Summary
Aging leads to a decrease in muscle function, mass, and strength in skeletal muscle of animals and humans. The transcriptome identified activation of the JAK/STAT pathway, a pathway that is associated with skeletal muscle atrophy, and endurance training has a significant effect on improving sarcopenia; however, the exact mechanism still requires further study. We investigated the effect of endurance training on sarcopenia. Six-month-old male SAMR1 mice were used as a young control group (group C), and the same month-old male SAMP8 mice were divided into an exercise group (group E) and a model group (group M). A 3–month running exercise intervention was performed on group E, and the other two groups were kept normally. Aging caused significant signs of sarcopenia in the SAMP8 mice, and endurance training effectively improved muscle function, muscle mass, and muscle strength in the SAMP8 mice. The expression of JAK2/STAT3 pathway factor was decreased in group E compared with group M, and the expression of SOCS3, the target gene of STAT3, and NR1D1, an atrophy-related factor, was significantly increased. Endurance training significantly improved the phenotypes associated with sarcopenia, and the JAK2/STAT3 pathway is a possible mechanism for the improvement of sarcopenia by endurance training, while NR1D1 may be its potential target.
Keywords: Sarcopenia, Endurance training, Janus kinase 2/signal transducer and activator of transcription 3 (JAK2/STAT3), Nuclear receptor subfamily 1, group D member 1 (Nr1d1)
Introduction
Sarcopenia is the progressive age-related decline of muscle mass and function. With the increasing demographic aging of society, sarcopenia has increasingly affected the independence and quality of life of older adults, not only raising this group’s risk of hospital admission and death, but also placing a heavy socioeconomic burden on society[1]. Currently, there are no Food and Drug Administration (FDA)-approved drugs for the global treatment of sarcopenia, and exercise is recognized as one of the most effective means to prevent and treat sarcopenia. Efficacy studies of exercise intervention in sarcopenia have been widely conducted in human and animal models, but studies of mechanisms related to exercise intervention in sarcopenia are more common in animal models. It has been reported that endurance training can improve mitochondrial metabolism and oxidative capacity in the skeletal muscle of aging mice by enhancing mitochondrial unfolded protein responses (UPRmt) [2], and long-term or habitual endurance training has significantly improved skeletal muscle function in SAMP8 mice with sarcopenia [3,4]
Skeletal muscle atrophy is associated with a variety of biological mechanisms—not only with an imbalance of protein synthesis and muscle degradation, but also with chronic inflammation and other factors. Some studies have found that an increase in atrophy-related factors is also an important cause of skeletal muscle atrophy. The Janus kinase 2/signal transducer and activator of transcription 3 (JAK2/STAT3) signaling pathways play important roles in various pathophysiological processes such as cell growth, differentiation, stress, proliferation, and apoptosis, and in regulating processes that include skeletal muscle atrophy. Suppressor of cytokine signaling (SOCS3) is a target gene of signal transducers and activators of transcription (STATs). SOCS3 directly inhibits the activation of STATs: activated STATs stimulate SOCS3 gene transcription, which causes SOCS3 protein to bind to phosphorylated JAK and receptors, thereby shutting down the pathway.
Nuclear receptor subfamily 1D group member 1 (Nr1d1) is highly expressed in skeletal muscle and is involved in the regulation of several physiological processes, including skeletal muscle mitochondrial biogenesis, oxidative capacity, skeletal myogenesis, and muscle fiber size through various pathways [5,6]. A single aerobic exercise session upregulates NR1D1 expression in skeletal muscle [7], and overexpression of NR1D1 in skeletal muscle can improve mitochondrial respiration and motility [8], demonstrating the important regulatory role of NR1D1 in skeletal muscle function.
Exercise has significantly ameliorated age-related myasthenia and can affect the expression of atrophy-related factors NR1D1 and JAK2/STAT3 pathway factors. We sought to explore the potential benefits of endurance training on sarcopenia by observing the effect of endurance training on NR1D1 and the JAK2/STAT3 pathway.
