Abstract
Suppression subtractive hybridization (SSH) is one of the most powerful and popular methods for isolating differentially expressed transcripts. However, SSH-generated libraries typically contain some background clones representing non-differentially expressed transcripts. To overcome this problem we developed a simple procedure that substantially decreases the number of background clones. This method is based on the following difference between target and background cDNAs: each kind of background molecule has only one orientation with respect to the two different flanking adapter sequences used in SSH, while truly differentially expressed target cDNA fragments are represented by both sequence orientations. The described method selects the molecules that arose due to hybridization of such mirror-orientated molecules. The efficiency of this method was demonstrated in both model and real experimental subtractions.
INTRODUCTION
A powerful approach for studying the genetic nature of many biological processes is to characterize genes that vary in expression level during this process. Suppression subtractive hybridization (SSH) (1,2) is a highly efficient and widely used (3–5) PCR-based method for identifying differentially expressed genes. A key feature of the SSH method is simultaneous subtraction and normalization that makes it possible to equalize abundance of target cDNAs in the subtracted population. As a result, rare differentially expressed transcripts can be enriched by ~1000-fold. Success of SSH application experiments is limited by factors including high-complexity cDNA samples and having a small number of differences (targets) between cDNA samples.
The major drawback of SSH is the presence of background clones representing non-differentially expressed (redundant) cDNA species in the subtracted libraries. In some difficult cases (e.g. subtraction of vertebrate brain samples), the number of background clones may considerably exceed the number of target clones in subtracted libraries. An especially challenging problem is the inclusion of so called ‘false positive’ clones that give a differential signal in a primary screening procedure but are not confirmed by further detailed analysis. To overcome this problem we developed a simple procedure that substantially decreases the number of background clones in the libraries generated by SSH.
MATERIALS AND METHODS
Telencephalons of E15.5 and E13.5 mouse embryos were surgically extracted. After removing pial membrane, cortices were separated from the rest of the brain tissue (basal ganglia, hippocampus and olfactory bulb). Total cortex RNA was purified as described (6). Human skeletal muscle polyA+ RNA was obtained from Clontech (CA) and ϕX174 DNA was obtained from Promega (WI). Double-strand cDNA synthesis was performed using the template switch technique (SmartTM PCR cDNA Synthesis Kit; Clontech).
The PCR-amplified cDNAs were then subtracted using the PCR-SelectTM cDNA Subtraction Kit (Clontech; detailed protocol in 7,8). We introduced some modifications in the SSH protocol that increased the efficiency of the method. Generally, for successful SSH outcome primary PCR should consist of no more than 27 cycles. This corresponds to ~1000 or more cDNA molecules taken for amplification (9,10). For our brain cDNA subtraction 30 cycle primary PCR was needed to amplify the subtracted sample up to 10–20 ng/µl concentration. To overcome this problem, 10 independent tubes of primary PCR were generated. The samples were then combined and 1000-fold diluted. The sample was subsequently amplified again up to 10–20 ng/µl in 10–12 cycle PCR using the same primer and conditions as described for primary PCR. This additional PCR step greatly decreased the portion of background molecules that could be amplified during or following secondary PCR. Such background cDNAs are flanked by one of the nested primers (NP1 or NP2R, see below) on both termini and originated from tester–tester homohybrids. Amplification of such symmetrically flanked molecules is inhibited in primary PCR due to the suppression PCR effect but is permitted in secondary PCR. This type of background is very dangerous for mirror orientation selection (MOS) because this procedure is based on selection of the cDNA molecules symmetrically flanked with NP2R primer. Secondary (nested) PCR was performed using the following primers: NP1, 5′-TCGAGCGGCCGCCCGGGCAGGT (XmaI restriction site underlined); NP2R, 5′-AGCGTGGTCGCGGCCGAGGT. PCR products were phenol/chloroform extracted and ethanol precipitated. The pellet was dissolved in NTE buffer (10 mM NaCl, 10 mM Tris–HCl, 0.1 mM EDTA) up to a concentration of 20–30 ng/µl of cDNA. To remove NP1 adapters, 5 µl of the cDNA sample was mixed with 2 µl of 10× XmaI restriction buffer, 12 µl H2O and 1 µl XmaI (10 U/µl). The reaction was allowed to proceed for 1 h at 37°C. The enzyme was then inactivated by adding 2 µl of 200 mM EDTA and incubated at 70°C for 10 min. One microliter of XmaI-digested cDNA (5–7 ng) was mixed with 1 µl of 4× hybridization buffer (2 M NaCl, 200 mM HEPES pH 8.3, 0.8 mM EDTA) and 2 µl of H2O [or 2 µl of driver skeletal muscle cDNA (300 ng/µl) in some model