Abstract
In patients with mitochondrial disease a continuously increasing number of mitochondrial DNA (mtDNA) mutations and polymorphisms have been identified. Most pathogenic mtDNA mutations are heteroplasmic, resulting in heteroduplexes after PCR amplification of mtDNA. To detect these heteroduplexes, we used the technique of denaturing high performance liquid chromatography (DHPLC). The complete mitochondrial genome was amplified in 13 fragments of 1–2 kb, digested in fragments of 90–600 bp and resolved at their optimal melting temperature. The sensitivity of the DHPLC system was high with a lowest detection of 0.5% for the A8344G mutation. The muscle mtDNA from six patients with mitochondrial disease was screened and three mutations were identified. The first patient with a limb-girdle-type myopathy carried an A3302G substitution in the tRNALeu(UUR) gene (70% heteroplasmy), the second patient with mitochondrial myopathy and cardiomyopathy carried a T3271C mutation in the tRNALeu(UUR) gene (80% heteroplasmy) and the third patient with Leigh syndrome carried a T9176C mutation in the ATPase6 gene (93% heteroplasmy). We conclude that DHPLC analysis is a sensitive and specific method to detect heteroplasmic mtDNA mutations. The entire automatic procedure can be completed within 2 days and can also be applied to exclude mtDNA involvement, providing a basis for subsequent investigation of nuclear genes.
INTRODUCTION
Energy production in cells by the process of oxidative phosphorylation (OXPHOS) is one of the important functions of mitochondria. Part of the proteins of the OXPHOS complex is encoded by the mitochondrial DNA (mtDNA). This is a maternally inherited 16 569 bp long, circular double-stranded molecule with 37 genes, encoding OXPHOS subunits, two rRNA genes and 22 tRNA genes. Mutations in the mtDNA as well as mutations in nuclear genes can cause a clinical phenotype of a mitochondrial disorder. In the past 12 years numerous mtDNA mutations have been described in patients with mitochondrial disease (1). These mutations were inherited maternally, or originated de novo. Except for the mtDNA mutations causing Leber hereditary optic neuropathy, most other pathogenic mutations are in general heteroplasmic (i.e. a mixture of wild-type and mutant mtDNA in the same tissue or cell) and the mtDNA mutations become clinically manifest when the percentage of a mutation increases above a tissue-specific threshold (2). Non-pathogenic polymorphisms are normally homoplasmic (3).
Many mtDNA disorders show a wide spectrum of clinical manifestations and variation in the mode of onset, course and progression of the disease (4). This is seen between families, but also within families. Maternal relatives can inherit different percentages of mutant mtDNAs and can present with different clinical manifestations (2). Patients with a mitochondrial disorder are usually screened for the presence of deletions and specific mtDNA mutations in blood or muscle, based on these symptoms. The protocols are becoming increasingly complex, as the number of mutations in mtDNA accumulate and the specificity of the symptoms lags behind. Furthermore, only known mutations are detected with these protocols and unknown mutations remain unresolved. As an alternative the entire mitochondrial genome can be screened, but these approaches remain laborious [denaturing gradient gel electrophoresis (DGGE) analysis], lack sufficient sensitivity (sequence analysis) or do not sufficiently discriminate between hetero- and homoplasmic variants [single stranded conformation polymorphism (SSCP) analysis]. Therefore, we applied the relatively new and automatable denaturing high performance liquid chromatography (DHPLC) technology (5–7), which specifically identifies heteroduplexes that result from heteroplasmic mutations. When known mutations were readily detected even at low levels of heteroplasmy, we developed a protocol to screen the mtDNA for mutations by DHPLC in a rapid and sensitive way. A group of six patients with mitochondrial disease was screened for mutations as a test case and three mutations were identified.
MATERIALS AND METHODS
Patients
Five patients with 4.5, 9, 18, 55 and 76% of the A3243G mutation, respectively, and three patients with 0.5, 7 and 19% of the A8344G substitution, respectively, were used to test the sensitivity and specificity of the DHPLC system. Genomic DNA was isolated from blood and (when available) muscle according to the protocol described by Mullenbach et al. (8). Heteroplasmy was determined, as described below, using PCR primers encompassing the mutations (Table 1). Another patient carried a 4 bp deletion in the cytochrome b gene (60% heteroplasmy). The mtDNA of six patients with mitochondrial (encephalo)myopathies, lactic acidosis, OXPHOS deficiencies and ragged red fibres was screened.
