Abstract
The CRISPR gene editing tool holds great potential for curing genetic disorders. However, the safe, efficient, and specific delivery of the CRISPR/Cas9 components into cells and tissues remains a challenge. While many currently available delivery methods achieve high levels of gene editing effects in vivo, they often result in genotoxicity and immunogenicity. Extracellular vesicles (EVs), which are cell‐derived lipid nanoparticles, are capable of transferring protein and nucleic acid cargoes between cells, making them a promising endogenous alternative to synthetic delivery methods. This review provides a comprehensive analysis of the currently available strategies for EV‐mediated delivery of CRISPR/Cas9. These strategies include cell‐based, passive loading obtained by overexpression of CRISPR/Cas9, active loading involving protein or RNA dimerization, and loading into already purified EVs. All these approaches suggest that EV‐based CRISPR/Cas9 delivery is useful for achieving both in vitro and in vivo gene editing. Despite that, substantial variations in cellular uptake and gene editing efficiencies indicate that further improvement and standardization are required for the therapeutic use of EVs as a CRISPR/Cas9 delivery vehicle. These improvements include, but is not limited to, the high‐yield purification of EVs, increased loading and release efficiencies, as well as improved tissue‐ or cell‐specific targeting specificities.
Keywords: Cas9, CRISPR, exosome, extracellular vesicles, gene editing, gene therapy, LNP
1. INTRODUCTION
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR‐associated 9 (CRISPR/Cas9) is a revolutionary genome editing (GE) technology with the potential to cure genetic disorders, cancers, and other diseases (Cong et al., 2013; Jinek et al., 2012; Liang et al., 2022; Xiang et al., 2021). Mediated by a programmable and single guide RNA (sgRNA), the Cas9 protein can introduce a double‐stranded DNA break (DSB) at a specific target site in genomes, which is subsequently repaired by the cellular DSB repair machineries. In mammalian cells, the DSB is predominately repaired by non‐homologous end joining (NHEJ), an error‐prone DNA repair process that often results in small insertions or deletions known as indels.
While multiple clinical trials are assessing different CRISPR/Cas9 therapies in humans, no drugs have yet been approved for broad clinical application (Henderson, 2023). The first GE medicine designed to cure sickle‐cell disease (SCD) or transfusion‐dependent beta‐thalassemia (TDT) is expected to receive FDA approval in December 2023. However, one of the main challenges facing the technology is the need for safe, efficient, and specific delivery of CRISPR/Cas9 to target cells and tissues (Horodecka & Düchler, 2021). Viral vectors have been the most commonly used in vivo delivery method. However, concerns regarding the inevitable side‐effects such as integration of the viral DNA into the host genome, genotoxicity, and immunogenicity for most viral vectors, have led to the urgent search for alternative and improved delivery methods (Duan et al., 2021; McAndrews et al., 2021). Strategies have been developed to mitigate random integration of viral vectors into host genomes, such as integration‐deficient lentiviral vectors. Adeno‐associated viral (AAV) vectors, which are the clinically approved, exhibit reduced genomic integration potential. However, limitations such as restricted cargo size remain. Currently, several non‐viral delivery methods are being explored, including extracellular vesicles (EVs), which are endogenous nanocarriers secreted by all cells (Cheng et al., 2021).
While EVs show great promise as delivery vehicles due to their intrinsic nature, they also face challenges in term of purification, cargo loading, and targeting (Liang et al., 2022). In this focused review, we comprehensively analyse the current development and potential use of EVs as a delivery method for CRISPR/Cas9. As such, we provide a comprehensive overview of the development of EV‐based CRISPR/Cas9 delivery methods, with a particular focus on methods for loading Cas9 and sgRNA into EVs. We also highlight the pros and cons of EV‐based CRISPR delivery, unmet technical challenges, potential solutions, and most importantly, the potential of EVs in gene therapy.
2. CRISPR GENE EDITING TECHNOLOGY
The CRISPR/Cas9 endonuclease system was originally discovered in bacteria and archaea as a part of their adaptive immune system (Bolotin et al., 2005; Ishino et al., 1987; Jansen et al., 2002; Mojica et al., 2005). Decades of research and understanding of CRISPR biology and functions in bacteria eventually led to the groundbreaking invention of a simple, efficient, and powerful CRISPR‐based gene editing tool (Jinek et al., 2012), which has now been successfully tested in all model organisms, including humans (Cheng et al., 2021; Cong et al., 2013; Esvelt et al., 2013; Hou et al., 2013; Jinek et al., 2012; Wang et al., 2013; Zhang et al., 2014). Over the last decade, the CRISPR gene editing tool has been substantially broadened to achieve epigenetic editing, gene activation, gene interference, base editing, nucleic acid detection, live cell imaging, and more recently prime editing. For a comprehensive overview of the genome editing revolution, we recommend readers consult the recent review by John van der Oost and Constantinos Patinios (van der Oost & Patinios, 2023). Computational tools are continuously being generated to facilitate the design of a single guide RNA (sgRNA) with high efficiency and specificity. For example, based on individually barcoded and array‐synthesized CRISPR sgRNA libraries, we have developed high‐throughput methods to measure on‐target and off‐target activities for over 10,000 sites simultaneously in cells. These large amounts of experimentally obtained data enable us to develop better prediction models for CRISPR on‐target and off‐target efficiency (Corsi et al., 2022; Pan et al., 2022; Xiang et al., 2021).
Most CRISPR/Cas9 gene editing systems to date are based on a two‐component system, consisting of a single guide RNA (sgRNA) and the Cas9 protein. The sgRNA is a chimeric RNA composed of the CRISPR RNA (crRNA) and trans‐activating CRISPR RNA (tracrRNA). Guided by the sgRNA, the Cas9 protein is directed to a specific site in the genome and cleaves the DNA at that site, thus generating double‐strand DNA breaks (DSBs) (McAndrews et al., 2021). The modifications to the genome rely on the endogenous repair of the DSBs (Demirci et al., 2022). Non‐homologous end joining (NHEJ) is the primary cellular DSB repair mechanism in mammalian cells. However, NHEJ is an error‐prone process and often leads to insertions or deletions (indels), resulting in gene knockout when DSBs are introduced in protein‐coding exons (Lino et al., 2018). Homology‐directed repair (HDR) is another DNA repair mechanism that occurs when a DNA donor template containing homologous sequences to the regions flanking the DSB site is provided. HDR can direct the insertion of desired sequences into the cleavage site, thereby enabling precise genetic modifications of the gene (Behr et al., 2021; Demirci et al., 2022). Beyond introducing DNA mutations, the CRISPR/Cas9 system can also be used for gene regulation by using an endonuclease‐dead Cas9 (dCas9) fused to a transcriptional activator or repressor domain (Lainšček et al., 2018). For instance, we previously demonstrated that fusing dCas9 to DNA methyltransferase domains can achieve RNA‐guided DNA methylation (Lin et al., 2018).
To achieve gene editing, it is essential to deliver the Cas9 and sgRNA components into cells and tissues efficiently and specifically. To date, the CRISPR/Cas9 system can be delivered to cells in three different types of cargo: (I) plasmid DNA encoding Cas9 and sgRNA, (II) messenger RNA (mRNA) for the translation of the Cas9 protein and a separate synthetic or in vitro transcribed sgRNA, or (III) a ribonucleoprotein (RNP) complex consisting of a Cas9 protein and an sgRNA (Cheng et al., 2021). The pros and cons of each type of cargo are depicted in Figure 1. Plasmid DNA is transcribed in the nucleus and is very stable, which could lead to long‐term expression as well as possible integration into the genome. In contrast, mRNA and RNP have a more rapid onset and are only transiently expressed and present in the cells due to degradation, which greatly limits the potential off‐target and other genotoxic events (Cheng et al., 2021).
FIGURE 1.
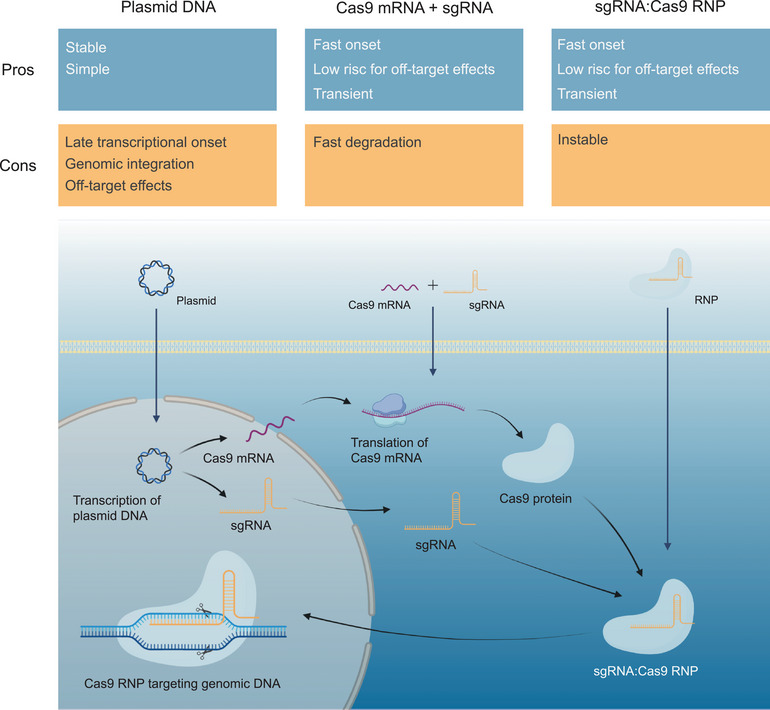
Overview of the three types of CRISPR/Cas9 cargo including the pros and cons of their application as well as the cellular transcription and translation. Created with BioRender.com.
3. CRISPR/CAS9 DELIVERY SYSTEMS
To exploit the CRISPR/Cas9 system for gene therapy, it must be transported to the target cells of a specific tissue and subsequently into the nucleus of those cells (Demirci et al., 2022). Delivery systems are required to ensure specific delivery, enhance cellular uptake and protect the cargo from enzymatic degradation in the extracellular space (Busatto et al., 2021). An ideal CRISPR/Cas9 delivery system is non‐immunogenic, non‐genotoxic, stable, efficient, and specific in delivering cargo to cells without causing off‐target effects (Wilbie et al., 2019). Moreover, the ideal delivery system is optimized for the specific target organ and target cell type. Distinct cell types exhibit variation in endocytic uptake efficiency as well as varying cytotoxic sensitivity towards chemical components and physical alterations (Wilbie et al., 2019). Available delivery systems can be roughly divided into three groups: physical, viral, and non‐viral as depicted in Figure 2.
FIGURE 2.
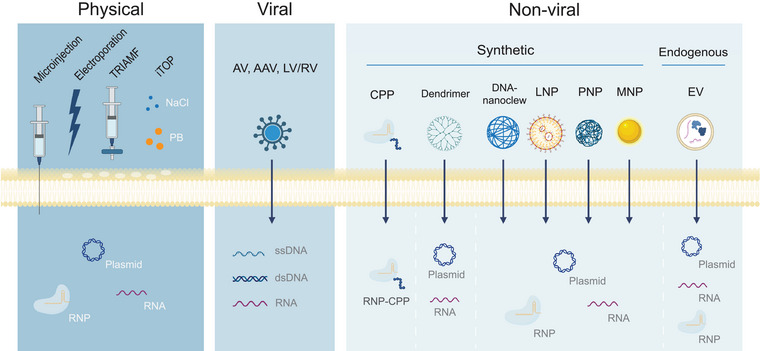
Cellular CRISPR/Cas9 delivery systems and their potential cargo divided into physical, viral and non‐viral subgroups. TRIAMF (transmembrane internalization assisted by membrane filtration), iTOP (induced transduction by osmocytosis and propane betaine (PB)), AV (Adenovirus), AAV (adeno‐associated virus), LV/RV (lentivirus/retrovirus), CPP (cell penetrating peptide), LNP (lipid nanoparticle), PNP (polymeric nanoparticle), MNP (metallic nanoparticle), EV (extracellular vesicle). Created with BioRender.com.
3.1. Physical delivery methods
Physical delivery methods include, for example, microinjection of the CRISPR components directly into individual cells, electroporation, and mechanical deformation, which forms pores in the cell membrane, allowing cargo to enter by diffusion (Cheng et al., 2021; Wilbie et al., 2019). These methods exhibit high loading efficacy ex vivo and are effective for cells that can be reintroduced into the body after loading (Cheng et al., 2021; Wilbie et al., 2019). TRIAMF (transmembrane internalization assisted by membrane filtration) and iTOP (induced transduction by Osmo cytosis and propane betaine) comprise two novel delivery methods, which allow the delivery of CRISPR RNPs exhibiting less cytotoxicity when compared to electroporation for ex vivo delivery (Wilbie et al., 2019; Yen et al., 2018). Downsides of the physical methods include the lack of in vivo applications and the potential disruption of cell integrity due to manipulation of the cell membrane (Cheng et al., 2021). On the other hand, hydrodynamic injection is a physical delivery method that can be used in vivo. In this approach, a solution containing DNA, RNA, and/or proteins is injected into a vein rapidly and in a large volume (Suda & Liu, 2007). This increases the hydrodynamic pressure in the capillaries thereby enhancing the permeability of endothelial and parenchymal cells. For selected cell types with limited endocytosis, physical delivery methods are preferred. For example, electroporation and TRIAMF are favoured ex vivo CRISPR delivery strategies for hematopoietic stem and progenitor cells in research (Yen et al., 2018).
