The importance of histones and chromatin structure in the regulation of eukaryotic gene transcription has become much more widely accepted over the past few years. It has been clear for a decade that histones contribute to the regulation of transcription both in vitro and in vivo (reviewed in references 14, 34, 50, 64, and 120). More recent studies have led to the striking observation that several protein complexes involved in transcription regulation can function, at least in part, by modifying histones or altering chromatin structure (for recent reviews, see references 3, 44, 49, 51, 52, 87, 100, and 119). While it is clear that many of these protein complexes have functions in addition to chromatin modification, they illustrate the importance of chromatin structure as a part of transcription regulation mechanisms.
The most widely characterized chromatin-modifying complexes studied to date can be classified into two major groups, based on their modes of action, as follows: (i) ATP-dependent complexes, which use the energy of ATP hydrolysis to locally disrupt or alter the association of histones with DNA, and (ii) histone acetyltransferase (HAT) and histone deacetylase (HDAC) complexes, which regulate the transcriptional activity of genes by determining the level of acetylation of the amino-terminal domains of nucleosomal histones associated with them.
This review will focus primarily on the ATP-dependent remodeling complexes. For recent reviews of HAT and HDAC complexes, see references 5, 20, 31, and 55. Here we provide an organized listing of the ATP-dependent chromatin-remodeling complexes described to date and illustrate the relationships between their subunits. We also review the data available with regard to their mechanisms of action and promoter targeting as well as regulation of their activity. Finally, we examine the relationship between these complexes and the HAT complexes.
CLASSIFICATION AND SUBUNIT COMPOSITION
All of the ATP-dependent chromatin-remodeling complexes contain an ATPase subunit that belongs to the SNF2 superfamily of proteins. Based on the identity of this subunit, they have been classified into two main groups, the SWI2/SNF2 group and the imitation SWI (ISWI) group (25). A third class of ATP-dependent complexes that contain a Snf2-like ATPase and also show deacetylase activity has been recently described (see below). Tables 1 to 3 classify all of the complexes described in the following paragraphs and includes lists of all of their known subunits organized by homology, starting with the ATPase subunit.
TABLE 1.
Known subunits of the SWI2/SNF2 group of ATP-dependent chromatin-remodeling complexesa
| Yeast SWI/SNF | Yeast RSC | Drosphila Brahma | Human SWI/SNF |
|---|---|---|---|
| Swi2/Snf2 | Sth1/Nsp1 | Brm | hBRG1 or hBRM |
| Swi1 | p270(?)/BAF250 | ||
| Snf5 | Sfh1 | Snr1 | hSNF5/INI1/BAF47 |
| Swi3 | Rsc8/Swh3 | Bap155/Moira | BAF170, BAF155 |
| Swp82 | |||
| Swp73/Snf12 | Rsc6 | Bap60 | BAF60 (a,b,c) |
| Swp61/Arp7 | Rsc11/Arp7 | Bap55 (?) | BAF53 (?) |
| Swp59/Arp9 | Rsc12/Arp9 | Bap55 (?) | BAF53 (?) |
| Snf6 | |||
| Swp29/TafII30 | |||
| Snf11 | |||
| Rsc1 or -2 | |||
| Rsc3-5,-7,-9,-10 | |||
| Rsc13-15 | |||
| Bap111 | BAF57 | ||
| β-Actin/Bap47 (*) | β-Actin (*) | ||
| Bap74 |
All the known subunits of the SWI2/SNF2 group of ATP-dependent chromatin-remodeling complexes cited in the text are listed. This list is clearly not complete, as additional subunits of some complexes have not yet been identified. Conserved subunits are grouped horizontally, with the ATPase subunit listed first. Question marks denote incomplete or conflicting information, and asterisks indicate tentative components that are still controversial.
TABLE 3.
Known subunits of the Mi-2 group of ATP-dependent chromatin-remodeling complexesa
| Human NURD/NuRD/NRD | Xenopus Mi-2 |
|---|---|
| Mi-2β/CHD4, Mi2α/CHD3 (?) | Mi-2 |
| HDAC1 (NURD63) | Rpd3 |
| HDAC2 (NURD59) | Rpd3 |
| RbAp48 (NURD56) | RbAp48/46 |
| RbAp46 (NURD55) | RbAp48/46 |
| MTA1/2 (NURD70) | MTA1-like |
| MBD3 | MBD3 and MBD3 LFb |
| p66 | |
| Sin3(*) |
Same as Table 1, but for the Mi-2 group of ATP-dependent chromatin-remodeling complexes.
LF, long form.
Apart from these eukaryotic remodeling complexes stands RapA, a bacterial protein that consistently copurifies with DNA-dependent RNA polymerase and shows sequence homology to the Swi2/Snf2 ATPase. It has been suggested that it might have a role in prokaryotic transcription analogous to that of the eukaryotic ATP-dependent remodeling complexes (101).
The SWI2/SNF2 group.
The SWI2/SNF2 group includes yeast SWI/SNF (ySWI/SNF), yeast RSC, the Drosophila Brahma complex, and the human BRM (hBRM) and BRG1 (hBRG1) complexes (Table 1). All of these contain a highly conserved ATPase subunit, which belongs to the Swi2/Snf2 subfamily of proteins: Swi2/Snf2, Sth1, Brm, hBRM, and BRG1, respectively. A mouse homologue, mBRG1, has also been described (90). The homology of these proteins extends beyond the ATPase domain, as they all contain a bromodomain in the C-terminal region and two other conserved regions of unknown function called domains 1 and 2 (58, 102).
The ySWI/SNF complex was the first remodeling complex to be described. It contains 11 known subunits, including Swi2/Snf2 (Table 1). Several of the subunits were initially identified genetically as gene products involved in the regulation of either the HO endonuclease gene or the SUC2 gene, which encodes invertase. HO is required for mating type switching, hence SWI, while SUC2 mutants are classified as sucrose nonfermenters, thus SNF (reviewed in references 37 and 83). The SWI/SNF genes were subsequently shown to be involved in the transcriptional regulation of a wider subset of yeast genes (36). Additionally, genetic studies provided a connection between the functions of the SWI/SNF complex and chromatin. Several mutations that suppressed swi/snf phenotypes corresponded to genes encoding histones and other chromatin proteins (54, 91). The relationship between the function of the ySWI/SNF complex and chromatin was strengthened when the complex was purified and found to alter nucleosome structure in an ATP-dependent mechanism (18, 84).
The highly related RSC complex contains many proteins that are homologues of SWI/SNF subunits. Actually, the two complexes share at least two identical subunits (Table 1). The RSC complex was initially identified by these sequence homologies and subsequently purified (10). The biochemical activities of the RSC complex that have been observed thus far are similar to those of ySWI/SNF (see below). However, the RSC complex is far more abundant than SWI/SNF in the yeast cell (thousands of molecules compared to 100 to 200 molecules of SWI/SNF). In addition, it contains several subunits that are essential for viability whereas none of the SWI/SNF subunits is essential (10). The 15 RSC subunits include a homologue of the Swi2/Snf2 ATPase, called Sth1, and homologues of Snf5, Swi3, and Swp73 (Sfh1, Rsc8/Swh3, and Rsc6, respectively).
In addition to these, SWI/SNF and RSC contain the same two actin-related proteins, Arp7 and Arp9 (9, 85). Arp7 is identical to Swp61 of the SWI/SNF complex and Rsc11 of the RSC complex, while Arp9 is identical to Swp59 and Rsc12. Yeast cells have many other known actin-related proteins (Arp1 to Arp10), many of which remain uncharacterized. It has been suggested that these actin-related proteins may link remodeling complexes to either actin-binding proteins or to nuclear proteins not previously thought to be associated with actin, such as components of the nuclear matrix or chromatin itself. Moreover, Baf53, an actin-related protein, and actin itself have been reported to be components of the mammalian SWI/SNF complexes (115). Finally, an actin-related protein, Bap55, and actin itself were also found in the Drosophila Brahma complex (81; see below).
