Abstract
Interest in the use of extracellular vesicles (EVs) as biomarkers of disease is rapidly growing. However, one main unsolved issue in the EV field is finding a technique able to eliminate non‐EV contaminants present in biofluid samples in a one‐step isolation protocol. Due to the expansion and value of size exclusion chromatography (SEC) as one of the best EV isolation methods, we have tested several agarose resins with different agarose percentages, bead sizes and crosslinking features to optimize EV isolation. For this optimization of SEC, we first employed conditioned media from a melanoma cell culture, a simpler sample in comparison to biological fluids, but which also contains abundant contaminants such as soluble protein and lipoproteins (LPPs). The distinct agaroses and the combinations of resins with different agarose percentages in the same column were tested. Soluble protein, EVs and LPPs levels from the different eluted fractions were quantitated by immunodetection or absorbance measurements. Samples were also analysed by NTA and TEM to verify the yield and the LPP contamination. Different percentages of agarose resins (2%, 4% and 6%) yielded samples with increasing LPP contamination respectively, which was not improved in the columns that combined them. Crosslinking of the agarose did not affect EV isolation yield nor the LPP contamination. In contrast, reducing the bead size greatly improved EV purity. We thus selected 4% Rapid Run Fine agarose beads as the resin that more efficiently isolated EVs with almost no contamination of other particles. Using blood plasma samples, this resin also demonstrated an improved capacity in the isolation of EVs from LPPs in comparison to the agaroses most commonly used in the field and differential ultracentrifugation.
Keywords: biofluids, disease biomarkers, extracellular vesicles, lipoproteins, plasma, purification, size exclusion chromatography
1. INTRODUCTION
Extracellular vesicles are secreted by virtually every cell type and have been recognized as a vehicle of intercellular communication since they transfer bioactive lipids, proteins and nucleic acids between cells. The term ‘extracellular vesicles’ comprehends apoptotic bodies, microvesicles and exosomes, which differ in their subcellular origin and biophysical characteristics (Yáñez‐Mó et al., 2015). They play important roles in physiological and pathological conditions, and there is an enormous interest in their potential use in the therapy of various pathologies, including cancer (Chen et al., 2022; Ma et al., 2021), and in the discovery of new biomarkers (Fais et al., 2016; Gutiérrez‐Fernández et al., 2021).
The main handicap in the field is the lack of a standardized or ideal isolation method for EVs that can combine a high yield alongside a high purity, with an easy translation to clinical practice. Several factors would determine which method is to be used for EV isolation: costs, the time required, equipment accessibility, sample origin and downstream analyses. The advantages and disadvantages of the most usually employed isolation methods have been previously reviewed (Clos‐Sansalvador et al., 2022; Monguió‐Tortajada et al., 2019).
Differential ultracentrifugation (dUC) was the first method proposed and thus is the most widely used isolation method (Gardiner et al., 2016; Théry et al., 2006) as it renders relatively high EV yields. However, samples obtained by this method are usually contaminated with protein aggregates, which co‐sediment with EVs. Moreover, the biological functionality of EVs isolated by this technique may be at least partially compromised due to the huge centrifugal forces to which samples are exposed (Guan et al., 2020). In addition to the requirement of special and expensive equipment that is not available for every researcher, it is highly time‐consuming, there is not a proper consensus protocol and the final yield is pretty much operator‐dependent.
Flotation in a density gradient ultracentrifugation is based on the separation of particles depending on their density. It is commonly employed, particularly in combination with other techniques, for EV isolation from biofluids (Karimi et al., 2018; Onódi et al., 2018; Van Deun et al., 2014), since it can resolve EVs from the majority but not all the lipoprotein nanoparticles (Karimi et al., 2018). Again, its principal disadvantages are that it is time‐consuming, expensive and has a low EV recovery rate. It also requires access to an ultra‐centrifuge, which again might not be available for every researcher. Finally, different compounds are commonly used to stablish the density gradients, such as sucrose and Optiprep, both displaying distinct results and having different impacts on the biological functionality of EVs (Kuipers et al., 2022).
Precipitation‐based methods are an option when looking for an easy, cheap and high‐yield method (Konoshenko et al., 2021; Ludwig et al., 2018). However, generated EV samples have low purity, due to the co‐precipitation of low‐solubility particles in the presence of a hydrophilic polymer. Thus, besides EVs, the resulting pellet will include proteins and other non‐soluble molecules. Therefore, for most applications, like protein composition or functional analysis, additional purification steps by other techniques may be required (Ludwig et al., 2018; Paolini et al., 2016).
Methods based on immunoaffinity take advantage of the expression of certain membrane markers of EVs. They are easy to use and specific, being a suitable option for EV isolation from biological samples due to their complexity and the reduced sample volume available. However, to date, no universal EV marker has been reported, being tetraspanins the most abundant membrane molecules and the most widely employed within the field (Campos‐Silva et al., 2021; Campos‐Silva et al., 2019). Tetraspanins conform a superfamily comprising 33 members in mammals which display differential expression levels in several cell types (Yáñez‐Mó et al., 2009; Zuidscherwoude et al., 2017). The most ubiquitous tetraspanins, and thus the most widely employed as EV markers, are CD9, CD63 and CD81. In any case, immunoaffinity methods would lead to the selection of a given subpopulation of EVs, depending on the antibody employed, and also, they are usually expensive (Clos‐Sansalvador et al., 2022; Kowal et al., 2016).
Different chromatographic methods have been also employed for extracellular vesicles isolation, from the most classical size exclusion chromatography (SEC) to more complex technologies like hydrophobic interaction chromatography using capillary‐channelled polymer fibre phase (Huang et al., 2021). The use of size exclusion chromatography for EV isolation has significantly grown during the last few years (Monguió‐Tortajada et al., 2019). This method allows proper elimination of soluble proteins, renders higher yields than density gradient ultracentrifugation and is not marker dependent, collecting all EVs without specific subpopulation enrichment. Importantly, SEC does not require expensive instrumentation and can be easily adapted to in‐field sample processing (Borgheti‐Cardoso et al., 2020; de Menezes‐Neto et al., 2015). SEC is based on the differential elution profiles of particles running through a porous matrix depending on their size. Due to their larger size, EVs would traverse the column avoiding the matrix pores, and elute soon after the void volume. Smaller particles, like proteins and most of the lipoprotein subpopulations will be able to enter the resin pores, being retained for a longer time in the column and thus eluting later than EVs. Therefore, a final advantage of SEC is that it allows the interrogation of different fractions of the sample (EVs vs. soluble protein) for biological activities (Calle et al., 2021; Carreras‐Planella et al., 2019).
The characteristics (pore size, bead size and crosslinking level) of the agarose resin employed would be determinant to obtain an optimal result. Pore size is defined by the agarose density, decreasing when the latter increases. This characteristic would determine the exclusion limit and the fractionation range of the agarose. Agarose bead size would define the column packing, decreasing the elution speed as size decreases. This can increase the peak resolution of sample components. The level of agarose crosslinking determines its chemical and physical stability, which is especially important for long gravity columns or ÄKTA™ systems.
These aspects are crucial when the goal is to isolate EVs from complex biofluid samples like serum or plasma. SEC can eliminate soluble proteins and most of the lipoprotein subpopulations present in that sample. Nevertheless, lipoproteins like chylomicrons present a broad range of sizes that overlap with EV size (Karimi et al., 2018) and LDL lipoproteins, being smaller than EVs, greatly overpass EVs in numbers so they are difficult to eliminate from the EV samples. With this goal in mind, we have tested different agarose resins to improve EV isolation from plasma samples. We first tested agaroses that differ in their pore size, crosslinking level and bead size to improve EV separation from lipoproteins of a melanoma cell‐conditioned culture medium. Finally, we have tested the selected agarose with a more complex sample like plasma and compared the performance with the classical differential ultracentrifugation method.
2. MATERIALS AND METHODS
2.1. Cell culture and plasma samples
Human melanoma cell line SK‐MEL‐147 was cultured in DMEM supplemented with 10% heat‐inactivated FBS (Sigma), penicillin (100 U/mL, Panreac) and streptomycin (100 μg/mL; Panreac), at 37°C in a 5% CO2 atmosphere. For EV production, 1.6·106 cells were grown in p150 plates (Corning) for 6 days, in DMEM (Gibco) supplemented with 5% EV‐depleted FBS, until they reached confluence (≈ 25·106 cells in a p150 plate). Cell viability was always over 95%. EV depletion was performed by ultracentrifugation at 120,000 g for 16 h.
Blood from healthy donors was collected in an EDTA tube, centrifuged at 2500 g for 15 min for the obtention of plasma and again centrifuged in the same conditions to completely remove contaminants. Plasma samples were aliquoted (500 μL) and frozen at −80°C. Before being used for SEC or UC isolation, samples were thawed at 4°C and centrifuged again at 2500 g for 15 min to eliminate possible aggregates formed during the defrosting process. SPREC code for the samples used would correspond to PL2‐EDG‐A‐A‐A‐G‐A (López‐Guerrero et al., 2023).
2.2. EV isolation by size exclusion chromatography (SEC)
Conditioned media from 1.5 p150 plates (≈23 mL) was centrifuged for 5 min at 500 g and 30 min at 3200 g to eliminate remaining cells, cellular debris and apoptotic bodies. Conditioned media was then concentrated using Amicon Ultra‐15 filters (100K, Millipore) to a final volume of ≈500 μL.