experiments] and incubated in a thermal cycler at 98°C for 1.5 min and then at 68°C for 3–12 h. It should be noted that theoretically, duration of hybridization could strongly affect the MOS outcomes. Too short a hybridization could result in enrichment of highly abundant cDNA species and loss of rare species due to second-order kinetics of re-annealing. Complexity of typical SSH samples is no more than 103 independent cDNA species. Such low-complexity samples can be almost completely re-annealed during a relatively short length of time. Our practice showed that 3 h hybridization is enough and following hybridization prolongation has little effect on the MOS efficiency. After hybridization the sample was mixed with 200 µl of dilution buffer (50 mM NaCl, 20 mM HEPES pH 8.3, 0.2 mM EDTA) and heated in a thermal cycler at 70°C for 7 min. One microliter of diluted cDNA was taken for subsequent PCR in a total volume of 20 µl. The PCR mixture contained 1× Advantage KlenTaq Polymerase Mix with the provided buffer (Clontech), 200 µM dNTPs and 0.6 µM adapter-specific primer NP2Rs (5′-GGTCGCGGCCGAGGT; this primer, which is shorter than NP2R, was designed to reduce the strong suppression PCR effect that occurs for short DNA fragments). The PCR mixture was incubated in a thermal cycler at 72°C for 2 min to extend the 3′-ends of DNA duplexes and was then immediately switched to the amplification program (25 cycles; Hybaid OmniGene thermocycler, tube control mode) 95°C, 7 s; 62°C, 20 s; 72°C, 2 min. Generally, for each particular sample the number of PCR cycles needed should be determined experimentally (usually this PCR consists of 18–23 cycles). Note that after XmaI digestion, a small portion of the NP1 adapter sequence remains intact. After hybridization, the target duplexes formed by annealing of DNA strands with opposite adapter orientation bear two unpaired 3′-terminal bases originating from the NP1 adapter. These bases do not impede the 3′-end extension due to the proofreading polymerase that is included in the Advantage KlenTaq Polymerase Mix.
The PCR product was cloned into pCR-Script Amp using the PCR-Script Cloning Kit (Stratagene, CA). Randomly selected clones were arrayed in 96-well microtiter dishes with 150 µl Luria–Bertrani broth with ampicillin and grown overnight on the shaker. One microliter of the bacterial cultures was used for PCR in 96-well PCR plates using the NP2Rs primer. PCR products were spotted on nylon membranes. Hybridization was performed with [32P]dATP-labeled subtracted cDNA from both forward and reverse subtractions as described (11).
RESULTS AND DISCUSSION
The SSH technique is based on the suppression PCR effect that is mediated by long inverted terminal repeats attached to the ends of DNA fragments (12). By incorporating this suppression effect in a PCR amplification scheme, the SSH method normalizes sequence abundance within the amplified cDNA population and prevents amplification of undesirable DNA fragments. The SSH scheme includes the following main steps: (i) subdivision of tester cDNA into two samples and ligation of these samples with two different suppression adapters; (ii) hybridization of tester with excess driver; and (iii) amplification of the tester cDNA molecules that are flanked only with different suppression adapters (this fraction contains the enriched and normalized target cDNA) (1,2,7,8).
We propose that there are the following two main sources of background amplification in SSH. (i) Long oligonucleotides from non-ligated suppression adapters can non-specifically anneal during subtractive hybridization to cDNA molecules having similar sequences. After DNA elongation such molecules can serve as a template for the SSH primary as well as for the secondary (nested) PCR. Also, some background can be generated due to non-specific annealing of PCR primers. (ii) Some redundant cDNA molecules can by chance evade elimination by hybridization with driver and be amplified in subsequent PCRs. For any given redundant cDNA species the latter explanation is extremely unlikely. We estimate that in most cases just a single molecule of each redundant cDNA species is present among several thousand other cDNA molecules that are used for PCR after the subtractive hybridization. However, a huge excess of redundant sequences relative to target cDNAs can cumulatively result in a high number of such background molecules.
Clones representing type (i) background can be easily revealed by differential screening because they do not produce a differential signal. On the contrary, type (ii) background clones show differential signals during screening with probes prepared from two reciprocal (forward and reverse) subtracted samples. Only northern blot and RT–PCR analysis can demonstrate the equal abundance of such sequences in the initial mRNA samples. As a consequence, the elimination of this type of background is the most difficult and time-consuming step in subtracted library analysis. As an alternative to screening with subtracted probes, it is possible to use the tester and driver cDNA as probes for differential screening, but in this case many clones representing rare transcripts give no signals.