Homologues of SWI/SNF proteins were previously identified in Drosophila melanogaster via genetic screens for suppressors of the transcriptional repressor Polycomb (102) and shown to form part of a large multisubunit complex called Brahma (23). This complex contains eight major proteins, including the ATPase subunit Brm. The proteins that copurify with Brm have been called BAPs for Brm-associated proteins (Table 1). Brm complex subunits Bap45/Snr1, Bap155/Moira, and Bap60 are conserved between yeast and humans (23, 81). Moira/Swi3D is a homologue of yeast Swi3 and the human proteins Baf155 and Baf170 (see below) and is reportedly identical to Bap155, a component of the Brahma complex purified by Papoulas et al. (19, 81). Moira is also highly homologous to a mouse Swi3-related protein, Srg3 (42). Moira is similar to Swi3, Baf155, Baf170, and Srg3 across three regions. The first region is rich in prolines and in hydrophobic and aromatic amino acids. The second region is a tryptophan-rich SANT domain. This name derives from its presence in Swi3, Ada2, N-CoR, and TFIIB (see below and reference 1). No function has been reported yet for the SANT domain, but it may be involved in Moira's association with Brm. The third region of homology is a leucine zipper motif, which is thought to be involved in the self-association ability of Moira (19). Interestingly, the Brm complex includes a protein that seems to be unique to higher eukaryotes, Bap111. This protein contains an HMG domain and is homologous to Baf57, present in human complexes but not in the related yeast RSC and SWI/SNF complexes. Other components are Bap74, a protein that has some sequence identity to Hsc4, a constitutive (non-heat-inducible) chaperone protein, and Bap47, which shares conserved regions with Act1 and Act2, two nonmuscle actins (81).
Kwon et al. reported the isolation of two Brg1-containing human complexes with ATP-remodeling abilities (56). Later, two SWI/SNF-like multisubunit complexes of approximately 2 MDa were purified from human cells (114, 115). These complexes contained different DNA-dependent ATPase/helicase subunits, BRG1 and hBRM, and are called hBRG1 and hBRM, respectively. These ATPases are more than 70% identical to one another and are highly homologous to Swi2/Snf2 across the entire gene (25). The proteins that are associated with BRG1 and hBRM in these two complexes, called BAFs or BRG1-associated factors, are also quite similar. In agreement with Kwon's data, it has been suggested that there might be multiple complexes, maybe in different cell types, each containing a different subset of BAF proteins and either Brg1 or hBrm as the catalytic subunit. One of the BAF proteins, p47, is a human homologue of yeast Snf5 (hSNF5/INI1/BAF47). Both complexes also contain BAF155 and BAF170, which are both homologues of the yeast Swi3 protein. These proteins, as well as Swi3 and Drosophila Moira (see above), contain a myb-like tryptophan repeat region called a SANT domain, which is proposed to be a DNA-binding domain (1). However, neither Swi3 nor BAF170 has been shown to bind DNA (89, 115). This type of conserved region is also found in the Ada2 subunit of the Gcn5-dependent HAT complexes and in the ISWI subunit of the Drosophila chromatin-remodeling complexes, suggesting that they might have a role in chromatin remodeling different from DNA binding. Both hBRG1 and hBRM complexes also contain a subunit that is homologous to the yeast Swp73 protein, BAF60. This protein exists in three forms, a, b and c, which might associate with the ATPase subunits in different subcomplexes. In fact, hBRG1 and hBRM have been found in many different cell lines from a wide range of tissues, and the complexes containing them might have slightly different subunit compositions (114). Interestingly, hSNF5/INI1 is also present in many types of cells as well, some of which contain neither hBRG1 nor hBRM.
The ISWI group.
The second group of ATP-dependent remodeling complexes contains the ISWI protein as the ATPase subunit (Table 2). The most extensively studied members of this group, ACF (ATP-utilizing chromatin assembly and remodeling factor), NURF (nucleosome-remodeling factor), and CHRAC (chromatin accessibility complex), were purified from Drosophila extracts using biochemical methods based on their ability to disrupt and/or generate regularly spaced nucleosomal arrays (40, 106, 108). All of these complexes contain the nucleosome-dependent ATPase ISWI, which has homology with Swi2/Snf2 exclusively over the region of the ATPase domain (26). The ISWI-containing complexes are smaller and have fewer subunits than their SWI/SNF counterparts. NURF has a molecular mass of approximately 500 kDa and contains four subunits, including ISWI, p215, and the WD repeat protein Nurf-55, a protein identical to the 55-kDa subunit of Drosophila chromatin assembly factor dCAF-1 (66). The smallest subunit of NURF, Nurf-38, was reported to be inorganic pyrophosphatase. Both recombinant Nurf-38 and purified NURF complex have inorganic pyrophosphatase activity; however, inhibition of this activity does not affect the ability of NURF to remodel chromatin (29). CHRAC has a molecular mass of approximately 670 kDa and contains five subunits, two of which were identified as ATPases, ISWI and topoisomerase II (108). ACF has a molecular mass of approximately 220 kDa and contains ISWI as the catalytic subunit (40). Recently, the protein Acf1 was described as a component of ACF (41). Purification of ACF from Drosophila revealed that it exists as two complexes. Both of them contain ISWI plus one of the two Acf1 forms, p170 or p185. By contrast, Acf1 did not copurify with NURF or CHRAC. Since the predicted size of ACF is only 220 kDa, it is believed that ACF exists as heterodimers of either form of Acf1 and ISWI (41).
TABLE 2.
Known subunits of the ISWI group of ATP-dependent chromatin-remodeling complexesa
| Drosophila NURF | Drosophila CHRAC | Drosophila ACF | Yeast ISWI | Human RSF |
|---|---|---|---|---|
| ISWI | ISWI | ISWI | ISW1,b ISW2c | hSNF2h |
| Nurf-55 | ||||
| p215 | ||||
| Nurf-38/iPPase | ||||
| Topoisomerase II | ||||
| p175 | ||||
| p20 | ||||
| p18 | ||||
| Acf1(p185 or p170) | ||||
| p74b | ||||
| p105b | ||||
| p110b | ||||
| p140c | ||||
| p325 |
Same as Table 1, but for the ISWI gorup of ATP-dependent chromatin-remodeling complexes.
Subunit of the ISW1 complex.
Subunit of the ISW2 complex.
ISW1 and ISW2, two ISWI-related proteins, were recently identified in yeast based on their sequence homology to the ATPase domain of Drosophila ISWI (105). In fact, the proteins are homologous across most of their sequences, including a SANT domain (see above). ISW1 and ISW2 are components of two distinct multisubunit complexes that possess diverse nucleosome-remodeling and spacing abilities. Using Flag epitope-tagged ISW1 and ISW2 and chromatographic techniques, it was shown that ISW1p forms a four-subunit complex with p110, p105, and p74, while ISW2p copurifies with p140 to form a two-subunit complex. To date, no further complexes containing ISW1p or ISW2p have been detected (105).
An ISWI-containing complex, RSF, was purified from human cells based on its ability to facilitate transcription from chromatin templates. It also contains the uncharacterized protein p325 (60).
To date, all of the eukaryotes analyzed, including yeast, plants, nematodes, flies, bovines, mice, and humans, have putative ISWI homologs (25, 76, 105). This suggests that these proteins have an important function conserved in evolution. It is also remarkable that all three of the species studied in detail to date (flies, humans, and yeast), contain several ATP-dependent remodeling complexes. The abundance of these complexes seems to be highly variable: some of them are present in a high concentration, such as yeast RSC, while others, such as ySWI/SNF, are present in much lower amounts. Finally, while some complexes seem to be dispensable, others contain essential proteins, suggesting that they are not totally redundant. It will be exciting to establish if these related complexes carry out different functions in the cell and to determine to what degree their properties overlap.
The Mi-2 group: chromatin-remodeling and deacetylase complexes (Table 3).
Similar complexes that possess both chromatin-remodeling and deacetylase activities were recently purified from human cells by several different groups and are known as NURD, NuRD, and NRD (103, 121, 124). The minor differences reported in their compositions might reflect the purification protocols used in the different cases. The complex, which we will generically refer to as hNURD, contains the HDACs HDAC1 and -2, the retinoblastoma protein (Rb)-associated proteins RbAp46 and -48, and the Swi2/Snf2 ATPase homologue CHD4, also known as Mi-2β. It is not clear if CHD3, a Mi-2α homolog, is contained in the same complex or in a similar one. These CHD/Mi-2 proteins are know to be self antigens in the human disease Mi-2 dermatomyositis, and they have, besides the Snf2 ATPase motif, two PhD zinc fingers and two chromodomains (118). It was shown that the complex has the ability to both deacetylate histones and remodel chromatin, presumably by means of the HDAC and Mi-2/CHD subunits, respectively. The complex was also shown to contain MTA1 and/or -2, proteins that are found in metastatic cells (121, 125). Recently obtained data suggest that MTA2 modulates the deacetylase activity of the hNURD complex (125).