Materials and Methods
Experimental animals and grouping
Male SAMR1 and SAMP8 mice (7 months of age) were purchased from Peking University School of Medicine (Department of Laboratory Animal Science) under license number SCXK (Beijing) 2016–0010. Animal feeding and training were carried out in the Animal Experiment Center of Chengdu Institute of Physical Education (license number: Ethics Committee of Chengdu Institute of Physical Education (2021) No. 39). The laboratory room temperature was controlled at 20 °C−25 °C, and a good ventilation environment and ambient humidity were guaranteed for 12 h. Turning the lights on and off simulated the alternation of day and night. The mice had free access to standard SPF-grade full-price solid nutrient feed and filtered warm water. The litter tray and animal laboratory were cleaned daily. The experimental groupings were as follows: SAMR1 mice and SAMP8 mice were raised in a standard environment to 28 weeks of age. The SAMR1 mice (eight mice) were used as the SAMR1 model control group (Group C), and 16 SAMP8 mice were randomly divided into a model group (Group M), endurance training group (Group E), and SAMR1 model control group (Group C) according to the digital table method. The specific exercise protocols were as follows: the endurance training group undertook 8 weeks of uniform horizontal treadmill exercise (once a day, five times a week).
Determination of sarcopenia
Sarcopenia was determined using the following tests: ① Grip strength test: grip strength as an indicator of muscle strength of SAMP8 mice was tested once per week using a rat and mouse grip strength tester (YLS-13A). ② Accelerating rotarod test: this test was performed once a week using a mouse rotarod instrument set to 300 s, 4 rpm to 40 rpm. The time when a mouse fell from the rotarod was recorded. Using the accelerating rotarod test of different groups of mice, the muscle endurance of SAMP8 mice was estimated.
Exercise protocol
Endurance training was performed using an animal experimental running table with an adaptive exercise intervention for 3 days according to the training model of Bedford et al. [9] The protocol was 10 min of training per day at a speed of 9 m/min with 2 days of rest. Formal training took place 5 days/week, 1 session/day, for a total of 8 weeks. Weeks 1–2 involved the running table at a running speed of 12 m/min for 30 min; week 3 involved the table at a speed of 15 m/min for 30 min; week 4 was 15 m/min for 45 min; and weeks 5–8 were 15 m/min for 60 min. The running table’s slope was 0°, and training took place every Monday to Friday, with 2 days’ rest. The animals were checked for health after each experiment, and the mice were weighed and their body weight recorded weekly during the training period.
Sample collection
After the last intervention, the mice in each group were fasted for 24 h (with free access to water). Intraperitoneal injection of 1 % pentobarbital sodium (0.1 mL/100 g) was administered under anesthesia. Surgery was performed in the laboratory of Chengdu Institute of Physical Education. The vastus lateralis (VL) and gastrocnemius (GA) muscles of the left and right sides were removed and stored at −80 °C. For the left triceps surae, after removal of fascia and fat, the tissue was fixed with 4 % paraformaldehyde for histomorphological examination. For the VL, after removing the fascia and fat, the tissue was cut into pea-shaped pieces and put in a cryopreservation tube. After numbering, the VL was temporarily stored in liquid nitrogen and divided into two parts. One part was wrapped in tin foil and then frozen in liquid nitrogen. This was stored in a −80 °C freezer for subsequent use in western blot and real-time quantitative PCR detection; another part was used for transcriptome sequencing.
Histopathology
For the muscle fiber cross-sectional area test, the main steps in measuring the cross-sectional area of the triceps surae with hematoxylin–eosin (HE) staining were as follows:
The triceps surae was fixed in 4 % paraformaldehyde, embedded in paraffin, and sliced into 4-μm-thick slices.
For dewaxing, the slices were first baked at 64 °C in an oven for 1 h and then dewaxed with xylene, three times in total, 30 min each time.
The slices were transferred to absolute ethanol and 95 % ethanol for dehydration in sequence and then dehydrated twice at each concentration for 5 min each time.
The slices were transferred to 80 % ethanol, 70 % ethanol, and 50 % ethanol in sequence for dehydration, dehydrated for 5 min at each concentration, and transferred to distilled water for 3 min after completion.
The slides were stained with hematoxylin for 1 min and briefly immersed in tap water to wash off excess staining solution.
The slides were immersed in 0.5 % hydrochloric acid alcohol for 10 s for differentiation (prepared with 70 % alcohol).
Then, the slides were returned to blue in ammonia water (0.05 % lithium carbonate) for 10 s, washed with running water for 20 min and continued to return to blue, and the transferred to 80 % ethanol for 5 min.