3.2. Viral vectors
Viral vectors are the most commonly used method for delivering CRISPR/Cas9 and other therapeutic genes due to their high transduction efficacy and targeting abilities (Cheng et al., 2021; Tong et al., 2019; Wilbie et al., 2019). The three widely used viral vectors include adenoviruses (AdVs), adeno‐associated viruses (AAVs), and lentiviruses/retroviruses (LV/RVs). These vectors utilize the viral shell loaded with CRISPR‐encoding genes either as dsDNA, ssDNA or ssRNA, respectively (Wilbie et al., 2019).
AAVs show relatively low immunogenicity and cytotoxicity, and they rarely cause chromosomal integration, making them the most preferred viral vector and ideal for in vivo use. However, a significant downside is the limited packaging capacity, as they cannot accommodate cargo larger than 5 kb. This limitation hinders the simultaneous encapsulation of both Cas9 and sgRNA. To address this challenge, multiple studies have employed strategies such as splitting up the cargo into multiple AAVs, truncating the cargo, or using smaller Cas proteins like S. aureus Cas9 (SaCas9) (Ahmadi et al., 2023; Cheng et al., 2021).
AdVs are larger in size and therefore capable of packaging both Cas9 and sgRNA. They do not integrate into the host genome, which reduces the risk of off‐target effects. However, they often trigger a strong immune response making them undesirable in many cases. LV/RVs are also larger in size and exhibit relatively low immunogenicity. A downside to LV/RVs is that they can integrate into the host genome, resulting in long‐lasting expression of Cas9, which can cause insertional mutagenesis and off‐target effects (Cheng et al., 2021).
AAVs are currently being used in a few clinical trials for CRISPR/Cas9 delivery as they have shown to be highly efficient for gene editing (Henderson, 2023). However, previous trials using AAVs for the delivery of other gene therapeutic vectors have resulted in the deaths of patients. For instance, four individuals in a trial on X‐linked myotubular myopathy suffered liver damage (Pagliarulo, 2021), and two patients were affected in separate studies on Duchenne Muscular Dystrophy (DMD) (Bigica, 2022; Bryson, 2022). These examples highlight the safety risks associated with viral vectors, as they can induce immune activation and toxicity (Cheng et al., 2021). Furthermore, the development of antibodies against viral proteins restricts their application to a single administration, and some individuals may already possess pre‐existing immunity against AAVs (Mirjalili Mohanna et al., 2022). Another significant concern with using viral vectors is the potential risk of random integration into the host genome, whether in recipient or non‐recipient cells, thus posing a risk of genotoxic off‐target effects or oncogenesis (Ye et al., 2020).
3.3. Non‐viral delivery methods (synthetic)
To overcome the limitations and drawbacks of physical and viral methods, new synthetic non‐viral delivery methods have been investigated to deliver CRISPR/Cas9 for gene therapy purposes (Cheng et al., 2021). These methods include transfection agents, cell penetrating peptides (CPP), dendrimers, DNA nano clews and nanoparticles such as lipid nanoparticles (LNPs), polymeric nanoparticles (PNPs), and metallic nanoparticles (MNPs) like gold nanoparticles (Chen et al., 2022; Duan et al., 2021; Han et al., 2021; Wilbie et al., 2019). These examples are just a selection, as new synthetic nanoparticles are continuously being developed.
CPPs can be coupled to either Cas9 or sgRNA, which facilitates direct uptake by recipient cells, often in combination with receptor‐mediated uptake as tandem peptides. This strategy has proven effective for targeting tumour cells in vivo (Song et al., 2021). LNPs and DNA nano clews encapsulate CRISPR/Cas9 DNA, RNA or RNP through electrostatic interactions and sequence complementarity, respectively. Meanwhile, polymeric and gold nanoparticles carry CRISPR molecules conjugated on their surface.
LNPs have gained a lot of attention, particularly after their successful utilization in COVID‐19 vaccines (Taha et al., 2022). They show great potential for CRISPR/Cas9 delivery in several studies and are currently being used in clinical trials in humans (Henderson, 2023; Taha et al., 2022). LNPs offer advantages such as high loading efficiency, flexible design allowing for surface modification and specific targeting (Han et al., 2021), as well as low immunogenicity and strong biocompatibility in the human body (Duan et al., 2021). However, their rapid blood clearance and susceptibility to endosomal degradation might contribute to lower in vivo gene editing efficiency (Demirci et al., 2022).
4. Extracellular vesicles (EVs)
EVs are nanovesicles secreted by cells into the extracellular space (Liang et al., 2022), representing a natural form of LNP. EVs play an important role in intercellular communication as they transfer molecular structures such as proteins and nucleic acids between cells (Du et al., 2022). They have significant implications in various physiological and pathological processes within the body including immune modulation, cancer development, and tissue repair. However, many aspects of their intrinsic functions remain unknown (Chen et al., 2019; Meng et al., 2020).
EVs are composed of a phospholipid bilayer membrane with various EV‐specific lipids and proteins, the latter including tetraspanins CD9 and CD63 as well as LAMP2B1 (Figure 3). EVs can be categorized into three subgroups according to their size and biogenesis (Moghadasi et al., 2021; Van Niel et al., 2018). Exosomes are the smallest (50–150 nm) and are generated through the inward budding of multivesicular endosomes (MVEs). The MVE escapes lysosomal degradation and releases the exosomes through fusion with the plasma membrane, facilitated by specific protein‐protein and protein‐lipid interactions (Aheget et al., 2020). On the other hand, microvesicles (100–1000 nm) and apoptotic bodies (50–5000 nm) are larger and are formed directly by outward budding of the plasma membrane.
FIGURE 3.
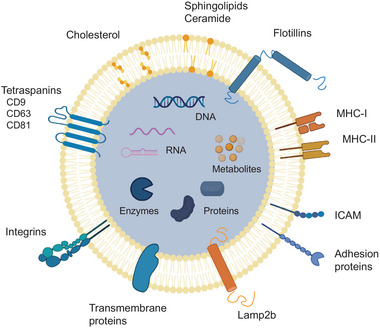
Endogenous EV composed of a lipid bilayer with a specific lipid composition and various surface proteins, some of them EV‐specific. EVs contain and transport various nucleic acids, metabolites and proteins including enzymes. Created with BioRender.com.
Several mechanisms are involved in the biogenesis of EVs, including cellular sorting of EV‐specific cargo and membrane proteins (Van Niel et al., 2018). In exosomes, both Endosomal Sorting Complex Required for Transport (ESCRT)‐dependent and ‐independent pathways contribute to their formation, with the latter involving tetraspanins that form membrane platforms. For microvesicles, the cytoskeleton and rearrangement of lipid and protein components of the plasma membrane are involved in EV formation and cargo sorting. For an in‐depth overview of EV biogenesis, we recommend readers refer to comprehensive EV‐focused reviews by Van Niel et al. (2018), and Hessvik and Llorente (2018). Despite variations in biogenesis, the three EV subgroups share common physiochemical properties (Beetler et al., 2022), and thus will be collectively referred to as EVs in this review.
EVs in the extracellular space can dock to the cell membrane of recipient cells and become internalized through either endocytosis, which is the primary uptake mechanism, or fusion with the plasma membrane (Van Niel et al., 2018). Upon endocytosis, EVs enter the endosomal pathway and become incorporated into MVEs. These MVEs can either fuse with lysosomes, leading to the degradation of EVs and their cargo. Alternatively, they can undergo retrograde fusion, resulting in the release of the EVs and their content to the cell cytoplasm (Van Niel et al., 2018). Although not fully comprehended, target cell specificity involves specific surface interactions, such as integrins on the EV surface and intercellular adhesion molecules (ICAMs) on the cell surface (Van Niel et al., 2018). To characterize isolated EVs, a combination of various methods is recommended, including transmission electron microscopy (TEM), scanning electron microscopy (SEM), atomic force microscopy (AFM), nanoparticle tracking analysis (NTA), dynamic light scattering (DLS), and flow cytometry. Furthermore, specific surface or cargo proteins of the EVs can be detected using immunoblotting (Meng et al., 2020).
EVs hold great potential as biomarkers, drug delivery agents and CRISPR/Cas9 delivery vehicles due to their remarkable molecular loading capabilities (Chung et al., 2020). Their endogenous origin makes them biocompatible, non‐immunogenic and stable in the body (Liang et al., 2022). Furthermore, they can cross biological barriers, evade phagocytosis, and easily enter cells (Liang et al., 2022; Majeau et al., 2022; Wang et al., 2022). Notably, certain EVs can cross the blood‐brain barrier, which is a challenge for many other delivery strategies like synthetic nanoparticles (Jiang et al., 2022). Currently there are multiple clinical trials using EVs for drug delivery, as well as trials using mesenchymal cell‐derived EVs for regenerative purposes (Han et al., 2021; Nikfarjam et al., 2020). Challenges of using EVs for CRISPR/Cas9 delivery include the successful large‐scale production, efficient loading, and specific targeting, all of which need to be addressed.
5. CRISPR/CAS9 DELIVERY WITH EV
In recent years, the use of EVs for delivering CRISPR/Cas9 has gained considerable attentions. Two primary loading strategies have been explored: non‐cell‐based and cell‐based loading (Han et al., 2021). Non‐cell‐based loading directly incorporating cargo into isolated EVs, while cell‐based loading entails donor cells producing EVs containing CRISPR/Cas9 cargo. Cell‐based loading can be accomplished through passive packaging via CRISPR/Cas9 overexpression or active packaging through protein‐protein or protein‐RNA dimerization. These loading methods have shown promise in achieving gene editing both in vitro and in vivo. However, there remains a need to improve and standardize these methods, including the purification of EVs, augmentation of loading and release efficiencies, and improvement of tissue‐ or cell‐specific targeting, for their effective application in gene therapy. Nevertheless, the potential of EVs in gene therapy, combined with their capacity to traverse biological barriers and evade phagocytosis, positions them as a promising candidate for delivering CRISPR/Cas9. A summary of the existing loading strategies is presented in Figure 4, with non‐cell‐based methods detailed in Table 1 and cell‐based methods outlined in Table 2.
FIGURE 4.
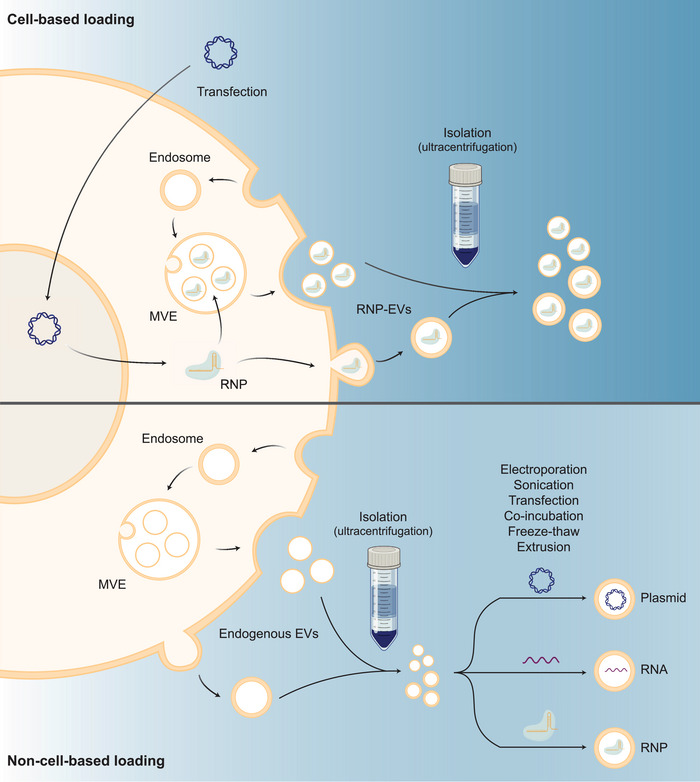
Cell‐based loading of CRISPR/Cas9 RNP (upper) and non‐cell‐based loading of CRISPR/Cas9 plasmid, RNA or RNP post‐EV‐purification (lower). Endogenous EVs are formed through inward budding of the multivesicular endosome (MVE), which are released by merging of the MVE with the cell membrane, or direct outward budding of the cell membrane. Created with BioRender.com.
TABLE 1.