A Mi-2 complex related to hNURD was identified in Xenopus egg extracts (109). This complex contains the deacetylase Rpd3, the histone-binding protein RbAp48/46 homologue, and a protein with homology to human Mi-2/CHD3/4. In contrast to the human complexes, Xenopus Mi-2 was reported to contain a variable amount of Sin3. The authors confirmed that this complex possesses both deacetylase and ATPase activities. A recent paper from the same group reports the characterization of the remaining subunits of the Mi-2 Xenopus complex (111). The 35-kDa band was shown to correspond to two alternatively spliced peptides that are homologous to mammalian MBD3 (methyl-CpG-binding domain-containing protein), which the authors call Xenopus laevis MBD3 and MBD3LF (long form). The other bands were identified as an MTA-like peptide and a novel 66-kDa protein that also has a human homologue. The authors suggest that the MBD3 peptide could recruit the histone deacetylase (HDAC) and remodeling activities of the Xenopus Mi-2 complex to methylated DNA.
In agreement with this, the hNURD complex was also found to contain two forms of MBD3 (125). Furthermore, the authors report that MBD3 directly interacts with several subunits of the NuRD complex, with the exception of Mi-2. In contrast to the Xenopus situation, in this case neither MBD3 nor NURD could bind to methylated DNA. However, MBD2, a homologue of MBD3, is able to bind both methylated DNA and the NURD complex. The authors suggest that the NURD complex can be tethered to methylated DNA via its interaction with MBD2. Interestingly, MBD2 is also associated with transformed cells (125).
The functional significance of these multifunctional complexes could be very interesting. While remodeling complexes are usually associated with derepression of transcription, deacetylases are related to repression. In this respect, it was proposed that the ATP-dependent remodeling activity of the NURD complexes might facilitate the deacetylation of the target histones. Furthermore, the presence of methyl-CpG-binding proteins in both the human and Xenopus Mi-2 complexes supports the idea that these activities might be specifically directed to methylated regions of the genome. In turn, this would lead to repression either via compaction of the chromatin structure or by allowing the binding of repressor proteins. Finally, it has been suggested that Mi-2 could be recruited to specific genes by repressors (45, 47). Thus, these discoveries establish links among chromatin remodeling, deacetylation, methylation, silencing, and cancer progression.
MECHANISM OF ACTION OF ATP-DEPENDENT REMODELING COMPLEXES
The mechanisms by which the ATP-dependent nucleosome-remodeling factors alter nucleosome and chromatin structure are not yet clear. However, pieces of information that illustrate both similarities and differences in the activities of these complexes are becoming available. For example, both the SWI/SNF and NURF complexes utilize the energy of ATP hydrolysis to alter nucleosome structure. However, while the Swi2/Snf2 ATPase of SWI/SNF is induced by either DNA or nucleosomes (18), the ISWI ATPase in NURF requires nucleosomes with intact histone amino-terminal tails for maximum stimulation (30).
Binding of remodeling complexes to DNA and nucleosomes.
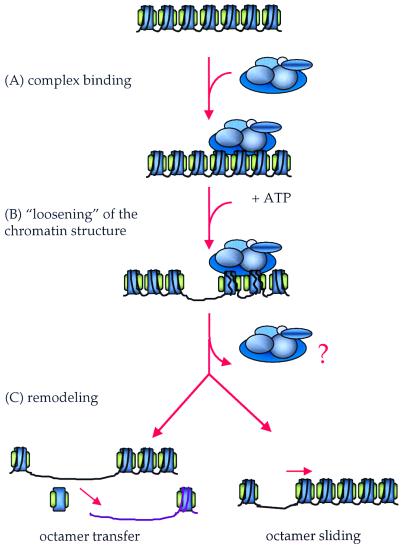
In order to remodel chromatin, remodeling complexes have to be able to recognize and bind to their substrate (Fig. 1A). The NURF complex has not been found to form a stable complex with nucleosomes or DNA in vitro. Instead, it appears that it may interact with nucleosome substrates in a manner dependent on the core histone tails (30, 106). Possibly, the ISWI subunit of NURF, CHRAC, and ACF binds DNA at least transiently. The topoisomerase II subunit of CHRAC represents another potential DNA-binding subunit (108). In addition, the fact that recombinant ISWI has some nucleosome-remodeling activity on its own (15, 33, 57) indicates that this subunit must be capable of interacting directly with nucleosome substrates.
FIG. 1.
Two-step model of SWI/SNF and RSC action in chromatin remodeling. The binding of the remodeling complex to chromatin is ATP independent (A). Upon ATP addition, the conformation of nucleosomes changes as a consequence of the alteration of histone-DNA interactions (B). This disruption results in remodeling of the chromatin (C), which might occur while the complex is still bound or might persist after it is released from the chromatin (indicated by the question mark). Remodeling that occurs may result in transfer of histone octamers to different DNA segments in trans or in sliding of the octamers in cis (i.e., to a different position in the same DNA molecule). The exact consequence of remodeling is likely dependent on the exact context of nucleosomes at a given promoter and can lead to either (i) activation of transcription or (ii) repression.
In contrast to the ISWI complexes, SWI/SNF and the related RSC complex clearly bind DNA and nucleosomes with high affinity (17, 63, 89). The ySWI/SNF complex is able to bind naked DNA in an ATP-independent manner, with a Kd in the nanomolar range (89). It is likely that this binding occurs through minor-groove interactions, since the complex can be displaced from DNA by distamycin A or chromomycin A3, two minor-groove-binding reagents (17, 89). The DNA-binding properties of the ySWI/SNF complex are similar to those of HMG box proteins, which bind nonspecifically to DNA in a length-dependent manner with a preference for four-way junctions and cruciforms. It has been proposed that the complex might bind DNA via one of the two subunits that contain an HMG box. The first one is INI1/hSNF5L1, which prefers to bind supercoiled DNA over relaxed circular DNA (69). The second subunit is BAF57, which can bind four-way junction DNA (113). Thus, it is possible that the SWI/SNF complex recognizes particular structural features in chromatin, such as crossovers in the linker DNA.
Extensive studies have not been able to demonstrate any DNA sequence specificity for the SWI/SNF complex. However, PYR (a mammalian complex related to SWI/SNF that was isolated from murine erythroleukemia cell nuclear extracts) prefers to bind to a 250-bp pyrimidine-rich DNA sequence located between the human fetal and adult β-globin genes. Furthermore, deletion of this DNA element resulted in delayed gene switching of human γ- to β-globin in transgenic mice (77). Therefore, it was proposed that the PYR complex could possibly promote globin gene switching by enhancing the binding of β-globin activators via its chromatin-remodeling functions.
The mode of binding of ySWI/SNF to nucleosomes appears to include interactions beyond those made with the minor groove of DNA, since this binding is not as easily disrupted upon addition of distamycin A (18). The affinity of SWI/SNF for nucleosomes is slightly higher than that for naked DNA. This is possibly due to additional interactions of SWI/SNF with the core histones. Depletion of the H2A/H2B dimer either in vivo (by mutations) or in vitro (by addition of H2A/H2B chaperones) has been demonstrated to bypass SWI/SNF requirements (12, 18, 35). Moreover, using site-directed mutagenesis, Recht and Osley have recently created SWI/SNF-independent mutations (sin) in the core histones that suppress SWI/SNF defects (91). These mutations occur in regions required for H2A/H2B dimerization and dimer-tetramer association and in the H2B amino terminus. These data argue for an important inhibitory role for the H2B N terminus in chromatin that can be antagonized by the SWI/SNF complex. There are data supporting the notion that histone N-terminal tails are required for the ability of SWI/SNF to catalytically remodel nucleosomal arrays (62). In contrast, human SWI/SNF (hSWI/SNF) is able to remodel tail-less mononucleosomes, as well as nucleosomal arrays, suggesting that the mechanism of nucleosome remodeling by hSWI/SNF is not dependent on the core histone tails (32).
ATP-dependent nucleosome disruption.
