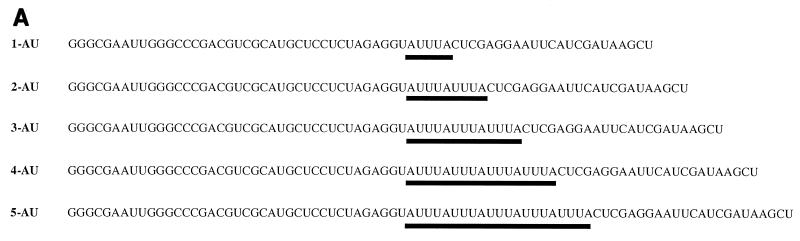Abstract
Apobec-1, the catalytic subunit of the mammalian apolipoprotein B (apoB) mRNA-editing enzyme, is a cytidine deaminase with RNA binding activity for AU-rich sequences. This RNA binding activity is required for Apobec-1 to mediate C-to-U RNA editing. Filter binding assays, using immobilized Apobec-1, demonstrate saturable binding to a 105-nt apoB RNA with a Kd of ∼435 nM. A series of AU-rich templates was used to identify a high-affinity (∼50 nM) binding site of consensus sequence UUUN[A/U]U, with multiple copies of this sequence constituting the high-affinity binding site. In order to determine whether this consensus site could be functionally demonstrated from within an apoB RNA, circular-permutation analysis was performed, revealing one major (UUUGAU) and one minor (UU) site located 3 and 16 nucleotides, respectively, downstream of the edited base. Secondary-structure predictions reveal a stem-loop flanking the edited base with Apobec-1 binding to the consensus site(s) at an open loop. A similar consensus (AUUUA) is present in the 3′ untranslated regions of several mRNAs, including that of c-myc, that are known to undergo rapid degradation. In this context, it is presumed that the consensus motif acts as a destabilizing element. As an independent test of the ability of Apobec-1 to bind to this sequence, F442A cells were transfected with Apobec-1 and the half-life of c-myc mRNA was determined following actinomycin D treatment. These studies demonstrated an increase in the half-life of c-myc mRNA from 90 to 240 min in control versus Apobec-1-expressing cells. Apobec-1 expression mutants, in which RNA binding activity is eliminated, failed to alter c-myc mRNA turnover. Taken together, the data establish a consensus binding site for Apobec-1 embedded in proximity to the edited base in apoB RNA. Binding to this site in other target RNAs raises the possibility that Apobec-1 may be involved in other aspects of RNA metabolism, independent of its role as an apoB RNA-specific cytidine deaminase.
Two forms of apolipoprotein B (apoB) are known to circulate in the plasma of mammals (reviewed by Young [56]). apoB100, a 512-kDa protein primarily synthesized in the liver as a structural component of very-low-density lipoprotein particles, is the product of an ∼14-kb nuclear mRNA. A truncated protein, apoB48, is synthesized in the small intestine and contains the amino-terminal 2,152 amino acids of the larger protein (56). This organ-specific partitioning of apoB production is the result of RNA editing of a common apoB gene. A site-specific C-to-U deamination in the nuclear transcript encoding mammalian apoB mRNA is responsible for the production of an in-frame UAA translational stop codon and results in translation of the truncated protein product (12, 14, 16, 43). apoB mRNA editing occurs in the small intestines of all mammals and in the livers of certain species, although notably not in humans (24, 30).
apoB mRNA editing is mediated by a multicomponent enzyme complex containing a catalytic subunit, Apobec-1, as well as other essential protein factors whose precise number and identities have yet to be established (25, 39, 49, 50, 54). Apobec-1 is a cytidine deaminase with homology to other members of a multigene family, particularly around the active site, which includes a zinc-coordinating motif (H/C)-(A/V)-E-(X)24-30-P-C-X-X-C (5, 46). In addition to functioning as a cytidine deaminase on a monomeric nucleoside substrate, Apobec-1 demonstrates RNA binding activity with specificity for AU-rich templates (1, 34, 37). The domains within Apobec-1 that coordinate this RNA binding activity have been localized to the amino-terminal half of the protein and involve residues flanking the active site (34, 37, 38). From a functional perspective, this activity assumes considerable importance, since mutations in Apobec-1 that interfere with apoB RNA binding consistently eliminate RNA editing (34, 37, 38, 51). Recent molecular modeling, based upon the crystal structure of the Escherichia coli cytidine deaminase, suggests that the dimeric interface formed between the paired monomers of Apobec-1 may allow entry of the apoB RNA substrate (38). Accordingly, information concerning the presentation and accessibility of cytidine nucleotides for deamination bears directly on the question of target site selection within an RNA template.
While uncertainty exists with respect to the identities of all the requisite trans-acting factors for apoB mRNA editing, the nucleotide requirements, by contrast, have been clearly established (1, 4, 8, 15, 17, 19, 37, 47). Prominent among these is an 11-nucleotide (nt) element located 4 nt downstream of the edited base, referred to as a mooring sequence (4, 19, 47). This element is absolutely required for editing of the C in apoB at position 6666 and will function to facilitate editing when inserted into a heterologous RNA context (3, 8). In addition, other sequence elements, located both 5′ and 3′ of the edited base, are required for maximal editing efficiency, as are the length and AU content of the flanking RNA sequence (26). In this context, it is noteworthy that the region flanking the edited base in apoB RNA is itself composed of ∼70% AU residues. RNA secondary-structure predictions, using a 55-nt apoB RNA flanking the edited site, suggest that the edited C is found within the loop of a highly conserved stem-loop structure (44). These predictions were very recently substantiated through RNase mapping of 40- and 55-nt apoB RNAs (44).
For the present report, we have performed RNA binding studies with synthetic AU-rich RNAs to determine a high-affinity binding site for Apobec-1. Secondary-structure predictions for these templates suggest that a U-rich sequence is required at an open loop for Apobec-1 binding. In addition, using circular-permutation analysis, we have directly demonstrated an Apobec-1 consensus binding site within a synthetic apoB RNA transcript that contains all the requisite elements for high-efficiency editing. We further demonstrate that the 3′ untranslated (3′ UTR) of c-myc mRNA, a cellular transcript known to be rapidly degraded, contains the high-affinity Apobec-1 binding site and binds to Apobec-1 in vitro. As evidence for the functional relevance of this binding site, the half-life of c-myc mRNA was extended in F442A cells overexpressing Apobec-1, suggesting a novel role for Apobec-1 in regulating RNA stability, mediated through binding to its high-affinity target.