Studies using non‐cell‐based loading strategies.
| Study | EV producing cell | EV validation | EV loading method | Cargo type | Target cells | Target genes | Results | Cell targeting mechanism and in vivo administration |
|---|---|---|---|---|---|---|---|---|
| (Wan et al., 2022) | Hepatic stellate cells (LX‐2) |
DLS TEM Markers: CD63+ TSG101+ GM130‐ |
Electroporation on isolated EVs (20% loading efficiency) |
RNP |
AML‐12 LX‐2 |
PUMA CcnE1 KAT5 |
In vitro gene editing: AML‐12(PUMA): 29.7% indels AML‐12(CcnE1): 25.9‐28.8% indels LX‐2 (KAT5): 20.3 % indels |
– |
| Hepatic cells of APAP‐induced liver injury mice | PUMA |
In vivo gene editing: 26.1 % indels |
Intravenous injection, Tropism |
|||||
| Hepatic cells of CCL4‐induced liver fibrosis mice |
CcnE1 |
In vivo gene editing: 9.7% indels |
||||||
| Hepatic cells of Huh‐7 cell‐induced hepatocellular carcinoma mice | KAT5 |
In vivo gene editing: 21.3% indels |
||||||
| (Kim et al., 2017) | SKOV3 cells (ovarian tumor cell line) |
DLS TEM AFM Markers: CD63+ TSG101+ |
Electroporation on isolated EVs (1.75% loading efficacy) |
Plasmid | SKOV3 cells | PARP‐1 |
In vitro gene editing: 27% indels |
‐ |
| SKOV3 xenograft mice | PARP‐1 |
In vivo gene editing: ‐ Reduced tumor weight and volume ‐ Reduced PARP‐1 protein levels |
Intratumoral or intravenous (tail) injection, Tropism |
|||||
| (Majeau et al., 2022) | Mouse serum |
DLS TEM hs‐FCM Markers: CD9+ |
Lipofectamine CRISPRMAX transfection on isolated EVs |
RNP | Anterior tibia muscle cells in reporter Ai9 mice | Ai9 (2 target sites) |
In vivo gene editing: 8.7% gene deletion |
Intramuscular injection |
| Anterior tibia muscle cells of hDMD/mdx mice with a point mutation in exon 23 of hDMD gene | DMD gene (both intron 22 and 24) |
In vivo gene editing: 13.8% deletion of exon 23+24 |
||||||
| (McAndrews et al., 2021) |
In vitro studies: HEK293T |
NTA FCM Markers: Alix+ CD9+ CD47+ CD63+ CD81+ |
Transfection with Exo‐Fect on isolated EVs | Plasmid | KPC689 cells (murine pancreatic cancer cell line with KRAS mutation) | KrasG12D |
In vitro gene editing: 58% reduced KrasG12D mRNA levels |
– |
| In vivo studies: MSCs | KPC689 cells injected into pancreas of mice |
In vivo gene editing: Statistically insignificantly reduced KrasG12D mRNA expression |
Intravenous and intratumoral injection | |||||
| (Zhuang et al., 2020) | HEK293T |
DLS TEM Markers: CD63+ TSG101+ |
Sonication or repeated freeze‐thaw cycles on isolated EVs (15.3% and 37.6% loading efficiency respectively) | RNP | Human liver organoids (PLOs) from human patient primary liver tumor tissue | WNT10B |
Ex vivo gene editing: TDN‐EVs‐RNP: approx. 28% indels EVs‐RNP: approx. 13% indels |
3D tetrahedral DNA nanostructures (TDNs) targeting cancer cell surface proteins were decorated in the surface of isolated EVs through cholesterol conjugation and a heat‐shock process. Gene editing was close to 5‐fold greater with TDN modified EVs compared to non‐modified EVs. In vivo: Intravenous injection |
| HepG2 xenograft liver tumor in mic |
In vivo gene editing: ‐ Ceased tumor development ‐ Decreased tumor volume Increased ALT enzyme levels with increasing TDN concentration suggesting liver toxicity |
|||||||
| Wang et al. (2021) | Human umbilical cord mesenchymal stem cells (HucMSCs) |
NTA TEM Markers: CD9+ CD63+ LaminA‐ |
Electroporation on isolated EVs | Plasmid | RAW264.7 (macrophages) | CCL2 gene (insertion of sTNFR1 gene by HDR) |
In vitro gene insertion: Increased sTNFR1 protein expression |
CAQK peptides were chemically crosslinked to the EV surface targeting activated immune cells at the SCI site. CAQK modified EVs mainly accumulated at the SCI site whereas nonmodified EVs accumulated mainly in the liver. In vivo: Intravenous injection |
| Activated immune cells of the spinal cord injury (SCI) in mice |
In vivo gene insertion: sTNFR1 levels increased by more than 5‐fold |
|||||||
| (Xu et al., 2020) |
Anti‐CD19‐CAR‐HEK293T HEK293T |
NTA TEM Markers: AnnexinV+ |
Electroporation on isolated EVs | Plasmid |
Raji cells (Burkitt lymphoma cell line, CD19+) Daudi cells |
MYC oncogene |
In vitro gene editing: Raji cells: 5.71% indels (more than 33.8% cells undergo apoptosis) Daudi cells: 3.85% indels |
AntiCD19‐CAR was incorporated into the EV membrane by plasmid transfection of EV‐donor cells. CD19‐specific CAR‐EVs increased CD19+ Raji cells targeting up to 2‐fold. In vivo: Intracardial or intratumoral injection |
| Raji xenograft NOD/SCID mice |
In vivo gene editing: 1.4‐1.8% indels (cells undergo apoptosis) |
|||||||
| (Usman et al., 2018) | Red blood cells (RBC) isolated from group O blood |
NTA TEM Markers: Alix+ TSG101+ Stomatin+ Calnexin‐ |
Electroporation on isolated EVs (18% loading efficiency for Cas9 mRNA) | Plasmid | 293T‐eGFP | EGFP |
In vitro gene editing: 10 % gene knockout |
– |
| Cas9 mRNA and sgRNA | NOMO1‐eGFP cells | EGFP |
In vitro gene editing: NOMO1: 32% of cells had complete loss of eGFP expression |
|||||
| MOLM13 (Leukemia cell line) | mir‐125b‐2 locus |
In vitro gene editing: MOLM13: 98% reduction of miR‐125b expression and 90% reduction of miR‐125a expression |
Density light scatter (DLS), Nanoparticle tracking analysis (NTA), Atomic force microscopy (AFM), Transmission electron microscopy (TEM), High‐sensitivity flow cytometry (hs‐FCM), Flow cytometry (FCM).
TABLE 2.
Studies using cell‐based EV loading strategies.
| Study | EV loading method | Donor cells | Cargo | EV validation | Target cells | Target genes/sequences | CRISPR Results | In vivo cell targeting mechanism |
|---|---|---|---|---|---|---|---|---|
| Passive packaging strategies | ||||||||
| (Luo et al., 2022) | Donor cells transfected with lentiviral particles containing plasmids encoding dCas‐VP64 (catalytically inactive Cas9 fused to a transcriptional activator), sgRNA, Lamp2b‐RBP4 and MS2‐P65. Cargo packaged into EVs by overexpression. | Hepatocyte AML12 cells | RNP (dCas9) |
NTA TEM Markers: CD63+, Lamp2B+ TSG101+ GM130‐ |
Hepatic stellate cells (HSC) |
HNF4α HGF1 FOXA2 |
In vitro gene activation: Conversion of HSCs to hepatocyte‐like cells (HLCs) |
Lamp2b‐RBP4 was incorporated into the EV membrane targeting HSCs In vivo: Intravenous injection (tail) |
| HSCs of CCL4‐induced hepatic fibrosis mouse model |
HNF4α HGF1 FOXA2 |
In vivo gene activation: ‐Conversion of HSC to HLCs ‐Decreased fibrosis ‐Reduced protein levels of profibr‐ ogenic mediators and collagen ‐Recovered liver counts |
||||||
| (Chen et al., 2019) | Donor cells transfected with CRISPR/Cas9 expression vectors targeting HPV (Human Papilloma Virus) or HBV (Hepatitis B Virus). Cargo packaged into EVs by overexpression. | HUH‐7 cells | RNP |
DLS TEM Markers: CD63+ CD81+ |
HUH‐7 cells transfected with HBV expression plasmid | HBV DNA (2 target sites) |
In vitro gene editing: Deletion in selected clones |
– |
| HeLa cells | RNP | HeLa cells transfected with HPV expression plasmid | HPV DNA (2 target sites) |
In vitro gene editing: Deletion in selected clones |
||||
| (He et al., 2020) |
Donor cells transfected with pLenti‐CRISPR‐V2 and pCMV‐VSV‐G. Cas9 packaged into EVs by overexpression. Loading of sgRNA expression plasmids by electroporation of isolated EVs. |
HEK293 cells Liver cancer HepG2 cells |
Cas9 protein + sgRNA plasmid |
DLS TEM Markers: CD40+ |
HepG2 cells | IQGAP1 |
In vitro gene editing: HepG2 EVs: 19.5% indels and 10.57% apoptosis HEK293 EVs: 15.9% indels and 12.07% apoptosis |
– |
| HepG2 xenograft mice | Iqgap1 |
In vivo gene editing: HepG2 EVs: Up to 25 % reduced IQGAP1 protein level HEK293 EVs: Up to 50% reduced IQGAP1 protein level |
Intratumoral injection, Tropism |
|||||
| (Lainšček et al., 2018) |
GEDEX: Donor cells transfected with plasmids encoding Cas9 or dCas9‐VPR (catalytically inactive Cas9 fused to the transcriptional activator VPR) and sgRNA. Cargo packaged into EVs by overexpression (100 ng Cas9 protein each 10 ug EVs). |
HEK293T cells | RNP (Cas9) |
NTA DLS TEM Markers: CD63+ TSG101+ Calnexin‐ |
HEK293 cells | MYD88 |
In vitro gene editing (Cas9): 9.2‐10.1% indels |
– |
| RNP (dCas9) |
HEK293 cells Mouse Neuro2A cells |
ACTC1 Actc1 |
In vitro gene activation (dCas9): HEK293 (ACTC1): 7‐fold increase in mRNA levels Neuro2A (Actc1): 5‐fold increase in mRNA levels. |
– | ||||
| BALB/c mouse model with alpha‐naphthylisothiocyanate (ANIT) induced hepatotoxicity. | Hgf |
In vivo gene activation (dCas9): ‐ 1.5‐fold increased HGF protein levels in liver ‐ Reduced protein levels of biochemical liver damage markers ‐ Reduced liver damage morphology |
Hydrodynamical intravenous injection (tail) | |||||
| (Montagna et al., 2018) |
VEsiCas: Donor cells transfected with plasmids encoding Cas9, pVAX‐T7‐sgRNA (for cytosolic transcription of sgRNA) and VSV‐G. Cargo packaged into EVs by overexpression. |
BSR‐T7/5 cells | RNP | – | HEK293T cells |
CXCR4 VEGFA site 3 |
In vitro gene editing: CXCR4: > 60% indels VEGFA site3: > 30% indels |
– |
| HEK293‐EGFP cells | EGFP (2 target sites) |
In vitro gene editing: 17% EGFP deletion |
– | |||||
| Cardiac muscle cells of EGFP transgenic mice. | EGFP (2 target sites) |
In vivo gene editing: 30% EGFP negative cardiomyocytes compared to 0 % in control mice. |
Intracardiac injection | |||||
| Active packaging strategies | ||||||||
| (Yao et al., 2021) |
ABP Com‐com: Donor cells transfected with plasmids encoding CD63 fused to Aptamer‐Binding Protein (ABP) Com (CD63‐Com), aptamer com‐sgRNA with Cas9 and VSV‐G. The Com‐com interaction increased loading of sgRNA:Cas9 into EVs by 2‐5‐fold. |
HEK293T cells | RNP |
NTA TEM Markers: CD9+ CD63+ RAB5B+ |
HEK293T GFP‐reporter cells | DMD exon 53 |
In vitro genome editing: 51.4% indels |
– |
| Tibialis anterior muscle cells of Del52hDMD/mdx mice | DMD exon 53 |
In vivo genome editing: 0.2 % indels |
Intramuscular injection | |||||
|
(Osteikoetxea et al., 2022) JEV |
CIBN‐CRY2: Donor cells transfected with plasmids encoding CIBN‐MysPalm and Cas9‐CRY2 with sgRNA. Blue light (488 nm) and FAD molecule‐induced CIBN‐CRY2 dimerization increased loading of sgRNA:Cas9 into EVs by up to 10‐fold. |
Expi293F cells | RNP |
NTA TEM Markers: Alix+ TSG101+ Flotilin+ Syntenin+ CD63+ CD81+ Calnexin‐ |
HEK293‐loxP‐GFP‐STOP‐LoxP‐RFP Cre reporter cells. | LoxP (excises GFP) |
In vitro gene editing: 42.0% RFP+ cells |
‐ |
| HepG2‐loxP‐tdTomato‐STOP‐loxP‐GFP Cre reporter cells. | LoxP (excises tdTomato) |
In vitro gene editing: 19.2% GFP+ cells |
||||||
| HEK293T cells | PCSK9 |
In vitro gene editing: 4.4% indels |
||||||
| (Ye et al., 2020) |
GFP‐GFPnanobody: Donor cells transfected with plasmids encoding CD63‐GFP, Cas9‐GFP‐nanobody and sgRNA. GFP and GFP‐nanobody binding increased loading of sgRNA into EVs 2‐fold. |
HEK293T cells | RNP |
NTA TEM Markers: CD63+ |
Lung adeno‐carcinoma cell line A549 with stop‐DsRed genomic sequence | 5′‐ and 3′‐ends of stop sequence (2 targets sites) |
In vitro genome editing: ‐ Deleted stop sequence ‐ Increased DsRed fluorescent cells |
– |
| (Gee et al., 2020) |
FKBP12‐FRB (NanoMEDIC): Donor cells transfected with plasmids encoding FRB‐Cas9, FKBP12‐GagHIV, and sgRNA flanked by self‐cleaving ribozymes and packaging signal that binds Gag. Addition of rapamycin analog facilitates dimerization of FKBP12 and FRB. The viral GAG peptide accumulates in EVs. Addition of rapamycin analog increased Cas9 loading by 2‐fold. |
HEK293T cells | RNP |
NTA TEM Markers: CD63+ CD81+ |
DMD patient‐derived Δex44 iPSC‐derived skeletal muscle cells | DMD exon 45 (both acceptor and donor splice sites) |
In vitro gene editing: 92% exon 45 skipping |
– |