Table 1. Primers used to amplify genes containing the A3243G, A8344G, A3302G, T3271C and T9176C mutations.
| Mutation | Position | Length (bp) | Sequence forward primer | Sequence reverse primer |
|---|---|---|---|---|
| A3243G | 3189–3383 | 195 | 5′-CAA CTT AGT ATT ATA CCC ACA C-3′ | 5′-TTT CGT TCG GTA AGC ATT AT-3′ |
| A8344G | 8211–8410 | 200 | 5′-TCG TCC TAG AAT TAA TTC CC-3′ | 5′-GGG GGT AAT TAT GGT GGG CC-3′ |
| A3302G | 3130–3383 | 254 | 5′-AGG ACA AGA GAA ATA AGG CC-3′ | 5′-TTT CGT TCG GTA AGC ATT AT-3′ |
| T3271C | 3130–3301 | 172 | 5′-AGG ACA AGA GAA ATA AGG CC-3′ | 5′-TAA GAA GAG GAA TTG AAC CTC TGA CCT TAA-3′ |
| T9176C | 9035–9203 | 169 | 5′-TCA TGC ACC TAA TTG GAA GCG-3′ | 5′-GTG TTG TCG TGC AGG TAC CAG CTT ACT-3′ |
Mismatches are underlined.
DHPLC analysis of specific mutations
Primers (Gibco BRL, Life Technologies, Cleveland, OH) used for the amplification of fragments containing the A3243G mutation [in tRNALeu(UUR)] and the A8344G mutation (in tRNALys) are displayed in Table 1. Reactions were performed in a 50 µl vol using 66 ng genomic DNA as template, 8.33 µM dNTP each (Pharmacia Biotech, Buckinghamshire, UK), 14 pmol of the forward and reverse primer, 1 U Taq DNA polymerase (PE Applied Biosystems, Foster City, CA) and OptiTaq buffer B (1.5 mM MgCl2, 50 mM KCl, 10 mM Tris–HCl, pH 8.4) for tRNALeu(UUR) and OptiTaq buffer G (50 mM NaCl instead of 50 mM KCl) for tRNALys. Using the GeneAmp“ PCR system 9700 (PE Applied Biosystems), PCR conditions for tRNALeu(UUR) were as follows: first, one cycle of 94°C for 5 min, followed by 32 cycles of 94°C for 1 min, 53°C for 1 min, 72°C for 45 s, and finally, one cycle of 72°C for 7 min followed by cooling to 4°C. PCR conditions for tRNALys were similar, except for the annealing temperature of 54°C and the number of cycles, which was 35. PCR products were tested by gel electrophoresis on a 2% agarose gel stained with ethidium bromide.
DHPLC analysis was performed on an automated DHPLC instrument (Transgenomic Inc., San Jose, CA). The stationary phase consisted of a DNA Sep“ column, which binds DNA during analysis. The mobile phase consisted of two eluents (pH 7.0). Buffer A contained triethylammonium acetate (TEAA), which interacts with the negatively charged phosphate groups on the DNA as well as with the surface of the column (http://www.transgenomic.com/Pages/Applicationnotes.shtml#101 ). Buffer B contained TEAA with 25% of the denaturing agent acetonitrile. Fragments were eluted with a linear acetonitrile gradient of 2% per min at a flow rate of 0.9 ml/min. Increasing the concentration of acetonitrile at a fixed temperature will denature the fragments. Temperatures for successful resolution of heteroduplexes were both calculated by the DHPLC Melt program (http://insertion.stanford.edu/cgi-bin/melt.pl ) and experimentally determined for the fragments containing the A3243G and A8344G mutation. For this latter purpose samples were analyzed at increasing column temperatures, until a significant decrease in retention time occurred.
DHPLC analysis of the complete mitochondrial genome
For DHPLC analysis of the mtDNA in small fragments, appropriate restriction sites were selected (Clone Manager 4.0 program, Scientific & Educational software) to yield fragments between 90 and 600 bp. These fragments were generated from longer PCR fragments using the primers shown in Table 2 (Gibco BRL, Life Technologies). Reactions were performed in a 50 µl vol using 330 ng genomic DNA as template, 8.33 µM dNTP each (Pharmacia Biotech), 14 pmol each primer, 2 U Taq DNA polymerase (PE Applied Biosystems) and OptiTaq buffer B. Fragment 2 was amplified using 14 pmol of the forward and 28 pmol of the reverse primer. Using the GeneAmp“ PCR system 9700 (PE Applied Biosystems), PCR conditions were as follows: first, one cycle of 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 57°C for fragments 1–10 and 67°C for fragments 11–13 for 1 min, 72°C for 2 min, and finally, one cycle of 72°C for 7 min followed by cooling to 4°C. PCR results were tested by gel electrophoresis on a 1% agarose gel stained with ethidium bromide.
Table 2. Primer sequences used to amplify the mtDNA in 13 fragments of 1–2 kb in length.