The original biochemical approach used to assay the ISWI complexes differed from that used for SWI/SNF and RSC, making it difficult to compare their activities. The ISWI complexes were tested both for the ability to disrupt or assemble spaced nucleosome arrays and for the ability to stimulate in vitro transcription (40, 60, 106, 108). In comparison, SWI/SNF and RSC were assayed for the ability to disrupt the rotational phasing of DNA sequences on nucleosome core particles. Alternatively, they were tested for the ability to stimulate the binding of transcription factors to nucleosome cores (10, 18, 38, 56, 114). However, NURF was also shown to disrupt rotational phasing of DNA on nucleosome cores but in a manner that appeared slightly different from that of SWI/SNF (106). The differences in the chromatin-remodeling activities of these complexes are now becoming more apparent.
In vitro studies have shown that the ySWI/SNF and hSWI/SNF complexes disrupt the rotational phasing of DNA on the surface of the histone octamer (Fig. 1B). In other words, the complex alters the histone-DNA contacts in an ATP-dependent manner, resulting in a pattern of DNase I digestion different from that of nucleosomal DNA (18, 38; reviewed in reference 51). Another consequence of this disruption is that it enhances the access of DNA-binding proteins to nucleosomal DNA. This has been shown with several types of sequence-specific DNA-binding proteins and with restriction endonucleases (18, 38, 56, 61, 107). Mutations in the ATP-binding domain of the Swi2/Snf2 subunit abolish SWI/SNF activity in these assays (18).
Studies of the human Swi2/Snf2 homologues BRG1 and hBRM have shown that these proteins can disrupt nucleosomes in the absence of the other subunits (86). The addition of three other hSWI/SNF subunits, INI1, BAF155, and BAF170, increases the nucleosomal disruption efficiency of BRG1 to that of the intact hSWI/SNF complex (86). In a similar fashion, recombinant ISWI has also been shown to be able to promote the binding of GAGA factor to its cognate site on DNA and chromatin and to increase the regularity of arrays of nucleosomes (15). Interestingly, each of the different ISWI-containing complexes can only perform a subset of these functions, suggesting that the activities of the ATPase subunit might be modulated by the different protein subunits present in each of them.
The mechanisms by which ATP-dependent nucleosome-remodeling complexes alter nucleosome structure remain vague. It is suspected that there are significant mechanistic differences between the action of the SWI/SNF and RSC complexes and that of the ISWI complexes. As mentioned above, the interactions of these complexes with nucleosomes differ and it is likely that their effects on nucleosome structure differ as well.
Data produced by several laboratories over the past several years are beginning to reveal important details and narrow the range of possibilities regarding nucleosome disruption by the SWI/SNF and RSC complexes. For example, the homology of the Swi2/Snf2 ATPase domain to that of helicases suggested that this protein might also function as a helicase. However, helicase activity was not observed in the purified complex (18) and single-stranded DNA was not detected within disrupted nucleosomes (17). Another model posited that SWI/SNF might facilitate the loss of H2A/H2B dimers from nucleosomes, since a histone chaperone stimulated the activity of SWI/SNF in facilitating transcription factor binding to nucleosomes (18). However, the fact that nucleosomes in which the histones were protein-protein cross-linked were still disrupted by SWI/SNF (as measured by the classic change in DNase I digestion patterns) indicated that displacement of histone dimers was not required for this alteration in the digestion pattern (4, 17). Nucleosome disruption by SWI/SNF also results in a change in the location of the histone H2A N-terminal tail on nucleosomal DNA. However, cross-linking of this tail to DNA, which impedes its movement, does not prevent nucleosome disruption (59).
The ATP-dependent disruption of nucleosome cores by a SWI/SNF or RSC complex generates a stably disrupted nucleosomal conformation that persists after the removal of ATP and detachment of the SWI/SNF or RSC complex (17, 39, 63, 94). Moreover, SWI/SNF or RSC can hasten the reverse of this process, converting the disrupted nucleosome conformation back to the original state (63, 94). Thus, these complexes can catalyze the interconversion between two nucleosome conformations (reviewed in reference 48). When nucleosome cores are used as the substrate of this reaction, the disrupted nucleosome conformation is relatively stable. However, if nucleosome arrays are used, their conversion back to the original nucleosome conformation appears to be more rapid (61).
Some features of the SWI/SNF- or RSC-disrupted nucleosome conformation are (i) loss of the rotational phasing of the DNA on the surface of the histone octamer, although the DNA remains at least partly associated with the histone octamer surface (17); (ii) increased accessibility of the nucleosomal DNA to transcription factors and restriction enzymes (17, 94); (iii) reduction of the total amount of DNA associated with the histone octamer as measured by electron spectroscopic imaging, suggesting that DNA at the edge of the nucleosome “peels off” the histone octamer (4); (iv) apparent association of disrupted nucleosome cores into a dinucleosomelike species, perhaps through interactions of loosened DNA with histone octamers from other nucleosomes (63, 94); (v) reduction of the stability of the nucleosome at elevated ionic strengths (63); and (vi) relocation of the histone H2A N-terminal tail from a position approximately 40 bp on either side of the nucleosome dyad (center) to a location near the nucleosome dyad (59).
The data obtained thus far suggest that the histone-DNA interactions have been reduced and significantly altered in the disrupted nucleosome conformation, diminishing the stability of histone binding to DNA and permitting increased affinity for transcription factors. This might be accomplished by simple loss of DNA associated at the edges of the nucleosome core. However, the observed relocation of the H2A amino termini (59) is most consistent with a conformational change in the histones themselves. For a detailed discussion of the conformational changes induced by SWI/SNF or RSC, see reference 49.
It is important to note that the many features of the mechanism described above may not apply to the actions of the ISWI complexes. As noted above, neither a stable complex between these complexes and nucleosomes nor a persistently altered nucleosome conformation has been observed. By contrast, mechanistic studies on the ISWI-containing NURF and CHRAC complexes suggest a gradual movement of nucleosomes along DNA in cis as a consequence of their action (33, 57). Nucleosome sliding may result from a more modest and more reversible form of “hit-and-run” nucleosome disruption by these complexes (see below).
Chromatin remodeling.
Chromatin remodeling is defined here as a stable alteration in the structure of nucleosomes and the distribution on DNA. Remodeling can be considered a consequence of nucleosome disruption. In this view, chromatin-remodeling complexes interact and disrupt nucleosome conformation, which may itself represent chromatin remodeling or may subsequently lead to nucleosome movement (Fig. 1C). Nucleosome movement is likely to occur during the interaction of the ISWI complexes (33, 57). In the case of the SWI/SNF and RSC complexes, where a stably disrupted nucleosome conformation has been observed, it is possible that disrupted nucleosomes move subsequently to the binding of these complexes. The interactions of sequence-specific transcription factors with DNA during chromatin remodeling results in the formation of DNase-hypersensitive sites (82, 112). This indicates either that the nucleosomes have been moved off those sequences or that they are now occupied by both factors and histones (reviewed in reference 97).
To understand the functions of ATP-dependent complexes in chromatin remodeling, it is necessary to consider what happens to the histones. First, it is important to realize that the consequences of nucleosome disruption by ATP-dependent complexes may differ at different promoters or enhancers. This has been clearly demonstrated in vitro, where several factors binding the human immunodeficiency virus type 1 enhancer form a stable complex with histones, whereas multiple Gal4 dimers facilitate histone displacement by SWI/SNF (98). Moreover, SWI/SNF and RSC can participate in repression, as well as activation, of transcription in vivo (22, 68, 72). Thus, chromatin remodeling need not always favor transcription activation.
There are two primary models for nucleosome rearrangement during chromatin remodeling: displacement of histones in trans and sliding of histone octamers along DNA in cis (Fig. 1). Displacement of histones in trans has been demonstrated for the RSC and SWI/SNF complexes (65, 79), and it was shown to require the presence of histone acceptors (65). Moreover, the binding of transcription factors, which further destabilize histone-DNA interactions, facilitates trans displacement (80). In this way, trans displacement can lead to the formation of DNase-hypersensitive sites at the sites of transcription factor binding. It has been suggested that the disrupted nucleosome conformation described above might fortuitously result from two histone octamers interacting with the same piece of DNA as a consequence of SWI/SNF or RSC action (65). However, electron spectroscopic imaging indicates a reduction of the DNA mass associated with the normal mass of histones in individual nucleosomes (4). Thus, association of two nucleosome cores or two histone octamers to form a particle the size of a dinucleosome does not appear to be required to observe features of the disrupted nucleosome.