Empty columns with both under and upper cellulose frits were packed with 10 mL of the desired resin and equilibrated with two column volumes of filtered PBS before SEC. The concentrated conditioned media or plasma samples (500 μL) were loaded and 25 fractions of 500 μL were collected by gravity elution.
Resins tested (Table 1) are composed in all cases by agarose (chains of galactose dimers: β‐D‐galactopyranose linked with 3,6‐anhydro‐α‐L‐galactopyranose via 1–4 bonds). Resins were from Agarose Bead Technologies (ABT): 2% BCL Agarose Bead Standard (2BCL), 4% BCL Agarose Bead Standard (4BCL), 6% BCL Agarose Bead Standard (6BCL), 4% B Agarose Bead Standard (4B), 4% Rapid Run Agarose Bead Standard (4RR), 4% Rapid Run Agarose Bead Fine (4RRF), 4% BCL Agarose Bead Macro (4 M); or Cytiva: Sepharose® CL‐2B (CL‐2B) and Sepharose® 4 Fast Flow (4FF).
TABLE 1.
Resins information.
| Resin name | N° CAT | Exclusion limit (Da) | Fractionation range (Da) | Bead size (μm) | Crosslinking level |
|---|---|---|---|---|---|
| 2% BCL Agarose Bead Standard—ABT | A‐1021S‐X | >4 × 10⁷ | 7 × 10⁴–4 × 10⁷ | 50–150 (D50 ∼90) | Yes |
| 4% BCL Agarose Bead Standard—ABT | A‐1041S‐X | >2 × 10⁷ | 7 × 10⁴–2 × 10⁷ | 50–150 (D50 ∼90) | Yes |
| 6% BCL Agarose Bead Standard—ABT | A‐1061S‐X | >4 × 10⁶ | 1 × 10⁴–4 × 10⁶ | 50–150 (D50 ∼90) | Yes |
| 4% B Agarose Bead Standard—ABT | A‐1040S‐X | >2 × 10⁷ | 7 × 10⁴–2 × 10⁷ | 50–150 (D50 ∼90) | No |
| 4% Rapid Run Agarose Bead Standard—ABT | 4RRS‐X | ∼3 × 10⁷ | 6 × 10⁴–3 × 10⁷ | 50–150 (D50 ∼90) | Highly crosslinked |
| 4% Rapid Run Agarose Bead Fine—ABT | 4RRF‐X | ∼3 × 10⁷ | 6 × 10⁴–3 × 10⁷ | ∼50 | Highly crosslinked |
| 4% BCL agarose Bead Macro—ABT | A‐1041 M‐X | >2 × 10⁷ | 7 × 10⁴–2 × 10⁷ | 150–350 | Yes |
| Sepharose CL‐2B—Cytiva | 17014001 | >4 × 10⁷ | 7 × 10⁴−4 × 10⁷ | 60–200 | Yes |
| Sepharose 4 Fast Flow—Cytiva | 17014901 | ∼3 × 10⁷ | 6 × 10⁴−3 × 10⁷ | D50 ∼90 | Yes |
2.3. EV isolation by differential ultracentrifugation (dUC)
Plasma samples (0.5 mL) were diluted to a final volume of 3 mL in filtered PBS and ultracentrifuged at 100.000 g for 2 h in a Beckman (TL‐100) ultracentrifuge using the angular rotor (TLA 100.3). Supernatant was discarded and pellet was resuspended in filtrated PBS to the same final sample volume than that obtained from samples isolated by SEC (1 mL).
We have submitted all relevant data of our experiments to the EV‐TRACK knowledgebase (EV‐TRACK ID: EV230602) (Van Deun et al., 2017).
2.4. Protein analyses
Protein concentration in samples was measured with Pierce™ BCA protein assay kit (Thermo Scientific, 23225) or by measuring Absorbance at 280 nm in a Nanodrop system.
For dot blot analysis, 1 μL of each SEC fraction or from the sample obtained by ultracentrifugation was directly spotted on a nitrocellulose membrane (GE Healthcare). Membranes were blocked with 5% skimmed‐milk in TBS/0.1% Tween‐20 for 30 min.
For western‐blot analyses, when indicated the EV enriched fractions were pooled and concentrated by centrifugation at 100,000 g for 2 h, in a Beckman Optima™ TLX ultracentrifuge with the angular rotor TLA 100.3. Pellets were lysed with 100 μL of 1% Triton in TBS, in presence of protease inhibitors. For total SK‐MEL‐147 cell lysates, confluent cultures were washed with PBS and lysed in 1% Triton (ACROS Organics) in TBS, in the presence of protease inhibitors (Roche). Loading was normalized to protein content or sample volume as indicated in the legend of the corresponding figures. Proteins were electrotransferred to PVDF membranes (BioRad) with a Transfer‐Blot Turbo system (BioRad). Membranes were blocked with 5% skimmed‐milk in TBS and 0.1% Tween‐20 for 30 min.
For EV detection by dot‐blot or western‐blot, we employed antibodies against tetraspanins (hybridoma supernatant, undiluted) CD9 (VJ/1.20), CD63 (Tea3.10) (Yáñez‐Mó et al., 1998), CD81 (5A6; provided by Dr. S Levy, Stanford, USA), or non‐tetraspanin EV markers TSG101 (Genetex, GTX118736) and Syntenin‐1 (Synaptic systems, 133003) and anti‐ARF‐6 (Sigma, A5230) antibodies, diluted 1:1000. To detect lipoproteins, anti‐ApolipoproteinB (Calbiochen, 178467) and anti‐ApolipoproteinE (Calbiochem, 178479) were used, diluted 1:500. As non‐EV markers we employed anti‐VDAC1 (Abcam, ab154856) and anti‐calnexin (Enzo, ADI‐SPA‐865‐F), diluted 1:1000. Anti‐mouse (31430, Thermo Fisher Scientific), anti‐rabbit (31460, Thermo Fisher Scientific) and anti‐goat (SAB3700316‐500 μg, Sigma) antibodies conjugated with HRP were used as secondary antibodies, diluted 1:10,000.
Membranes were revealed with Super Signal West Femto HRP substrate (Thermo Scientific). Images were acquired with a LAS 4000 mini system (General Electrics) and processed with Fiji ImageJ.
2.5. Nanoparticle tracking analysis (NTA)
The size distribution of EVs and the number of particles after SEC were analysed by NTA with NanoSight NS300 (Malvern Instruments Ltd.), equipped with a 532 nm laser. Pools of SEC EV‐enriched fractions were diluted 100 times for the analyses. Results were analysed with NTA 3.0 software. Five videos of 60 s of duration were analysed for each sample, using camera level at 11 and analysis threshold at 5.
2.6. Transmission electron microscopy (TEM)
SEC isolated EVs were diluted 1/2 or 1/3 in filtrated PBS, adsorbed on carbon‐coated nickel grids and contrasted with 2% uranyl acetate. Samples were visualized in a transmission electron microscopy JEM1400 Flash (Jeol). The morphology, size and relative number of isolated EVs were analysed with TEM Exosome Analyzer software (Kotrbová et al., 2019).
3. RESULTS
3.1. Comparison between standard agarose beads with different pore size
Starting from the seminal description of SEC as an efficient method to isolate EVs from biofluids (Böing et al., 2014), the agarose resins that are more commonly employed are 2% and 4% agaroses. The percentage of agarose in the beads determines the exclusion size of the vesicles eluted in the main fractions, being centred around 35 nm diameter for 4% and around 70 nm of diameter for 2%. These differences would, of course, impact the yield of EVs and also the levels of contamination of the samples with different lipoproteins, which cover the range from 5 to 1200 nm of diameter. For this reason, we decided to start testing standard agarose resins of different densities (2% from two different vendors, 4% and 6%) in a 10 mL column format. As samples, we used a starting volume of ≈23 mL of a melanoma cell line conditioned media, concentrated to ≈0.5 mL (final volume loaded on the column). Twenty‐five fractions of 0.5 mL were collected. The elution profile of EVs, LPPs and total soluble protein was determined and quantitated by dot blot directly spotting 1 μL of each fraction on the nitrocellulose membrane (Figure 1a and S1a) or by measuring absorbance at 280 nm of the eluate fractions (Figure 1a). The three agaroses from Agarose Beads Technologies (ABT) presented a similar elution profile of EVs, as detected by dot‐blot with antibodies against different tetraspanins (CD9, CD81 and CD63), whereas the elution profile of LPPs and soluble protein varied as the pore of the agarose decreased. The co‐isolation of LPPs and part of the soluble protein in EV‐enriched fractions was higher using agaroses with smaller pores, with significant differences in LPPs contamination between 6% agarose and the other resins tested (Figure 1b). Although EV yield also increased slightly with the reduction of the pore size, with significant differences between 2% and 6% resins (Figure 1c), EV purity index is clearly compromised in 6% agarose columns (Figure 1d). A different 2% agarose from a different vendor (Sepharose® CL‐2B, Cytiva) presented some differences with ABT 2%BCL in the EV elution profile (Figure 1a); showing a wider peak and a considerably smaller yield of EV isolation (Figure 1c).
FIGURE 1.