The slides were stained for 1 min with eosin staining solution (95 % ethanol).
They were then were transferred to 80 % ethanol, 85 % ethanol, 90 % ethanol, and 95 % ethanol for dehydration, dehydrated for 5 min at each concentration, and then dehydrated in absolute ethanol twice for 5 min each time.
Transparency of the slides was ensured by keeping them in xylene for 5 min, and they were then sealed with neutral resin after drying.
Transcriptome sequencing
Four biological replicates of transcriptome sequencing were performed on muscle samples from groups M and E after modeling intervention. Total RNA from muscle tissue was assessed using an RNA Nano 6000 Assay Kit from the Bioanalyzer 2100 system (Agilent Technologies, CA, USA) and RNeasy RNA purification kit (DNase). Total RNA was used as input material for the RNA sample preparations. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads, after which fragmentation was carried out using divalent cations under an elevated temperature in First Strand Synthesis Reaction Buffer (5×). First-strand cDNA was synthesized using a random hexamer primer and M-MuLV Reverse Transcriptase (RNase H), while second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of the 3′ ends of DNA fragments, adaptors with a hairpin loop structure were ligated to prepare for hybridization. To select cDNA fragments that were preferentially 370–420 bp in length, the library fragments were purified with an AMPure XP system (Beckman Coulter, Brea, CA, USA). Then, PCR was performed with Phusion High-Fidelity DNA polymerase, universal PCR primers, and Index (X) Primer. Lastly, PCR products were purified (using an AMPure XP system), and library quality was assessed on an Agilent Bioanalyzer 2100 system.
After the library check was qualified, the different libraries were pooled according to the requirements of effective concentration and target data volume and sequenced with the Illumina NovaSeq 6000; a total of 150 bp paired-end reads were generated. The basic principle of the sequencing was synthesis. Four fluorescently labeled dNTPs, DNA polymerase, and adapter primers were added to the sequencing flow cell for amplification. When each sequencing cluster extended the complementary chain, each fluorescently labeled dNTP released the corresponding fluorescence; the sequencer captured the fluorescent signal and converted the light signal into a sequencing peak using computer software to obtain the sequence information of the fragment to be detected.
Differential expression of genes and enrichment analysis
For samples with biological replicates, differential expression analysis between the two comparison sets was performed using DESeq2 software (1.20.0). DESeq2 provides statistical procedures for determining differential expression in digital gene expression data using models based on negative binomial distribution. The resulting P-value (padj) was adjusted using the Benjamini–Hochberg correction to control for the false-discovery rate. The criteria padj ≤ 0.05 and |log2(fold change)| ≥1 were set as the threshold for significantly differential expression. Gene Ontology (GO) enrichment analysis of differentially expressed genes was achieved using clusterProfiler (3.8.1) software, in which gene length bias was corrected. GO terms with corrected P-values lower than 0.05 were considered significantly enriched by differentially expressed genes.
Western blotting
The removed VL tissue was homogenized in a high-speed cryogenic triturator (Servicebio, China) using RIPA lysis buffer (Servicebio), and the lysates were separated using SDS-PAGE and transferred to PVDF membranes. The membranes were incubated with primary anti-JAK2, anti-STAT3, anti-Socs3, and anti-NR1D1 antibodies, and the indicated secondary antibodies (Abcam, Cambridge, UK). GAPDH was used as an internal control. Subsequently, using a chemiluminescent substrate (Thermo Fischer Scientific Inc., MA, USA), the treated PVDF membranes were scanned using Tianneng GIS chassis control software V2.0 to identify the bands and visualize the proteins.
RNA isolation and real-time quantitative PCR analysis
Muscle tissues (50 mg) were ground in liquid nitrogen, and then 1 ml of pre-chilled TRIzol was added; the muscle tissues were then centrifuged for 10 min (10,000 g, 2°C–8°C). Then, 75℅ ethanol precipitation was used to isolate RNA, followed by centrifugation for 3 min (2400 g, 2°C–8°C). The supernatant was discarded and total RNA remained. The concentration and purity of RNA were analyzed using an ultra-micro UV spectrophotometer and agarose gel electrophoresis, and cDNA was synthesized by reverse transcription. PIKORed 96 (ThermoFisher) was used to measure the expression levels of Bmal1 and Clock mRNA via real-time fluorescence quantitative PCR. The primers used to measure the expression were designed and synthesized by Shanghai Shengong Bioengineering Management Technology Development Service Co., Ltd., and purified by ULTRAPAGE. The primer sequence is shown in Table 1. Real-time quantitative PCR was performed, and the cycle threshold values of each sample were analyzed using Thermo Scientific PikoReal software. The relative mRNA content level was calculated via 2−ΔΔCT (Table 1).