| Fragment | Position | Length (bp) | Sequence forward primer | Sequence reverse primer |
|---|---|---|---|---|
| 1 |
656–2490 |
1835 |
5′-TGG TCC TAG CCT TTC TAT TAG C-3′ |
5′-GGG TAA GAT TTG CCG AGT TCC-3′ |
| 2 |
2433–4224 |
1792 |
5′-CAG GCA TGC TCA TAA GGA AAG G-3′ |
5′-GGA GAC ATA TCA TAT AAG TAA TGC-3′ |
| 3 |
4152–5735 |
1584 |
5′-CGA CCA ACT CAT ACA CCT CC-3′ |
5′-GAG AAG TAG ATT GAA GCC AG-3′ |
| 4 |
5470–6908 |
1439 |
5′-CGC TAC TCC TAC CTA TCT CC-3′ |
5′-AGA TCA TTT CAT ATT GCT TCC GT-3′ |
| 5 |
6789–8000 |
1212 |
5′-GGA ATA GAC GTA GAC ACA CGA G-3′ |
5′-CAA CGT CAA GGA GTC GCA GGT-3′ |
| 6 |
7699–8738 |
1040 |
5′-CCT GTA TGC CCT TTT CCT AAC- 3′ |
5′-ATA AGA GAT CAG GTT CGT CCT T-3′ |
| 7 |
8577–10407 |
1831 |
5′-ACC CGC CGC AGT ACT GAT CAT-3′ |
5′-CCA ATT CGG TTC AGT CTA ATC C-3′ |
| 8 |
10233–11249 |
1017 |
5′-GCT ATT ACC TTC TTA TTA TTT GAT C-3′ |
5′-GTG CGA TGA GTA GGG GAA GG-3′ |
| 9 |
10866–12420 |
1554 |
5′-TCA TCC CTC TAC TAT TTT TTA ACC-3′ |
5′-TTT GTT AGG GTT AAC GAG GG-3 |
| 10 |
12175–13708 |
1534 |
5′-TGA CAA CAG AGG CTT ACG ACC-3′ |
5′-CCA GGC GTT TAA TGG GGT TTA GT-3′ |
| 11 |
13354–14458 |
1105 |
5′-TTT ATG TGC TCC GGG TCC ATC AT-3′ |
5′-GAT GGC TAT TGA GGA GTA TCC T-3′ |
| 12 |
14399–15593 |
1195 |
5′-ACA CTC ACC AAG ACC TCA ACC-3′ |
5′-ATC GGA GAA TTG TGT AGG CGA AT-3′ |
| 13 | 15498–711 | 1783 | 5′-GCG ACC CAG ACA ATT ATA CCC T-3′ | 5′-AAC GGG GAT GCT TGC ATG TGT-3′ |
Fragments were denatured after PCR at 95°C for 10 min, reannealed at 65°C for 10 min and slowly (1°C/s) cooled to 4°C to form heteroduplexes. PCR fragments were then cleaved in smaller fragments varying in size from 90 to 560 bp by the enzymes shown in Table 3. For each reaction ~300 ng of PCR product and 0.8–15 U of the appropriate enzyme (Roche Diagnostics, Mannheim, Germany and New England Biolabs, Beverly, MA) were used in a volume of 50 µl, according to the conditions given by the manufacturer, leaving out the dithiothreitol, which may be harmful to the DNA Sep“ column. Digestion results were tested by gel electrophoresis on a 3% agarose gel stained with ethidium bromide.
Table 3. Enzymes used for restriction digestion (in a volume of 50 µl) and DHPLC analysis temperatures.
| Fragment | Enzyme | Restriction site | Enzyme used (U) | Incubation temperature (°C) | Fragments after restriction | Temperatures for DHPLC analysis (°C) |
|---|---|---|---|---|---|---|
| 1 | BfaI | C↓TAG | 10 | 37 | 132 187 260 365 401 484 | 55, 57, 58, 59, 60 |
| 2 | NlaIII | CATG↓ | 10 | 37 | 120 295 382 460 527 | 55, 58, 59, 60, 61 |
| 3 | HpaII | C↓CGG | 5 | 37 | 135 396 493 560 | 55, 56, 58, 59, 60 |
| 4 | HaeIII | GG↓CC | 5 | 37 | 123 190 233 369 524 | 55, 57, 59, 60, 61 |
| 5 | HaeIII | GG↓CC | 5 | 37 | 113 170 240 300 389 | 55, 57, 58, 59, 60 |
| 6 | AluI | AG↓CT | 10 | 37 | 229 377 434 | 54, 57, 58, 59, 60 |
| 7 | TaqI | T↓CGA | 5 | 65 | 159 227 270 308 381 486 | 50, 55, 58, 59, 60 |
| 8 | HinfI | G↓ANTC | 15 | 37 | 141 205 278 393 | 53, 56, 57, 58, 60 |
| 9 | HphI | GGTGA(N)8↓ | 7.5 | 37 | 284 330 413 528 | 55, 57, 58, 59, 60 |
| 10 | SfaNI | GCATC(N)5↓ | 0.8 | 37 | 292 347 410 485 | 55, 57, 58, 59, 60 |
| 11 | HaeIII | GG↓CC | 5 | 37 | 500 593 | 57, 58, 59, 60, 61 |
| 12 | DpnII | GA↓TC | 5 | 37 | 191 235 297 472 | 55, 57, 59, 60, 61 |
| 13 | AluI | AG↓CT | 10 | 37 | 90 130 218 280 482 548 | 53, 57, 58, 59, 60 |
The optimal conditions for DHPLC analysis of fragments created by digestion were calculated for each individual fragment using the DHPLC Melt program (http://insertion.stanford.edu/cgi-bin/melt.pl ) and the WAVEmaker Utility Software (Transgenomic Inc.). The fragments were first analyzed under non-denaturing conditions (50°C) to test for the presence of aspecific fragments and the resolution. Each vial of 100 µl digestion product is sufficient for analysis at five temperatures using 10–20 µl per injection. The injection volume can be adjusted to the signal of the product on agarose.