In contrast to the observed trans displacement of histones by the SWI/SNF and RSC complexes, the ISWI complexes NURF and CHRAC have thus far been found to facilitate nucleosome movement in cis. This process is referred to as nucleosome sliding (33, 57). Nucleosome sliding induced by CHRAC or NURF appears to occur without displacement of the histone octamer from the DNA and appears to be progressive, suggesting a series of catalyzed small steps rather than any large leaps forward (33, 57). These results raise the obvious question of whether the ISWI complexes catalyze chromatin remodeling by a mechanism completely different from that of SWI/SNF and RSC. However, further analysis of the SWI/SNF complex has shown that it too can catalyze nucleosome sliding (116). In these experiments, SWI/SNF preferred to slide histone octamers along DNA rather than displace them in trans. However, when a barrier was present that prevented nucleosome sliding by SWI/SNF, the complex was still able to displace histones in trans (116). Neighboring positioned nucleosomes may also provide a barrier to sliding, favoring the displacement pathway (79). The emerging picture is that the ISWI complexes and SWI/SNF (and presumably RSC as well) can facilitate nucleosome sliding on DNA in cis. However, when sliding is not possible, SWI/SNF can also facilitate nucleosome displacement in trans. Histone displacement in trans has not been observed with a CHRAC or NURF complex, despite attempts to do so (33, 57).
What special characteristic of SWI/SNF provides it with this displacement activity? One possibility mentioned above is the fact that SWI/SNF binds DNA with high affinity, which has not been observed with the ISWI complexes. DNA binding by SWI/SNF may provide an extra push, facilitating dissociation of the histones either onto other DNA in trans or to histone chaperones. The possibility that DNA binding participates in trans displacement is consistent with the observation that binding of GAL4 dimers to nucleosomal DNA further stimulates displacement in trans by SWI/SNF (79, 80). Thus, the binding of SWI/SNF and the binding of transcription factors may contribute to histone displacement in trans by further destabilizing histone-DNA interactions within the disrupted nucleosome. This raises the formal possibility that the ISWI complexes might also be able to participate in trans displacement of histones if sequence-specific transcription factors (e.g., GAGA factor) provided a DNA-binding activity to complete with histone-DNA interactions. This experiment has not yet been reported.
ATP-DEPENDENT CHROMATIN-REMODELING COMPLEXES AND THE CELL CYCLE
One aspect of ATP-dependent chromatin-remodeling complexes that has not been studied extensively is their function in the different phases of the cell cycle. These complexes have been implicated in the regulation of cellular growth and proliferation. For example, hBRG1 and Sfh1p have been shown to play a role in cell cycle progression (11, 46), while BRM knockout mice show increased cell proliferation (92). In addition, human complexes were identified as coregulators of genes involved in cellular transformation (reviewed in reference 71). Conceivably, these effects could be indirect, i.e., mediated by the ability of ATP-dependent remodeling complexes to remodel promoters of genes involved in cell cycle control.
However, the scenario seems to be more complex. Several studies done with human cell lines have shown that BRG1 and hbrm, the ATPase subunits of the hSWI/SNF complexes, can physically interact with Rb. Furthermore, these studies showed that hBRG1/hBRM can function as tumor suppressor genes and induce the formation of growth-arrested cells in an Rb-dependent manner (24, 99). Rb has a central role in the control of fundamental cellular processes such as proliferation and differentiation. Therefore, these studies suggest that the ATPase subunits of hSWI/SNF complexes cooperate with Rb to regulate cell fate.
In addition to associating with Rb, hBRG1 has been shown to coimmunoprecipitate with cyclin E. In turn, cyclin E associates with the cycle-dependent kinase Cdk2 to control the G1/S checkpoint of the cell cycle. Other SWI/SNF subunits were also coimmunoprecipitated in this study, suggesting that cyclin E/Cdk2 might interact with the SWI/SNF complex as a whole. Furthermore, the cdk2-cyclin E complex can phosphorylate both hBRG1 and BAF155. The authors propose that this phosphorylation might regulate the activity of hSWI/SNF (95).
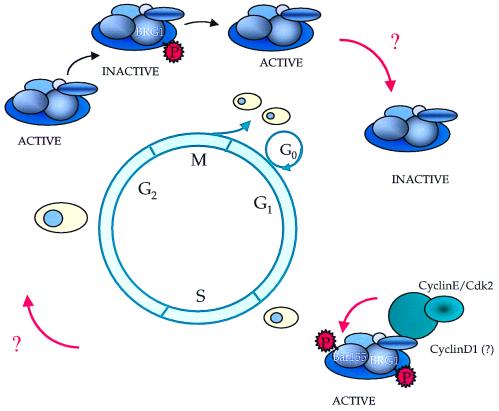
Several lines of evidence suggest that phosphorylation might regulate the function of ATP-dependent remodeling complexes during the cell cycle (Fig. 2). For example, the phosphorylation state of Sfh1p, a component of yeast RSC, oscillates during the cell cycle, an event that might be related to its role in G2/M transition (11). Other experiments have shown that hBRM and hBRG1 are phosphorylated during mitosis, concomitantly with their exclusion from condensed chromosomes. In addition, hbrm was partially degraded in mitotic cells. These events were suggested to cause the inactivation of SWI/SNF during cell division (70). Recent experiments by the Kingston group confirmed these data (96). Furthermore, these experiments showed that the phosphorylation state of another SWI/SNF subunit, hSWI3, followed that of Brg1. In addition, the mitotic complexes lost their ability to disrupt nucleosomes. As cells exited mitosis, the SWI/SNF complexes recovered the ability to disrupt nucleosomes and simultaneously became dephosphorylated (96). Finally, it has been shown that the BAF complex (or hSWI/SNF) is rapidly targeted to chromatin via the phosphoinositol pathway upon lymphocyte activation (126).
FIG. 2.
Cell cycle regulation of the SWI/SNF complex. The activity of hSWI/SNF is regulated, at least in part, by phosphorylation of some of its subunits. The complex is activated after G1 by a cyclin E/cdk2-dependent phosphorylation event of BAF155 and BRG1. Phosphorylation toward the end of G2, which might also occur at the level of BRG1, inactivates it, while a dephosphorylation even occurring late in G2 also seems to have an activating role in SWI/SNF function. The question marks denote the lack of information concerning how these two apparently contradictory sets of data are related.
These studies, taken together, argue that the activity of the ATP-dependent remodeling complexes in both yeast and mammalian cells is regulated by the cell cycle machinery through multiple mechanisms. Among these are the reversible phosphorylation of several subunits, targeted protein degradation, and modulation of the localization of the complex in the cell. Some of the complexes also seem to interact with proteins that are essential to the regulation of the cell cycle, suggesting that their role is more complex than initially suspected.
TARGETING TO PROMOTERS
Why do a subset of cellular genes specifically require the function of the SWI/SNF complexes for full activation? This question has been unclear for many years. One possibility is that the strength of a particular promoter plays a role in its dependence on chromatin-modifying complexes. An otherwise weak promoter may require the complex for full activity, while a strong promoter may not. In support of this hypothesis, the removal of two of the four Gal4-binding sites in the Gal1-Gal10 upstream activation sequence causes the normally SWI/SNF-independent Gal1 promoter to become SWI/SNF-dependent (8, 28). Another possibility is that remodeling complexes are only required for the transcription of promoters that possess positioned nucleosomes, which leave transcription factor sites unavailable for binding.
How are chromatin-remodeling complexes recruited to the specific promoters of the genes they regulate in living cells? Since there are only about 100 SWI/SNF molecules per yeast cell (10), the concentration of the complex near a target gene needs to be increased to allow remodeling of nucleosomes at specific promoters. An early report suggested that gene-specific activators might interact with components of the hSWI/SNF complex. In the presence of hSWI/SNF, Gal4-VP16 bound more strongly to nucleosomal templates than either the DNA-binding domain of Gal4 [Gal4(1-94)] or Gal4-AH (the AH domain is a weaker activation domain than that of VP16) (56). Since the DNA-binding domain of each of these fusion proteins is the same, Kwon et al. implied that the activation domain might play a role in the enhanced binding.