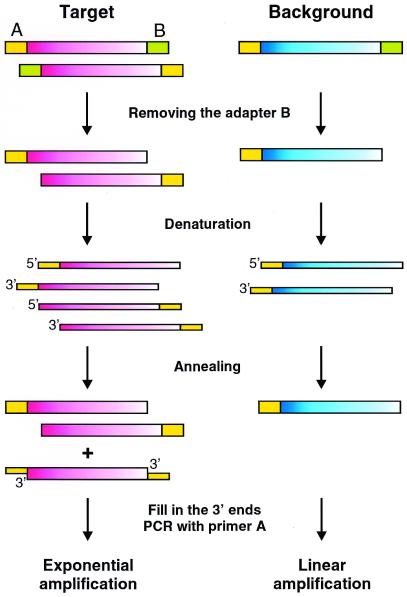
We developed a special procedure to decrease the portion of background clones in the subtracted samples. This technique is based on the rationale that after PCR, each species of background molecule has only one orientation relative to the adapter sequences. This directionality corresponds to the orientation of the progenitor molecule. On the contrary, the target cDNA fragments are involved in PCR amplification due to efficient enrichment in the SSH procedure. As a result, each specific sequence has many progenitors and is represented by both sequence orientations. We call our method MOS because this difference between target and background populations is used for specific amplification of target molecules (Fig. 1). The procedure includes removing one adapter (adapter B in Fig. 1) by restriction endonuclease, heat-denaturation and re-annealing of the SSH sample. Some of the newly formed hybrids from target cDNAs bear adapter A at both termini. Such molecules are generated as a result of hybridization of molecules with mirror orientation of adapters A and B. Thus, they can only be derived from target cDNA fraction. Next, the 3′-ends are filled in and PCR with primer corresponding to adapter A is performed. In this PCR only molecules bearing adapter A at both termini can be amplified exponentially. Thus, the final PCR product is enriched for target sequences.
Figure 1.
Schematic representation of the MOS method. Rectangles represent DNA molecules (broad rectangles, double-stranded DNA; narrow rectangles, single-stranded DNA). Yellow rectangles, adapter A; green rectangles, adapter B. Pink molecules, target cDNA; blue, background cDNA. The gradient of filling shows the orientation of the cDNA molecules.
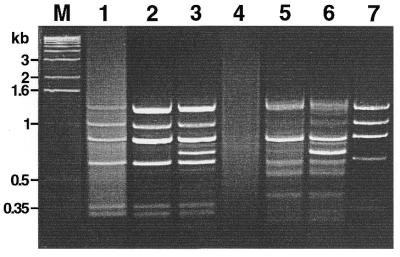
This scheme was verified in model experiments. To create artificial tester samples, we added various amounts of bacteriophage ϕX174 DNA as a target for subtraction in human skeletal muscle double-stranded cDNA. The amount of ϕX174 DNA added was 0.01% or 0.001% of the human cDNA. The same cDNA without viral DNA was used as driver. Both the tester and the driver were digested by HaeIII and used in SSH and subsequently in MOS procedures. Figure 2 shows the electrophoretic analysis of final PCR products. After subtraction of the tester with 0.01% of the target, the pattern of bands corresponding to ϕX174 was clearly visible but a bright smear representing background cDNA was also present (lane 1; compare with control ϕX174 DNA in lane 7). MOS application reduced the background level as shown by the disappearance of the smear in lane 2. The efficiency of MOS was even more demonstrative in subtraction of the tester with 0.001% of target. In this case the SSH product was an even smear with no apparent bands (lane 4) and obviously included a very low portion of the target DNA. However, after MOS the sample contained bright bands (lane 5) corresponding to some ϕX174 fragments. In these model experiments we also tested the addition of excess driver (as described for the SSH method) in the hybridization mixture during MOS (lanes 3 and 6). It is apparent that the addition of driver did not enrich the target but did lead to the appearance of background bands. Therefore, we did not use this modification in further experiments.
Figure 2.
Application of the MOS technique to the model subtraction (gel-electrophoresis of the PCR products). Lane M, 1 kb ladder (Gibco BRL). Lanes 1–3, results of applying of SSH and MOS procedures to tester cDNA containing 0.01% ϕX174 DNA; lanes 4–6, tester cDNA containing 0.001% ϕX174 DNA. Lanes 1 and 3, samples after SSH. Lanes 2 and 5, samples after MOS. Lanes 3 and 6, samples after MOS with addition of excess driver. Lane 7, amplification product of ϕX174 DNA digested with HaeIII and ligated with adapters A and B. Small divergences in length of the fragments presenting in SSH-generated and control ϕX174/HaeIII samples on the one hand and in MOS-generated samples on the other hand are due to the different PCR primers used.