Sequence analysis
PCR fragments were purified with the QIA quick PCR purification kit protocol (Qiagen, Valencia, CA) and cycle sequenced in a volume of 20.0 µl using 8.0 µl BigDye Terminator Ready Reaction Kit (PE Applied Biosystems), 3.2 pmol of the forward or reverse primer and 6.0 µl of PCR product. Using a GeneAmp“ PCR system 9700 (PE Applied Biosystems), cycle sequence conditions were as follows: 25 cycles of 96°C for 10 s, 50°C for 5 s, 60°C for 4 min, and finally cooling to 4°C. Then, DNA was precipitated by mixing 20 µl of the product with 74 µl 70% ethanol and 0.5 mM MgCl2, incubated at room temperature for 15 min and centrifuged at 10 000 g for 15 min. Samples were loaded on a denaturing Long Ranger gel (Pharmacia Biotech), on the ABI Prism 377 automatic sequencer/genetic analyzer and analyzed with the Sequence“ 2.1 analysis software (PE Applied Biosystems).
Determination of heteroplasmy
The percentage heteroplasmy of the A3243G, A8344G, A3302G, T3271C and T9176C mutations was determined by analyzing fluorescently labeled PCR products on a 4% polyacrylamide gel under non-denaturing conditions. Primers (Gibco BRL, Life Technologies) used for the amplification of fragments containing these five mutations are displayed in Table 1. PCR amplification, as described for the A3243G mutation, was followed by a single last cycle, using fluorescent dUTPs (dTTP:fdUTP, 250:1). The PCR fragment with the A3243G mutation was digested with ApaI (New England Biolabs), yielding two fragments in the presence of the mutation (140 and 60 bp) and one fragment in the wild-type (200 bp). The PCR fragment with the A8344G mutation was digested with BglI (New England Biolabs), yielding two fragments in the presence of the mutation (133 and 42 bp) and one fragment in the wild-type (175 bp). The PCR fragment containing the A3302G mutation was digested with DdeI (New England Biolabs), yielding three fragments (106, 85 and 63 bp) in the patient and two fragments (191 and 63 bp) in the wild-type. The PCR fragment containing the T3271C mutation was generated with a mismatch primer, creating an AflII site (New England Biolabs), yielding two fragments in the patient (26 and 146 bp) and one fragment (171 bp) in the wild-type. The PCR fragment containing the T9176C mutation was also generated with a mismatch primer, creating a BstXI site (New England Biolabs), yielding two fragments (148 and 21 bp) in the patient and one fragment (169 bp) in the wild-type. Digestion products were then analyzed on an ABI Prism 377 automatic genetic analyzer with the Genescan“ 2.1 analysis software (PE Applied Biosystems). The ratio of the mutation peaks versus wild-type was determined. In case not all peaks could be quantified, a correction was made for the size of the fragments and the related incorporation of fdUTP.
RESULTS
The sensitivity of DHPLC analysis
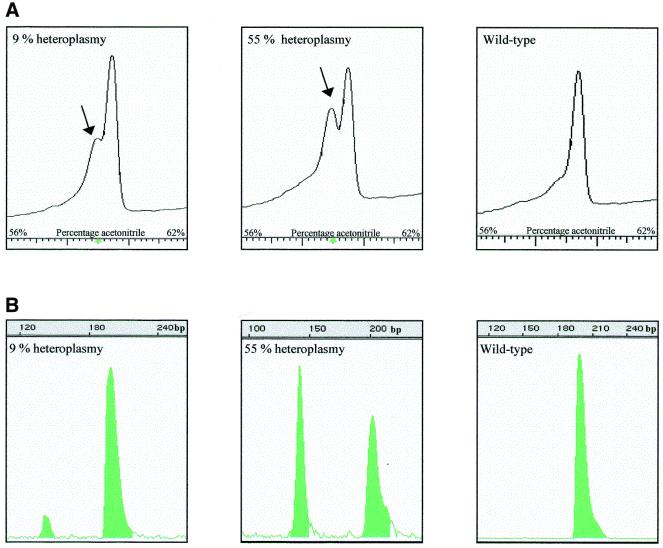
DHPLC analysis was performed on a variety of percentages of the A3243G and A8344G mutations in PCR fragments of 200 bp (Fig. 1). In the absence of heteroplasmic mutations, the chromatogram shows only a single homoduplex peak, whereas a heteroplasmic mutation leads to two peaks, the first one representing the heteroduplexes and the second one the homoduplexes. In the case of a low percentage of heteroplasmy, heteroduplexes are represented as a ‘shoulder’ in the peak. The A3243G mutation was detected in samples with heteroplasmy ranging from 4.5 to 76% (Fig. 1B). Figure 1A shows 9 and 55% mutated mtDNA compared to 0% heteroplasmy (wild-type). The A8344G mutation was also detected in samples with 0.5, 7 and 19% heteroplasmy (Fig. 1D). Figure 1C shows 0.5 and 19% mutated mtDNA compared to 0% heteroplasmy. The sensitivity of the method is thus high for the examples given.