MATERIALS AND METHODS
Expression of GST–Apobec-1.
Apobec-1 cDNA was cloned into plasmid pGEX-4T3 and expressed as a glutathione S-transferase (GST) fusion protein as previously described (34). The fusion protein was purified from bacterial lysates by using glutathione-agarose beads (Sigma, St. Louis, Mo.) in a buffer containing 50 mM Tris-HCl (pH 8.0)–10 mM reduced glutathione. The material was further purified by Sephadex G-75 chromatography, and the dominant peak was dialysed against a buffer containing 20 mM HEPES (pH 7.9), 0.1 M KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol (DTT), and 20% glycerol. The purity was determined to be greater than 85% by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis and Western blotting using both an antipeptide antiserum (21) and an affinity-purified antibody generated against the fusion protein. Mutant forms of Apobec-1, deficient in RNA binding activity (F66,87L and H61R) were created by a two-step PCR-based method as described previously (34). For the F66,87L mutation, a single F66L mutant was used for a second round of mutagenesis. After being sequenced, clones containing the desired mutations were selected and subcloned for eukaryotic expression. The primers used for mutagenesis were H61R sense primer, 5′-CCAACAAACGCGTTGAAG-3′ (nt 173 to 191); H61R antisense primer, 5′-GACTTCAACGCGTTTGTTGGTG-3′ (nt 192 to 171); F66L sense primer, 5′-GAAGTCAATTACATAGAAAAATTTA-3′ (nt 228 to 252); F66L antisense primer, 5′-ATTTTTCTATGTAATTGACTTC-3′ (nt 249 to 228); F87L sense primer, 5′-CATTACCTGGTTGCTGTCCTGGAG-3′ (nt 290 to 313); F87L antisense primer, 5′-CTCCAGGACAGCAACCAGGTAATG-3′ (nt 313 to 290); Apobec-1 outside sense primer, 5′-GTAGGATCCATGAGTTCCGAGACAGGC-3′ (BamHI); and Apobec-1 outside antisense primer, 5′-AGTGTCGACTTTCAACCCTGTGGCCCACAG-3′ (SalI). The underlined nucleotides represent restriction sites engineered for cloning purposes, while the boldface underlined sequences represent mutations. For eukaryotic expression, the wild-type or mutant Apobec-1 cDNA was subcloned into pRC/CMV (InVitrogen, Carlsbad, Calif.).
Plasmid construction and in vitro transcription.
The constructions of plasmids p1-AU to p5-AU, pM1 to pM8, p3′ c-myc, p3′IL-2, and p3′TNF-α were described earlier (6, 7), and the plasmids were a gift from T. Lindsten, University of Chicago. These sequences were cloned into plasmid pGEM7Zf(+). pRB105 contains a 105-bp fragment of rat apoB cDNA (nt 6639 to 6743), and pCB150 contains a 160-bp fragment of chicken apoB cDNA (nt 6608 to 6768) cloned into plasmid pGEM3Zf(+) (1). The plasmids were linearized and used as templates for in vitro transcription with T7 RNA polymerase. In vitro transcription reactions were conducted in the presence of [α-32P]UTP (specific activity, 3,000 Ci/mmol), and the resultant RNA product was gel purified through 8% PAGE—8 M urea. The transcripts were labeled to a specific activity of approximately 3 × 108 cpm/μg.
RNA-protein UV cross-linking and EMSA.
A 32P-labeled cRNA template (50,000 cpm) was incubated with 500 ng of GST–Apobec-1 in a binding buffer containing 20 mM HEPES (pH 7.9), 100 mM KCl, and 1 mM DTT for 20 min at room temperature and then treated sequentially with RNase T1 (1-U/ml final concentration) and heparin (5-mg/ml final concentration). The mixture was then subjected to UV cross-linking on ice in a Stratalinker (Stratagene, La Jolla, Calif.) with an energy of 250 mJ/cm2 and resolved by SDS–10% PAGE under reducing conditions. For electrophoretic mobility shift assay (EMSA), the mixture was immediately analyzed (after RNase T1 and heparin addition) by electrophoresis in a 5% native PAGE (37.5:1) using 45 mM Tris borate–0.1 mM EDTA, pH 8.6. Competitor RNA was added where indicated at 250 ng per reaction mixture. In the supershift assay, 1 μl of undiluted anti-Apobec-1 antibody was added, and incubation continued for a further 20 min before treatment with RNase T1 and heparin.
Nitrocellulose filter-binding assays.
GST–Apobec-1 was diluted to the desired concentration in 10 μl of binding buffer and added to 20 μl of binding buffer containing 1.5 nM labeled cRNA (15,000 cpm). The reaction mixture was incubated for 20 min at room temperature. The mixture was then passed through a nitrocellulose filter (HAWP; 0.45-μm pore size) (Millipore) that was preequilibrated with binding buffer. The filter was washed extensively with the binding buffer solution, dried, and analyzed by scintillation spectroscopy (Model 1500; Packard, Downers Grove, Ill.) using Ultima Gold scintillation cocktail (Packard). Each point in the binding curve represents the average of binding reactions performed in triplicate. Dissociation constants were determined as previously described (11). Control experiments revealed that GST alone exhibited no apoB RNA binding activity (references 1 and 2 and data not shown).
In vitro conversion and primer extension assay.
Twenty femtomoles of either the 105- or a 470-nt rat apoB cRNA (nt 6512 to 6982) was incubated with 10 μg of chicken intestinal S-100 extracts and 500 ng of GST–Apobec-1 in a buffer containing 20 mM HEPES (pH 7.9), 100 mM KCl, and 1 mM DTT. Control reactions contained 250 ng of tRNA, and where indicated, 250 ng of competitor RNA was substituted for tRNA. The mixture was incubated for 3 h at 30°C. The RNA was extracted, annealed to 100 pg of a 32P-end-labeled primer extension primer (nt 6705 to 6671), and reverse transcribed with 20 U of Moloney murine leukemia virus reverse transcriptase at 42°C for 1 h in a buffer containing 50 mM (each) dATP, dCTP, and dTTP and 250 mM ddGTP. The extended products were then fractionated by gel electrophoresis in an 8 M urea–8% polyacrylamide gel. The gel was dried and analyzed by autoradiography on XAR-5 film.