In our practice, we repeatedly used the MOS technique successfully, including the following example of a real MOS application. The structural heterogeneity of neural tissues and the consequent highly complex gene expression profiles presents a great challenge in isolating genes that are developmentally regulated in the mammalian brain. In order to identify genes that are involved in the establishment of cellular identity in murine cortical neurons, we compared two cDNA samples (E13 and E15) prepared from cerebral cortex on 13- and 15-day embryos. A detailed description of this comparison and the isolated sequences will be published elsewhere. The statistical analysis of this experiment, presented below, illustrates the utility of MOS in cases where subtraction yields a small portion of target clones (Table 1). Subtraction was performed by the SSH method in both directions: using E13 as a tester and E15 as a driver (E13–E15) and vice versa (E15–E13). Subtracted sample E15–E13 was cloned and 192 clones from this library were analyzed by differential screening with two subtracted cDNA probes mentioned above. Screening revealed 17 differential clones (9% of analyzed clones), and further analysis (by means of Southern blot hybridization with initial E13 and E15 amplified cDNA samples) confirmed only four of these clones (24% of putative differential clones) to have differential expression patterns. After application of the MOS technique to subtracted samples, the primary screening of 480 clones from the E15-specific library revealed 87 differentially expressed clones (18% of analyzed clones), and 71 of these (82% of putative differential clones) were confirmed by further analysis. In this case MOS increased the portion of truly differential clones 7.5-fold and decreased the portion of false positive clones in the enriched sample 4-fold. It should be noted that the complexity of confirmed differential clones in the MOS library is rather high, 62 out of 71 clones represent different cDNA species. So, one of the main advantages of the SSH method, simultaneous isolation of many differentially expressed sequences, is conserved.
Table 1. Comparison of the enriched cDNA libraries generated by the SSH and MOS techniques.
| Analyzed clones | Putative differential clones | Confirmed differential clones | Different target cDNA species | |
|---|---|---|---|---|
| SSH |
192 |
17 (9%)a |
4 (24%)b |
4 |
| MOS | 480 | 87 (18%)a | 71 (82%)b | 62 |
aPercentage of all analyzed clones.
bPercentage of putative differential clones.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Prof. Eugene Sverdlov (Institute of Bioorganic Chemistry RAS), Dr Paul Siebert (Clontech Laboratories Inc.) for fruitful discussion, Dr Luda Diatchenko (Clontech Laboratories Inc.) for critical reading of the manuscript and Eric Machleder (Clontech Laboratories Inc.) for help in the manuscript preparation. This work was supported by Clontech Laboratories Inc. and the Russian Foundation for Fundamental Research (grant no. 98-04-48508).
REFERENCES
- 1.Diatchenko L., Lau,Y.F.C., Campbell,A.P., Chenchik,A., Moqadam,F., Huang,B., Lukyanov,S., Lukyanov,K., Gurskaya,N., Sverdlov,E.D. and Siebert,P.D. (1996) Proc. Natl Acad. Sci. USA, 93, 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurskaya N.G., Diatchenko,L., Chenchik,A., Siebert,P.D., Khaspekov,G.L., Lukyanov,K.A., Vagner,L.L., Ermolaeva,O.D., Lukyanov,S.A. and Sverdlov,E.D. (1996) Anal. Biochem., 240, 90–97. [DOI] [PubMed] [Google Scholar]
- 3.von Stain O.D., Thies,W.-G. and Hofmann,M. (1997) Nucleic Acids Res., 25, 2598–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokomizo T., Izumi,T., Chang,K., Takuwa,Y. and Shimizu,T. (1997) Nature, 387, 620–624. [DOI] [PubMed] [Google Scholar]
- 5.Yamagishi H., Garg,V., Matsuoka,R., Thomas,T. and Srivastava,D. (1999) Science, 283, 1158–1161. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski P. and Sacchi,N. (1987) Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 7.Diatchenko L., Lukyanov,S., Lau,Y.F.C. and Siebert,P.D. (1999) Methods Enzymol., 303, 349–380. [DOI] [PubMed] [Google Scholar]
- 8.Chenchik A., Zhu,Y., Diatchenko,L., Li,R., Hill,J. and Siebert,P. (1998) In Siebert,P.D. and Larrick,J.W. (eds), Gene Cloning and Analysis by RT–PCR. BioTechniques Books, Natick, MA, pp. 305–319.
- 9.Lukyanov K.A., Matz,M.V., Bogdanova,E.A., Gurskaya,N.G. and Lukyanov,S.A. (1996) Nucleic Acids Res., 24, 2194–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukyanov K.A., Diachenko,L., Chenchik,A., Nanisetti,A., Siebert,P.D., Usman,N.Y., Matz,M.V. and Lukyanov,S.A. (1997) Biophys. Biochem. Res. Commun., 230, 285–288. [DOI] [PubMed] [Google Scholar]
- 11.Jin H., Cheng,X., Diatchenko,L., Siebert,P.D. and Huang,C.C. (1997) Biotechniques, 23, 1084–1086. [DOI] [PubMed]
- 12.Siebert P.D., Chenchik,A., Kellogg,D.E., Lukyanov,K.A. and Lukyanov,S.A. (1995) Nucleic Acids Res., 23, 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]