| Gastrocnemius muscle cells in GAG‐Luc2‐hDMD Ex45(stop) KI mice | DMD exon 45 (both acceptor and donor splice sites) |
In vivo gene editing: 6.9% exon 45 skipping |
Intramuscular injection | |||||
| Anterior tibialis muscle cells in NOG‐mdx mouse with nonsense mutation in Ex23 of hDMD gene | DMD exon 23 (both acceptor and donor splice sites) |
In vivo gene editing: 1.1% gene editing leading to 1.6% exon 23 skipping |
||||||
| (Ilahibaks et al., 2023) |
FKBP12‐FRB (TOP‐EV): Donor cells transfected with plasmids encoding sgRNA, VSV‐G, FRB‐Cas9, FKBP12 with N‐myristoylation sequence. Rapamycin induces FKBP12‐FRB dimerization. VSV‐G and rapamycin together increased Cas9 protein packaging 9.5‐fold whereas VSV‐G itself increased packaging 7.8‐fold. |
HEK293FT cells | RNP |
NTA TEM Markers: Alix+ Syntenin+ Calnexin‐ |
Cas9 stoplight reporter cells | Linker region in stoplight cassette (1 and 2 nt indels result in eGFP expression) |
In vitro gene editing: Rapamycin only: 0–1% eGFP signal VSV‐G only: 11.6% eGFP signal VSV‐G + rapamycin: 29.4% eGFP signal |
– |
| (Campbell et al., 2019) |
DmrC‐DmrA (Gesicles): Donor cells transfected with plasmids encoding Cas9‐DmrC, Cherrypicker Red‐DmrA, A/C dimerization molecule, VSV‐G and sgRNA. DmrC‐DmrA dimerization increased loading of Cas9 into the gesicles by 2‐fold. |
HEK293‐FT cells | RNP |
NTA TEM |
HIV‐NanoLuc CHME‐5 microglial cells containing HIV provirus with NanoLuciferase Reporter | HIV LTR (long terminal repeat) 5′ region |
In vitro gene editing: 8% indels |
– |
| Wang, Yu et al. (2018) |
ARMMs: Donor cells transfected with plasmids encoding 4WW‐Cas9‐anti‐GFP‐sgRNA and ARRDC1. The WW domains coupled to Cas9 interacts with the PPXY motifs of ARRDC1 which increased sgRNA loading significantly, compared to Cas9 alone. |
HEK293T | RNP |
NTA TEM Markers: Flotillin + Vinculin ‐ |
GFP positive U2OS cells (1 gene copy) | GFP |
In vitro gene editing: The GFP negative cells increased from 4.9% to 13.4%. |
– |
| Li, Zhou et al. (2019) |
ARE‐HuR: Donor cells transfected with plasmids encoding dCas9‐ARE, CD9‐HuR (Human antigen R) and sgRNA. The addition of the ARE motif increased dCas9 mRNA loading by up to 10‐fold. |
HEK293T |
sgRNA and dCas9 mRNA |
DLS TEM Markers: CD9+ CD63+ TSG101+ Lamp2B+ GM130‐ |
Adipogenic stem cells | CEBPA |
In vitro gene repression (dCas9): More than 2‐fold decreased mRNA levels |
– |
| Liver cells of C56BL/6 mice |
In vivo gene repression (dCas9): Up to 2‐fold decrease in mRNA levels |
Intravenous injection (tail) | ||||||
| Li, Zhang et al. (2023) |
pTA‐CasRx: Donor cells transfected with a plasmid encoding pTA‐Cas13‐HA and sgRNA. The pTA extracellular secretion signal increased Cas13 loading into EVs by 8‐fold. |
HEK293T | RNP (Cas13) |
NTA TEM IEM Markers: CD63 |
LPS (lipopolysaccharide)‐activated bone marrow‐derived macrophages from mice |
Il‐6 Tnf Il‐1β |
In vitro mRNA inactivation (Cas13): Reduced NOS2 (marker for macrophage activation) mRNA levels by 4‐fold |
– |
| Lung tissue of LPS‐induced acute lung injury mice |
In vivo mRNA inactivation (Cas13): Reduced IL‐6, TNF‐α and IL‐1β mRNA levels by 2‐fold |
Four intratracheal injections | ||||||
Density light scatter (DLS), Nanoparticle tracking analysis (NTA), Transmission electron microscopy (TEM), Immunogold electron microscopy (IEM).
5.1. Non‐cell‐based loading methods
Non‐cell‐based loading methods require a pure EV sample (Han et al., 2021) prior to loading, which can be achieved using techniques such as electroporation, sonication, freeze–thaw cycles, and transfection (Table 1). Ultracentrifugation is commonly used for EV isolation, but it is time‐consuming and challenging to obtain a substantial yield of pure EVs, and there is currently no standardized approach for this (Meng et al., 2020). Electroporation is primarily used for loading EVs with plasmid DNA (Kim et al., 2017; Wang et al., 2022; Xu et al., 2020). Although very few studies have used electroporation for Cas9 protein (Wan et al., 2022) or mRNA (Usman et al., 2018) loading, this method can result in partial membrane disruption and cargo aggregation, potentially compromising EV integrity and gene editing outcomes. This issue is especially concerning for RNPs, as their tertiary structure must be maintained to function properly, a requirement not applicable to plasmid DNA (Man et al., 2020). Conversely, using plasmid DNA as EV cargo carries the risk of integration into the genome, resulting in potential gene disruption and off‐target effects due to sustained CRISPR/Cas9 expression. Therefore, RNPs and (m)RNA, which are only transiently present in target cells, are preferred cargo options. For gene regulation by dCas9, higher quantities and repeated administration may be necessary to achieve the desired effect.
Usman et al performed direct loading of Cas9 mRNA and sgRNA into isolated EVs through electroporation and observed successful gene editing upon addition of these EVs to isolated leukaemia cells (Usman et al., 2018). Loading RNA into EVs via electroporation has primarily been applied to small RNAs like miRNA and siRNA, and as far as we are aware, only a handful of studies have effectively achieved such loading (Aslan et al., 2021; Wang et al., 2018). Therefore, further investigation into efficient direct mRNA loading of EVs is required.
As an alternative method for EV packaging, Zhuang et al. found that repeated freeze‐thaw cycles are more effective than sonication for loading RNPs into EVs, facilitating subsequent ex vivo gene editing in recipient cells (Zhuang et al., 2020). To date, two studies have reported the successful transfer of Cas9 RNPs to EVs via electroporation and freeze‐thaw cycles, respectively, which was previously very difficult to achieve (Wan et al., 2022; Zhuang et al., 2020). Both Lipofectamine and Exo‐Fect can be used for efficient plasmid loading of EVs, resulting in gene editing in recipient cells (Majeau et al., 2022; McAndrews et al., 2021). However, the lipid composition of the transfection agent may integrate into the EV membrane, forming an EV‐lipid hybrid. The cytotoxic and immunogenic effects of this phenomenon should be considered for in vivo administration (Majeau et al., 2022).
The successful non‐cell‐based loading of CRISPR/Cas9 into EVs is evident in relatively high in vitro gene editing outcomes across various cell lines, targeting different genes, with reported editing efficiencies reaching up to 30%. However, the therapeutic effect in vivo in mice varies not only due to various target tissues and delivery strategies. For instance, Wan et al. observed in vivo indel frequencies of up to 26.1% when targeting hepatic cells through intravenous injections (Wan et al., 2022). In contrast, Xu et al. observed a maximum of 1.8% editing efficiency in lymphoma cells through intracardial and intratumoral injections (Xu et al., 2020). The latter result was presumably lower due to apoptosis and hence negative selection of the edited cells. Majeau et al. achieved a 13.4% exon deletion in muscle cells through intramuscular injections (Majeau et al., 2022). Given that indel formation via NHEJ can lead to knockout of genes and insertions through HDR can result in genomic integration of genes, CRISPR efficiency can also be measured by assessing the gene product. Downstream effects and altered phenotypes due to gene editing were observed in cells in vitro or in vivo in the form of reduced or increased mRNA and protein levels or GFP fluorescence signal (Kim et al., 2017; McAndrews et al., 2021; Wang et al., 2022; Zhuang et al., 2020).
The non‐cell‐based loading methods can also be applied to EVs isolated from body fluids like blood plasma (Majeau et al., 2022; Usman et al., 2018), making large‐scale production more feasible. Furthermore, the EVs produced by red blood cells, which lack a nucleus, are devoid of genetic material, preventing the risk of horizontal gene transfer (Horodecka & Düchler, 2021; Usman et al., 2018).
5.2. Cell‐based loading methods—Passive strategies
The cell‐based loading methods rely on transfecting donor cells with plasmid DNA encoding the CRISPR/Cas9 system. This results in the production of EVs containing both Cas9 protein and sgRNA (Han et al., 2021). Various approaches have been developed for enriching EVs with Cas9 RNPs (summarized in Table 2). The straightforward approach of overexpressing Cas9 protein and sgRNA in donor cells leads to the packaging of the RNP into released EVs through a concentration‐dependent sorting mechanism (Osteikoetxea et al., 2022). This efficient passive method has been extensively applied (Chen et al., 2019; He et al., 2020; Lainšček et al., 2018; Montagna et al., 2018; Yao et al., 2021).
By passively loading CRISPR/Cas9 into EVs, in vitro gene editing efficiencies ranging from 9.2% to 60.0% have been achieved in recipient cells, along with downstream in vivo effects. It is worth noting that the target cell type and target gene may influence the CRISPR efficiencies. Moreover, Luo et al. and Lainšček et al. exploited intravenously injected EVs delivering deactivated Cas9 (dCas9) for the successful in vivo activation of genes in mouse models (Lainšček et al., 2018; Luo et al., 2022). He et al. investigated passive loading of Cas9 protein and subsequent direct loading of sgRNA encoding plasmids by electroporation after EV purification. This approach resulted in promising indel frequencies of 15.9% to 19.5% in HepG2 cells in vitro (He et al., 2020). Another highly effective passive loading strategy exploited by Montagna et al. takes advantage of the T7 RNA polymerase for transcription of the sgRNA in the cytoplasm, achieving at least 30% and 60% gene editing efficiencies in HEK293T cells in vitro while targeting VEGFA and CXCR4, respectively (Montagna et al., 2018). The high gene editing efficiencies obtained by bypassing the transcription of sgRNA in the nucleus of donor cells (He et al., 2020; Montagna et al., 2018) might indicate that sgRNA availability is a limiting factor for passive packaging of Cas9‐sgRNA RNPs. In addition, both studies incorporated vesicular stomatitis virus glycoprotein (VSV‐G) into the membrane. VSV‐G, a viral surface protein, plays a vital role in the endosomal escape of membrane‐encapsulated virus particles (Montagna et al., 2018). The overexpression of VSV‐G increases protein packaging into EVs and improves the protein delivery efficiency1, which likely contributed to the high gene editing frequencies observed (He et al., 2020; Montagna et al., 2018). Notably, both studies report significant downregulation of gene products because of gene knockout upon locally administered CRISPR‐EVs in vivo (He et al., 2020; Montagna et al., 2018). A key challenge for passive packaging strategies is to obtain a high quantity of RNPs within the EVs.
5.3. Cell‐based loading methods‐active strategies
Another cell‐based loading strategy leverages active packaging to increase EV loading. This is achieved by fusing sgRNA with a protein‐binding RNA motif or fusing the Cas9 protein with a protein, peptide or lipid that is specific to the EV membrane or naturally accumulates in EVs through a dimerization system (Campbell et al., 2019; Gee et al., 2020; Li et al., 2019; Osteikoetxea et al., 2022; Yao et al., 2021; Ye et al., 2020). Donor cells are transfected with plasmids encoding Cas9 and sgRNA, with either component fused to a specific protein domain or RNA sequence, respectively. The dimerization partner is then fused to an EV‐specific protein (as illustrated in Figure 5).
