol., 17, 788–792. [DOI] [PubMed] [Google Scholar]
- 5.Kwiatkowski M., Fredriksson,S., Isaksson,A., Nilsson,M. and Landegren,U. (1999) Nucleic Acids Res., 27, 4710–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oroskar A.A., Rasmussen,S.E., Rasmussen,H.N., Rasmussen,S.R., Sullivan,B.M. and Johansson,A. (1996) Clin. Chem., 42, 1547–1555. [PubMed] [Google Scholar]
- 7.Yang M., Kong,R.Y.C., Kazmi,N. and Leung,A.K.C. (1998) Chem. Lett., 257–258. [Google Scholar]
- 8.Mitra R.D. and Church,G.M. (1999) Nucleic Acids Res., 27, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamture J.B., Beattie,K.L., Burke,B.E., Eggers,M.D., Ehrlich,D.J., Fowler,R., Hollis,M.A., Kosicki,B.B., Reich,R.K., Smith,S.R., Varma,R.S. and Hogan,M.E. (1994) Nucleic Acids Res., 22, 2121–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beattie W.G., Meng,L., Turner,S.L., Varma,R.S., Dao,D.D. and Beattie,K.L. (1995) Mol. Biotechnol., 4, 213–225. [DOI] [PubMed] [Google Scholar]
- 11.Beattie K.L., Beattie,W.G., Meng,L., Turner,S.L., Coral-Vazquez,R., Smith,D.D., McIntyre,P.M. and Dao,D.D. (1995) Clin. Chem., 41, 700–706. [PubMed] [Google Scholar]
- 12.Joos B., Kuster,H. and Cone,R. (1997) Anal. Biochem., 247, 96–101. [DOI] [PubMed] [Google Scholar]
- 13.Rogers Y.H., Jiang-Baucom,P., Huang,Z.J., Bogdanov,V., Anderson,S. and Boyce-Jacino,M.T. (1999) Anal. Biochem., 266, 23–30. [DOI] [PubMed] [Google Scholar]
- 14.Guo Z., Guilfoyle,R.A., Thiel,A.J., Wang,R. and Smith,L.M. (1994) Nucleic Acids Res., 22, 5456–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilham P.T. (1968) Biochemistry, 7, 2809–2813. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh S.S. and Musso,G.F. (1987) Nucleic Acids Res., 15, 5353–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf S.F., Haines,L., Fisch,J., Kremsky,J.N., Dougherty,J.P. and Jacobs,K. (1987) Nucleic Acids Res., 15, 2911–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lund V., Schmid,R., Rickwood,D. and Hornes,E. (1988) Nucleic Acids Res., 16, 10861–10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gingeras T.R., Kwoh,D.Y. and Davis,G.R. (1987) Nucleic Acids Res., 15, 5373–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasmussen S.R., Larsen,M.R. and Rasmussen,S.E. (1991) Anal. Biochem., 198, 138–142. [DOI] [PubMed] [Google Scholar]
- 21.Chrisey L.A., Lee,G.U. and O’Ferrall,C.E. (1996) Nucleic Acids Res., 24, 3031–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Donnell M.J., Tang,K., Koster,H., Smith,C.L. and Cantor,C.R. (1997) Anal. Chem., 69, 2438–2443. [DOI] [PubMed] [Google Scholar]
- 23.Maskos U. and Southern,E.M. (1992) Nucleic Acids Res., 20, 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beier M. and Hoheisel,J.D. (1999) Nucleic Acids Res., 27, 1970–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]