Circular-permutation analysis.
RNA was synthesized by in vitro transcription as described above except that a fivefold molar excess of 5′ GMP was added to the reaction to generate RNA with 5′ monophosphate (22, 40, 41). The RNA was then circularized with T4 RNA ligase in a buffer containing 50 mM Tris HCl (pH 7.6), 10 mM MgCl2, 10 mM β-mercaptoethanol, 0.2 mM ATP, 0.1 mg of bovine serum albumin/ml, and 15% dimethyl sulfoxide for 2 h at 37°C. Alkaline lysis was performed with ∼0.7 μM purified circular RNA by boiling the mixture for 1 min in buffer containing 1 mM glycine and 0.4 mM MgSO4, pH 9.5. The hydrolysis mixture was neutralized by the addition of 120 mM Tris HCl (pH 7.5)–1 mM EDTA, renatured by heating it to 95°C for 2 min, and cooled quickly on ice. The population of RNA molecules was then bound to Apobec-1 and filtered through a nitrocellulose filter membrane as described above. The Apobec-1-bound RNA molecules were eluted from the membrane and ethanol precipitated in 50 mM potassium acetate and 0.2 M KCl. To determine the 5′ ends of the circularly permutated RNAs, the eluted RNAs were annealed with a molar excess of 32P-labeled primer (5′-GATTCTATCAATAATCTG-3′) and reverse transcribed with Moloney murine leukemia virus reverse transcriptase in a buffer containing 1 mM deoxynucleoside triphosphates for 1 h at 42°C. The products were analyzed on a 6% denaturing urea gel and autoradiographed as described above.
Cell culture and RNA turnover studies.
F442A preadipocyte cells were obtained from Reed Graves (University of Chicago) and maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics (50 μg of penicillin/ml, 50 μg of streptomycin/ml, and 2 μg of gentamycin/ml). The cells were transfected with Apobec-1 cloned into a eukaryotic expression plasmid, pRC/CMV (InVitrogen), using Lipofectamine (Life Technologies, Gaithersburg, Md.), and individual colonies were selected in the presence of 800 μg of G418/ml. The clones were screened by Northern blot analysis of total RNA, which was normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA (24). Protein extracts were prepared from positive clones, resolved through denaturing SDS–12.5% PAGE, and transferred to Immobilon-P membranes. The blots were probed with anti-Apobec-1 antibody followed by secondary anti-rabbit immunoglobulin G (IgG) conjugated to horseradish peroxidase. The immunoreactive Apobec-1 was detected by enhanced chemiluminescence (Amersham Life Sciences, Arlington Heights, Ill.). For decay studies with stable vector and Apobec-1-expressing cell lines, 106 cells were seeded in a 10-cm-diameter dish and grown for 48 h in DMEM–10% FBS and then washed three times in phosphate-buffered saline and serum starved in DMEM–0.5% FBS for 48 h prior to stimulation with DMEM–15% FBS. After 1 h, actinomycin D (10-μg/ml final concentration) was added. For studies involving mutant apobec-1, namely, H61R and F66,87L, a transient-transfection strategy was used. Twenty-four hours after a 35-mm-diameter dish was seeded with 5 × 104 cells, 3 μg of appropriate plasmid DNA was transfected using FuGene (Roche Biochemicals, Nutley, N.J.). Forty-eight hours posttransfection, fresh medium containing 15% FBS was added to the cells, and the mixture was incubated for 1 h followed by treatment with actinomycin D as detailed above. At the indicated time points, total cellular RNA was isolated from the cells using TRIzol reagent (Life Technologies). Ten micrograms of total RNA was size fractionated in a 1% agarose–2% formaldehyde gel and blotted onto a Nytran-Plus membrane (Schleicher and Schuell, Keene, N.H.). Hybridization was performed in QuikHyb solution (Stratagene) with a full-length mouse c-myc cDNA probe (a kind gift from K. N. Subramanian, University of Illinois) followed by a mouse β-actin cDNA probe (Ambion, Austin, Tex.). Hybridization intensity was quantitated by PhosphorImager scanning (model PSI; Molecular Dynamics, Sunnyvale, Calif.).
RESULTS
GST–Apobec-1 binds with low affinity to apoB RNA.
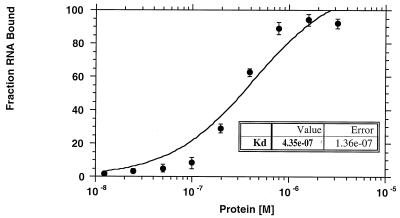
Previous work has determined that the ability of Apobec-1 to bind apoB RNA is required for RNA editing (34, 37), a function independent of its cytidine deaminase activity (34). To determine the binding affinity of Apobec-1 to the apoB mRNA, nitrocellulose filter binding was performed using a radiolabeled 105-nt rat apoB RNA in the presence of increasing concentrations of GST–Apobec-1. This apoB RNA template supports optimal in vitro editing, with greater than 85% C-to-U conversion (data not shown), suggesting that the requisite cis-acting elements are present, as expected, within this fragment (4, 15, 19). As seen in Fig. 1, Apobec-1 binds this apoB RNA saturably, with a calculated Kd of 435 ± 136 nM. By comparison, the binding affinity of apobec-1 to a 150-nt chicken apoB RNA was determined to be 570 ± 107 nM. This represents the lower limit of detection, a finding consistent with previous data (1) demonstrating that this template does not bind to apobec-1 in UV cross-linking assays.
FIG. 1.
Binding affinity of GST–Apobec-1 to rat apoB RNA. Increasing amounts of GST–Apobec-1 (1.2 × 10−8 to 3.2 × 10−6 M) were bound to 15,000 cpm of 32P-radiolabeled 105-nt rat apoB transcript (RB105) and incubated at room temperature for 20 min. The mixture was then filtered through a nitrocellulose membrane, and the retained material was analyzed by scintillation spectroscopy. The data are plotted as the fraction of RNA bound (mean ± standard deviation) to the indicated amount of GST–Apobec-1 (Protein [M]). Each point represents data from three independent experiments.