In the two primary models of SWI/SNF targeting that exist, (i) remodeling complexes are targeted to promoters via interactions with sequence-specific transcription factors and (ii) the SWI/SNF complex is recruited to promoters through association with RNA polymerase II. Studies showing association of SWI/SNF with the yeast RNA polymerase II holoenzyme (117) and mammalian RNA polymerase II (13, 75) support the second model. SWI/SNF might be recruited to a promoter with RNA polymerase II, and/or SWI/SNF could recruit the transcription machinery to promote and enhance the transcription of a gene via its interaction with the holoenzyme. The Swi2/Snf2, Swi3, Snf5, and Snf11 proteins have been reported to be integral components of the mediator complex, which is tightly associated with the C-terminal domain of RNA polymerase II (117); however, these results have been questioned (10). Consistent with these possibilities, it has been shown that hSWI/SNF can cause the activator-dependent release of paused RNA polymerase II (6).
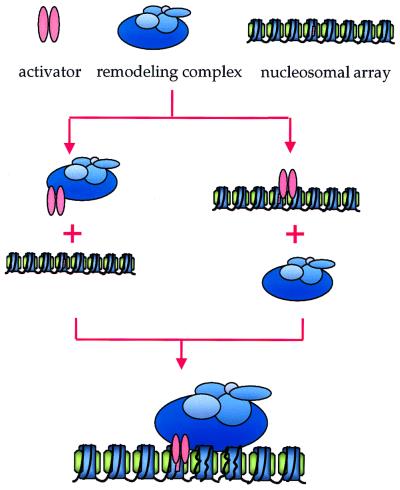
Many in vivo studies involving nuclear hormone receptors favor the first model, i.e., targeting of chromatin-modifying complexes via gene-specific activators (Fig. 3). Using coimmunoprecipitation studies, Yoshinaga et al. detected an interaction between a region of the glucocorticoid receptor (GR) and Swi3 (a component of the SWI/SNF complex) (122). However, the interaction does not occur in Δswi1 or Δswi2 strains, suggesting that the GR interacted with Swi3 in the context of the SWI/SNF complex. Östlund Farrants and colleagues later argued that the binding of the receptor to mononucleosomes containing GR response elements potentiated the nucleosome disruption ability of the SWI/SNF complex (78). Others have shown the presence of hSWI/SNF components, as well as transcriptional coactivators, in an activated GR complex (27) and interactions between Brg1 and the progesterone receptor (67).
FIG. 3.
Targeting of the SWI/SNF complex. The remodeling complexes are directed to sites in chromatin via their interactions with transcriptional regulatory factors. The available data do not distinguish between the two models presented. It is possible that the factor binds the DNA first and then acts as a docking pad for the remodeling complex. Alternatively, it is possible that the interaction between the factor and the remodeling complex takes place in solution and the DNA-binding domain of the transcription factor directs the remodeling complex to chromatin in a later step.
Additional studies that do not involve nuclear hormone receptors also support the activator-mediated targeting model. An association of the NURD complex (containing the Mi-2 DNA-dependent ATPase) with the Ikaros DNA-binding proteins in lymphoid cells was demonstrated (47). In addition, the erythroid transcription factor EKLF has been shown to require E-RC1 (a SWI/SNF-related complex) to activate transcription in vitro (2). Finally, it has been shown that heat shock factor 1 can direct chromatin disruption in the transcribed region of the hsp70 gene (7).
Direct evidence of an association between activator and remodeling complexes has been limited until recently. Several laboratories have recently demonstrated that the ySWI/SNF complex can directly interact with acidic transcriptional activators. Mutation of the acidic activation domains of herpesvirus VP16 and yeast Gcn4 affects their interaction with SWI/SNF (73, 74). By contrast, glutamine-rich and proline-rich activation domains do not interact directly with ySWI/SNF (74, 123). The acidic activators were able to interact with SWI/SNF in the context of yeast whole-cell extract. Furthermore, using an immobilized DNA template assay, it was shown that Gal4-AH and Gal4-VP16 were able to recruit SWI/SNF out of yeast nuclear extracts in a manner that was independent of RNA polymerase II holoenzyme and TATA-binding protein (123). Acidic activators can also recruit SWI/SNF remodeling activity to increase restriction enzyme accessibility and mononucleosome disruption by DNase I, while a proline-rich activator cannot (123; A. H. Hassan and J. L. Workman, unpublished results).
Targeting of the SWI/SNF complex has also been linked to gene regulation. Activator-dependent transcriptional stimulation of nucleosomal arrays upon ySWI/SNF targeting to specific promoters was demonstrated in vitro (74). An in vivo function for activator interactions was shown as well (73). The current models for activator-mediated targeting are shown in Fig. 3. It is not known whether the activator associates with DNA and/or chromatin before or after it binds the SWI/SNF complex, but it is likely that both situations occur in vivo.
All of the above studies support a positive role for SWI/SNF in transcriptional regulation via its interaction with activator proteins. However, a recent study suggests a role for gene-specific repressors in the targeting of the SWI/SNF complex. Yeast SWI/SNF components coimmunoprecipitated with the corepressors Hir1p and Hir2p, which negatively regulate the transcription of the yeast histone HTA1-HTB1 gene locus (22). Another possible connection between a SWI/SNF complex and repression was suggested previously. The human proteins hBRM and BRG-1 interact with Rb, which in turn interacts with E2F1 (24, 104). The binding of hbrm to E2F1 via Rb represses the activity of E2F1 and induces complete cell cycle arrest.
CHROMATIN REMODELING AND HAT COMPLEXES
It has been suggested that chromatin-remodeling complexes might act together with HATs to antagonize chromatin-mediated transcriptional repression of some promoters. The subset of yeast genes which are regulated by the yeast SWI/SNF complex, including HO, SUC2, INO1, and Ty insertions, overlaps, to a certain degree, those controlled by Gcn5-containing yeast HATs (36). In addition, both types of complexes are required for the function of the yeast Gcn4 activator and the mammalian GR. Pollard and Peterson reported that several previously proposed SWI/SNF components were, in fact, subunits of the Gcn5-containing HATs. Moreover, the authors were unable to recover Ada/Swi double mutants except in combination with mutations in chromatin components (88). Similarly, a different group isolated a SWI/SNF mutant that displayed a synthetic lethal phenotype with HAT components (93). This phenotype suggests that the two types of complexes perform overlapping functions related to the transcription of essential yeast genes in a chromatin context. It is possible that the two types of chromatin-modifying complexes function in parallel pathways, so that when both are lost, the cell dies. Alternatively, this phenotype might indicate that the complexes interact functionally in the cell so that each functions best in the presence of the other.
Both HATs and HDACs have been found to be associated with remodeling complexes (93, 103, 110). Thus, it is likely that HATs, HDACs, and ATP-dependent remodeling complexes can work together on some promoters that are sensitive to nucleosomal structure. However, the question of how the chromatin-remodeling complexes and the HATs might act in concert to activate transcription still remains unanswered. It is possible that acetylation occurs first and that this modification, functioning either as a flag or by directly opening the chromatin structure, allows ATP complexes to remodel nucleosomes more easily. In support of this hypothesis, it has been shown that bromodomains, which are contained in some SWI subunits (and also in nuclear HATs), interact specifically with acetylated lysines in histone tail peptides (21). Conversely, it is possible that the remodeling complexes act first. Two recent papers support this second option. Analyzing in vivo cross-linking of proteins to the HO promoter in yeast, Cosma et al. have shown that the binding of the Swi5 transcription activator to its upstream activating sequence site is required for the subsequent interaction of SWI/SNF. In turn, SWI/SNF is required for the interaction of Gcn5-containing HAT complexes. Both complexes, but not the continued presence of Swi5p, are required for the maintenance of an active chromatin state, allowing the binding of the activator SBF and, hence, full transcriptional enhancement (16). In the second study of the HO promoter, Krebs et al. have shown that the interactions of Gcn5, SWI/SNF, and Swi5p result in the cell cycle-regulated acetylation of approximately 1 kb of upstream regulatory sequences. This includes factor-binding sites and the TATA box, but not the open reading frame. The acetylation event precedes transcription and possibly SBF binding. These authors have also shown that this acetylation can be reversed through the action of the Sin3p/Rpd3 deacetylase complex (53).