Table 1.
PCR primer sequences
| Primer name | Upstream | Downstream |
|---|---|---|
| β-actin | 5′-ctacctcatgaagatcctgacc-3′ | 5′-cacagcttctctttgatgtcac-3′ |
| Socs3 | 5′-gagagcggattctactggag-3′ | 5′-agcttgagtacacagtcgaag-3′ |
| Nr1d1 | 5′-cgtcatcctcttcatcctcctcctc-3′ | 5′-cttggtaatgttgcttgtgcccttg-3′ |
Statistical analyses
Data are presented as mean ± standard deviation. Before statistical analysis, all data were assessed for a normal distribution using the one-sample Kolmogorov–Smirnov test (10). Statistical analyses were undertaken using Prism 9.0 (GraphPad, San Diego, CA, USA). Depending on the characteristics of the variables, either the Student t-test (two-sided) or Mann–Whitney U-test was used to compare the measured parameters in 7-month-old and 9-month-old mice. Differences in the relative grip strength, accelerating rotarod test, and mean cross-sectional fiber area of the GA among groups C, M, and E were assessed using one-way analysis of variance (ANOVA) or Kruskal–Wallis H-tests, depending on whether the variables had a normal distribution. Intergroup differences in levels of JAK2, STAT3, SOCS3, and NR1D1 were assessed by one-way ANOVA followed by Tukey’s post hoc test. P<0.05 was considered significant.
Results
Endurance training improves age-related decline in skeletal muscle
To verify age-related muscle changes and phenotypic characteristics of sarcopenia, and to assess the ameliorative effect of endurance training on sarcopenia in SAMP8 mice, we tested the relative grip strength of each group of mice before and after endurance training, and further recorded accelerating rotarod test durations, changes in muscle cross-sectional area, and changes in muscle ultrastructure of the gastrocnemius muscle.
Grip strength and accelerating rotarod test
Muscle strength and endurance of SAMP8 and SAMR1 mice decreased from 7 months of age, with the relative grip strength and accelerating rotarod test duration of mice in group C being significantly higher than those in group M (P<0.01) (Fig. 1A,B). By 9 months of age, the relative grip strength and accelerating rotarod test duration of the M group had decreased even more significantly (P<0.0001) (Fig. 1C,D). SAMP8 mice exhibited more obvious sarcopenic features than SAMR1 mice. After endurance training, relative grip strength (P<0.05) and accelerating rotarod test duration of mice (P<0.05) significantly increased in group E compared to group M (P<0.05) (Fig. 1A–1D); endurance training significantly improved age-related decreases in muscle strength and endurance in SAMP8 mice (Fig. 1E,F).
Fig. 1.
Changes in muscle mass and function of mice in each group prior to the intervention. A: Relative grip strength of mice in each group at 7 months. B: Accelerating rotarod test duration in each group of mice at 7 months. C: Relative grip strength of mice in each group at 9 months. D: Accelerating rotarod test duration in each group of mice at 9 months. E: Changes in relative grip strength of mice in each group from 7 to 9 months. F: Accelerating rotarod test duration in each group from 7 to 9 months.
Cross-sectional area of GA and skeletal muscle mass
We also tested the mice’s cross-sectional muscle area to assess muscle function (Fig. 2A). Hematoxylin and eosin (HE) staining showed that the gastrocnemius cross-sectional area in group M was significantly reduced compared to that of group C (P<0.0001). Further, group E’s cross-sectional muscle area was significantly increased compared to that of group M (P<0.0001) (Fig. 2B). Muscle mass and relative grip strength were significantly lower in group M than group C (P<0.001), and then showed significant improvement after endurance training (P<0.01) (Fig. 2C).
Fig. 2.