Figure 1.
DHPLC analysis of two known mutations with different amounts of mutated mtDNA. (A and C) DHPLC elution peaks of characterized percentages of the A3243G and A8344G mutation (B and D) using increasing amounts of acetonitrile (x-axis). The y-axis represents the intensity of the peaks. The arrow indicates the heteroduplexes. (B and D) The same samples using last-cycle PCR followed by restriction digestion to determine the exact percentage of the mutation. On the x-axis the length of the fragments after digestion is displayed.
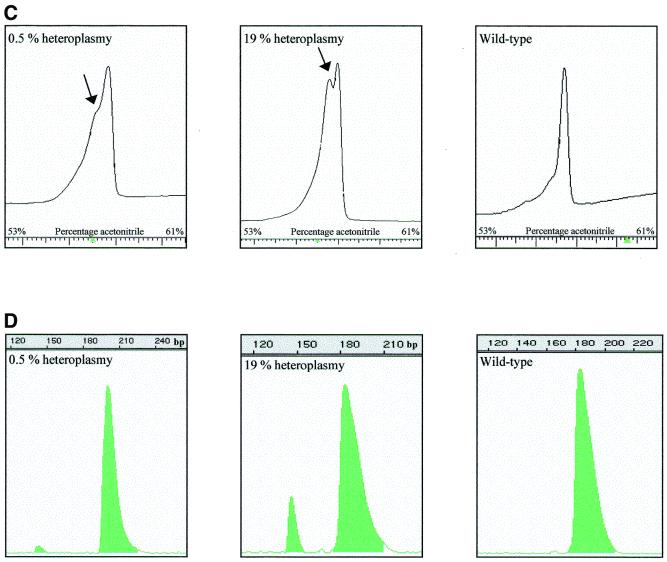
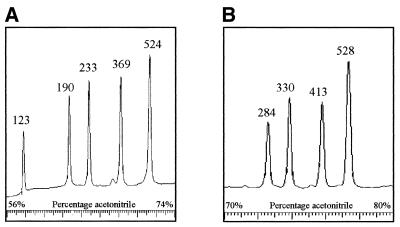
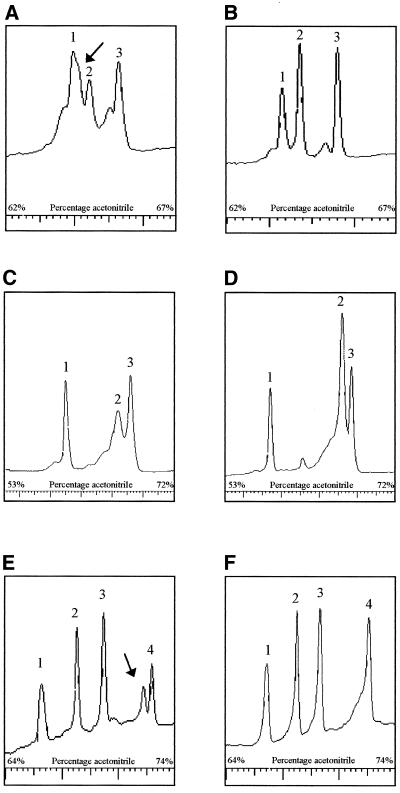
Next we amplified the entire mtDNA in 13 fragments (numbered 1–13, primers in Table 2). PCR products were optimized and restriction enzymes were chosen to generate fragments in the DHPLC size range without overlap (Table 3). Figure 2 shows chromatograms of the digestion products of fragments 4 and 9 under non-denaturing conditions. Fragment 4 yields HaeIII fragments of 123, 190, 233, 369 and 524 bp in length and fragment 9 HphI fragments of 284, 330, 413 and 528 bp. The resolution was good and no additional fragments were observed. Because polymorphisms in the mtDNA can occur within the restriction sites of the enzymes, sometimes less and sometimes more fragments are observed. Five such polymorphisms were identified (Table 4). At higher temperatures the number of fragments decreased, depending on the melting temperature of the individual fragments. It should be noted that separation of the fragments is based on melting characteristics and that it is not always the largest fragment which is finally last eluted. Additional digestion reactions were performed in case of ambiguity to determine which fragment each of the peaks represents. Figure 3A displays the A3243G mutation with 55% heteroplasmy detected in peak 2 (460 bp) of fragment 2. Figure 3C shows the A8344G mutation with 7% heteroplasmy at 57°C. Figure 3E shows the heteroplasmic 14787delTTAA mutation (60%) in the cytochrome b gene, present in peak 4 (472 bp) of fragment 12 at 57°C. The corresponding wild-type fragments are shown in Figure 3B, 3D and 3F, respectively. All mutations were readily detected in the digestion products, although the patterns can be quite complex because heteroduplexes of one fragment can overlap with homoduplexes of another fragment. Any alteration, which is either the presence of a heteroduplex or the relative decrease in intensity of the homoduplexes, is an indication for a mutation.