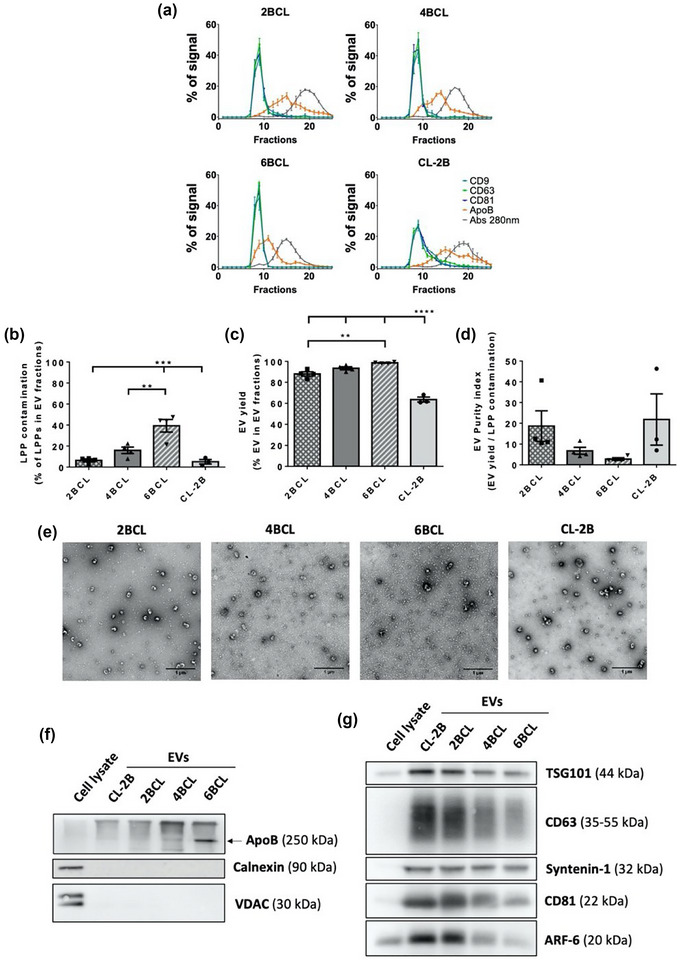
Comparison between BCL agaroses (2BCL, 4BCL, 6BCL and CL‐2B). (a) Elution profile of EVs, LLPs and soluble protein. Densitometry of dot blot analyses for EVs markers (CD9, CD63 and CD81) and LPPs markers (ApoB), and the absorbance at 280 nm for total protein analysis is plotted for each fraction as the % of a given marker signal in each fraction (being the 100% of the signal the sum for each marker of the signal in all fractions eluted from the column). The mean ± SEM of at least three independent experiments is shown. (b) LPPs contamination in EVs fractions, represented as the % of LPPs that co‐isolate in the three fractions with a higher level of EVs (usually F8‐10). The means ± SEM of at least three independent experiments are shown. Significant differences in the One‐way Anova statistical test are indicated with ** (p ≤ 0.01), *** (p ≤ 0.001). (c) EV yield, represented as the % of EVs in the three fractions with a higher level of EVs. The means ± SEM of at least three independent experiments are shown. Significant differences in the One‐way Anova statistical test are indicated with ** (p ≤ 0.01), **** (p ≤ 0.0001). (d) EV purity index, represented as the ratio between EV yield and LPP contamination in EV fractions. The means ± SEM of at least three independent experiments are shown. (e) Representative TEM images of negatively stained samples from the pools of EV‐enriched fractions. Bars = 1 μm. (f) Western blot analyses of non‐EV markers (ApoB, Calnexin or VDAC) or (g) EVs markers (TSG101, CD63, Syntenin‐1, CD81 and ARF6) in pools of EV‐enriched fractions. Protein content was measured by BCA and a total of 10 μg was loaded in each lane. The same amount of total cell lysate was included.
EVs contained in the three most enriched fractions from each column were pooled and characterized according to MISEV recommendations (Théry et al., 2018). Transmission electron microscopy (TEM) analyses from negatively stained samples revealed the presence of EVs with the characteristic cup‐shape (Figure 1e). We could also confirm the increasing amount of LPPs when agarose pore decreases. LPPs were detected as electron‐clear regular‐size small particles with sizes compatible with LDL. TEM images analyses confirmed a slight but significant increase in the mean size of EVs obtained with CL‐2B in comparison with 4%BCL and 6%BCL ABT agaroses (Figure S1b), although this difference could not be detected in NTA analyses (Figure S1c). The number of particles detected by NTA (Figure S1d) and the size profile (Figure S1e) was similar in all cases, with a characteristic peak around 150 nm (Figure S1e). It should be pointed out, however, that most LPPs in these samples (≈30 nm in diameter as measured by TEM) lie below the size resolution limit of NTA (60–70 nm) and would not be detected/counted by this technique.
Negative (Figure 1f) and positive (Figure 1g) markers of EVs were also analysed by western blot, proving EV isolation and the increasing co‐isolation of lipoproteins employing agaroses with smaller pores. Some EV markers were almost undetectable in total cell lysates, supporting the high enrichment of EVs in EV samples.
3.2. Combination of agaroses with different pore size for EV isolation by SEC
A previous work recently demonstrated that the combination of resins with different pore sizes allows the separation of particles with different diameters (Kaddour et al., 2020) when the gradient column, with particle purification liquid chromatography (PPLC), was integrated into a turbidimetry‐enabled system.
To test whether this combinatorial approach could be a good solution for EV SEC, we combined our 2%, 4% and 6% BCL agaroses in both gradient configurations (Figure 2a). Elution profiles using both columns were similar, with no significant differences in LLPs contamination in EV fractions (Figure 2b,c), which was corroborated by TEM images (Figure S2a). The yield of EVs was similar for both combinations (Figure 2d) and thus there were no significant differences in EV Purity index (Figure 2e). No significant differences were observed in the mean size of particles by TEM (Figure S2b) or NTA analysis (Figure S2e), in the size distribution (Figure S2c) or the concentration of particles by NTA (Figure S2d). Western blot analysis demonstrated the isolation of EVs, with a similar level of LPPs contamination between them (Figure S2f,g). Thus, as evidenced from their elution profiles, these gradient columns did not improve the isolation of EVs over that obtained with 2% or 4% BCL simple agarose columns (Figures 1a and 2b).
FIGURE 2.
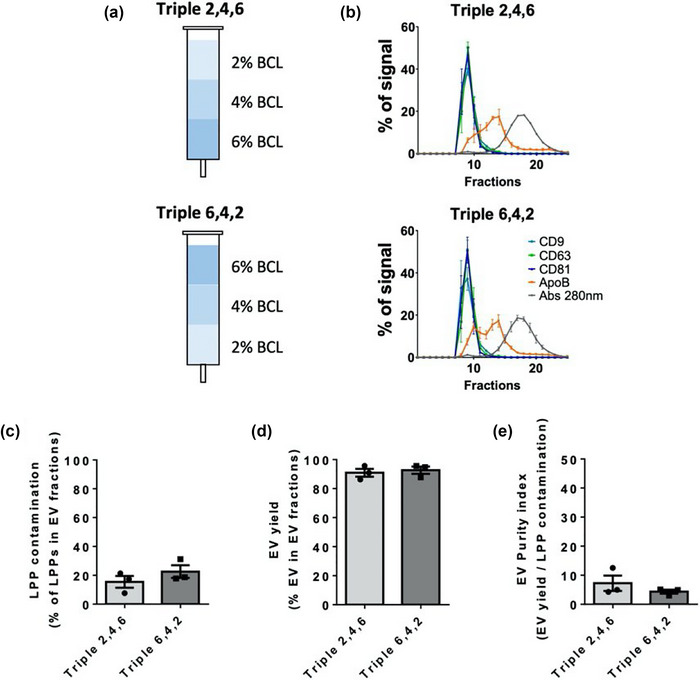
SEC with columns that combine three different BCL agaroses. (a) Scheme of the combination of BCL agaroses in triple columns. (b) Elution profile of EVs, LLPs and soluble protein. Densitometry of dot blot analyses for EVs markers (CD9, CD63 and CD81) and LPPs markers (ApoB), and representation of the absorbance at 280 nm in fractions for total protein analysis as the % of a given marker signal in each fraction (being the 100% of the signal the sum for each marker of the signal in all fractions eluted from the column). The mean ± SEM of at least three independent experiments is shown. (c) LPPs contamination in EVs fractions, represented as the % of LPPs that co‐isolate in the three fractions with a higher level of EVs (usually F8‐10). The means ± SEM of at least three independent experiments are shown. (d) EV yield, represented as the % of EVs in the three fractions with a higher level of EVs. The means ± SEM of at least three independent experiments are shown. (e) EV purity index, represented as the ratio between EV yield and LPP contamination in EV fractions. The means ± SEM of at least three independent experiments are shown.
3.3. Modification of bead size and crosslinking level of agaroses
Next, we decided to look for an agarose that would improve the isolation of EVs with a lower level of LPPs contamination in comparison with 2%BCL or 4%BCL results. Since the 4% agarose has a smaller size exclusion limit that would incorporate in the sample the whole range of small EVs, we tested columns with 4% of agarose, but whose agarose beads differ in their crosslinking level or bead size.
First, we evaluated the elution profiles of agaroses with increasing levels of crosslinking (4B, 4BCL and 4RR) (Figure 3a, top panels). Cross‐linking is a chemical process to enhance the chemical and mechanical stability of the agarose beads. We observed no significant differences amongst their profiles in LPPs contamination within EV fractions (Figure 3a,b), in EV yield (Figure 3c) or in the EV purity index (Figure 3d). Another crosslinked agarose from a different vendor (GE Healthcare Sepharose® 4 Fast Flow; 4FF) also rendered similar results to 4RR agarose, in terms of LPPs co‐isolation, EV yield and EV purity index. These data suggest that chemical crosslinking does not change the size exclusion limit or performance of the resins.