A: Cross-sectional HE staining of triceps muscle fibers in each group of mice. (C) HE staining of muscle fibers in the model control group [200×, scale bar 10 um]. M) HE staining of muscle fibers in the non-exercise group [200×, scale bar 10 um]. E) HE staining of muscle fibers in the endurance training group [200×, scale bar 10 um]). Changes in skeletal muscle fiber cross-section and muscle mass in each group of mice following 8 weeks of endurance training. B: Changes in muscle fiber cross-sectional area of triceps in each group of mice. C: Changes in relative muscle mass (muscle wet weight/body weight) of triceps in each group of mice. (**p<0.01, ***p<0.001, ****p<0.0001).
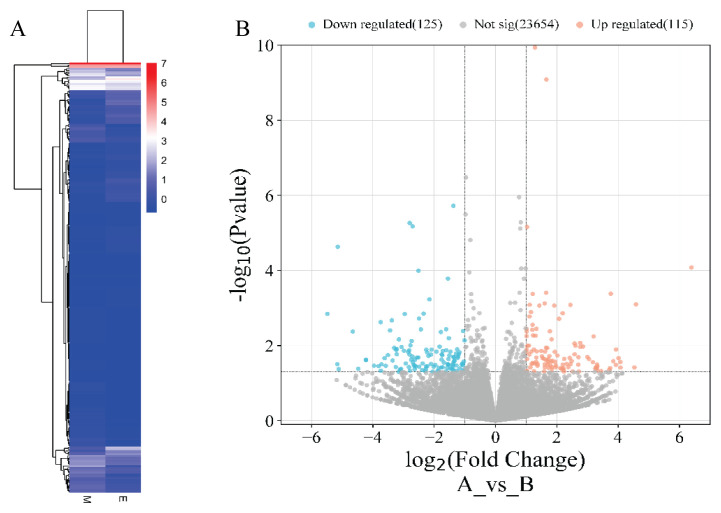
Bioinformatics analysis of the Nr1d1 in SAMP8 mice
Screening for highly relevant factors in sarcopenia using transcriptome sequencing yielded the results that follow (Fig. 3B). Based on the transcriptome differential gene expression matrix, compared to group M, a total of 115 mRNA expressions were upregulated and 125 mRNA expressions were downregulated in group E (FC≥0, P<0.05) (Fig. 3B); NR1D1 mRNA was upregulated in group E compared to group M (log2FoldChange = 1.65, P value = 0.0000000008).
Fig. 3.
A: Heat map of differentiated gene clustering. B: Volcano map.
Endurance training inhibits the JAK2/STAT3 pathway
We also examined the effects of aging and endurance training on the expression of the JAK2/STAT3 pathway and its inhibitory regulator SOCS3. JAK2 and STAT3 protein expression was significantly greater in group M compared with group C (P<0.05) (Fig. 4B,C). SOCS3 protein and mRNA expression in skeletal muscle were significantly lower in group M compared with group C. After endurance training, SOCS3 protein and mRNA expression were significantly upregulated in group E compared with group M (P<0.05) (Fig. 4D,E), while JAK2 and STAT3 protein expression was significantly downregulated (P<0.05) (Fig. 4B,C).
Fig. 4.
Western blot and RT-qPCR were used to detect changes in the protein and mRNA expression of JAK2/STAT3 pathway factors. A: Western blot bands: Western blot of JAK2, STAT3, and SOCS3 proteins. B: Changes in JAK2 protein expression. C: Changes in STAT3 protein expression. D: Changes in SOCS3 protein expression. E: Changes in SOCS3 mRNA expression (**p<0.01, ***p<0.001, ****p<0.0001).
Endurance exercise induces changes in NR1D1 expression in skeletal muscle in sarcopenia
We verified the changes in the expression of the target factors using western blot and real-time quantitative PCR. NR1D1 protein expression showed that group E protein expression was significantly higher than group M (P<0.05) (Fig. 5B). Compared with group C, Nr1d1 mRNA expression levels were significantly lower in group M (P<0.01) (Fig. 5C). Compared with group M, Nr1d1 mRNA expression levels were significantly higher in group E (P<0.05) (Fig. 5C).
Fig. 5.
Western blot and RT-qPCR were performed to detect changes in the protein and mRNA expression of the target factor NR1D1. A: Western blot bands: Western blot of NR1D1 proteins. B: Changes in NR1D1 protein expression. C: Changes in NR1D1 mRNA expression (**p<0.01, ***p<0.001, ****p<0.0001).