Figure 2.
DHPLC analysis of fragments 4 and 9 after digestion with HaeIII and HphI, respectively. Fragment 4 is cleaved into fragments of 123, 190, 233, 369 and 524 bp (A) and fragment 9 into fragments of 284, 330, 413 and 528 bp (B). Intensities of the elution peaks are an indication of the size of the fragment. In case of ambiguity, additional digestion reactions were performed to determine which fragment each of the peaks represents. Fragments 4 and 9 are representative for all other fragments.
Table 4. Polymorphisms creating (+) or deleting (–) restriction sites of the enzymes used.
| Polymorphism | Gene | Restriction site | Result | No. of patients (total 6) |
|---|---|---|---|---|
| A2706G | 16S rRNA | + | 295 bp → 146 + 149 bp | 4 |
| G6260A | COX I | – | 233 + 123 bp → 356 bp | 2 |
| C8958A | ATPase6 | – | 381 + 486 bp → 867 bp | 1 |
| T11299C | ND4 | + | 528 → 139 + 389 bp | 1 |
| A15607G | Cyt b | + | 280 → 170 + 110 bp | 2 |
Figure 3.
DHPLC analysis of known mutations in digestion products compared with wild-types. The A3243G mutation (55% heteroplasmy) in tRNALeu(UUR) (A) is compared with the wild-type fragment (B), representing the 382 (peak 1), 460 (peak 2) and 527 bp (peak 3) digestion products of fragment 2 at 58°C. The mutation is present in peak 2, which decreases in intensity. The arrow indicates the heteroduplexes merging into peak 1. The A8344G mutation (7% heteroplasmy) in tRNALys (C) is compared with the wild-type fragment 6 (D), representing the 229 (peak 1), 434 (peak 2) and 377 bp (peak 3) products of fragment 6 at 57°C. The mutation is present in peak 2, which decreases in intensity and broadens, whereas the size of peaks 1 and 3 remains the same. The 14787delTTAA (60% heteroplasmy) in cytochrome b (E) is compared with the wild-type fragment 12 (F), showing four homoduplex peaks at 57°C representing the 191 (peak 1), 235 (peak 2), 297 (peak 3) and 472 bp (peak 4) products. The mutation is present in peak 4. The arrow indicates the heteroduplexes.
DHPLC analysis of patients with unknown mutations
The muscle mtDNA of six patients with evident mitochondrial disease were screened by DHPLC. One patient with a limb-girdle-type myopathy showed heteroduplexes in the fragment containing the tRNALeu(UUR) gene (Fig. 4A). Sequence analysis revealed a heteroplasmic A3302G substitution. Heteroplasmy was determined to be ~70% in muscle. A second patient also showed a heteroduplex in the tRNALeu(UUR) gene (Fig. 4C), which by sequence analysis turned out to be the T3271C mutation. The percentage of the mutation was 80% in muscle from this patient with mitochondrial myopathy and cardiomyopathy. The pattern is different from the previous patient because of an NlaIII polymorphism. A third patient with signs of Leigh syndrome showed heteroduplexes in the largest individual fragment of 867 bp (Fig. 4E). Sequence analysis revealed a heteroplasmic T9176C substitution in the ATPase6 gene. The percentage of the mutation in muscle was ~93%. No comparable wild-type pattern is available because of a polymorphism in a TaqI site in this patient. The other three patients did not carry a heteroplasmic mutation in the mtDNA.
Figure 4.
DHPLC analysis of fragments 2 and 7 of three patients with unknown mutations. Fragment 2 of patient 1 (A) was compared with the wild-type fragment 2 (B), showing three homoduplex peaks at 55°C representing the 382 (peak 1), 460 (peak 2) and 527 bp (peak 3) products. Patient 1 showed heteroduplexes in peak 2 (460 bp) as indicated by the arrow. Sequence analysis revealed the presence of an A3302G substitution. Fragment 2 of patient 2 showed a shoulder in peak 3 (460 bp) (C), as indicated by the arrow, compared to the wild-type at 58°C (D). The four peaks represent the 295 (peak 1), 382 (peak 2), 460 (peak 3) and 527 bp (peak 4) fragments. In these two samples the polymorphic restriction site, seen in the other patients, was absent, showing the fragment of 295 bp as the fourth peak. Sequence analysis revealed the presence of a T3271C substitution. Patient 3 showed heteroduplexes in the 867 bp digestion product of fragment 7 (E) as indicated by the arrow. No similar wild-type fragment was available because of a TaqI polymorphism. Sequence analysis revealed the presence of a T9176G substitution.