FIGURE 5.
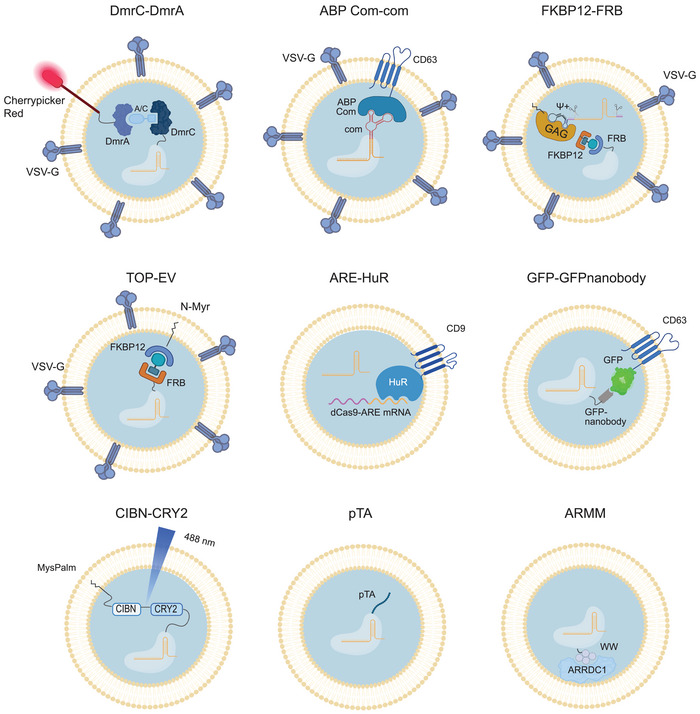
Cell‐based active loading of CRISPR/Cas9 into EVs by nine different protein‐protein or RNA‐RNA dimerization strategies. Further details on the dimerization systems incl. references in Table 2. Created with BioRender.com.
Osteikoetxea et al. used the CIBN‐CRY2 heterodimerization system to enhance active Cas9 packaging into EVs. This is achieved by fusing CIBN to a short amino sequence that ensures lipid modification with the EV‐specific lipid anchor MysPalm in cells, and fusing Cas9 to CRY2 (Osteikoetxea et al., 2022). The dimerization increased Cas9 loading by ten‐fold and resulted in gene editing efficiencies ranging from 4.4% to 42.0% in vitro across various cell lines targeting different genes. Importantly, the heterodimerization is light‐induced and hence reversible, which facilitates the release of Cas9 RNP in recipient cells.
CD63‐mediated loading of CRISPR/Cas9 is another active loading strategy, exploited by several studies (Osteikoetxea et al., 2022; Yao et al., 2021; Ye et al., 2020). By fusing the aptamer binding protein (ABP) Com with CD63 and combining the aptamer Com system with an sgRNA, Yao et al. observed a two‐ to five‐fold increase in Cas9 protein loading and enhanced gene editing, although it only yielded an indel frequency of 0.2% in vivo in muscle cells of Del52hDMD/mdx mice targeted through intramuscular injections (Yao et al., 2021). Coupling GFP to CD63 and a GFP‐specific nanobody to Cas9, Ye et al. achieved a two‐fold increase in RNP loading and in vitro target gene deletion (Ye et al., 2020).
Despite the increased loading of Cas9 RNP achieved by active packaging, the dimerization interaction between sgRNA or Cas9 and components of the EV membrane can impede endosomal escape in target cells. This might explain the low levels of gene editing efficiencies obtained both in vitro and in vivo despite employing active packaging methods (Wang et al., 2018; Yao et al., 2021; Ye et al., 2020). This could potentially provide an explanation to the observation by Yao et al., who observed that CD63‐mediated loading of sgRNA required co‐expression of VSV‐G to achieve a therapeutic effect (Yao et al., 2021). Additionally, Osteikoetxea et al. only observed a significant therapeutic effect when fusing Cas9 to MysPalm and not CD63 (Osteikoetxea et al., 2022). This suggests that active loading, mediated by Cas9 or sgRNA interaction with or direct fusion to the CD63 tetraspanin, could potentially hinder their release from endosomes.
Another packaging strategy utilizes arrestin domain containing protein 1 (ARRDC1)‐mediated microvesicles (ARMMs). ARRDC1 recruits ESCRT to the cell membrane, initiating budding and thereby increasing microvesicle production. Wang and colleagues discovered that the interaction between ARRDC1 and 4 WW domains coupled with Cas9 significantly increased the loading of anti‐GFP sgRNA into EVs resulting in a reduced number of GFP‐positive cells (GFP knockout) in U2OS cell cultures in vitro (Wang et al., 2018).
The Gesicle system is a commercialized CRISPR/Cas9 delivery method that utilizes the A/C‐induced heterodimerization of DmrA and DmrC domains, linked to the transmembrane Cherrypicker red protein and Cas9, respectively. The advantage of this system is that the dimerization is reversible due to the dilution of the A/C molecule in recipient cells and VSV‐G is further applied to enhance Gesicle formation. Campbell et al. successfully increased Cas9 loading by two‐fold using the Gesicle production system and obtained an 8% indel frequency in the target HIV LTRs in vitro (Campbell et al., 2019).
Nanoblades are a novel technology designed to efficiently deliver a genomic cleaving agent, such as the Cas9 protein complexed with guide RNAs, into specific cells, particularly human T cells, B cells, and hematopoietic stem and progenitor cells (HSPCs). We have previously demonstrated the effectiveness of these nanoblades, which are essentially modified virus‐like particles (VLPs), derived from murine leukemia virus (MLV) or HIV, where the viral structural protein Gag has been fused to Cas9 (Gutierrez‐Guerrero et al., 2021). This fusion facilitates their targeted entry into cells, enabling precise gene editing purposes with high efficiency and minimal toxicity.
For RNA‐RNA interaction‐mediated loading of mRNA, Li et al. exploited the RNA binding protein HuR (Human antigen R) fused to CD9 and dCas9 mRNA with ARE signals, which increased dCas9 mRNA loading up to 10‐fold (Li et al., 2019). Intravenously injected dCas9‐loaded EVs into mice resulted in a 2‐fold reduction of target gene expression in recipient cells of the liver.
Gee et al. explored active loading of both Cas9 and sgRNA (Gee et al., 2020). Here, the FKBP12‐FRB dimerization system, including a rapamycin analogue, was coupled to the viral EV membrane‐anchoring protein GagHIV and Cas9, respectively, and sgRNA was flanked by self‐cleaving ribozymes and coupled to an HIV Ψ packaging signal with affinity for GagHIV. This induced high levels of in vitro gene editing targeting DMD slicing acceptor and donor sites, leading to 92% exon skipping, and 1.1% in vivo gene editing, leading to 1.6% exon skipping in NOG‐mdx mice using intramuscular injections for targeting (Gee et al., 2020). The combined active packaging strategy significantly increased the gene editing compared to sgRNA active loading alone. Nevertheless, active enrichment of EVs with either sgRNA or Cas9, which is the most used strategy, also ensures effective packaging of the Cas9:sgRNA complex due to the intrinsic affinity between sgRNA and Cas9 (Osteikoetxea et al., 2022; Yao et al., 2021; Ye et al., 2020; ).
Other strategies for the FKBP12‐FRB dimerization system include linking FKBP12 to the EV membrane through an N‐myristoylation sequence (Ilahibaks et al., 2023). Ilahibaks and colleagues found that VSV‐G overexpression in donor cells was necessary to achieve in vitro gene editing in recipient cells, and that the addition of rapamycin for FKBP12‐FRB dimerization increased the gene editing efficiency from 11.6% to 29.4% 66. Importantly, the reversible dimerization of FKBP12 and FRB by rapamycin or analogs allows for endosomal escape due to the dilution of rapamycin in recipient cells (Gee et al., 2020; Ilahibaks et al., 2023).
A recent strategy involves the use of the pTA extracellular secretion signal, which is not linked to EV packaging but concentrates the protein on the extracellular membrane. Li and colleagues found that tagging Cas13 (an RNA editing CRISPR system) with pTA increased its loading into EVs by eightfold (Li et al., 2023). Upon intratracheal injection of CRISPR‐EVs into an LPS‐induced inflammatory lung tissue mouse model, Cas13 was delivered to macrophages and lung tissues, where it reduced mRNA levels of inflammatory target genes. This approach shows promise for the efficient delivery of RNA editing systems using EVs.
In summary, active loading strategies that rely on interaction with EV membrane lipid‐interacting molecules, such as the lipid Myristoylation anchor or the viral GAGHIV protein, as well as incorporation of VSV‐G, hold promise for efficient endosomal escape and selective packaging of RNPs into EVs (Gee et al., 2020; Osteikoetxea et al., 2022; Yao et al., 2021). Additionally, reversible dimerization systems allow for Cas9 dissociation in recipient cells (Campbell et al., 2019; Osteikoetxea et al., 2022). These active loading strategies selectively package RNPs into EVs. However, Cas9 mRNA may become co‐packaged into the EVs (Luo et al., 2022; Yao et al., 2021; Ye et al., 2020) or be selectively packaged (Li et al., 2019). Compared to delivery of plasmid DNA for CRISPR/Cas9 editing, the RNP delivery strategies, including active loading, result in far less off‐target editing (Campbell et al., 2019; Gee et al., 2020; Yao et al., 2021; ). This difference might suggest limited packaging of plasmid DNA in EVs in donor cells, thereby minimizing the risk of DNA integration in recipient cells. Ilahibaks and colleagues investigated the risk of plasmid DNA delivery via EVs from donor cells and found that EVs were not contaminated with expression plasmids, which were overexpressed in donor cells (Ilahibaks et al., 2023). Moreover, several of the passive loading strategies were also found to avoid co‐packaging of plasmid DNA (Chen et al., 2019; Lainšček et al., 2018; Montagna et al., 2018). However, it should be noted that multiple studies using EVs for drug delivery have reported that donor cells can co‐package plasmids into EVs (Han et al., 2021). Therefore, to create a plasmid DNA‐free system, the transfer of plasmid DNA via EVs should always be thoroughly tested.
While cell‐based loading methods are scalable and allow for engineering of EVs with specific proteins, they require a high number of genetically modified, cultured cells for therapeutic use (Man et al., 2020; Ng et al., 2022). A combination of 3D culture and bioreactor might offer a potential solution to mitigate some of these challenges.
5.4. HYBRID EVs
A novel approach to loading CRISPR/Cas9 into EVs involves fusing EVs with liposomes, polymers, or other synthetic nanoparticles to create hybrids. Hybrid EVs are capable of accommodating significantly larger molecules in higher quantities than EVs alone. These hybrids also exhibit improved cellular uptake while maintaining the surface properties of EVs (Busatto et al., 2021; Liang et al., 2022; Rodríguez & Vader, 2022). To form hybrid EVs, multiple methods have been reported. The most applied strategies include extrusion, co‐incubation, freeze‐thaw, and electroporation (Li et al., 2023; Ou et al., 2021).
The EV‐liposome hybrid system was tested by Liang et al., who used co‐incubation to fuse chondrocyte‐targeting EVs (CAP‐EVs) with liposomes loaded with plasmids encoding Cas9 and sgRNA (Liang et al., 2022). The in vitro knockout efficiency of the hybrids was significantly higher compared to EVs or liposomes alone. The hybrids caused minimal cytotoxicity when compared to liposomes, whereas EVs alone did not induce cytotoxicity. Lin and colleagues found that EV‐liposome hybrids generated by co‐incubation can deliver CRISPR/Cas9 expression plasmids to mesenchymal stem cells, unlike EVs and liposomes alone, although with a similar cytotoxic effect as liposomes (Lin et al., 2018). Thus, hybrids exhibit promising packaging abilities and can be cost‐effectively created on a large scale from EVs isolated from body fluids. However, the cytotoxicity associated with currently available hybrids needs to be addressed for clinical therapeutics (Busatto et al., 2021).
In addition to therapeutics, the potential applications of the EV‐liposome hybrid system are enormous. For instance, they can be applied in rapid and sensitive diagnosis. Ning and colleagues reported a strategy for the detection of SARS‐CoV‐2 RNA in plasma (Ning et al., 2021). The assay is developed by fusing plasma SARS‐CoV‐2 RNA‐containing EVs with RT‐RPA‐CRISPR‐loaded liposomes, which triggers RT‐RPA‐targeted amplification of the SARS‐CoV‐2 RNA and CRISPR (Cas12a)‐mediated cleavage of a SARS‐CoV‐2 RNA targeting quenched fluorescent probe. The presence and concentration of SARS‐CoV‐2 RNA are quantified in proportion to the fluorescent signal. These collective findings demonstrated the great potential and broad applications of CRISPR delivery by EVs.