Our knowledge of the mechanisms by which ATP-dependent complexes and HATs might function together to potentiate transcription is still highly speculative and awaits the resolution of their individual modes of action. Moreover, the in vivo dependence of yeast genes on SWI/SNF or Gcn5-containing HAT complexes differs dramatically and for some of genes, one or the other complex may suffice. The order of recruitment seems clearest at the HO promoter, but it may differ significantly at other genes or under different inducing conditions. This issue is further complicated by the presence of the ISWI-containing remodeling complexes and additional HAT complexes.
CONCLUSIONS
Considerable progress has been made in the past few years in the identification and characterization of ATP-dependent chromatin-remodeling complexes. They are conserved in evolution, and there is more than one type of complex in each cell. Interestingly, although their mechanism of action is still not fully understood, it seems that the different types of complexes might also have different ways of disrupting the histone-DNA contacts. Very recently obtained data offer insight into their possible regulation. In addition, the data show how they might be directed to the subset of cellular genes that require their function for activation. All of these points warrant further study. Therefore, they are being actively pursued by several groups.
Some important questions that remain to be addressed include the extent of modification that remodeling complexes exert on any given gene. It will be interesting to determine whether only the nucleosomes that are positioned on the promoter are remodeled or whether these modifications extend beyond the regulatory region, maybe also facilitating elongation by the RNA polymerase. Another important aspect of this is the effect that ATP-dependent remodeling complexes might have on higher-order structure.
Finally, the possible concerted action of ATP-dependent remodeling complexes, HATs, and HDACs to regulate the transcription of certain genes is very intriguing. Although several recent studies have started to suggest how these complexes might work together, further studies are necessary to establish the possible redundancy or synergism in the function of these different chromatin-modifying machineries. Discovery of the ATP-dependent chromatin-remodeling complexes and other chromatin-modifying activities linked to transcription (e.g., HATs, HDACs, etc.) has revolutionized our view of eukaryotic transcription. We are finally beginning to envision how gene expression might be regulated in the context of chromatin structure.
ACKNOWLEDGMENTS
We thank members of the Workman laboratory for reading and commenting on the manuscript. We are particularly grateful to the referees of the original version for their detailed and thoughtful comments and suggestions that have improved the quality of this review.
Work done in the authors' laboratory was supported by a grant from NIGMS. J.L.W. is an HHMI Associate Investigator.
REFERENCES
- 1.Aasland R, Stewart A F, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional corepressor NCoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- 2.Armstrong J A, Bieker J J, Emerson B M. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong J A, Emerson B M. Transcription of chromatin: these are complex times. Curr Opin Genet Dev. 1998;8:165–172. doi: 10.1016/s0959-437x(98)80137-8. [DOI] [PubMed] [Google Scholar]
- 4.Bazett-Jones D P, Côté J, Landel C C, Peterson C L, Workman J L. The SWI/SNF complex creates loop domains in DNA and polynucleosome arrays and can disrupt DNA-histone contacts within these domains. Mol Cell Biol. 1999;19:1470–1478. doi: 10.1128/mcb.19.2.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger S L. Gene activation by histone and factor acetyltransferases. Curr Opin Cell Biol. 1999;11:336–341. doi: 10.1016/S0955-0674(99)80046-5. [DOI] [PubMed] [Google Scholar]
- 6.Brown S A, Imbalzano A N, Kingston R E. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 7.Brown S A, Kingston R E. Disruption of downstream chromatin directed by a transcriptional activator. Genes Dev. 1997;11:3116–3121. doi: 10.1101/gad.11.23.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns L G, Peterson C L. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cairns B R, Erdjument-Bromage H, Tempst P, Winston F, Kornberg R D. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 10.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 11.Cao Y, Bairns B R, Kornberg R D, Laurent B C. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol Cell Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Li B, Workman J L. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO J. 1994;13:380–390. doi: 10.1002/j.1460-2075.1994.tb06272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho H, Orphanides G, Sun X, Yang X-J, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark-Adams C D, Norris D, Osley M A, Fassler J S, Winston F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 1988;2:150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- 15.Corona D F V, Langst G, Clapier C R, Bonte E J, Ferrari S, Tamkun J W, Becker P B. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- 16.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 17.Côté J, Peterson C L, Workman J L. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc Natl Acad Sci USA. 1998;95:4947–4952. doi: 10.1073/pnas.95.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 19.Crosby M A, Miller C, Alon T, Watson K L, Verrijzer C P, Goldman-Levi R, Zak N B. The trithorax group gene moira encodes a Brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol Cell Biol. 1999;19:1159–1170. doi: 10.1128/mcb.19.2.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davie J R. Covalent modifications of histones: expression from chromatin templates. Curr Opin Genet Dev. 1998;8:173–178. doi: 10.1016/s0959-437x(98)80138-x. [DOI] [PubMed] [Google Scholar]
- 21.Dhalluin C, Carlson J E, Zeng L, He C, Aggarwal A K, Zhou M-M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 22.Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley M A. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell. 1999;4:75–83. doi: 10.1016/s1097-2765(00)80189-6. [DOI] [PubMed] [Google Scholar]
- 23.Dingwall A K, Beek S J, McCallum C M, Tamkun J W, Kalpana G V, Goff S P, Scott M P. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunaief J L, Strober B E, Guha S, Khavari P A, Ålin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 25.Eisen J A, Sweder K S, Hanawalt P C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elfring L K, Deuring R, McCallum C M, Peterson C L, Tamkun J W. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 28.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Hörz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 29.Gdula D A, Sandaltzopoulos R, Tsukiyama T, Ossipow V, Wu C. Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes Dev. 1998;12:3206–3216. doi: 10.1101/gad.12.20.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georgel P T, Tsukiyama T, Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grant P A, Berger S L. Histone acetyltransferase complexes. Semin Cell Dev Biol. 1999;10:169–177. doi: 10.1006/scdb.1999.0298. [DOI] [PubMed] [Google Scholar]
- 32.Guyon J R, Narlikar G J, Sif S, Kingston R E. Stable remodeling of tailless nucleosomes by the human SWI-SNF complex. Mol Cell Biol. 1999;19:2088–2097. doi: 10.1128/mcb.19.3.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamiche A, Sandaltzopoulos R, Gdula D A, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 34.Han M, Grunstein M. Nucleosome loss activates downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 35.Hirschhorn J N, Bortvin A L, Ricupero-Hovasse S L, Winston F. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol Cell Biol. 1995;15:1999–2009. doi: 10.1128/mcb.15.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengarner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 37.Imbalzano A N. Energy-dependent chromatin remodelers: complex complexes and their components. Crit Rev Eukaryot Gene Exp. 1998;8:225–255. doi: 10.1615/critreveukargeneexpr.v8.i3-4.10. [DOI] [PubMed] [Google Scholar]
- 38.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 39.Imbalzano A N, Schnitzler G R, Kingston R E. Nucleosome disruption by human SWI/SNF is maintained in the absence of continued ATP hydrolysis. J Biol Chem. 1996;271:20726–20733. doi: 10.1074/jbc.271.34.20726. [DOI] [PubMed] [Google Scholar]
- 40.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 41.Ito T, Levenstein M E, Fyodorov D V, Kutach A K, Kobayashi R, Kadonaga J T. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeon S H, Kang M G, Kim Y H, Jin Y H, Lee C, Chung H-Y, Kwon H, Park S D, Seong R H. A new mouse gene, SRG3, related to SWI3 of Saccharomyces cerevisiae, is required for apoptosis induced by glucocorticoids in a thymoma cell line. J Exp Med. 1997;185:1827–1836. doi: 10.1084/jem.185.10.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 44.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 45.Kehle J, Beuchle D, Treuheit S, Christen B, Kennison J A, Bienz M, Muller J. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- 46.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 47.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 48.Kingston R E. A shared but complex bridge. Nature. 1999;399:199–200. doi: 10.1038/20302. [DOI] [PubMed] [Google Scholar]