DISCUSSION
PCR reactions were optimized to amplify the entire mtDNA in 13 overlapping fragments using two different PCR conditions. Restriction digestion conditions were adjusted for each individual fragment. Testing the digestion products on agarose revealed five different polymorphisms (9–11). These changes in restriction digestion sites are important to know prior to the analysis, because the melting temperatures from these new fragments may differ from the original fragments. All fragments showed good resolution and no aspecific products were detectable. Digestion of fragment 9 yielded products that eluted after 10 min. It is possible to start at a higher acetonitrile concentration to decrease the time required for analysis. However, if the analysis time is too short, the risk of fragments eluting into the first injection peak exists. The optimal temperatures for DHPLC analysis and acetonitrile gradients of all individual fragments can be determined experimentally or using the DHPLC Melt program (http://insertion.stanford.edu/cgi-bin/melt.pl ). The predicted temperatures did not differ by more than 1–2°C from the experimentally determined temperatures. When fragments are being analyzed at 1°C below and above the recommended temperature, this 1–2°C difference is within the range of reliable detection of heteroduplexes. For each fragment of 1–2 kb, five temperatures are recommended for analysis, ranging from 50 to 60°C. Digestion products should be first analyzed for wild-type fragments at each individual temperature to determine the normal melting behavior. When screening for unknown mutations it is important to be able to discriminate between peaks, which appear as a consequence of the melting process and peaks caused by the presence of heteroduplexes.
Samples with 4.5, 9, 18, 55 and 76% of the A3243G mutation and 0.5, 7 and 19% of the A8344G mutation were used to test the sensitivity and accuracy of the DHPLC. All heteroplasmy percentages were detectable, making DHPLC analysis a very sensitive technology. Mutations were represented as two peaks and mutations with low percentages, such as 0.5% heteroplasmy, as a ‘shoulder’ in the peak. Apart from the heteroduplex peak, which under some conditions can overlap with (melting) homoduplexes, the reduction in intensity of a peak in comparison with the other peaks is a strong indicator for the presence of a mutation in that specific fragment. Transgenomic‘ describes the resolution of hetero- and homoduplexes to be optimal, when all four hetero- and homoduplex species are fully resolved (http://www.transgenomic.com/Pages/Applicationnotes.shtml#101 ). In the DHPLC analysis performed here, only two species were consistently visible. This was also reported by others using DHPLC as a mutation detection system (6,7,12). It has also been described that different mutations show different profiles (7). The stability of heteroduplexes and the degree of partial denaturation surrounding it varies, depending on the nature of the mutation and the flanking base pairs. This may explain why some mutations show a better resolution of hetero- and homoduplexes than other mutations (7).
DHPLC has been described as a very sensitive method in mutation screening of nuclear genes. Sensitivity and specificity of this method have been evaluated in a blind analysis performed on exon H of the factor IX gene and exon 16 of the neurofibromatosis type 1 gene. In this analysis 55/55 (100%) individuals carrying 48 unique mutations were correctly identified, as were 55/55 individuals with wild-type alleles (7). DHPLC has also been used to analyze sequence variation in the BRCA2 gene, identifying 82 sequence variants (13). In another study 37 out of 40 (92.5%) PCR products containing defined sequence variation were identified and no alterations among 196 PCR products containing homozygous normal sequence were indicated (6). The few exceptions encountered may relate to the extreme G-C content of the tested fragments. This high sensitivity indicates that the absence of heteroduplexes after DHPLC analysis of the mtDNA most likely excludes the presence of heteroplasmic mutations in the mtDNA, making the involvement of such a mtDNA mutation as the cause for the disease unlikely. This provides a strong basis for further investigation of nuclear genes. This is also an important issue for counselling as mtDNA is transmitted maternally, whereas nuclear gene defects are predominantly autosomal recessive.
The entire mitochondrial genome of six patients showing clinical signs of a mitochondrial disorder was screened with DHPLC. In one patient, the A3302G mutation in tRNALeu(UUR) was found with ~70% heteroplasmy in muscle. This mutation involves the aminoacyl stem of tRNALeu(UUR) and has been described before, associated with abnormal mitochondrial RNA processing (14). A second patient carried the T3271C mutation in the anticodon stem of the tRNALeu(UUR) gene, which was previously reported in MELAS patients (15). In a third patient, with signs of Leigh syndrome, the T9176C mutation in the ATPase6 gene was found with ~93% heteroplasmy in muscle. This mutation was first described in two patients with familial bilateral striatal necrosis and later described in patients with Leigh disease (16–18). A high percentage of this mutation in blood and muscle is associated with poor prognosis (16,17). We feel that the power of the method has convincingly been demonstrated to rapidly identify mutations in the mtDNA.
Methods to screen part of or the complete mtDNA sequence for mutations have been reported previously (19,20). Most of these methods have been based on SSCP analysis, sequence analysis and DGGE (21–24). DHPLC is superior to the first two, because these methods do not discriminate between pathogenic mutations and polymorphisms and lack sufficient sensitivity to detect low percentages of mutations. A two-dimensional DGGE method has a similar sensitivity and detects heteroduplexes, but can be less easily automated (24). The protocol described here can be automated and can be completed for a single patient within 2 days.