6. TARGETING EVs TO SPECIFIC CELLS AND TISSUES
To achieve cell‐specific gene editing and avoid off‐target events in EV‐based CRISPR therapeutic applications, it is crucial that EVs are exclusively taken up by the desired target cells. Targeting mechanisms of EVs can prevent accumulation in the liver, spleen, and lungs after intravenous or intraperitoneal administration (Majeau et al., 2022). Li, Zhang and colleagues found that EVs accumulated in the liver and spleen (60%) and to some extent in the lungs, kidneys and intestinal tract (10%) for 24 h upon intraperitoneal injection of a single dose of EVs (1.0 × 1012) in mice (Li et al., 2023).
It is worth noting that EVs tend to target the cells from which they are derived due to specific surface protein interactions, that is, tropism (He et al., 2020; Kim et al., 2017; Wan et al., 2022). Kim et al. found that EVs purified from cancer cells tend to accumulate in the tumour after systemic administration (Kim et al., 2017). However, cancer‐derived EVs should be used with caution since they can contain different molecules that can enhance tumour growth and lead to metastasis (Majeau et al., 2022). He et al. found that cancer‐derived EVs tend to accumulate in the tumour to a higher degree than HEK293T‐derived EVs, however resulting in a limited reduction of tumour size and gene editing, presumably due to their tumour‐stimulating content (He et al., 2020). Liver targeting tropism has proven to be efficient (Li et al., 2019; Wan et al., 2022), which is no surprise since EVs naturally tend to accumulate in the liver and spleen, and this may not apply to other organs or tissues.
Alternatively, EVs can be modified with specific surface ligands by transfecting donor cells with expression plasmids. Surface ligands enable EVs to enter target cells through receptor binding, such as coating with anti‐CD19‐CAR to target CD19+ cells (Xu et al., 2020), fusion of the EV surface LAMP2B protein with RBP4 or chondrocyte‐affinity peptide (CAP) to target HSC (Luo et al., 2022) and chondrocytes (Liang et al., 2022), respectively. Another targeting mechanism is to apply surface modification post EV isolation. Zhuang et al. fused tetrahedral DNA nanostructures (TDNs) with cancer protein‐specific DNA aptamers and cholesterols for anchoring to the EV membrane using a modified heat‐shock process, which resulted in reduced tumour size due to gene editing, however increased liver toxicity (Zhuang et al., 2020). Wang et al. chemically crosslinked a CAQK peptide to the EV membrane which increased accumulation at the spinal cord injury (SCI) site (Wang et al., 2022).
Targeted administration of EVs, such as intramuscularly for muscle targeting (Gee et al., 2020; Majeau et al., 2022; Yao et al., 2021), or intratumorally for tumour targeting (He et al., 2020; McAndrews et al., 2021), provides more specific alternatives to intravenous administration, albeit limited to selected parts of the body. Both Kim et al. and Xu et al. have found that intratumoral injections are superior for targeting tumours compared to intravenous and intracardial injections, respectively (Kim et al., 2017; Xu et al., 2020).
In summary, the targeting of EVs to specific cells and tissues is crucial for effective CRISPR/Cas9 delivery. Tropism and surface modification with ligands or aptamers are promising strategies for achieving this goal. Targeted administration methods offer more specific alternatives to systemic delivery. However, it is essential to carefully assess the potential cytotoxicity of modified EVs and the potential risks associated with using cancer‐derived EVs.
7. APPLICATIONS OF EVS AS A CRISPR/CAS9 DELIVERY TOOL IN THERAPEUTICS
The strategies developed so far for delivering CRISPR/Cas9 vary greatly in terms of gene editing effects and cell targeting specificity. In addition to differences in EV loading and targeting methods, injection routes, the type of donor cells, EV particle number, selection of target genes or reporter systems, gRNA design, gene editing analysis methods, cargo type, and selected target cells also vary among the strategies applied, making direct comparisons difficult. It is important to note that the variation in gene editing efficiencies could be attributed to either the CRISPR/Cas9 system or the EV delivery method.
Despite these challenges, the current strategies suggest that CRISPR/Cas9‐carrying EVs are safe, non‐immunogenic, and effective for gene editing in mice, showing great promise for their use in humans. Both passive and active cell‐based loading studies have reported little to no plasmid DNA present in the EVs, as well as minimal off‐target editing, indicating a low risk of genotoxicity associated with EV‐mediated delivery of CRISPR/Cas9 RNA or RNP cargo. In comparison to the delivery of CRISPR/Cas9 using viral vectors, RNP‐ or RNA‐loaded EVs are expected to hold a therapeutic advantage in ensuring minimal genotoxicity and furthermore, a minimal immunogenic reaction. Nevertheless, preclinical studies on EVs as a CRISPR delivery vehicle are required to shed light on this, as viral vectors have been the preferred delivery system so far due to their intrinsic gene delivery efficacies (Henderson, 2023).
A key challenge moving forward is to develop loading methods that are non‐destructive, effective, and scalable to obtain a high yield of cargo‐loaded EVs. Standardized comparisons between loading methods will enable a thorough evaluation of genotoxic and cytotoxic collateral effects, which is necessary for the safe, efficient, and specific application of CRISPR/Cas9‐loaded EVs in curing human diseases.
Large quantities of EVs can be obtained through purification from body fluids or culture medium, but the subsequent loading of EVs may compromise their intrinsic characteristics. Enhancing cargo loading by modifying EVs or fusing them with liposomes to overcome packaging and targeting challenges could potentially alter the natural, non‐immunogenic and nontoxic advantages of EVs. For example, while VSV‐G has shown promise for efficient EV loading and release from donor cells (Ilahibaks et al., 2023), its status as a viral protein in the EV membrane raises concerns about the safety of therapeutic applications. Although modifying EVs using viral proteins has demonstrated utility in increasing EV formation and gene editing (Yao et al., 2021; Gee et al., 2020), the exogenous nature of these viral proteins necessitates comprehensive testing for toxicity and immune system activation prior to clinical applications is required.
Newer approaches using biomimetic vesicles or exosome‐like nanocarriers, which are synthetic vesicles designed to mimic EV properties, offer easier mass production for therapeutic use (Duan et al., 2021; Jiang et al., 2022; Tenchov et al., 2021). However, but the available data on these approaches is limited. An alternative to EVs is synthetic LNPs, which can efficiently package CRISPR components and are easy to produce in large quantities circumventing the need for producer cells (Duan et al., 2021; Wilbie et al., 2019). In contrast to EVs, LNPs have been investigated in a preclinical study that exhibited promising results for delivering CRISPR RNA to liver cells in patients suffering from transthyretin amyloidosis (Gillmore et al., 2021; Henderson, 2023). Importantly, only mild adverse events were observed, in line with the biocompatibility of LNPs. LNPs have shown great promise for targeting the liver, where they accumulate due to opsonization by apolipoprotein E in the blood (Wilbie et al., 2019). However, this characteristic also limits their use for targeting other organs as they are rapidly degraded in the liver. EVs, on the other hand, naturally target the cell type from which they are derived (tropism) and thus hold an advantage over LNPs. Moreover, EVs not only serve as a vehicle but may also provide additional therapeutic benefits. For instance, MSC‐derived EVs have been shown to mitigate inflammation and stimulate regeneration at injury sites partly due to their paracrine activity (Mendt et al., 2019; Wang et al., 2022). However, EVs can also naturally carry non‐beneficial cargo from donor cells, such as those from cancer cells, which is challenging to control in comparison to synthetically engineered LNPs.
8. CONCLUSIONS
Altogether, EVs hold great potential as a safe, efficient, and specific delivery tool for CRISPR/Cas9‐mediated gene editing therapies. However, further improvements are needed to optimize gene editing efficiency and cell specificity. Currently, two main approaches exist for EV loading: direct loading of purified EVs and cell‐based loading, either passively through CRISPR/Cas9 overexpression or actively through protein or RNA interactions. Challenges for both methods include high‐scale purification, effective cargo loading, gene editing efficiency, selective cell targeting, and a clear overview of the collateral effects. Direct loading may lead to cargo aggregation, while cell‐based methods face challenges such as low yield of EVs and low endosomal escape. Solutions like viral proteins and EV hybrids are being developed to overcome these challenges.
The optimal CRISPR vehicle for therapeutics might involve combining natural systems like EVs with viral or synthetic nanoparticles. Furthermore, EVs can be targeted using tropism, surface modifications, or direct injection. But additional research is necessary to explore their potential applications in treating various diseases. Given that the CRISPR technology has demonstrated great promise in curing and treating multiple genetic and infectious diseases, the development of safe, efficient, and specific delivery tools is essential for its successful use in therapeutics.
AUTHOR CONTRIBUTIONS
Anne Højberg Berggreen: Conceptualization; data curation; investigation; visualization; writing—original draft; writing—review and editing. Julie Lund Petersen: Conceptualization; data curation; formal analysis; investigation; supervision; visualization; writing—original draft; writing—review and editing. Lin Lin: Investigation; supervision; validation; writing—review and editing. Karim Benabdel: Investigation; supervision; writing—review and editing. Yonglun Luo: Conceptualization; funding acquisition; investigation; project administration; resources; supervision; visualization; writing—review and editing
CONFLICT OF INTEREST STATEMENT
The authors have no conflicts of interest to disclose.
ACKNOWLEDGMENTS
This project is partially supported by the M‐ERA.Net3 and the Innovation Fund Denmark (9355 PIECRISCI), FEDER/Junta de Andalucia‐Consejería de Transformación Económica, Industria, Conocimiento y Universidades/Project (ProyExcel_00875), and Consejeria de Salud y Familia (CSyF) (PECART‐O027‐2020)) and based on work from COST Action Gene Editing for the treatment of Human Diseases, CA21113, supported by COST (European Cooperation in Science and Technology). K.B. held Nicolas Monardes contracts from CSyF.
Berggreen, A. H. , Petersen, J. L. , Lin, L. , Benabdellah, K. , & Luo, Y. (2023). CRISPR delivery with extracellular vesicles: Promises and challenges. Journal of Extracellular Biology, 2, e111. 10.1002/jex2.111
Anne Højberg Berggreen and Julie Lund Petersen contributed equally to this work.