FIGURE 3.
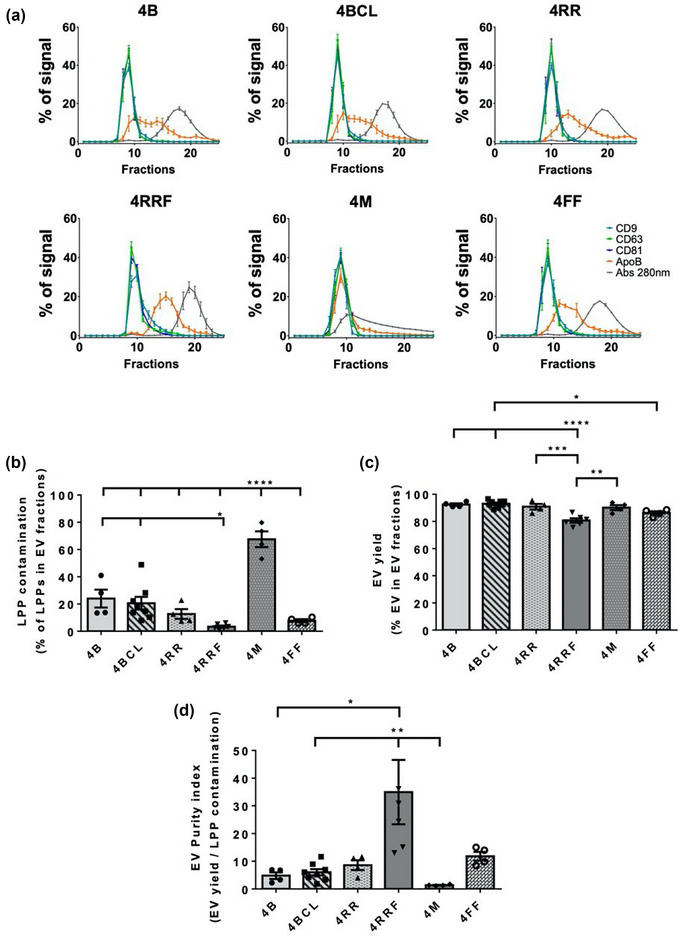
Analysis of the repercussion of agarose crosslinking or bead size on SEC performance. (a) Elution profile of EVs, LLPs and soluble protein. Densitometry of dot blot analyses for EVs markers (CD9, CD63 and CD81) and LPPs markers (ApoB), and representation of the absorbance at 280 nm in fractions for total protein analysis as the % of a given marker signal in each fraction (being the 100% of the signal the sum for each marker of the signal in all fractions eluted from the column). The mean ± SEM of at least three independent experiments are shown. (b) LPPs contamination in EVs fractions, represented as the % of LPPs that co‐isolate in the three fractions with a higher level of EVs (usually F8‐10). The means ± SEM of at least three independent experiments are shown. Significant differences in the One‐way Anova statistical test are indicated with *** (p ≤ 0.001) or **** (p ≤ 0.0001). (c) EV yield, represented as the % of EVs in the three fractions with a higher level of EVs. The means ± SEM of at least three independent experiments are shown. Significant differences in the One‐way Anova statistical test are indicated with * (p ≤ 0.05), ** (p ≤ 0.01), ***(p ≤ 0.001), **** (p ≤ 0.0001). (d) EV purity index, represented as the ratio between EV yield and LPP contamination in EV fractions. The means ± SEM of at least three independent experiments are shown. Significant differences in the One‐way Anova statistical test are indicated with * (p ≤ 0.05), ** (p ≤ 0.01).
Second, agaroses with different bead sizes were tested: 4 M, a 4% macro beads agarose, and 4RRF, a fine beads agarose. Since small bead sizes are more prone to deformation, this agarose had also a high level of crosslinking. As shown in Figure 3a lower panels and Figure 3b, the use of 4 M agarose results in highly overlapping elution profiles of EVs, LPPs and soluble protein, with major contamination of LPPs in EV fractions. On the contrary, 4RRF presented better resolution between EVs, LPPs and soluble protein peaks than any other agarose previously tested (Figure 3a). These columns render EVs with very low levels of LPPs contamination (around 3.5% of LPPs in EV fractions), significantly lower than those from 4B and 4BCL agaroses (about 24% and 20%, respectively) (Figure 3b), which was corroborated in TEM analyses (Figure 4a). Although a decrease in LPP contamination led to slightly reduced EV yields (Figure 3c), EV purity index obtained for 4RRF column is clearly higher than for the other agaroses tested (Figure 3d).
FIGURE 4.
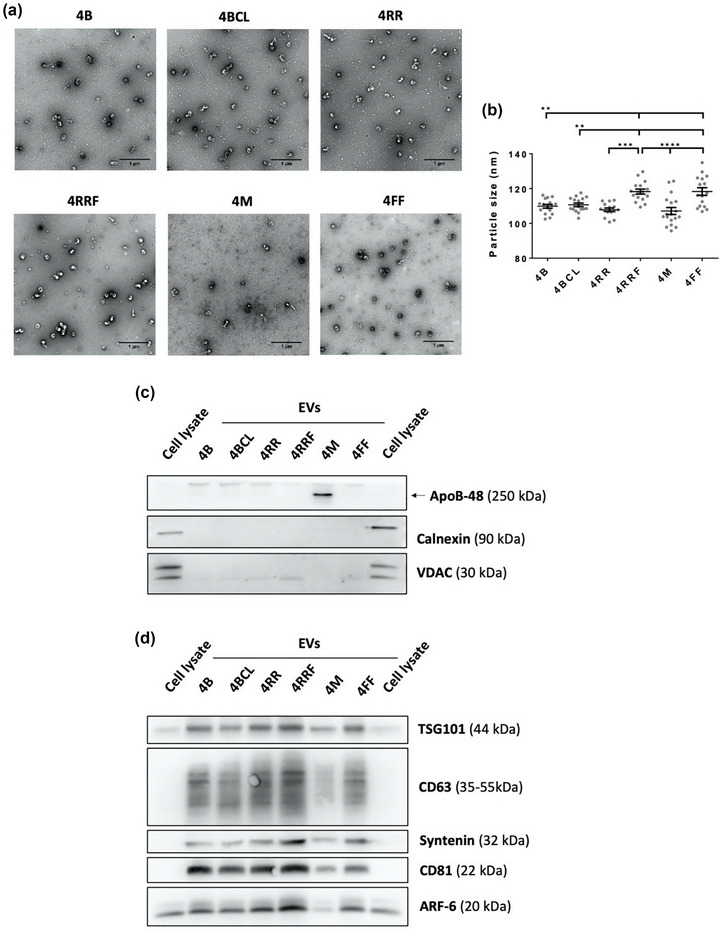
TEM and Western blot analysis of EVs isolated by SEC with different resins. (a) Representative TEM images of negatively stained samples from EV‐enriched fraction pools isolated with the different resins. Bars = 1 μm. (b) Mean EV diameter of EVs derived from TEM Exosome Analyser on TEM images. Sixteen images from two independent experiments were analyzed and the data was shown as means ± SEM. Significant differences in a One‐way Anova statistical test are indicated with ** (p ≤ 0.01), *** (p ≤ 0.001) or **** (p ≤ 0.0001). Western blot analyses of (c) non‐EV markers (ApoB, Calnexin or VDAC) or (d) EVs markers (TSG101, CD63, Syntenin‐1, CD81 and ARF6) in EV‐fraction pools isolated with the different resins. Protein content was measured by BCA and a total of 10 μg was loaded in each lane. The same amount of total cell lysate was included.
Detailed analyses revealed that EVs obtained with 4RRF and 4FF columns in comparison to those obtained with 4B, 4BCL, 4RR and 4 M columns had a slightly higher mean size when TEM images were analysed (Figure 4b), but those differences were small enough not to be detected by NTA (Figures S3a–c).
Western blot profiling confirmed proper EV isolation with all agaroses, with the exception of 4 M agarose, which presented a very low proportion of EVs versus protein/lipoprotein (Figure 4c,d). A higher proportion of EVs versus protein/lipoprotein was confirmed in samples obtained with the 4RRF column, confirming that this resin renders a better elimination of these contaminants and an enrichment in EVs in the sample.
3.4. EV isolation from plasma samples with 4RRF resin columns
Once we determined that the 4RRF column allows for better isolation of EVs with reduced LPPs and protein contamination from cell culture‐ conditioned media, we moved on to test this agarose with a more complex sample, such as human plasma. For these analyses we compared 4RRF with both 2%BCL and 4%BCL agaroses.
The elution profile of EVs and LPPs was determined by western blot (Figure 5a). Quantification of these results, together with absorbance at 280 nm for total protein measurement, rendered the elution profiles represented in Figure 5b. 4RRF column was again the best performer in terms of EV isolation, with lower contamination of LPPs in the pools of EV‐enriched fractions (Figure 5c,d). The presence of ApoE‐positive LPPs (Figure 5c) using this column was significantly lower (∼12%) than with the other columns employed (∼20%). Likewise, the percentage of ApoB‐LPPs obtained with the 4RRF column (∼18%) was clearly lower compared to that obtained with the 4BCL (∼33%) or 2BCL (∼24%) columns. However, a slight reduction in the amount of EVs isolated within EV‐enriched fractions was also observed in parallel with the decrease of LPP contamination (Figure 5e).
FIGURE 5.