GST–Apobec-1 binds to AU-rich sequence elements within the 3′ UTR of c-myc, IL-2, TNF-α, and GM-CSF RNA.
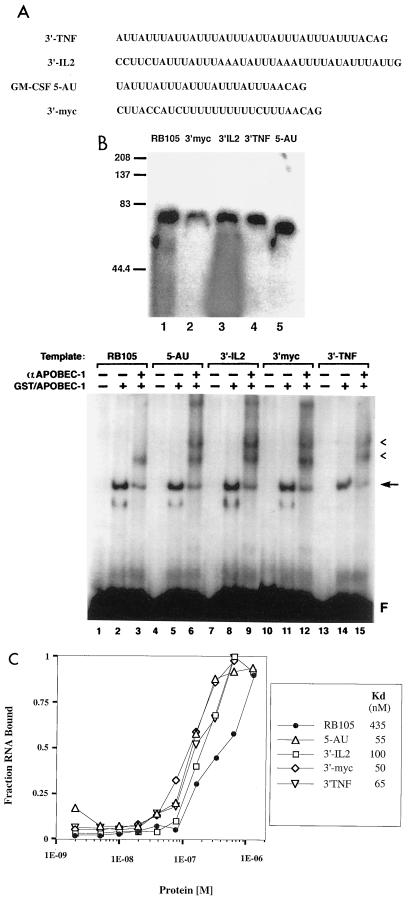
To further define the optimal binding site for Apobec-1, we examined the binding of Apobec-1 to transcripts containing different configurations of AU sequences found, respectively, in the 3′ UTRs of c-myc, interleukin 2 (IL-2), tumor necrosis factor alpha (TNF-α), and granulocyte/macrophage colony stimulating factor (GM-CSF). c-myc contains a series of U nucleotides and four dispersed AUUUA sequence elements, while IL-2, TNF-α, and GM-CSF have at least three tandem repeats of the sequence AUUUA (Fig. 2A). Radiolabeled RNA transcripts were synthesized and incubated with GST–Apobec-1, and the UV cross-link pattern was examined. In all cases, RNA-bound protein was observed with a mobility of ∼75 kDa (Fig. 2B, top). EMSA further confirmed the interaction of Apobec-1 with these various RNAs (Fig. 2B, bottom). Inclusion of anti-Apobec-1 antibody in the reaction led to a further shift in mobility of the bound RNA, confirming the specific interaction of Apobec-1 with these RNAs (Fig. 2B, bottom). The supershift with the rat apoB template showed a single band, whereas that observed for c-myc, IL-2, TNF-α, and GM-CSF showed multiple supershifted bands (Fig. 2B, bottom). In view of the presence of multiple copies of AU-rich sequence elements within these cytokine mRNAs, the most plausible explanation for this supershift pattern is that binding of Apobec-1 may have occurred at multiple sites, leading to formation of a higher-order complex in the presence of antibody. This supposition, however, will require experimental confirmation. Nitrocellulose filter binding assays were performed with these cytokine mRNAs, and the binding affinities were determined to be 50, 55, 65, and 100 nM for c-myc, GM-CSF, TNF-α, and IL-2, respectively (Fig. 2C). This suggests that Apobec-1 binds to the AU-rich sequences in the 3′ UTRs of several cytokine mRNAs with almost 10-fold-higher affinity than to apoB mRNA. The functional implications of this suggestion were examined and will be presented in a later section of this report.
FIG. 2.
RNA-protein complex formation between GST–Apobec-1 and AU-rich sequences present in the 3′ UTRs of rapidly degraded RNAs. (A) AU-rich sequences present in 3′ UTRs of TNF-α, IL-2, GM-CSF, and c-myc. These sequences were cloned into plasmid pGem-3Zf(+), linearized, and transcribed in the presence of [α-32P]UTP with T7 RNA polymerase. (B) Top: UV cross-linking. Five hundred nanograms of GST–Apobec-1 was incubated with 50,000 cpm of the indicated radiolabeled transcript followed by incubation with RNase T1 and heparin, subjected to UV cross-linking for 1.5 min, and analyzed by SDS–10% PAGE. The migration of the molecular mass markers is shown. Bottom: EMSA. GST–Apobec-1 was incubated with the indicated 32P-labeled RNA templates in the presence (+) or absence (−) of affinity-purified rabbit anti (α)-Apobec-1 IgG, and the resulting complexes were analyzed by nondenaturing polyacrylamide gel electrophoresis. Migration of the free probe (F), the GST–Apobec-1–RB105 complex (arrow), and the supershifted bands (arrowheads) is indicated. (C) Affinity of GST–Apobec-1 for AU-rich templates. Increasing amounts of GST–Apobec-1 (1.2 × 10−8 to 3.2 × 10−6 M) were bound to 15,000 cpm of the indicated 32P-radiolabeled transcript followed by filtration through a nitrocellulose membrane, and the retained material was analyzed by scintillation spectroscopy. The data are plotted as the fraction of RNA bound (mean ± standard deviation) to the indicated amount of GST–Apobec-1 (protein [M]). Each point represents data from three independent experiments.
At least two tandem repeats of an AUUU sequence are required for GST–Apobec-1 binding.
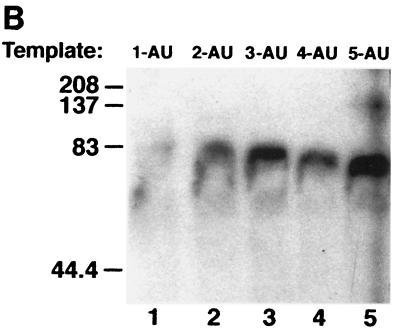
Since GST–Apobec-1 binds to AU-rich transcripts, especially AUUU multimers, we wished to determine the number of tandem AUUU repeats required for GST–Apobec-1 binding. Accordingly, UV cross-linking assays were undertaken with transcripts containing one to five tandem repeats of AUUU (Fig. 3A). GST–Apobec-1 efficiently cross-linked to transcripts containing 2-AU to 5-AU but did not cross-link to the 1-AU template (Fig. 3B). The overall AU content of the various templates ranged from 48 to 58%, lower than that of the apoB RNA discussed above (68%). These findings imply a structural requirement for apobec-1 binding, beyond the AU content of the template.