Acknowledgments
ACKNOWLEDGEMENT
We would like to thank Petra Lux from the Department of Biochemistry, Maastricht University, Maastricht, The Netherlands for her assistance on the DHPLC instrument.
REFERENCES
- 1.Servidei S. (2000) Neuromusc. Disord., 10, IX–XIII. [PubMed] [Google Scholar]
- 2.DiMauro S.M.D. and Moreas,D.T. (1993) Arch. Neurol., 50, 1197–1208. [DOI] [PubMed] [Google Scholar]
- 3.Zeviani M. and Antozzi,C. (1997) Mol. Hum. Reprod., 3, 133–148. [DOI] [PubMed] [Google Scholar]
- 4.Rose M.D. (1998) Arch. Neurol., 55, 17–24. [DOI] [PubMed] [Google Scholar]
- 5.Underhill P.A., Jin,L., Lin,A.A., Mehdi,S.Q., Jenkins,T., Vollrath,D., Davis R.W., Cavalli-Sforza,L.L. and Oefner,P.J. (1997) Genome Res., 7, 966–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W., Smith,D.I., Rechtzigel,K.J., Thibodeau,S.N. and James,C.D. (1998) Nucleic Acids Res., 26, 1396–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Donovan M.C., Oefner,P.J., Roberts,S.C., Austin,J., Hoogendoorn,B., Guy,C., Speight,G., Upadhyaya,M., Sommer,S.S. and McGuffin,P. (1998) Genomics, 52, 44–49. [DOI] [PubMed] [Google Scholar]
- 8.Mullenbach R., Lagoda,P.J. and Welter,C. (1989) Trends Genet., 5, 391. [PubMed] [Google Scholar]
- 9.Wallace D.C., Singh,G., Lott,M.T., Hodge,J.A., Schurr,T.G., Lezza,A.M., Elsas,L.J. and Nikoskelainen,E.K. (1988) Science, 242, 1427–1430. [DOI] [PubMed] [Google Scholar]
- 10.Rieder M.J., Taylor,S.L., Tobe,V.O. and Nickerson,D.A. (1998) Nucleic Acids Res., 26, 967–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howell N., Kubacka,I., Halvorson,S., Howell,B., McCullough,D.A. and Mackey,D. (1995) Genetics, 140, 285–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giordano M., Oefner,P.J., Underhill,P.A., Cavalli Sforza,L.L., Tosi,R. and Richiardi,P.M. (1999) Genomics, 56, 247–253. [DOI] [PubMed] [Google Scholar]
- 13.Wagner T.M.U., Hirtenlehner,K., Shen,P., Moeslinger,R., Muhr,D., Fleishmann,E., Concin,H., Doeller,W., Haid,A., Lang,A.H. et al. (1999) Hum. Mol. Genet., 8, 413–423. [DOI] [PubMed] [Google Scholar]
- 14.Bindoff L.A., Howell,N., Poulton,J., McCullough,D.A., Morten,K.J., Lightowlers,R.N., Turnbull,D.M. and Weber,K. (1993) J. Biol. Chem., 268, 19559–19564. [PubMed] [Google Scholar]
- 15.Goto Y., Nonaka,I. and Horai,S. (1991) Biochim. Biophys. Acta, 1097, 238–240. [DOI] [PubMed] [Google Scholar]
- 16.Thyagarajan D., Shanske,S., Vazquez-Memije,M., De Vivo,D. and DiMauro,S. (1995) Ann. Neurol., 38, 468–472. [DOI] [PubMed] [Google Scholar]
- 17.Campos Y., Martin,M.A., Rubio,J.C., Solana,L.G., Garcia-Benayas,C., Terradas,J.L. and Arenas,J. (1997) Neurology, 49, 595–597. [DOI] [PubMed] [Google Scholar]
- 18.Dionisi-Vici C., Seneca,S., Zeviani,M., Fariello,G., Rimoldi,M., Bertini,E. and De Meirleir,L. (1998) J. Inherit. Metab. Dis., 21, 2–8. [DOI] [PubMed] [Google Scholar]
- 19.Wong L.J.C. and Senadheera,D. (1997) Clin. Chem., 43, 1857–1861. [PubMed] [Google Scholar]
- 20.Fauser S. and Wissinger,B. (1997) Biotechniques, 22, 964–968. [DOI] [PubMed] [Google Scholar]
- 21.Suomalainen A., Ciafaloni,E., Koga,Y., Peltonen,L., DiMauro,S. and Schon,E.A. (1992) J. Neurol. Sci., 111, 222–226. [DOI] [PubMed] [Google Scholar]
- 22.Levin B.C., Cheng,H. and Reeder,D.J. (1999) Genomics, 55, 135–146. [DOI] [PubMed] [Google Scholar]
- 23.Barros F., Lareu,M.V., Salas,A. and Carracedo,A. (1997) Electrophoresis, 18, 52–54. [DOI] [PubMed] [Google Scholar]
- 24.Van Orsouw N.J., Zhang,X., Wei,J.Y., Johns,D.R. and Vijg,J. (1998) Genomics, 52, 27–36. [DOI] [PubMed] [Google Scholar]