Discussion
Sarcopenia is a progressive age-related loss of muscle mass and function, and recent studies have shown the importance of muscle strength for the development of skeletal muscle and for predicting adverse health events. In 2018, the European Working Group on Sarcopenia in Older People (EWGSOP) proposed muscle strength as the main parameter for the diagnosis of sarcopenia, and as a classification criterion to evaluate the severity of sarcopenia. In the present study, we explored the effect of endurance training to improve sarcopenia and its underlying mechanisms. Our study showed that endurance training was effective in improving muscle strength, muscle endurance, and muscle cross-sectional area in sarcopenia. The underlying mechanism may be that endurance training upregulates NR1D1, which inhibits the activation of the JAK2/STAT3 signaling pathway by upregulating SOCS3.
Sarcopenia onset may be related to various factors, such as low frequency of exercise, age-related decline in neuromuscular function, decreased immune function, and nutrition, but its exact pathogenesis remains unclear. The JAK/STAT signaling pathway is initiated by the binding of cytokine ligands to cytokine receptors on the cell surface, which triggers the activation of JAK. The activated JAK can phosphorylate key tyrosine residues on the receptor, which recruit specific STAT through its SH2 (Src Homology 2) structural domain to form a dimer. It is transported by importin-α into the nucleus and binds to specific DNA targets, thus exerting a series of pathophysiological effects such as regulation of cell growth, differentiation, proliferation, and apoptosis [10]. STAT has several protein families, including STAT1, STAT2, STAT3, STAT5A, STAT5B, and STAT6, of which the protein and gene expression of STAT3 are key to muscle atrophy[11]. In studies of cancer cachexia-associated muscle atrophy, certain inflammatory factors were found to activate JAK2/STAT3 pathway expression, while inhibition of expression delayed the process of myasthenia[11]. We found by transcriptome analysis that the JAK2/STAT3 pathway is highly enriched in sarcopenic muscles and is closely associated with the process of sarcopenia-related muscle atrophy.
Skeletal muscle aging in humans is a slow and long-term process. There have been numerous studies of exercise interventions in human sarcopenia, but they have been too short in duration to draw more informative conclusions about the entire sarcopenia process [12]. The use of shorter-lived, faster-aging animal models to simulate the muscle characteristics of the human aging process for exercise intervention studies could be an alternative. The accelerated-accelerated mouse prone 8 (SAMP8) model has age-related changes in muscle mass, contractile properties, metabolic characteristics, and muscle fiber types similar to the development of sarcopenia in humans, making it an ideal animal model for the study of sarcopenia [13,14]. It has been shown that the onset of sarcopenia in SAMP8 mice occurs between 7 and 8 months of age [15]. Based on the results of the model testing in this study, which showed that the mice started to develop the characteristics of sarcopenia at the age of 7 months. We carried out a 2-month endurance training intervention after this to observe the process of change in sarcopenia and the effect of intervention on endurance training.
Studies have concretely observed the beneficial effect of exercise on sarcopenia; exercise is widely recognized by scientific and clinical workers to help with age-related muscle atrophy[16–18]. Specifically, endurance exercise has been shown to be effective in preventing muscle loss and reducing the risk of sarcopenia in middle-aged and elderly populations [19], helping to improve age-related skeletal muscle dysfunction, aerobic capacity, and mitochondrial function in the elderly[16], and to enhance muscle mass and strength[20]. Locomotor adaptation of aging skeletal muscle is mainly related to: muscle hypertrophy; mitochondrial biogenesis; antioxidant and fiber conversion signals, such as mitogen-activated protein kinase (MAPK) signaling [21]; ATP cycling, and adenosine monophosphate-activated protein kinase (AMPK) signaling[22]; reduced nicotinamide adenine dinucleotide (NADH)/nicotinamide adenine dinucleotide (NAD+)/ratio and sirtuins (SIRT)[23]; mammalian target of rapamycin (mTOR) activation[24]; hypoxia-inducible factor 1 (HIF-1)[25]; mitochondrial biogenesis pathway; and peroxisome proliferator-activated receptor gamma coactivator l-alpha (PGC-1α) and 1-beta (PGC-1β).