FIG. 3.
Two tandem repeats of AUUU sequence are required for Apobec-1 binding. (A) Radiolabeled cRNAs were prepared containing one to five copies of an AUUUA motif (labeled 1-AU to 5-AU) and used in the in vitro UV cross-linking assays with GST–Apobec-1. The AUUU repeat in each transcript is underlined. Except for variations in AUUU iterations, each transcript contained the same flanking nucleotide sequence. (B) GST–Apobec-1 was incubated with the radiolabeled transcripts, subjected to UV cross-linking, and analyzed by SDS–10% PAGE. The migration of molecular mass markers is indicated.
Refinement of the binding site of GST–Apobec-1 to an AU-rich target.
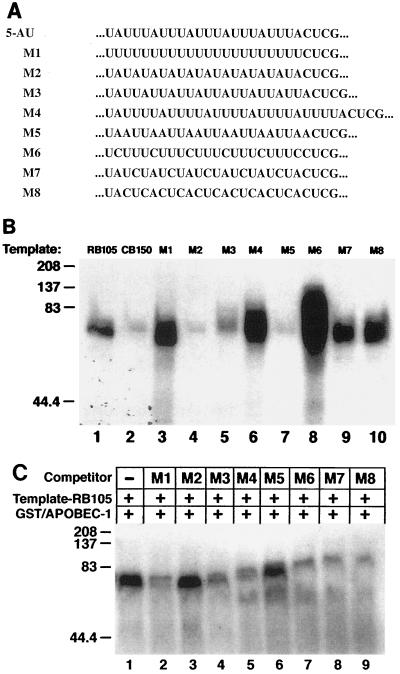
In order to further refine the sequence requirement for Apobec-1 binding, RNA transcripts were prepared from plasmids containing different AU-rich sequences in place of the 5-AU cassette described above (Fig. 4A). In this approach, the lengths and flanking sequences of these various AU-rich RNA templates were the same as in the 5-AU construct. Cross-linking experiments with these radiolabeled transcripts and GST–Apobec-1 (Fig. 4B) show that M1 (UUUU), M4 (UUUUA), and M6 (UUUC) exhibit more-prominent cross-linking activity than rat apoB (RB105). This impression was formally examined using immobilized GST–Apobec-1, revealing that the Kds of M1, M4, and M6 were in the range of 100 to 210 nM (data not shown). Transcripts M3 (UUA), M7 (UCUA), and M8 (CUCA) demonstrated weak cross-linking activity to GST–Apobec-1. In contrast, M2 (AUAU) and M5 (AAUU) templates, similarly to chicken apoB (CB150), showed no cross-linking to GST–Apobec-1 (Fig. 4B). By way of confirmation, a 1,000-fold excess of unlabeled M2 and M5 templates failed to compete for GST–Apobec-1 cross-linking to a radiolabeled RB105 template while the remaining transcripts generally competed in proportion to their apparent binding affinities (Fig. 4C). Modeling predictions suggest that RNA templates demonstrating cross-linking activity with GST–Apobec-1 contain at least 6 nt within a loop structure (data not shown). These predictions for M2 and M5 templates indicate that each contains a small loop with less than 6 nt (data not shown). Using the structural predictions alluded to above and the experimentally determined hierarchy of binding to GST–Apobec-1, a consensus binding site is proposed as shown in Fig. 5. The sequences are listed in descending order of observed cross-linking affinity. Based upon this sequence alignment, the hexamer UUUN(A/U)U emerges as a candidate Apobec-1 binding motif. This sequence is present in all transcripts demonstrating strong cross-linking with GST–Apobec-1.
FIG. 4.
GST–Apobec-1 binding to different AU-rich templates as a means to identify a consensus Apobec-1 binding site. (A) The AUUU sequence in construct 5-AU (shown underlined in Fig. 3A) was replaced with the indicated cassette, labeled M1 to M8. The flanking sequences in the various transcripts were identical. (B) UV cross-linking experiments were performed with GST–Apobec-1 and the indicated radiolabeled M transcripts, and the complex was analyzed by SDS–10% PAGE. RB105 (105-nt rat apoB cRNA; positive control) and CB150 (150-nt chicken apoB RNA; negative control) templates were also used in the cross-linking reaction. The migration of molecular mass markers is shown on the left. (C) Competition by the indicated M transcripts for GST–Apobec-1 binding to rat apoB RNA. Excess unlabeled M template cRNA was added to binding reaction mixtures containing radiolabeled RB105 and GST–Apobec-1. Following incubation, the reaction mixture was subjected to UV cross-linking followed by separation by SDS–10% PAGE and autoradiography. The migration of the molecular mass markers is shown on the left. +, present; −, absent.
FIG. 5.
Identification of a consensus Apobec-1 binding motif. UV cross-linking experiments to determine GST–Apobec-1 binding activity were performed (as shown in Fig. 4), and the sequences were arranged based on cross-linking efficiency. A consensus binding motif (UUUN[A/U]U) was derived from the alignment (shaded area).
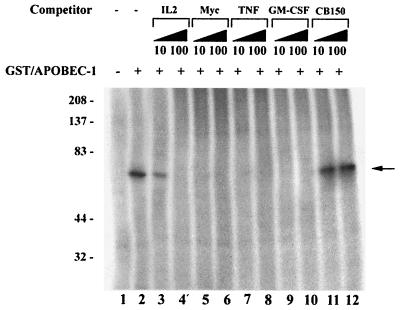
The results further suggest that binding occurs with higher affinity when multiple overlapping copies of this motif are present. Specifically, the IL-2 template, which contains two scattered UUUN(A/U)U motifs, binds to immobilized Apobec-1 with lower affinity than the c-myc and GM-CSF templates (100 vs. 50 to 55 nM) (Fig. 2C). Additionally, cross-competition experiments were conducted using a 10- or 100-fold excess of cold unlabeled IL-2, c-myc, GM-CSF, and TNF-α RNA templates in UV cross-linking assays containing GST–Apobec-1 and radiolabeled RB105. As shown in Fig. 6, a 100-fold but not 10-fold excess of the cold IL-2 template was required to inhibit GST–Apobec-1 from cross-linking to the RB105 template. On the other hand, a 10-fold excess of c-myc, GM-CSF, or TNF-α RNA was sufficient to inhibit the cross-linking reaction (Fig. 6). Furthermore, even a 100-fold excess of chicken apoB RNA—which lacks the consensus binding site but contains a similarly high AU content to the mammalian apoB RNA—failed to compete Apobec-1 binding to the rat apoB template (Fig. 6, lanes 11 and 12). Taken together, these data strongly suggest that multiple tandem repeats of the sequence UUUN(A/U)U may constitute a high-affinity binding site for Apobec-1. The findings also serve as a plausible basis for the low binding affinity of GST–Apobec-1 to the 105-nt rat apoB RNA (Fig. 1), which contains only a single copy of this motif.