REFERENCES
- Aheget, H. , Mazini, L. , Martin, F. , Belqat, B. , Marchal, J. A. , & Benabdellah, K. (2020). Exosomes: Their role in pathogenesis, diagnosis and treatment of diseases. Cancers (Basel), 13, 10.3390/cancers13010084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi, S. E. , Soleymani, M. , Shahriyary, F. , Amirzargar, M. R. , Ofoghi, M. , Fattahi, M. D. , & Safa, M. (2023). Viral vectors and extracellular vesicles: Innate delivery systems utilized in CRISPR/Cas‐mediated cancer therapy. Cancer Gene Therapy, 30, 936–954. 10.1038/s41417-023-00597-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan, C. , Kiaie, S. H. , Zolbanin, N. M. , Lotfinejad, P. , Ramezani, R. , Kashanchi, F. , & Jafari, R. (2021). Exosomes for mRNA delivery: A novel biotherapeutic strategy with hurdles and hope. BMC Biotechnology, 21, 10.1186/s12896-021-00683-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beetler, D. J. , Di Florio, D. N. , Bruno, K. A. , Ikezu, T. , March, K. L. , Cooper, L. T. , Wolfram, J. Jr , & Fairweather, D. (2022). Extracellular vesicles as personalized medicine. Molecular Aspects Medicine, 101155, 10.1016/j.mam.2022.101155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr, M. , Zhou, J. , Xu, B. , & Zhang, H. (2021). In vivo delivery of CRISPR‐Cas9 therapeutics: Progress and challenges. Acta Pharm Sin B, 11, 2150–2171. 10.1016/j.apsb.2021.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigica, A. (2022). Cure Rare Disease Announces Patient Death in CRISPR Gene Therapy Trial of Duchenne, https://www.neurologylive.com/view/cure‐rare‐disease‐announces‐patient‐death‐crispr‐gene‐therapy‐trial‐of‐duchenne
- Bolotin, A. , Quinquis, B. , Sorokin, A. , & Ehrlich, S. D. (2005). Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Reading), 151, 2551–2561. 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- Bryson, S. (2022). Pfizer Reports Death of Participant in DMD Gene Therapy Trial, https://musculardystrophynews.com/news/pfizer‐reports‐death‐duchenne‐md‐gene‐therapy‐trial‐participant/
- Busatto, S. , Iannotta, D. , Walker, S. A. , Di Marzio, L. , & Wolfram, J. (2021). A Simple and Quick Method for Loading Proteins in Extracellular Vesicles. Pharmaceuticals (Basel), 14, 10.3390/ph14040356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, L. A. , Coke, L. M. , Richie, C. T. , Fortuno, L. V. , Park, A. Y. , & Harvey, B. K. (2019). Gesicle‐mediated delivery of CRISPR/Cas9 ribonucleoprotein complex for inactivating the HIV provirus. Molecular Therapy, 27, 151–163. 10.1016/j.ymthe.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Zhu, D. , Liu, X. , & Peng, L. (2022). Amphiphilic dendrimer vectors for RNA delivery: State‐of‐the‐art and future perspective. Accounts of Materials Research, 3, 484–497. 10.1021/accountsmr.1c00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R. , Huang, H. , Liu, H. , Xi, J. , Ning, J. , Zeng, W. , Shen, C. , Zhang, T. , Yu, G. , Xu, Q. , Chen, X. , Wang, J. , & Lu, F. (2019). Friend or foe? Evidence indicates endogenous exosomes can deliver functional gRNA and Cas9 protein. Small, 15, e1902686. 10.1002/smll.201902686 [DOI] [PubMed] [Google Scholar]
- Cheng, H. , Zhang, F. , & Ding, Y. (2021). CRISPR/Cas9 delivery system engineering for genome editing in therapeutic applications. Pharmaceutics, 13, 10.3390/pharmaceutics13101649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, I. M. , Rajakumar, G. , Venkidasamy, B. , Subramanian, U. , & Thiruvengadam, M. (2020). Exosomes: Current use and future applications. Clinical Chimica Acta, 500, 226–232. 10.1016/j.cca.2019.10.022 [DOI] [PubMed] [Google Scholar]
- Cong, L. , Ran, F. A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. , Hsu, P. D. , Wu, X. , Jiang, W. , Marraffini, L. A. , & Zhang, F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi, G. I. , Qu, K. , Alkan, F. , Pan, X. , Luo, Y. , & Gorodkin, J. (2022). CRISPR/Cas9 gRNA activity depends on free energy changes and on the target PAM context. Nature Communications, 13, 3006. 10.1038/s41467-022-30515-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci, S. , Essawi, K. , Germino‐Watnick, P. , Liu, X. , Hakami, W. , & Tisdale, J. F. (2022). Advances in CRISPR delivery methods: Perspectives and challenges. Crispr Journal, 5, 660–676. 10.1089/crispr.2022.0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, R. , Wang, C. , Zhu, L. , & Yang, Y. (2022). Extracellular vesicles as delivery vehicles for therapeutic nucleic acids in cancer gene therapy: Progress and challenges. Pharmaceutics, 14, 10.3390/pharmaceutics14102236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, L. , Ouyang, K. , Wang, J. , Xu, L. , Xu, X. , Wen, C. , Xie, Y. , Liang, Y. , & Xia, J. (2021). Exosomes as targeted delivery platform of CRISPR/Cas9 for therapeutic genome editing. Chembiochem, 22, 3360–3368. 10.1002/cbic.202100359 [DOI] [PubMed] [Google Scholar]
- Duan, L. , Ouyang, K. , Xu, X. , Xu, L. , Wen, C. , Zhou, X. , Qin, Z. , Xu, Z. , Sun, W. , & Liang, Y. (2021). Nanoparticle delivery of CRISPR/Cas9 for genome editing. Frontiers in Genetics, 12, 673286. 10.3389/fgene.2021.673286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt, K. M. , Mali, P. , Braff, J. L. , Moosburner, M. , Yaung, S. J. , & Church, G. M. (2013). Orthogonal Cas9 proteins for RNA‐guided gene regulation and editing. Nature Methods, 10, 1116–1121. 10.1038/nmeth.2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee, P. , Lung, M. S. Y. , Okuzaki, Y. , Sasakawa, N. , Iguchi, T. , Makita, Y. , Hozumi, H. , Miura, Y. , Yang, L. F. , Iwasaki, M. , Wang, X. H. , Waller, M. A. , Shirai, N. , Abe, Y. O. , Fujita, Y. , Watanabe, K. , Kagita, A. , Iwabuchi, K. A. , Yasuda, M. , … Xu, H. (2020). Extracellular nanovesicles for packaging of CRISPR‐Cas9 protein and sgRNA to induce therapeutic exon skipping. Nature Communications, 11, 1334. 10.1038/s41467-020-14957-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmore, J. D. , Gane, E. , Taubel, J. , Kao, J. , Fontana, M. , Maitland, M. L. , Seitzer, J. , O'Connell, D. , Walsh, K. R. , Wood, K. , Phillips, J. , Xu, Y. , Amaral, A. , Boyd, A. P. , Cehelsky, J. E. , McKee, M. D. , Schiermeier, A. , Harari, O. , Murphy, A. , … Kyratsous, C. A. (2021). CRISPR‐Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. New England Journal of Medicine, 385, 493–502. 10.1056/nejmoa2107454 [DOI] [PubMed] [Google Scholar]
- Gutierrez‐Guerrero, A. , Abrey Recalde, M. J. , Mangeot, P. E. , Costa, C. , Bernadin, O. , Périan, S. , Fusil, F. , Froment, G. , Martinez‐Turtos, A. , Krug, A. , Martin, F. , Benabdellah, K. , Ricci, E. P. , Giovannozzi, S. , Gijsbers, R. , Ayuso, E. , Cosset, F. L. , & Verhoeyen, E. (2021). Baboon Envelope Pseudotyped “Nanoblades” Carrying Cas9/gRNA Complexes Allow Efficient Genome Editing in Human T, B, and CD34(+) Cells and Knock‐in of AAV6‐Encoded Donor DNA in CD34(+) Cells. Frontiers in Genome Ed, 3, 604371. 10.3389/fgeed.2021.604371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y. , Jones, T. W. , Dutta, S. , Zhu, Y. , Wang, X. , Narayanan, S. P. , Fagan, S. C. , & Zhang, D. (2021). Overview and update on methods for cargo loading into extracellular vesicles. Processes (Basel), 9, 10.3390/pr9020356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C. , Jaffar Ali, D. , Xu, H. , Kumaravel, S. , Si, K. , Li, Y. , Sun, B. , Ma, J. , & Xiao, Z. (2020). Epithelial cell ‐derived microvesicles: A safe delivery platform of CRISPR/Cas9 conferring synergistic anti‐tumor effect with sorafenib. Exerimental Cell Research, 392, 112040. 10.1016/j.yexcr.2020.112040 [DOI] [PubMed] [Google Scholar]
- Henderson, H. (2023). CRISPR Clinical Trials: A 2023 Update, https://innovativegenomics.org/news/crispr‐clinical‐trials‐2023/
- Hessvik, N. P. , & Llorente, A. (2018). Current knowledge on exosome biogenesis and release. Cellular and Molecular Life Sciences, 75, 193–208. 10.1007/s00018-017-2595-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horodecka, K. , & Düchler, M. (2021). CRISPR/Cas9: Principle, applications, and delivery through extracellular vesicles. International Journal of Molecular Sciences, 22, 10.3390/ijms22116072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Z. , Zhang, Y. , Propson, N. E. , Howden, S. E. , Chu, L. F. , Sontheimer, E. J. , & Thomson, J. A. (2013). Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proceedings of National Academy of Sciences U S A, 110, 15644–15649. 10.1073/pnas.1313587110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilahibaks, N. F. , Ardisasmita, A. I. , Xie, S. , Gunnarsson, A. , Brealey, J. , Vader, P. , de Jong, O. G. , de Jager, S. , Dekker, N. , Peacock, B. , Schiffelers, R. M. , Sluijter, J. P. G. , & Lei, Z. (2023). TOP‐EVs: Technology of protein delivery through extracellular vesicles is a versatile platform for intracellular protein delivery. Journal of Controlled Release, 355, 579–592. 10.1016/j.jconrel.2023.02.003 [DOI] [PubMed] [Google Scholar]
- Ishino, Y. , Shinagawa, H. , Makino, K. , Amemura, M. , & Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of Bacteriology, 169, 5429—5433. 10.1128/jb.169.12.5429-5433.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R. , Embden, J. D. , Gaastra, W. , & Schouls, L. M. (2002). Identification of genes that are associated with DNA repeats in prokaryotes. Molecular Microbiology, 43, 1565–1575. 10.1046/j.1365-2958.2002.02839.x [DOI] [PubMed] [Google Scholar]
- Jiang, X. C. , Zhang, T. , & Gao, J. Q. (2022). The in vivo fate and targeting engineering of crossover vesicle‐based gene delivery system. Advanced Drug Delivery Reviews, 187, 114324. 10.1016/j.addr.2022.114324 [DOI] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J. A. , & Charpentier, E. (2012). A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. M. , Yang, Y. , Oh, S. J. , Hong, Y. , Seo, M. , & Jang, M. (2017). Cancer‐derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism‐dependent targeting. Journal of Controlled Release, 266, 8–16. 10.1016/j.jconrel.2017.09.013 [DOI] [PubMed] [Google Scholar]
- Lainšček, D. , Kadunc, L. , Keber, M. M. , Bratkovič, I. H. , Romih, R. , & Jerala, R. (2018). Delivery of an Artificial Transcription Regulator dCas9‐VPR by Extracellular Vesicles for Therapeutic Gene Activation. ACS Synth Biol, 7, 2715–2725. 10.1021/acssynbio.8b00192 [DOI] [PubMed] [Google Scholar]
- Li, T. , Zhang, L. , Lu, T. , Zhu, T. , Feng, C. , Gao, N. , Liu, F. , Yu, J. , Chen, K. , Zhong, J. , Tang, Q. , Zhang, Q. , Deng, X. , Ren, J. , Zeng, J. , Zhou, H. , & Zhu, J. (2023). Engineered extracellular vesicle‐delivered CRISPR/CasRx as a novel RNA editing tool. Advanced Science, 2206517. 10.1002/advs.202206517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Zhou, X. , Wei, M. , Gao, X. , Zhao, L. , Shi, R. , Sun, W. , Duan, Y. , Yang, G. , & Yuan, L. (2019). In vitro and in vivo RNA Inhibition by CD9‐HuR functionalized exosomes encapsulated with miRNA or CRISPR/dCas9. Nano Letters, 19, 19–28. 10.1021/acs.nanolett.8b02689 [DOI] [PubMed] [Google Scholar]
- Liang, Y. , Liang, Y. , Iqbal, Z. , Wang, J. , Xu, L. , Xu, X. , Ouyang, K. , Zhang, H. , Lu, J. , Duan, L. , & Xia, J. (2022). Cell‐derived extracellular vesicles for CRISPR/Cas9 delivery: Engineering strategies for cargo packaging and loading. Biomaterial Science, 10, 4095–4106. 10.1039/d2bm00480a [DOI] [PubMed] [Google Scholar]
- Liang, Y. , Xu, X. , Xu, L. , Iqbal, Z. , Ouyang, K. , Zhang, H. , Wen, C. , Duan, L. , & Xia, J. (2022). Chondrocyte‐specific genomic editing enabled by hybrid exosomes for osteoarthritis treatment. Theranostics, 12, 4866–4878. 10.7150/thno.69368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, L. , Liu, Y. , Xu, F. , Huang, J. , Daugaard, T. F. , Petersen, T. S. , Hansen, B. , Ye, L. , Zhou, Q. , Fang, F. , Yang, L. , Li, S. , Fløe, L. , Jensen, K. T. , Shrock, E. , Chen, F. , Yang, H. , Wang, J. , Liu, X. , … Xu, X. (2018). Genome‐wide determination of on‐target and off‐target characteristics for RNA‐guided DNA methylation by dCas9 methyltransferases. Gigascience, 7, 1–19. 10.1093/gigascience/giy011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. , Wu, J. , Gu, W. , Huang, Y. , Tong, Z. , Huang, L. , & Tan, J. (2018). Exosome‐Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs. Advance Science (Weinh), 5, 1700611. 10.1002/advs.201700611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino, C. A. , Harper, J. C. , Carney, J. P. , & Timlin, J. A. (2018). Delivering CRISPR: a review of the challenges and approaches. Drug Deliv, 25, 1234–1257. 10.1080/10717544.2018.1474964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, N. , Zhong, W. , Li, J. , Zhai, Z. , Lu, J. , & Dong, R. (2022). Targeted activation of HNF4α/HGF1/FOXA2 reverses hepatic fibrosis via exosome‐mediated delivery of CRISPR/dCas9‐SAM system. Nanomedicine (Lond), 17, 141100–142700. 10.2217/nnm-2022-0083 [DOI] [PubMed] [Google Scholar]