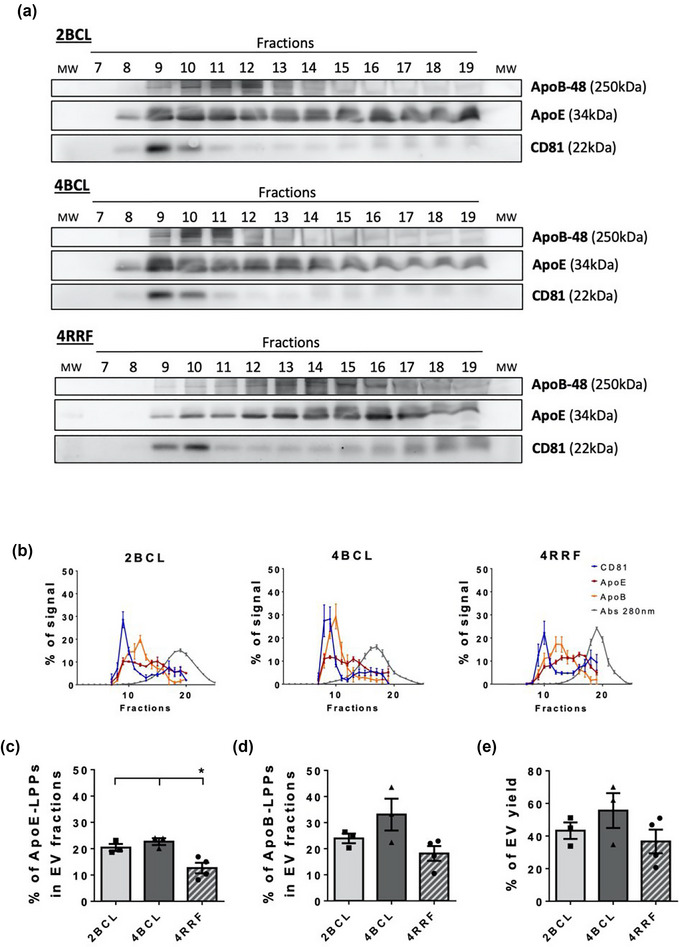
SEC of plasma samples. (a) Western blot analysis of EV marker (CD81) and LPPs (ApoB and ApoE). 20 μL of each fraction was loaded per lane. (b) Elution profile of EVs, LLPs and soluble protein. Densitometry of Western blot analysis for EVs markers (CD81) and LPPs markers (ApoB and ApoE), and representation of the absorbance at 280 nm in fractions for total protein analysis as the % of a given marker signal in each fraction (being the 100% of the signal the sum for each marker of the signal in all fractions analysed from the column). The mean ± SEM of at least three independent experiments is shown. (c) ApoE and (d) ApoB positive LPPs contamination in EV fractions, represented as the % of ApoE‐LPPs and ApoB‐LPPs that respectively co‐isolate in the two fractions with a higher level of EVs (usually F9‐10). The means ± SEM of at least three independent experiments are shown. Significant differences in the One‐way Anova statistical test are indicated with * (p ≤ 0.05). (e) Amount of EV yield, represented as the % of EVs recovered in the two fractions with a higher level of EVs (being the 100% of the signal the sum of the signal in all fractions analysed from the column).
No significant differences were observed in the size profile (Figure S4a) or concentration of particles (Figure S4b) obtained by NTA analysis of EV‐enriched samples. In TEM analyses, although LPPs were visible in EV‐enriched fractions, co‐isolation was clearly higher using 4BCL columns than with 4RRF columns (Figure S4c).
Serial ultracentrifugation has been the gold standard for EV isolation for many years. However, EV integrity and function is somewhat perturbed by the high centrifugation forces. In addition, higher level of protein aggregates contaminants are expected when compared to SEC. To directly confirm these differences, we compared the isolation of EVs using SEC with the resins that provided fair isolation results (2BCL and 4RRF) with dUC of human plasma samples. The same sample starting volume (500 μL) was employed for EV isolation and after isolation, samples were brought to the same final volume for comparative analyses. Our results confirmed the best performance of 4RRF resins in EV yield (as assessed by western‐blot with anti‐CD81), with lower LPP and protein contamination (Figure 6a,b). dUC samples showed a lower LPP content but also a very poor EV yield when compared to SEC samples. In contrast, total protein content was very elevated (Figure 6b), which points to a significant co‐isolation of proteins within the dUC sample. This protein co‐isolation was confirmed in TEM images: dUC samples where completely full of protein aggregates, being only possible to visualize a few EVs at very high dilutions (Figure 6c). Moreover, when measuring the yield as particles by NTA per millilitre of initial plasma, it was almost two orders of magnitude lower in dUC than in SEC samples (Figure 6d). The size profile observed for dUC was more heterogeneous than that of SEC samples consistent with a higher degree of vesicles and protein aggregation (Figure 6e).
FIGURE 6.
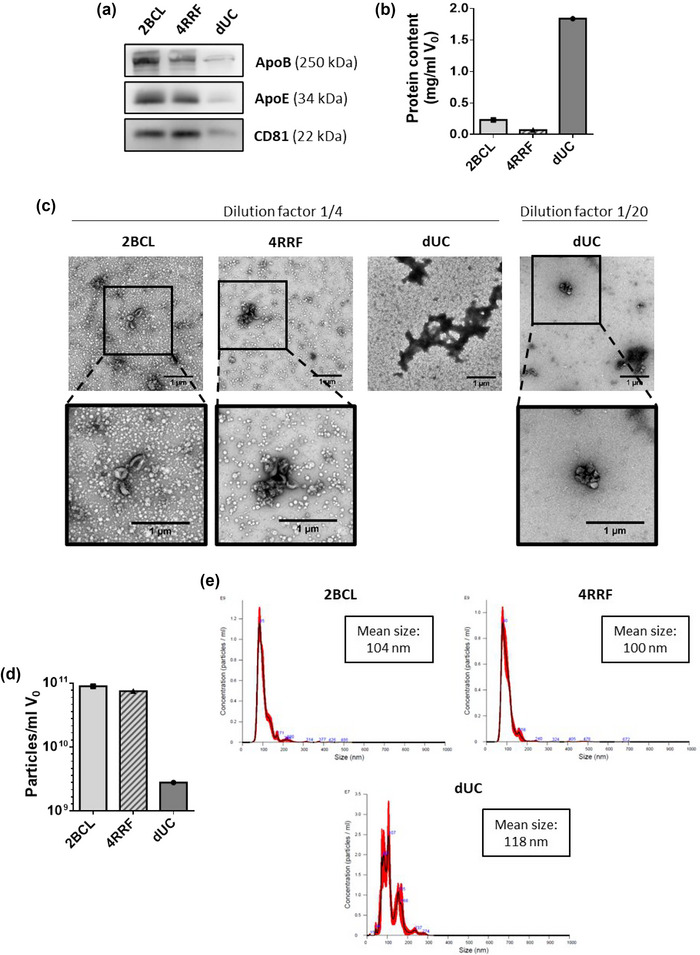
Comparison of EV isolation from plasma samples by SEC (with 2BCL or 4RRF resins) and dUC. (a) Western blot analyses of EV (CD81) and LPP (ApoB and ApoE) markers in EV samples. 20 μL of each sample were loaded per gel for immunoblot detection. (b) Protein content represented as mg of protein in isolated samples per milliliter of initial volume of plasma. (c) Representative TEM images of negatively stained samples isolated by SEC or dUC. Samples were previously diluted 1/4 (2BCL, 4RRF and dUC) or 1/20 (dUC). Bars = 1 μm. (d) Concentration of particles measured by NTA in EV samples isolated by SEC or dUC, represented as the number of particles per mL of initial volume of plasma. (e) Size profile of particles in EV isolated samples measured by NTA.
4. DISCUSSION
EV research is a rapidly expanding field because of their relevance in intercellular communication in pluricellular organisms, and even in cross‐kingdom biological communication during infection or commensalism. Among the bioactive molecules that EVs can exchange, genetic material is key not only for its crucial functions, but also because it can be readily detected following amplification. Thus, EVs have been incorporated into the opportunities that the liquid biopsy offers for non‐invasive diagnosis, prognosis or follow up of different diseases (Fais et al., 2016; Gutiérrez‐Fernández et al., 2021). Biological markers carried within EVs also represent an advantage compared to soluble markers, since upon EV isolation, those molecules are concentrated and separated from the characteristic high molecular background of the biological samples, easing EV‐borne biomarker identification and quantification. Other particles like LPPs (Vickers et al., 2011) or soluble protein complexes (Arroyo et al., 2011) may also carry RNA and other potential biomarkers in body fluids. A proper isolation of EVs from those carriers would lead to an enrichment of EV‐material and an improvement in sensitivity of detection of EV‐borne biomarkers (Lässer, 2019). Taking this into consideration, our aim was to move forward a one‐step method of EVs isolation that would eliminate almost all contaminants from biological samples.
During the last years there have been considerable advances in the isolation of EVs from different samples, particularly in the use of size exclusion chromatography (SEC) for this end. Several types of agaroses and other resins (Yoshitake et al., 2022; Kaddour et al., 2021) have been shown to achieve isolation of EVs from conditioned media or biological samples in a more efficient manner that other methods (Veerman et al., 2021). Whereas some studies have mainly focused on the selection of a particular agarose that improves EV yield (Guo et al., 2021), we wanted to exhaustively study the level of contaminants in EV‐enriched fractions for the selection of a superior agarose for EV isolation, and to test which are the main parameters in agarose resins to improve isolation yield and purity.