FIG. 6.
Multiple tandem repeats of the consensus sequence make up the Apobec-1 high-affinity binding site. UV cross-linking experiments were performed with GST–Apobec-1 and radiolabeled RB105 RNA in the presence of (+) of 10- or 100-fold excess of unlabeled c-myc (Myc), TNF-α (TNF), IL-2 (IL-2), GM-CSF, or chicken apoB (CB150) transcripts and subjected to UV cross-linking. Following cross-linking, the reaction mixture was separated in an SDS–10% PAGE and autoradiographed. The migration of molecular mass markers (kilodaltons) is shown on the left. The location of the GST–Apobec-1–RNA cross-linked complex is indicated by an arrow. This is a representation of two independent tests.
Mapping the Apobec-1 binding site in apoB mRNA through circular-permutation analysis.
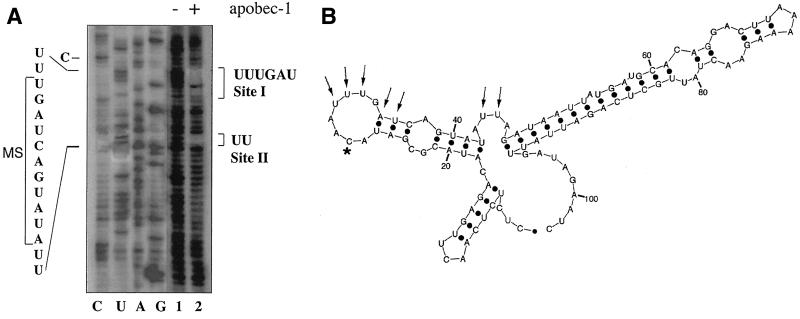
The consensus binding motif identified above (UUUN[A/U]U) is present in rat apoB RNA at a single site (UUUGAU) immediately 3′ to the edited base (Fig. 5). Circular-permutation analysis was performed in order to determine directly whether Apobec-1 interacts with this sequence element in apoB RNA. As detailed above (see Materials and Methods), a mixture of circularly permutated RNAs was generated by partial alkaline hydrolysis of a circularized 105-nt apoB RNA, which was then bound to GST–Apobec-1. The Apobec-1-bound RNA molecules were purified, annealed to excess primer, and subjected to reverse transcription and direct sequencing of the 5′ ends. The sequence information was then compared with the sequence determined using the circular RNA as a template. As shown in Fig. 7A, two distinct regions were identified as Apobec-1 binding sites. One site (site I) is the predicted UUUGAU sequence, located immediately downstream of the edited base. A second site (site II) for Apobec-1 binding was located downstream of the first site and is part of a UAUAUU sequence. Site II was previously determined to be an Apobec-1 binding site based on deletion analysis and UV cross-linking (37). Structural predictions suggest that site I is located within a distinct loop structure containing 8 nt while site II is located within a bulge containing 4 nt (Fig. 7B). The model for the region containing site I is predicted to position the edited C within the same 8-nt loop (Fig. 7B).
FIG. 7.
Direct determination of Apobec-1 binding sites in rat apoB RNA. (A) A circular-permutation assay (CPA) was performed with GST–Apobec-1 and a 105-nt RB105 cRNA. RB105 RNA was circularized and subjected to partial alkaline hydrolysis under denaturing conditions to generate a complete population of linear circular-permutated (CP) isoforms. The CP isomers were mixed with GST–Apobec-1, and the bound isomers were recovered following filtration through a nitrocellulose membrane. Bound RNAs were identified by primer extension with Moloney murine leukemia virus reverse transcriptase using a 32P-labeled primer located at the 3′ end of the apoB sequence. Total CP isomers (lane 1) and GST–Apobec-1-bound isomers (lane 2), along with reverse transcriptase sequencing of the circular form of RB105 (lanes C, U, A, and G), were separated in an 8 M urea–8% polyacrylamide gel and autoradiographed. The sequence of interest is shown on the left, and the 11-nt mooring sequence motif (MS) is bracketed. The two Apobec-1 binding sites (sites I and II) are shown on the right. (B) Schematic representation of structure of a 105-nt apoB RNA, including the binding sites for Apobec-1 (determined by CPA). RNA folding was determined by using the RNA mfold program. RB105 forms a three-branch structure, and the Watson-Crick (A:U and G:C) and the weaker G:U wobble pairs are shown as black dots. Nucleotides required for Apobec-1 binding and the edited C are shown by arrows and an asterisk, respectively.
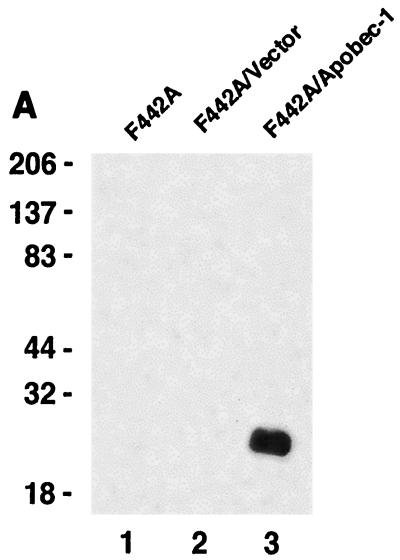
Binding of Apobec-1 to its consensus site within the 3′ UTR of c-myc mRNA alters RNA stability.