- 49.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 50.Knezetic J A, Luse D S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 51.Kornberg R D, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 52.Kornberg R D, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 53.Krebs J E, Kuo M-H, Allis C D, Peterson C L. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kruger W, Peterson C L, Sil A, Coburn C, Arents G, Moudrianakis E N, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 55.Kuo M-H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 56.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 57.Längst G, Bonte E J, Corona D F V, Becker P B. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 58.Laurent B C, Treich I, Carlson M. The yeast SNF2/SWI2 protein has a DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 59.Lee K-M, Sif S, Kingston R E, Hayes J J. hSWI/SNF disrupts interactions between the H2A N-terminal tail and nucleosomal DNA. Biochemistry. 1999;38:8423–8429. doi: 10.1021/bi990090o. [DOI] [PubMed] [Google Scholar]
- 60.LeRoy G, Orphanides G, Lane W S, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 61.Logie C, Peterson C L. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 1997;16:6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Logie C, Tse C, Hansen J C, Peterson C L. The core histone N-terminal domains are required for multiple rounds of catalytic chromatin remodeling by the SWI/SNF and RSC complexes. Biochemistry. 1999;38:2514–2522. doi: 10.1021/bi982109d. [DOI] [PubMed] [Google Scholar]
- 63.Lorch Y, Cairns B R, Zhang M, Kornberg R D. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 64.Lorch Y, LaPointe J W, Kornberg R D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 65.Lorch Y, Zhang M, Kornberg R D. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- 66.Martinez-Balbas M A, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci USA. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McKenna N J, Nawaz Z, Tsai S Y, Tsai M J, O'Malley B W. Distinct steady-state nuclear receptor coregulator complexes exist in vivo. Proc Natl Acad Sci USA. 1998;95:11697–11702. doi: 10.1073/pnas.95.20.11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moreira J M, Holmberg S. Transcriptional repression of the yeast CHA1 gene requires the chromatin-remodeling complex RSC. EMBO J. 1999;18:2836–2844. doi: 10.1093/emboj/18.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morozov A, Yung E, Kalpana G V. Structure-function analysis of integrase interactor 1/hSNF5L1 reveals differential properties of two repeat motifs present in the highly conserved region. Proc Natl Acad Sci USA. 1998;95:1120–1125. doi: 10.1073/pnas.95.3.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muchardt C, Reyes J-C, Bourachot B, Legouy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SWI/SNF complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- 71.Muchardt C, Yaniv M. The mammalian SWI/SNF complex and the control of cell growth. Semin Cell Dev Biol. 1999;10:189–195. doi: 10.1006/scdb.1999.0300. [DOI] [PubMed] [Google Scholar]
- 72.Murphy D J, Hardy S, Engel D A. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol Cell Biol. 1999;19:2724–2733. doi: 10.1128/mcb.19.4.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Natarajan K, Jackson B M, Zhou H, Winston F, Hinnebusch A G. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/Mediator. Mol Cell. 1999;4:657–664. doi: 10.1016/s1097-2765(00)80217-8. [DOI] [PubMed] [Google Scholar]
- 74.Neely K E, Hassan A H, Wallberg A E, Steger D J, Cairns B R, Wright A P H, Workman J L. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 75.Neish A S, Anderson S F, Schlegel B P, Wei W, Parvin J D. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okabe I, Bailey L C, Attree O, Srinivasan S, Perkel J M, Laurent B C, Carlson M, Nelson D L, Nussbaum R L. Cloning of human and bovine homologues of SNF2/SWI2: a global activator of transcription in yeast S. cerevisiae. Nucleic Acids Res. 1992;20:4649–4655. doi: 10.1093/nar/20.17.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O'Neill D, Yang J, Erdjument-Bromage H, Bornschlegel K, Tempst P, Bank A. Tissue-specific and developmental stage-specific DNA binding by a mammalian SWI/SNF complex associated with human fetal-to-adult globin gene switching. Proc Natl Acad Sci USA. 1999;96:349–354. doi: 10.1073/pnas.96.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Östlund Farrants A-K, Blomquist P, Kwon H, Wrange Ö. Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol Cell Biol. 1997;17:895–905. doi: 10.1128/mcb.17.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Owen-Hughes T, Utley R T, Côté J, Peterson C L, Workman J L. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 80.Owen-Hughes T, Workman J L. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 1996;15:4702–4712. [PMC free article] [PubMed] [Google Scholar]
- 81.Papoulas O, Beek S J, Moseley S L, McCallum C M, Sarte M, Shearn A, Tamkun J W. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 82.Pazin M J, Bhargava P, Geiduschek E P, Kadonaga J T. Nucleosome mobility and the maintenance of nucleosome positioning. Science. 1997;276:809–812. doi: 10.1126/science.276.5313.809. [DOI] [PubMed] [Google Scholar]
- 83.Peterson C L. Multiple SWItches to turn on chromatin? Curr Opin Genet Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- 84.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 85.Peterson C L, Zhao Y, Chait B T. Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J Biol Chem. 1998;273:23641–23644. doi: 10.1074/jbc.273.37.23641. [DOI] [PubMed] [Google Scholar]
- 86.Phelan M L, Sif S, Narlikar G J, Kingston R E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 87.Pollard K J, Peterson C L. Chromatin remodeling: a marriage between two families? Bioessays. 1998;20:771–780. doi: 10.1002/(SICI)1521-1878(199809)20:9<771::AID-BIES10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 88.Pollard K J, Peterson C L. Role of ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 90.Randazzo F M, Khavari P, Crabtree G, Tamkun J W, Rossant J. brg1: a putative murine homolog of the Drosophila brahma gene, a homeotic gene regulator. Dev Biol. 1994;161:229–242. doi: 10.1006/dbio.1994.1023. [DOI] [PubMed] [Google Scholar]
- 91.Recht J, Osley M A. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast. EMBO J. 1999;18:229–240. doi: 10.1093/emboj/18.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reyes J C, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schnitzler G, Sif S, Kingston R E. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 95.Shanahan F, Seghezzi W, Parry D, Mahony D, Lees E. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SFN complex, and alters the ability of BRG1 to induce growth arrest. Mol Cell Biol. 1999;19:1460–1469. doi: 10.1128/mcb.19.2.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sif S, Stukenberg P T, Kirschner M W, Kingston R E. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Steger D J, Workman J L. Remodeling chromatin structures for transcription: what happens to the histones? BioEssays. 1996;18:875–884. doi: 10.1002/bies.950181106. [DOI] [PubMed] [Google Scholar]
- 98.Steger D J, Workman J L. Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. EMBO J. 1997;16:2463–2472. doi: 10.1093/emboj/16.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Strober B E, Dunaief J L, Guha S, Goff S P. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996;16:1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 101.Sukhodolets M V, Jin D J. RapA, a novel RNA polymerase-associated protein, is a bacterial homolog of SWI2/SNF2. J Biol Chem. 1998;273:7018–7023. doi: 10.1074/jbc.273.12.7018. [DOI] [PubMed] [Google Scholar]
- 102.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SWI2/SNF2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 103.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 104.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsukiyama T, Palmer J, Landel C C, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 107.Utley R T, Côté J, Owen-Hughes T, Workman J L. SWI/SNF stimulates the formation of disparate activator-nucleosome complexes but is partially redundant with cooperative binding. J Biol Chem. 1997;272:12642–12649. doi: 10.1074/jbc.272.19.12642. [DOI] [PubMed] [Google Scholar]
- 108.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 109.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 110.Wade P A, Pruss D, Wolffe A P. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 111.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 112.Wall G, Varga-Weisz P D, Sandaltzopoulos R, Becker P B. Chromatin remodeling by GAGA factor and heat shock factor at the hypersensitive Drosophila hsp26 promoter in vitro. EMBO J. 1995;14:1727–1736. doi: 10.1002/j.1460-2075.1995.tb07162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang W, Chi T, Xue Y, Zhou S, Kuo A, Crabtree G R. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci USA. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang W, Côté J, Xue Y, Zhou S, Khavari P, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 115.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 116.Whitehouse I, Flaus A, Cairns B R, White M F, Workman J L, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 117.Wilson C J, Chao D M, Imbalzano A N, Schnitzer G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 118.Woodage T, Basrai M A, Baxevanis A D, Hieter P, Collins F S. Characterization of the CHD family of proteins. Proc Natl Acad Sci USA. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 120.Workman J L, Roeder R G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 121.Xue Y, Wong J, Moreno G T, Young M K, Côté J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 122.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 123.Yudkovsky N, Logie C, Hahn S, Peterson C L. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 125.Zhang Y, Ng H-H, Edrjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao K, Wang W, Rando O J, Xue Y, Swiderek K, Kuo A, Crabtree G R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]