- Majeau, N. , Fortin‐Archambault, A. , Gérard, C. , Rousseau, J. , Yaméogo, P. , & Tremblay, J. P. (2022). Serum extracellular vesicles for delivery of CRISPR‐CAS9 ribonucleoproteins to modify the dystrophin gene. Molecular Therapy, 30, 2429–2442. 10.1016/j.ymthe.2022.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man, K. , Brunet, M. Y. , Jones, M. C. , & Cox, S. C. (2020). Engineered extracellular vesicles: Tailored‐made nanomaterials for medical applications. Nanomaterials (Basel), 10, 10.3390/nano10091838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAndrews, K. M. , Xiao, F. , Chronopoulos, A. , LeBleu, V. S. , Kugeratski, F. G. , & Kalluri, R. (2021). Exosome‐mediated delivery of CRISPR/Cas9 for targeting of oncogenic Kras(G12D) in pancreatic cancer. Life Science Alliance, 4, 10.26508/lsa.202000875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendt, M. , Rezvani, K. , & Shpall, E. (2019). Mesenchymal stem cell‐derived exosomes for clinical use. Bone Marrow Transplantation, 54, 789–792. 10.1038/s41409-019-0616-z [DOI] [PubMed] [Google Scholar]
- Meng, W. , He, C. , Hao, Y. , Wang, L. , Li, L. , & Zhu, G. (2020). Prospects and challenges of extracellular vesicle‐based drug delivery system: considering cell source. Drug Delivery, 27, 585–598. 10.1080/10717544.2020.1748758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirjalili Mohanna, S. Z. , Djaksigulova, D. , Hill, A. M. , Wagner, P. K. , Simpson, E. M. , & Leavitt, B. R. (2022). LNP‐mediated delivery of CRISPR RNP for wide‐spread in vivo genome editing in mouse cornea. Journal of Controlled Release, 350, 401–413. 10.1016/j.jconrel.2022.08.042 [DOI] [PubMed] [Google Scholar]
- Moghadasi, S. , Elveny, M. , Rahman, H. S. , Suksatan, W. , Jalil, A. T. , Abdelbasset, W. K. , Yumashev, A. V. , Shariatzadeh, S. , Motavalli, R. , Behzad, F. , Marofi, F. , Hassanzadeh, A. , Pathak, Y. , & Jarahian, M. (2021). A paradigm shift in cell‐free approach: The emerging role of MSCs‐derived exosomes in regenerative medicine. Journal of Translational Medicine, 19, 302. 10.1186/s12967-021-02980-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mojica, F. J. , Diez‐Villasenor, C. , Garcia‐Martinez, J. , & Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of Molecular Evolution, 60, 174–182. 10.1007/s00239-004-0046-3 [DOI] [PubMed] [Google Scholar]
- Montagna, C. , Petris, G. , Casini, A. , Maule, G. , Franceschini, G. M. , Zanella, I. , Conti, L. , Arnoldi, F. , Burrone, O. R. , Zentilin, L. , Zacchigna, S. , Giacca, M. , & Cereseto, A. (2018). VSV‐G‐enveloped vesicles for traceless delivery of CRISPR‐Cas9. Molecular Therapy Nucleic Acids, 12, 453–462. 10.1016/j.omtn.2018.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, C. Y. , Kee, L. T. , Al‐Masawa, M. E. , Lee, Q. H. , Subramaniam, T. , Kok, D. , Ng, M. H. , & Law, J. X. (2022). Scalable Production of Extracellular Vesicles and Its Therapeutic Values: A Review. International Journal of Molecular Science, 23, 10.3390/ijms23147986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikfarjam, S. , Rezaie, J. , Zolbanin, N. M. , & Jafari, R. (2020). Mesenchymal stem cell derived‐exosomes: a modern approach in translational medicine. Journal of Translational Medicine, 18, 449. 10.1186/s12967-020-02622-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, B. , Huang, Z. , Youngquist, B. M. , Scott, J. W. , Niu, A. , Bojanowski, C. M. , Zwezdaryk, K. J. , Saba, N. S. , Fan, J. , Yin, X. M. , Cao, J. , Lyon, C. J. , Li, C. Z. , Roy, C. J. , & Hu, T. Y. (2021). Liposome‐mediated detection of SARS‐CoV‐2 RNA‐positive extracellular vesicles in plasma. Nature Nanotechnology, 16, 1039–1044. 10.1038/s41565-021-00939-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteikoetxea, X. , Silva, A. , Lázaro‐Ibáñez, E. , Salmond, N. , Shatnyeva, O. , Stein, J. , Schick, J. , Wren, S. , Lindgren, J. , Firth, M. , Madsen, A. , Mayr, L. M. , Overman, R. , Davies, R. , & Dekker, N. (2022). Engineered Cas9 extracellular vesicles as a novel gene editing tool. Journal of Extracellular Vesicles, 11, e12225. 10.1002/jev2.12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, Y. H. , Liang, J. , Czarny, B. , Wacker, M. G. , Yu, V. , Wang, J. W. , & Pastorin, G. (2021). Extracellular Vesicle (EV) biohybrid systems for cancer therapy: Recent advances and future perspectives. Semin Cancer Biolilogy, 74, 45–61. 10.1016/j.semcancer.2021.02.006 [DOI] [PubMed] [Google Scholar]
- Pagliarulo, N. (2021). Fourth trial volunteer dies in Astellas gene therapy study, https://www.biopharmadive.com/news/astellas‐gene‐therapy‐trial‐death‐fourth‐audentes/606508/
- Pan, X. , Qu, K. , Yuan, H. , Xiang, X. , Anthon, C. , Pashkova, L. , Liang, X. , Han, P. , Corsi, G. I. , Xu, F. , Liu, P. , Zhong, J. , Zhou, Y. , Ma, T. , Jiang, H. , Liu, J. , Wang, J. , Jessen, N. , Bolund, L. , … Yang, H. (2022). Massively targeted evaluation of therapeutic CRISPR off‐targets in cells. Nature Communicatins, 13, 4049. 10.1038/s41467-022-31543-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez, D. A. , & Vader, P. (2022). Extracellular vesicle‐based hybrid systems for advanced drug delivery. Pharmaceutics, 14. 10.3390/pharmaceutics14020267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X. , Liu, C. , Wang, N. , Huang, H. , He, S. , Gong, C. , & Wei, Y. (2021). Delivery of CRISPR/Cas systems for cancer gene therapy and immunotherapy. Advanced Drug Delivery Reviews, 168, 158–180. 10.1016/j.addr.2020.04.010 [DOI] [PubMed] [Google Scholar]
- Suda, T. , & Liu, D. (2007). Hydrodynamic Gene Delivery: Its Principles and Applications. Molecular Therapy, 15, 2063–2069. 10.1038/sj.mt.6300314 [DOI] [PubMed] [Google Scholar]
- Taha, E. A. , Lee, J. , & Hotta, A. (2022). Delivery of CRISPR‐Cas tools for in vivo genome editing therapy: Trends and challenges. Journal of Controlled Release, 342, 345–361. 10.1016/j.jconrel.2022.01.013 [DOI] [PubMed] [Google Scholar]
- Tenchov, R. , Bird, R. , Curtze, A. E. , & Zhou, Q. (2021). Lipid nanoparticles─from liposomes to MRNA vaccine delivery, a landscape of research diversity and advancement. ACS Nano, 15, 16982–17015. 10.1021/acsnano.1c04996 [DOI] [PubMed] [Google Scholar]
- Tong, S. , Moyo, B. , Lee, C. M. , Leong, K. , & Bao, G. (2019). Engineered materials for in vivo delivery of genome‐editing machinery. Nature Reviews Materials, 4, 726–737. 10.1038/s41578-019-0145-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usman, W. M. , Pham, T. C. , Kwok, Y. Y. , Vu, L. T. , Ma, V. , Peng, B. , Chan, Y. S. , Wei, L. , Chin, S. M. , Azad, A. , He, A. B. , Leung, A. Y. H. , Yang, M. , Shyh‐Chang, N. , Cho, W. C. , Shi, J. , & Le, M. T. N. (2018). Efficient RNA drug delivery using red blood cell extracellular vesicles. Nature Communications, 9, 2359. 10.1038/s41467-018-04791-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost, J. , & Patinios, C. (2023). The genome editing revolution. Trends Biotechnogy, 41, 396–409. 10.1016/j.tibtech.2022.12.022 [DOI] [PubMed] [Google Scholar]
- Van Niel, G. , D'Angelo, G. , & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology, 19, 213–228. 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- Wan, T. , Zhong, J. , Pan, Q. , Zhou, T. , Ping, Y. , & Liu, X. (2022). Exosome‐mediated delivery of Cas9 ribonucleoprotein complexes for tissue‐specific gene therapy of liver diseases. Scitific Advances, 8, eabp9435. 10.1126/sciadv.abp9435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Chang, M. , Zhang, R. , Wo, J. , Wu, B. , Zhang, H. , Zhou, Z. , Li, Z. , Zhang, F. , Zhong, C. , Tang, S. , Yang, S. , & Sun, G. (2022). Spinal cord injury target‐immunotherapy with TNF‐α autoregulated and feedback‐controlled human umbilical cord mesenchymal stem cell derived exosomes remodelled by CRISPR/Cas9 plasmid. Biomaterials Advances, 133, 112624. 10.1016/j.msec.2021.112624 [DOI] [PubMed] [Google Scholar]
- Wang, H. , Yang, H. , Shivalila, C. S. , Dawlaty, M. M. , Cheng, A. W. , Zhang, F. , & Jaenisch, R. (2013). One‐step generation of mice carrying mutations in multiple genes by CRISPR/Cas‐mediated genome engineering. Cell, 153, 910–918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.‐H. , Forterre, A. V. , Zhao, J. , Frimannsson, D. O. , Delcayre, A. , Antes, T. J. , Efron, B. , Jeffrey, S. S. , Pegram, M. D. , & Matin, A. C. (2018). Anti‐HER2 scFv‐directed extracellular vesicle‐mediated mRNA‐based gene delivery inhibits growth of HER2‐positive human breast tumor xenografts by prodrug activation. Molecular Cancer Therapeutics, 17, 1133–1142. 10.1158/1535-7163.mct-17-0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Yu, J. , Kadungure, T. , Beyene, J. , Zhang, H. , & Lu, Q. (2018). ARMMs as a versatile platform for intracellular delivery of macromolecules. Nature Communications, 9, 10.1038/s41467-018-03390-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbie, D. , Walther, J. , & Mastrobattista, E. (2019). Delivery Aspects of CRISPR/Cas for in Vivo Genome Editing. Acc Chem Res, 52, 1555–1564. 10.1021/acs.accounts.9b00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, X. , Corsi, G. I. , Anthon, C. , Qu, K. , Pan, X. , Liang, X. , Han, P. , Dong, Z. , Liu, L. , Zhong, J. , Ma, T. , Wang, J. , Zhang, X. , Jiang, H. , Xu, F. , Liu, X. , Xu, X. , Wang, J. , Yang, H. , … Bolund, L. (2021). Enhancing CRISPR‐Cas9 gRNA efficiency prediction by data integration and deep learning. Nature Communicatins, 12, 3238. 10.1038/s41467-021-23576-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, X. , Zhao, X. , Pan, X. , Dong, Z. , Yu, J. , Li, S. , Liang, X. , Han, P. , Qu, K. , Jensen, J. B. , Farup, J. , Wang, F. , Petersen, T. S. , Bolund, L. , Teng, H. , Lin, L. , & Luo, Y. (2021). Efficient correction of Duchenne muscular dystrophy mutations by SpCas9 and dual gRNAs. Molecular Therapy Nucleic Acids, 24, 403–415. 10.1016/j.omtn.2021.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q. , Zhang, Z. , Zhao, L. , Qin, Y. , Cai, H. , Geng, Z. , Zhu, X. , Zhang, W. , Zhang, Y. , Tan, J. , Wang, J. , & Zhou, J. (2020). Tropism‐facilitated delivery of CRISPR/Cas9 system with chimeric antigen receptor‐extracellular vesicles against B‐cell malignancies. Journal of Controlled Release, 326, 455–467. 10.1016/j.jconrel.2020.07.033 [DOI] [PubMed] [Google Scholar]
- Yao, X. , Lyu, P. , Yoo, K. , Yadav, M. K. , Singh, R. , Atala, A. , & Lu, B. (2021). Engineered extracellular vesicles as versatile ribonucleoprotein delivery vehicles for efficient and safe CRISPR genome editing. Journal of Extracellular Vesicles, 10, e12076. 10.1002/jev2.12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Y. , Zhang, X. , Xie, F. , Xu, B. , Xie, P. , Yang, T. , Shi, Q. , Zhang, C. Y. , Zhang, Y. , Chen, J. , Jiang, X. , & Li, J. (2020). An engineered exosome for delivering sgRNA:Cas9 ribonucleoprotein complex and genome editing in recipient cells. Biomaterials Science, 8, 2966–2976. 10.1039/d0bm00427h [DOI] [PubMed] [Google Scholar]
- Yen, J. , Fiorino, M. , Liu, Y. , Paula, S. , Clarkson, S. , Quinn, L. , Tschantz, W. R. , Klock, H. , Guo, N. , Russ, C. , Yu, V. W. C. , Mickanin, C. , Stevenson, S. C. , Lee, C. , & Yang, Y. (2018). TRIAMF: A new method for delivery of Cas9 ribonucleoprotein complex to human hematopoietic stem cells. Scientific Reports, 8, 10.1038/s41598-018-34601-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, J. , Wei, P. , Zhang, B. , Gou, F. , Feng, Z. , Mao, Y. , Yang, L. , Zhang, H. , Xu, N. , & Zhu, J. K. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnology Journal, 12, 797–807. 10.1111/pbi.12200 [DOI] [PubMed] [Google Scholar]