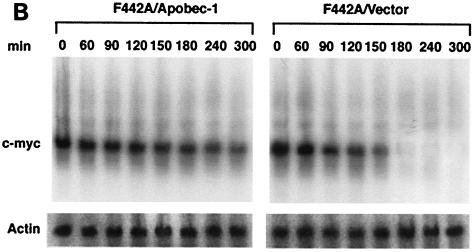
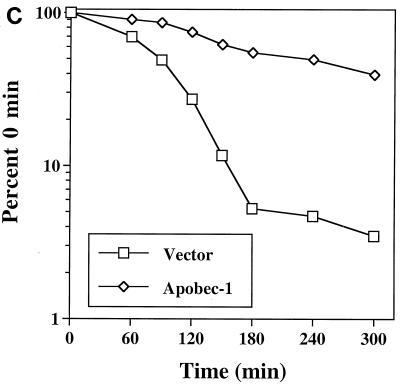
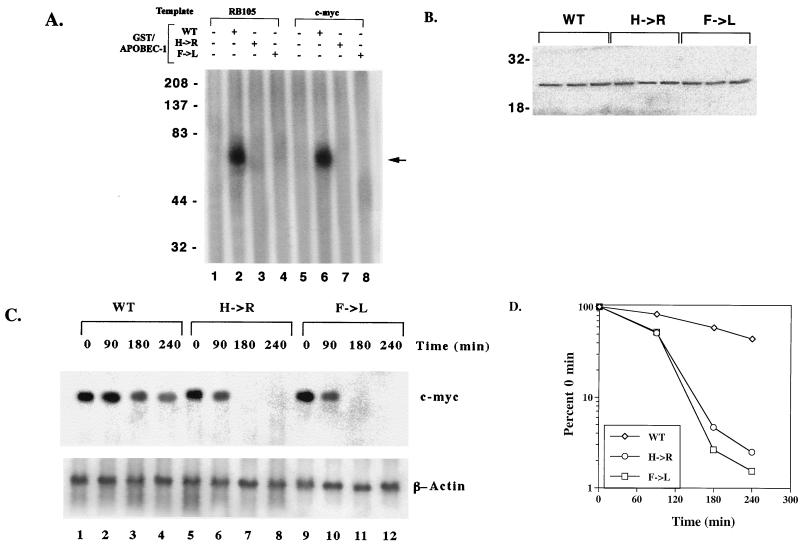
AU-rich sequences within the 3′ UTR of c-myc mRNA are known to contain RNA-destabilizing elements, the presence of which triggers rapid degradation of the transcript (10, 27, 28). The presence of a consensus binding site for Apobec-1 that appears to be embedded within the 3′ UTR of c-myc RNA led us to examine the hypothesis that Apobec-1 binding to this target may play a role in RNA stability. In order to determine the role of Apobec-1 in mRNA stability, we used a preadipocyte cell line, F442A, since these cells express neither endogenous apoB mRNA (data not shown) nor Apobec-1 (Fig. 8A). Accordingly, F442A cells were stably transfected with rat Apobec-1, and high levels of protein expression were confirmed in S-100 extracts probed with affinity-purified anti-Apobec-1 IgG (Fig. 8A). To determine the effects of overexpression of Apobec-1 on c-myc mRNA stability, total cellular RNA was isolated at various time points following the addition of actinomycin D, and c-myc mRNA abundance was determined by Northern blot analysis. The half-life of c-myc mRNA increased from ∼90 min in vector-transfected control cells to ∼240 min in cells expressing Apobec-1 (Fig. 7B and C). These findings suggest that Apobec-1 binding to its target consensus site within c-myc mRNA results in stabilization of the transcript. In order to determine whether the effects of Apobec-1 expression on c-myc mRNA stability require its RNA binding activity, further experiments were conducted using Apobec-1 mutants defective in RNA binding. Two such mutants were selected, a histidine61-to-arginine mutant and a phenylalanine66,87-to-leucine mutant, each of which is defective in apoB RNA binding and exhibits no C-to-U RNA-editing activity (34, 37). To confirm that these mutants do not bind c-myc RNA, wild-type GST–Apobec-1 and the H61R (denoted H→R) and F66,87L (F→L) mutants were incubated with either the rat apoB RNA (RB105) or c-myc RNA, and UV cross-linking was performed. As shown in Fig. 9A, the wild-type protein, but not the mutant proteins, cross-linked to both templates. This suggested that both the H→R and F→L mutants would be good candidates for determining the RNA binding requirement of Apobec-1 in c-myc mRNA stability. Accordingly, each of these mutants was transiently transfected into F442A cells, in parallel with the wild-type parental construct, and c-myc mRNA turnover was examined following actinomycin D treatment, as before. Comparable levels of expression of each of the wild-type and mutant Apobec-1 proteins were observed, indicating that transfection and expression efficiencies were similar (Fig. 9B). Expression of the wild-type Apobec-1 resulted in stabilization of c-myc mRNA, as observed earlier (compare Fig. 9 and Fig. 8), while transfection of the RNA binding-defective Apobec-1 mutants resulted in c-myc mRNA turnover that was indistinguishable from control vector-transfected cells (Fig. 8). Taken together, these data strongly suggest that it is the RNA binding activity of Apobec-1 that results in stabilization of the c-myc transcript.
FIG. 8.
Binding of Apobec-1 to AU-rich sequence in 3′ UTR of c-myc mRNA increases c-myc mRNA stability. (A) Wild-type Apobec-1 cDNA was transfected into F442A cells, and stable transfectants were selected with G418 (F442A/apobec-1). As a control, vector alone was transfected and colonies were isolated (F442A/vector). Cytosolic S100 extracts were subjected to 30% ammonium sulfate precipitation, and aliquots were separated in an SDS–12% PAGE and blotted on a polyvinylidine difluoride membrane. The blot was probed with affinity-purified rabbit anti-Apobec-1 IgG, and bands were visualized by enhanced chemiluminescence. Molecular mass markers (kilodaltons) are indicated on the left. (B) Northern blot analysis of c-myc mRNA. Cells were grown to ∼90% confluence, and actinomycin D (final concentration, 10 μg/ml) was added. At the indicated time points, RNA was extracted, size separated in a formaldehyde-agarose gel, and blotted onto Nytran-Plus membranes. The blots were probed sequentially with c-myc and mouse β-actin cDNAs. (C) Hybridization was quantitated with a PhosphorImager and normalized to β-actin. Data from five independent experiments were averaged (mean ± standard deviation) and are presented as a percentage of c-myc mRNA remaining, relative to that at time zero. Error bars fall within the symbol for the mean.