For the optimization of SEC and resin parameters, we decided to employ conditioned media from a melanoma cell culture, before moving on to more complex biological samples. The majority of researchers are obliged to work with media supplemented with FBS for the maintenance of the proper cell culture conditions (Lehrich et al., 2021). Therefore, although the conditioned media composition is considered to be simpler than that of many biological fluids, when supplemented with EV‐depleted FBS, culture media typically contain a high concentration of soluble proteins as well as subpopulations of LPPs (which ultimately can partially co‐elute with the EVs isolated by SEC).
First, we compared the EVs isolation capacity of standard agarose resins with different percentages of agarose. We confirmed a higher co‐elution of LPPs and soluble protein as the exclusion limit of the agarose decreases. On the other hand, as we raised the exclusion limit (lowering the percentage of agarose), LPP elimination decreases accompanied by a slight decrease in EV yield. When comparing 2% agaroses from two different suppliers, that from ABT allowed a similar depletion of LPPs in EVs fractions but with an improvement in EVs yield. Sepharose CL‐2B loses a subpopulation of small EVs in LPPs fractions, as revealed by size analysis of TEM micrographs from EV‐enriched fractions, when compared to 4BCL and 6BCL. An increment in the D10 particle size (diameter of the 10% smaller particles; data not shown) was also detected to be significant when comparing to 4BCL. These results were not confirmed by NTA analysis, probably due to the range of detection of this technique (over 60–70 nm diameter) (Bachurski et al., 2019).
In a recent study, Dr. Okeoma´s group isolated particles with different sizes by PPLC using a combination of dextran beads with different pore sizes, achieving an effective isolation of EVs from membraneless condensates (Kaddour et al., 2020). Although it was not directly addressed in that study, the exclusion limit of the resins employed would not allow the isolation of EVs from LPPs in that case. Here, to address whether a similar strategy could improve our results, we combined different percentage agarose beads in the same column. However, this approach did not improve peak resolution in comparison to simple columns of 2% or 4% agarose.
Since 2% agarose resins seem to lose the lower size range of EVs, we decided to focus on improving results using 4% resins, that should render the whole size range of EVs. When we tested different crosslinking levels on the agarose beads, we could not observe significant differences between them (4B, 4BCL or 4RR), although a slight tendency towards lower LPPs contamination is observed as crosslinking level is increased. This could be due to an increment in stiffness and hence, in stability of agarose, being exclusion limits and fractionation ranges better preserved. The major differences were observed when the agarose bead sizes were changed. With larger agarose beads (4 M agarose) we observed an increment in elution speed while columns with smaller beads (4RRF agarose) eluted slower in gravity conditions, rendering a superior peak resolution and thus better EV purity index in comparison to any of the agaroses previously tested in this study. Thus, our data reveal that bead size and not crosslinking may be the main determinant of EV purity in SEC isolation procedures.
It should be noted, however, that the mean size of EVs isolated with 4RRF is very slightly higher than EVs isolated with the other 4% agaroses, although this difference was only significant in TEM, but not NTA analysis. The tiny tail of EVs observed in the elution profile would also suggest a little bleed through of the smaller EVs into the LPPs fractions, which was confirmed by statistical analysis of EV yield based on dot blot results. Nevertheless, D10 particle size of these EVs was not increased either in NTA or TEM analyses (data not shown), although a slight increase in D90 by NTA and TEM analysis was observed, but only reached significance when compared with 4RR in TEM analysis. Importantly, total EVs yield obtained with 4RRF column was not affected in NTA analysis of number of particles, suggesting that the loss of small particles in LPPs fractions is really negligible in comparison to the improvement on EV purity obtained.
Promising results were also obtained for EVs isolation from plasma samples employing 4RRF agarose columns, with clearly lower LPP contaminants than with 2% and 4% agaroses, which are usually employed in EVs isolation. ApoB48‐positive LPPs elimination was not as good as that of ApoE‐positive ones, maybe because of the exclusive presence of ApoB‐48 in chylomicrons and chylomicrons remnants (Isherwood et al., 1997), which present a range of particle size that overlaps with EVs (Karimi et al.). Using 4RRF agarose columns, the amount of LPPs in the final sample decreased down to 10% (ApoE‐LPPs) or 18% (ApoB48‐LPPs), although these particles could be still detected in TEM images. In this regard, the initial concentration of LPPs in plasma samples has been determined to be around 1015 particles/mL for LPPs (Caulfield et al., 2008), which is several orders of magnitude over the estimated number for EVs, around ≈1010 particles/mL. EV numbers in the literature greatly vary depending on the isolation or quantification method employed, from 106–1013 particles per ml of plasma (Johnsen et al., 2019; Coumans et al., 2017). Those differences could be explained for the low or high presence of contaminants in the isolated EV samples. In our hands, concentration of particles on isolated EVs are around 1010 particles per ml of plasma.
For a complete elimination of LPPs a combination of methods would be needed (Karimi et al., 2018). Apart from the combination of SEC with density gradient ultracentrifugation, methods based on cation exchange chromatography have also been tested (Van Deun et al., 2020). Even so, LPPs elimination could be probably improved selecting the appropriate moment for blood collection (fasting conditions for a lower number of chylomicrons) and a more suitable pre‐processing protocol to obtain plasma samples. A careful optimization and consistency in preanalytical parameters are thus fundamental to ensure proper biomarker discovery and large cohorts analyses. SPREC codification could be a useful approach for this aspect (López‐Guerrero et al., 2023). Regarding this point, it is also important to consider an optimum elimination of platelets in plasma samples, as their presence in the sample would increase platelet‐derived EVs, which would mask results for downstream experiments (Mitchell et al., 2016). We have employed here the protocol published by the International Society on Thrombosis and Haemostasis (ISTH) (Lacroix et al., 2013), although other protocols have been recently proposed for improved results (Bettin et al., 2022).
A key factor that should be taken into account when choosing a method for EV isolation, is a careful consideration of the downstream experiments that are to be carried out. SEC is an easy and accessible technique for any laboratory, which provides isolated EVs samples with high purity. With our improved resins, for most downstream analyses LPP contamination could be considered negligible, allowing proper biomarker analyses without the need of combination with a different purification method, thus easing the implantation in a clinical setting. Moreover, one of the main advantages of the improved separation of EVs from LPPs and soluble proteins of our optimized agarose, is that we can separately interrogate these three main fractions (EVs, LPPs and soluble proteins) in the search of biomarkers or functional components.
AUTHOR CONTRIBUTIONS
Beatriz Benayas: Data curation; Formal analysis; Investigation; Methodology; Writing—original draft. Joaquín Morales: Investigation; Methodology. Carolina Egea: Project administration; Resources. Pilar Armisén: Formal analysis; Methodology; Project administration; Resources; Supervision. María Yáñez‐Mó: Conceptualization; Data curation; Formal analysis; Methodology; Project administration; Supervision; Writing—review & editing.
CONFLICTS OF INTEREST STATEMENT
C.E. is general manager of ABT, P.A. is Business and technical development manager at ABT and B.B. is employed by ABT in the context of an industrial doctorate. The remaining authors declare no conflict of interests.
Supporting information
Figure S1. Characterization of EV‐enriched fractions obtained by SEC with standard agarose resins.
Figure S2. Characterization of EV‐enriched fractions obtained by SEC with columns that combined agarose resins of different percentages.
Figure S3. NTA characterization of EV‐enriched fractions obtained by SEC with 4% agarose resins of different crosslinking or bead size.
Figure S4. NTA and TEM characterization of EV‐enriched fractions obtained by SEC of plasma samples.
ACKNOWLEDGEMENTS
We want to thank Dr Cabañas and Víctor Toribio for critical reading of this manuscript, Dr Dotti for the use of Nanosight equipment and the TEM Facility at CBMSO for their assistance in electron microscopy. This work has been supported by grants PID2020‐ 119627GB‐I00, DTS21/00134 and RED2018‐102411‐T TeNTaCLES from Ministerio Español de Ciencia e Innovación (Spain) to M.Y.‐M. B.B. is supported by predoctoral industrial contract IND2019_BMD‐17100 and J.M. by a INV‐CAM‐03 Yo Investigo contract, both from Comunidad Autónoma de Madrid.