NR1D1 plays a role in oxidative capacity, mitochondrial biogenesis, atrophy genes, and muscle fiber size in skeletal muscle[26]. NR1D1 has been shown to regulate muscle mass and fiber size by repressing the expression of genes of the ubiquitin-proteasome system degradation pathway, as well as the upstream forkhead box O1 (FoxO1) and forkhead box O3a (FoxO3a) regulators [5]. The JAK2/STAT3 pathway is closely related to several malignant diseases and skeletal muscle atrophy; researchers have observed changes in JAK2/STAT3 pathway-related molecule expression after overexpression and silencing of NR1D1 in ovarian cancer cells, and they discovered the inverse regulation of JAK2/STAT3 by NR1D1. In this study, through transcriptome enrichment screening analysis and molecular experiments, we found that NR1D1 inhibits the JAK2/STAT3 pathway. Based on these results, we hypothesize that there may be a potential regulatory role of NR1D1 on the JAK2/STAT3 signaling pathway in endurance training that improves sarcopenia. Future research should focus on using cell cultures, overexpression, silencing, and other molecular biology methods to determine the specific regulatory role and mechanisms of NR1D1 on the JAK2/STAT3 pathway.
On its own, NR1D1’s functions are also closely related to the mechanisms of sarcopenia. It plays an important role as a nuclear receptor in the transcriptional regulation of the molecular circadian clock, linking circadian rhythm to metabolism, and it is associated with bodily inflammatory processes [27,28]. Therefore, focusing on the role of NR1D1 in skeletal muscle function has important implications for the future treatment of aging skeletal muscle diseases.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81904318). Yu Xia was supported by the Sports Medicine Key Laboratory Project of Sichuan Province (GS21ZX01). The authors have no competing financial interests to declare.
Footnotes
Conflict of Interest: There is no conflict of interest.
References
- 1.Cruz-Jentoft AJ, Bahat G, Bauer J, et al. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing. 2019;48(1):16–31. doi: 10.1093/ageing/afy169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordeiro AV, Brícola RS, Braga RR, et al. Aerobic Exercise Training Induces the Mitonuclear Imbalance and UPRmt in the Skeletal Muscle of Aged Mice. J Gerontol A Biol Sci Med Sci. 2020;75(12):2258–2261. doi: 10.1093/gerona/glaa059. [DOI] [PubMed] [Google Scholar]
- 3.Liang J, Zhang H, Zeng Z, et al. MicroRNA profiling of different exercise interventions for alleviating skeletal muscle atrophy in naturally aging rats. Journal of Cachexia, Sarcopenia and Muscle. 2023;14(1):356. doi: 10.1002/jcsm.13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoki K, Konno M, Honda K, et al. Habitual Aerobic Exercise Diminishes the Effects of Sarcopenia in Senescence-Accelerated Mice Prone8 Model. Geriatrics (Basel, Switzerland) 2020;5(3) doi: 10.3390/geriatrics5030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayeuf-Louchart A, Thorel Q, Delhaye S, et al. Rev-erb-α regulates atrophy-related genes to control skeletal muscle mass. Sci Rep. 2017;7:14383. doi: 10.1038/s41598-017-14596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welch RD, Guo C, Sengupta M, et al. Rev-Erb co-regulates muscle regeneration via tethered interaction with the NF-Y cistrome. Mol Metab. 2017;6(7):703–714. doi: 10.1016/j.molmet.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rovina RL, da Rocha AL, Marafon BB, et al. One Bout of Aerobic Exercise Can Enhance the Expression of Nr1d1 in Oxidative Skeletal Muscle Samples. Front Physiol. 2021;12:626096. doi: 10.3389/fphys.2021.626096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woldt E, Sebti Y, Solt LA, et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19(8):1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tg B, Cm T, Nc W, Ra O, Cv G. Maximum oxygen consumption of rats and its changes with various experimental procedures. Journal of applied physiology: respiratory, environmental and exercise physiology. 1979;47(6) doi: 10.1152/jappl.1979.47.6.1278. [DOI] [PubMed] [Google Scholar]
- 10.Hu X, li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct Target Ther. 2021;6:402. doi: 10.1038/s41392-021-00791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eskiler GG, Bezdegumeli E, Ozman Z, et al. IL-6 mediated JAK/STAT3 signaling pathway in cancer patients with cachexia. Bratisl Lek Listy. 2019;66(11):819–826. doi: 10.4149/BLL_2019_136. [DOI] [PubMed] [Google Scholar]
- 12.Shen Y, Shi Q, Nong K, et al. Exercise for sarcopenia in older people: A systematic review and network meta-analysis. J Cachexia Sarcopenia Muscle. 2023;14(3):1199–1211. doi: 10.1002/jcsm.13225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romanick M, Brown-Borg HM. Murine models of atrophy, cachexia, and sarcopenia in skeletal muscle. Biochim Biophys Acta. 2013;1832(9):1410–1420. doi: 10.1016/j.bbadis.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishikawa T, Takahashi JA, Matsushita T, et al. Tubular aggregates in the skeletal muscle of the senescence-accelerated mouse; SAM. Mech Ageing Dev. 2000;114(2):89–99. doi: 10.1016/S0047-6374(00)00088-9. [DOI] [PubMed] [Google Scholar]
- 15.Guo AY, Leung KS, Siu PMF, et al. Muscle mass, structural and functional investigations of senescence-accelerated mouse P8 (SAMP8) Exp Anim. 2015;64(4):425–433. doi: 10.1538/expanim.15-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moghadam BH, Bagheri R, Ashtary-Larky D, et al. The effects of concurrent training order on satellite cell-related markers, body composition, muscular and cardiorespiratory fitness in older men with sarcopenia. J Nutr Health Aging. 2020;24(7):796–804. doi: 10.1007/s12603-020-1431-3. [DOI] [PubMed] [Google Scholar]
- 17.Park J, Kwon Y, Park H. Effects of 24-Week Aerobic and Resistance Training on Carotid Artery Intima-Media Thickness and Flow Velocity in Elderly Women with Sarcopenic Obesity. J Atheroscler Thromb. 2017;24(11):1117–1124. doi: 10.5551/jat.39065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen HT, Chung YC, Chen YJ, Ho SY, Wu HJ. Effects of different types of exercise on body composition, muscle strength, and IGF-1 in the elderly with sarcopenic obesity. J Am Geriatr Soc. 2017;65(4):827–832. doi: 10.1111/jgs.14722. [DOI] [PubMed] [Google Scholar]
- 19.Kim SH, Kim TH, Hwang HJ. The relationship of physical activity (PA) and walking with sarcopenia in Korean males aged 60 years and older using the Fourth Korean National Health and Nutrition Examination Survey (KNHANES IV-2, 3), 2008–2009. Arch Gerontol Geriatr. 2013;56(3):472–477. doi: 10.1016/j.archger.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Garatachea N, Pareja-Galeano H, Sanchis-Gomar F, et al. Exercise Attenuates the Major Hallmarks of Aging. Rejuvenation Res. 2015;18(1):57–89. doi: 10.1089/rej.2014.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez-Cabrera MC, Domenech E, Viña J. Moderate exercise is an antioxidant: upregulation of antioxidant genes by training. Free Radic Biol Med. 2008;44(2):126–131. doi: 10.1016/j.freeradbiomed.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Jäger S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc Natl Acad Sci U S A. 2007;104(29):12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchis-Gomar F. Sestrins: novel antioxidant and AMPK-modulating functions regulated by exercise? J Cell Physiol. 2013;228(8):1647–1650. doi: 10.1002/jcp.24338. [DOI] [PubMed] [Google Scholar]
- 24.Bodine SC, Stitt TN, Gonzalez M, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3(11):1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 25.Schmutz S, Däpp C, Wittwer M, et al. Endurance training modulates the muscular transcriptome response to acute exercise. Pflugers Archiv: Eur J Physiol. 2006;451(5) doi: 10.1007/s00424-005-1497-0. [DOI] [PubMed] [Google Scholar]
- 26.Woldt E, Sebti Y, Solt LA, et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19(8):1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fontaine C, Rigamonti E, Pourcet B, et al. The nuclear receptor rev-erbα is a liver x receptor (LXR) target gene driving a negative feedback loop on select LXR-induced pathways in human macrophages. Mol Endocrinol. 2008;22(8):1797–1811. doi: 10.1210/me.2007-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salminen A, Vihko V. Endurance training reduces the susceptibility of mouse skeletal muscle to lipid peroxidation in vitro. Acta Physiol Scand. 1983;117(1):109–113. doi: 10.1111/j.1748-1716.1983.tb07184.x. [DOI] [PubMed] [Google Scholar]