Benayas, B. , Morales, J. , Egea, C. , Armisén, P. , & Yáñez‐Mó, M. (2023). Optimization of extracellular vesicle isolation and their separation from lipoproteins by size exclusion chromatography. Journal of Extracellular Biology, 2, e100. 10.1002/jex2.100
REFERENCES
- Arroyo, J. D. , Chevillet, J. R. , Kroh, E. M. , Ruf, I. K. , Pritchard, C. C. , Gibson, D. F. , Mitchell, P. S. , Bennett, C. F. , Pogosova‐Agadjanyan, E. L. , Stirewalt, D. L. , Tait, J. F. , & Tewari, M. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proceedings of the National Academy of Sciences of the United States of America, 108(12), 5003–5008. 10.1073/pnas.1019055108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachurski, D. , Schuldner, M. , Nguyen, P.‐H. , Malz, A. , Reiners, K. S. , Grenzi, P. C. , Babatz, F. , Schauss, A. C. , Hansen, H. P. , Hallek, M. , & Pogge von Strandmann, E. (2019). Extracellular vesicle measurements with nanoparticle tracking analysis‐An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. Journal of Extracellular Vesicles, 8(1), 1596016. 10.1080/20013078.2019.1596016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettin, B. , Gasecka, A. , Li, B. , Dhondt, B. , Hendrix, A. , Nieuwland, R. , & van der Pol, E. (2022). Removal of platelets from blood plasma to improve the quality of extracellular vesicle research. Journal of Thrombosis and Haemostasis, 20(11), 2679–2685. 10.1111/jth.15867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böing, A. N. , van der Pol, E. , Grootemaat, A. E. , Coumans, F. A. W. , Sturk, A. , & Nieuwland, R. (2014). Single‐step isolation of extracellular vesicles by size‐exclusion chromatography. Journal of Extracellular Vesicles, 3, 23430. 10.3402/jev.v3.23430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgheti‐Cardoso, L. N. , Kooijmans, S. A. A. , Chamorro, L. G. , Biosca, A. , Lantero, E. , Ramírez, M. , Avalos‐Padilla, Y. , Crespo, I. , Fernández, I. , Fernandez‐Becerra, C. , Del Portillo, H. A. , & Fernàndez‐Busquets, X. (2020). Extracellular vesicles derived from Plasmodium‐infected and non‐infected red blood cells as targeted drug delivery vehicles. International Journal of Pharmaceutics, 587, 119627. 10.1016/j.ijpharm.2020.119627 [DOI] [PubMed] [Google Scholar]
- Calle, A. , Toribio, V. , Yáñez‐Mó, M. , & Ramírez, M. Á. (2021). Embryonic trophectoderm secretomics reveals chemotactic migration and intercellular communication of endometrial and circulating MSCs in embryonic implantation. International Journal of Molecular Sciences, 22(11), 5638. 10.3390/ijms22115638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos‐Silva, C. , Cáceres‐Martell, Y. , López‐Cobo, S. , Rodriguez, M. J. , Jara, R. , Yáñez‐Mó, M. , & Valés‐Gómez, M. (2021). An immunocapture‐based assay for detecting multiple antigens in melanoma‐derived extracellular vesicles. Methods in Molecular Biology (Clifton, N.J.), 2265, 323–344. 10.1007/978-1-0716-1205-7_24 [DOI] [PubMed] [Google Scholar]
- Campos‐Silva, C. , Suárez, H. , Jara‐Acevedo, R. , Linares‐Espinós, E. , Martinez‐Piñeiro, L. , Yáñez‐Mó, M. , & Valés‐Gómez, M. (2019). High sensitivity detection of extracellular vesicles immune‐captured from urine by conventional flow cytometry. Scientific Reports, 9, 2042. 10.1038/s41598-019-38516-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreras‐Planella, L. , Monguió‐Tortajada, M. , Borràs, F. E. , & Franquesa, M. (2019). Immunomodulatory effect of MSC on B cells is independent of secreted extracellular vesicles. Frontiers in Immunology, 10, 1288. 10.3389/fimmu.2019.01288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulfield, M. P. , Li, S. , Lee, G. , Blanche, P. J. , Salameh, W. A. , Benner, W. H. , Reitz, R. E. , & Krauss, R. M. (2008). Direct determination of lipoprotein particle sizes and concentrations by ion mobility analysis. Clinical Chemistry, 54(8), 1307–1316. 10.1373/clinchem.2007.100586 [DOI] [PubMed] [Google Scholar]
- Chen, J. , Tan, Q. , Yang, Z. , & Jin, Y. (2022). Engineered extracellular vesicles: Potentials in cancer combination therapy. Journal of Nanobiotechnology, 20(1), 132. 10.1186/s12951-022-01330-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clos‐Sansalvador, M. , Monguió‐Tortajada, M. , Roura, S. , Franquesa, M. , & Borràs, F. E. (2022). Commonly used methods for extracellular vesicles’ enrichment: Implications in downstream analyses and use. European Journal of Cell Biology, 101(3), 151227. 10.1016/j.ejcb.2022.151227 [DOI] [PubMed] [Google Scholar]
- Coumans, F. A. W. , Brisson, A. R. , Buzas, E. I. , Dignat‐George, F. , Drees, E. E. E. , El‐Andaloussi, S. , Emanueli, C. , Gasecka, A. , Hendrix, A. , Hill, A. F. , Lacroix, R. , Lee, Y. , van Leeuwen, T. G. , Mackman, N. , Mäger, I. , Nolan, J. P. , van der Pol, E. , Pegtel, D. M. , Sahoo, S. , … Nieuwland, R. (2017). Methodological guidelines to study extracellular vesicles. Circulation Research, 120(10), 1632–1648. 10.1161/CIRCRESAHA.117.309417 [DOI] [PubMed] [Google Scholar]
- de Menezes‐Neto, A. , Sáez, M. J. F. , Lozano‐Ramos, I. , Segui‐Barber, J. , Martin‐Jaular, L. , Ullate, J. M. E. , Fernandez‐Becerra, C. , Borrás, F. E. , & del Portillo, H. A. (2015). Size‐exclusion chromatography as a stand‐alone methodology identifies novel markers in mass spectrometry analyses of plasma‐derived vesicles from healthy individuals. Journal of Extracellular Vesicles, 4, 27378. 10.3402/jev.v4.27378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fais, S. , O'Driscoll, L. , Borras, F. E. , Buzas, E. , Camussi, G. , Cappello, F. , Carvalho, J. , Cordeiro da Silva, A. , Del Portillo, H. , El Andaloussi, S. , Ficko Trček, T. , Furlan, R. , Hendrix, A. , Gursel, I. , Kralj‐Iglic, V. , Kaeffer, B. , Kosanovic, M. , Lekka, M. E. , Lipps, G. , … Giebel, B. (2016). Evidence‐based clinical use of nanoscale extracellular vesicles in nanomedicine. ACS Nano, 10(4), 3886–3899. 10.1021/acsnano.5b08015 [DOI] [PubMed] [Google Scholar]
- Gardiner, C. , Vizio, D. D. , Sahoo, S. , Théry, C. , Witwer, K. W. , Wauben, M. , & Hill, A. F. (2016). Techniques used for the isolation and characterization of extracellular vesicles: Results of a worldwide survey. Journal of Extracellular Vesicles, 5, 32945. 10.3402/jev.v5.32945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, S. , Yu, H. , Yan, G. , Gao, M. , Sun, W. , & Zhang, X. (2020). Characterization of urinary exosomes purified with size exclusion chromatography and ultracentrifugation. Journal of Proteome Research, 19(6), 2217–2225. 10.1021/acs.jproteome.9b00693 [DOI] [PubMed] [Google Scholar]
- Guo, J. , Wu, C. , Lin, X. , Zhou, J. , Zhang, J. , Zheng, W. , Wang, T. , & Cui, Y. (2021). Establishment of a simplified dichotomic size‐exclusion chromatography for isolating extracellular vesicles toward clinical applications. Journal of Extracellular Vesicles, 10(11), e12145. 10.1002/jev2.12145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez‐Fernández, M. , de la Cuesta, F. , Tallón, A. , Cuesta, I. , Fernández‐Fournier, M. , Laso‐García, F. , Gómez‐de Frutos, M. C. , Díez‐Tejedor, E. , & Otero‐Ortega, L. (2021). Potential roles of extracellular vesicles as biomarkers and a novel treatment approach in multiple sclerosis. International Journal of Molecular Sciences, 22(16), 9011. 10.3390/ijms22169011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. , Ji, X. , Jackson, K. K. , Lubman, D. M. , Ard, M. B. , Bruce, T. F. , & Marcus, R. K. (2021). Rapid separation of blood plasma exosomes from low‐density lipoproteins via a hydrophobic interaction chromatography method on a polyester capillary‐channeled polymer fiber phase. Analytica Chimica Acta, 1167, 338578. 10.1016/j.aca.2021.338578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isherwood, S. G. , Williams, C. M. , & Gould, B. J. (1997). Apolipoprotein B‐48 as a marker for chylomicrons and their remnants: Studies in the postprandial state. Proceedings of the Nutrition Society, 56(1B), 497–505. 10.1079/PNS19970050 [DOI] [PubMed] [Google Scholar]
- Johnsen, K. B. , Gudbergsson, J. M. , Andresen, T. L. , & Simonsen, J. B. (2019). What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood‐borne biomarkers of cancer. Biochimica Et Biophysica Acta. Reviews on Cancer, 1871(1), 109–116. 10.1016/j.bbcan.2018.11.006 [DOI] [PubMed] [Google Scholar]
- Kaddour, H. , Lyu, Y. , Shouman, N. , Mohan, M. , & Okeoma, C. M. (2020). Development of novel high‐resolution size‐guided turbidimetry‐enabled particle purification liquid chromatography (PPLC): Extracellular vesicles and membraneless condensates in focus. International Journal of Molecular Sciences, 21(15), 5361. 10.3390/ijms21155361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddour, H. , Tranquille, M. , & Okeoma, C. M. (2021). The past, the present, and the future of the size exclusion chromatography in extracellular vesicles separation. Viruses, 13(11), 2272. 10.3390/v13112272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, N. , Cvjetkovic, A. , Jang, S. C. , Crescitelli, R. , Hosseinpour Feizi, M. A. , Nieuwland, R. , Lötvall, J. , & Lässer, C. (2018). Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cellular and Molecular Life Sciences, 75(15), 2873–2886. 10.1007/s00018-018-2773-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konoshenko, M. Y. , Lekchnov, E. A. , Bryzgunova, O. E. , Kiseleva, E. , Pyshnaya, I. A. , & Laktionov, P. P. (2021). Isolation of extracellular vesicles from biological fluids via the aggregation–precipitation approach for downstream miRNAs detection. Diagnostics, 11(3), 384. 10.3390/diagnostics11030384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotrbová, A. , Štěpka, K. , Maška, M. , Pálenik, J. J. , Ilkovics, L. , Klemová, D. , Kravec, M. , Hubatka, F. , Dave, Z. , Hampl, A. , Bryja, V. , Matula, P. , & Pospíchalová, V. (2019). TEM ExosomeAnalyzer: A computer‐assisted software tool for quantitative evaluation of extracellular vesicles in transmission electron microscopy images. Journal of Extracellular Vesicles, 8(1), 1560808. 10.1080/20013078.2018.1560808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal, J. , Arras, G. , Colombo, M. , Jouve, M. , Morath, J. P. , Primdal‐Bengtson, B. , Dingli, F. , Loew, D. , Tkach, M. , & Théry, C. (2016). Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proceedings of the National Academy of Sciences of the United States of America, 113(8), E968–E977. 10.1073/pnas.1521230113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers, M. E. , Koning, R. I. , Bos, E. , Hokke, C. H. , Smits, H. H. , & Nolte‐‘t Hoen, E. N. M. (2022). Optimized protocol for the isolation of extracellular vesicles from the parasitic worm Schistosoma mansoni with improved purity, concentration, and yield. Journal of Immunology Research, 2022, e5473763. 10.1155/2022/5473763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix, R. , Judicone, C. , Mooberry, M. , Boucekine, M. , Key, N. S. , & Dignat‐George, F. (2013). Standardization of pre‐analytical variables in plasma microparticle determination: Results of the International Society on Thrombosis and Haemostasis SSC Collaborative workshop. Journal of Thrombosis and Haemostasis : JTH, 11, 1190–1193. 10.1111/jth.12207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lässer, C. (2019). Mapping extracellular RNA sheds lights on distinct carriers. Cell, 177(2), 228–230. 10.1016/j.cell.2019.03.027 [DOI] [PubMed] [Google Scholar]
- Lehrich, B. M. , Liang, Y. , & Fiandaca, M. S. (2021). Foetal bovine serum influence on in vitro extracellular vesicle analyses. Journal of Extracellular Vesicles, 10(3), e12061. 10.1002/jev2.12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Guerrero, J. A. , Valés‐Gómez, M. , Borrás, F. E. , Falcón‐Pérez, J. M. , Vicent, M. J. , & Yáñez‐Mó, M. (2023). Standardising the preanalytical reporting of biospecimens to improve reproducibility in extracellular vesicle research‐A GEIVEX study. Journal of Extracellular Biology, 2(4), e76. 10.1002/jex2.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, A.‐K. , De Miroschedji, K. , Doeppner, T. R. , Börger, V. , Ruesing, J. , Rebmann, V. , Durst, S. , Jansen, S. , Bremer, M. , Behrmann, E. , Singer, B. B. , Jastrow, H. , Kuhlmann, J. D. , El Magraoui, F. , Meyer, H. E. , Hermann, D. M. , Opalka, B. , Raunser, S. , Epple, M. , … Giebel, B. (2018). Precipitation with polyethylene glycol followed by washing and pelleting by ultracentrifugation enriches extracellular vesicles from tissue culture supernatants in small and large scales. Journal of Extracellular Vesicles, 7(1), 1528109. 10.1080/20013078.2018.1528109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Dong, S. , Li, X. , Kim, B. Y. S. , Yang, Z. , & Jiang, W. (2021). Extracellular vesicles: An emerging nanoplatform for cancer therapy. Frontiers in Oncology, 10, 606906. https://www.frontiersin.org/articles/10.3389/fonc.2020.606906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, A. J. , Gray, W. D. , Hayek, S. S. , Ko, Y.‐A. , Thomas, S. , Rooney, K. , Awad, M. , Roback, J. D. , Quyyumi, A. , & Searles, C. D. (2016). Platelets confound the measurement of extracellular miRNA in archived plasma. Scientific Reports, 6, 32651. 10.1038/srep32651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monguió‐Tortajada, M. , Gálvez‐Montón, C. , Bayes‐Genis, A. , Roura, S. , & Borràs, F. E. (2019). Extracellular vesicle isolation methods: Rising impact of size‐exclusion chromatography. Cellular and Molecular Life Sciences, 76(12), 2369–2382. 10.1007/s00018-019-03071-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onódi, Z. , Pelyhe, C. , Terézia Nagy, C. , Brenner, G. B. , Almási, L. , Kittel, Á. , Manček‐Keber, M. , Ferdinandy, P. , Buzás, E. I. , & Giricz, Z. (2018). Isolation of high‐purity extracellular vesicles by the combination of iodixanol density gradient ultracentrifugation and bind‐elute chromatography from blood plasma. Frontiers in Physiology, 9, 1479. 10.3389/fphys.2018.01479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini, L. , Zendrini, A. , Noto, G. D. , Busatto, S. , Lottini, E. , Radeghieri, A. , Dossi, A. , Caneschi, A. , Ricotta, D. , & Bergese, P. (2016). Residual matrix from different separation techniques impacts exosome biological activity. Scientific Reports, 6, 23550. 10.1038/srep23550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Amigorena, S. , Raposo, G. , & Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current Protocols in Cell Biology, 30(1), 3.22.1–3.22.29. 10.1002/0471143030.cb0322s30 [DOI] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J.‐M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deun, J. , Jo, A. , Li, H. , Lin, H.‐Y. , Weissleder, R. , Im, H. , & Lee, H. (2020). Integrated dual‐mode chromatography to enrich extracellular vesicles from plasma. Advanced Biosystems, 4(12), 1900310. 10.1002/adbi.201900310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deun, J. , Mestdagh, P. , Agostinis, P. , Akay, Ö. , Anand, S. , Anckaert, J. , Martinez, Z. A. , Baetens, T. , Beghein, E. , Bertier, L. , Berx, G. , Boere, J. , Boukouris, S. , Bremer, M. , Buschmann, D. , Byrd, J. B. , Casert, C. , Cheng, L. , Cmoch, A. , … Hendrix, A. (2017). EV‐TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nature Methods, 14(3), 228–232. 10.1038/nmeth.4185 [DOI] [PubMed] [Google Scholar]
- Van Deun, J. , Mestdagh, P. , Sormunen, R. , Cocquyt, V. , Vermaelen, K. , Vandesompele, J. , Bracke, M. , De Wever, O. , & Hendrix, A. (2014). The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. Journal of Extracellular Vesicles, 3, 24858. 10.3402/jev.v3.24858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerman, R. E. , Teeuwen, L. , Czarnewski, P. , Güclüler Akpinar, G. , Sandberg, A. , Cao, X. , Pernemalm, M. , Orre, L. M. , Gabrielsson, S. , & Eldh, M. (2021). Molecular evaluation of five different isolation methods for extracellular vesicles reveals different clinical applicability and subcellular origin. Journal of Extracellular Vesicles, 10(9), e12128. 10.1002/jev2.12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers, K. C. , Palmisano, B. T. , Shoucri, B. M. , Shamburek, R. D. , & Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high‐density lipoproteins. Nature Cell Biology, 13(4), 423–433. 10.1038/ncb2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Alfranca, A. , Cabañas, C. , Marazuela, M. , Tejedor, R. , Angeles Ursa, M. , Ashman, L. K. , de Landázuri, M. O. , & Sánchez‐Madrid, F. (1998). Regulation of endothelial cell motility by complexes of tetraspan molecules CD81/TAPA‐1 and CD151/PETA‐3 with α3β1 integrin localized at endothelial lateral junctions. The Journal of Cell Biology, 141(3), 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Barreiro, O. , Gordon‐Alonso, M. , Sala‐Valdés, M. , & Sánchez‐Madrid, F. (2009). Tetraspanin‐enriched microdomains: A functional unit in cell plasma membranes. Trends in Cell Biology, 19(9), 434–446. 10.1016/j.tcb.2009.06.004 [DOI] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Siljander, P. R.‐M. , Andreu, Z. , Zavec, A. B. , Borràs, F. E. , Buzas, E. I. , Buzas, K. , Casal, E. , Cappello, F. , Carvalho, J. , Colás, E. , Silva, A. C. , Fais, S. , Falcon‐Perez, J. M. , Ghobrial, I. M. , Giebel, B. , Gimona, M. , Graner, M. , Gursel, I. , … De Wever, O. (2015). Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles, 4, 27066. 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitake, J. , Azami, M. , Sei, H. , Onoshima, D. , Takahashi, K. , Hirayama, A. , Uchida, K. , Baba, Y. , & Shibata, T. (2022). Rapid isolation of extracellular vesicles using a hydrophilic porous silica gel‐based size‐exclusion chromatography column. Analytical Chemistry, 94(40), 13676–13681. 10.1021/acs.analchem.2c01053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuidscherwoude, M. , Worah, K. , van der Schaaf, A. , Buschow, S. I. , & van Spriel, A. B. (2017). Differential expression of tetraspanin superfamily members in dendritic cell subsets. PLoS ONE, 12(9), e0184317. 10.1371/journal.pone.0184317 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Characterization of EV‐enriched fractions obtained by SEC with standard agarose resins.
Figure S2. Characterization of EV‐enriched fractions obtained by SEC with columns that combined agarose resins of different percentages.
Figure S3. NTA characterization of EV‐enriched fractions obtained by SEC with 4% agarose resins of different crosslinking or bead size.
Figure S4. NTA and TEM characterization of EV‐enriched fractions obtained by SEC of plasma samples.