FIG. 9.
RNA binding mutants of Apobec-1 do not stabilize c-myc mRNA. (A) Wild-type (WT) and mutant (H→R and F→L) Apobec-1 cDNAs were expressed as GST fusion proteins and affinity purified over glutathione-agarose beads. Two hundred fifty nanograms of the indicated fusion protein was incubated with either radiolabeled RB105 or c-myc transcript, subjected to UV cross-linking, and analyzed by SDS–10% PAGE. The location of the GST–Apobec-1–RNA cross-linking complex is indicated by an arrow. The migration of molecular mass markers (kilodaltons) is indicated. This is a representation of two independent tests. (B) WT and mutant (H→R and F→L) Apobec-1 cDNAs were transiently transfected into F442A cells. Total cellular extracts were prepared, and aliquots were separated in an SDS–12% PAGE and transferred to a polyvinylidine difluoride membrane. The blot was probed with affinity-purified rabbit anti-Apobec-1 IgG, and the bands were visualized by enhanced chemiluminescence. The migration of molecular mass markers (kilodaltons) is indicated on the left. (C) WT and mutant (H→R and F→L) Apobec-1 cDNAs were transfected into F442A cells. Seventy-two hours after transfection, actinomycin D was added, the cells were incubated for the indicated times, and RNA was isolated. The RNA was subjected to Northern blot hybridization and probed with mouse c-myc and β-actin cDNAs. (D) The blots were scanned by PhosphorImager, and hybridization to the c-myc transcript was normalized to that of β-actin. The data from three independent transfections were averaged (mean ± standard deviation) and are presented as a percentage of c-myc mRNA remaining, relative to that at time zero. Error bars fall within the symbol for the mean.
DISCUSSION
C-to-U deamination of nuclear mRNA is a highly restricted process in mammalian tissues. The operational constraints, however, are poorly understood. In the context of mammalian apoB RNA, the prototype for this category of RNA editing, the enzymatic modification targets a single C in a region that is enriched in A+U residues, suggesting that a defined structural conformation of the RNA may direct the editing machinery to the selected site and simultaneously make other potential sites less favorable. This suggestion was strengthened through studies that demonstrated Apobec-1 binding to apoB RNA as well as to a variety of AU- or U-rich targets (1, 37). In addition, functional analyses of recombinant apobec-1 protein have demonstrated that mutations which abolish RNA binding activity of the protein also eliminate its RNA-editing activity (34, 37, 38). Taken together, these earlier findings strongly implicate RNA binding in regulating apoB RNA editing, although details of the structural and more specific sequence requirements were left unanswered. The current studies sought to delineate the RNA binding activity of Apobec-1 in some detail, both in the context of apoB RNA and also other potential targets.
One of the central observations of this report concerns Apobec-1 binding to a 105-nt apoB RNA that is now shown to contain a high-affinity consensus binding site embedded within the region flanking the edited base. The binding affinity of Apobec-1 for the apoB RNA, ∼430 nM, is considerably lower than that of the consensus site (∼50 nM), a finding that has important functional implications. The presence of a high-affinity binding site for Apobec-1, contained in proximity to and 3′ of the edited base, may be of importance in positioning the canonical C within the active site of the homodimeric interface (38). Such optimal positioning with respect to the enzyme and its substrate may be of importance in regulating the C-to-U RNA editing activity of Apobec-1. This suggestion is broadly consistent with the observations of others that scrambling mutants of apoB RNA in which the region immediately 3′ to the edited base (containing the site I consensus) do not support C-to-U editing in vitro (4, 47). However, it is recognized that other explanations, including structural alterations in RNA folding, are equally plausible (44). A key consideration in assigning significance to the structural predictions and binding affinities currently reported is whether the enzymatic (i.e., editing) activity of Apobec-1 is favored or inhibited by high-affinity binding to a target RNA. This is of particular relevance, since Apobec-1 is one of a few well-characterized genes with both RNA binding and enzymatic activities (reviewed in reference 16). The demonstration that Apobec-1 binds to a 105-nt apoB RNA with low affinity is consistent with a scanning model in which Apobec-1 exhibits loose interactions with RNA targets enriched in AU residues. The model further predicts that upon recognition of an appropriate secondary-structural context, Apobec-1 undergoes selective, high-affinity binding, mediated through the consensus motif. This sequential-scanning-high-affinity-binding model may then facilitate a conformational adaptation that allows C-to-U deamination at the appropriate site. It bears emphasizing that, supplemental to its cytidine deaminase activity on a monomeric substrate, Apobec-1 exhibits an obligate requirement for an additional protein cofactor(s) for RNA-editing activity. These cofactors, in turn, have the ability to bind both apoB RNA (19, 25, 29, 31, 39) and apobec-1 (23, 31, 35, 36). As an example, one of these potential cofactors, p60, has been shown to bind to a UGAU sequence located downstream of the edited C, a motif that is contained within the Apobec-1 consensus site I (39). Previous studies suggest that the binding of a protein factor(s) present in chicken intestinal S100 extracts (a source of complementation activity but not Apobec-1 [50]) to the apoB RNA template is not competitive with the binding of Apobec-1 (2). Nevertheless, it remains a distinct possibility that the apparent binding affinity of recombinant GST–Apobec-1 alone may differ considerably from its affinity when examined in the context of the holoediting enzyme. Formal proof of this possibility will await identification and expression of the remaining components of the editing enzyme. By corollary, the importance of the consensus motif for Apobec-1 in regulating the specificity of the cytidine nucleotide selected for deamination by the apoB editing enzyme will require formal evaluation in the context of other targets.
The structural predictions with respect to apoB RNA binding, based upon modeling of a 105-nt RNA, are consistent with recent results from Richardson and colleagues, who used RNase mapping to delineate an RNA secondary structure for a 46-mer apoB RNA flanking the editing site (44). Both studies, using independent approaches, suggest that apoB RNA forms a stable hairpin stem-loop structure with the edited C located in an 8-nt loop (44). Using circular-permutation assays, the present studies indicate the presence of a binding site, UUUGAU (site I), spanning nt 6669 to 6674 (Fig. 7). This binding site is of potential functional significance, since elimination of the UGAU motifs flanking the edited C in chimeric editing constructs—one of which is contained within the site I binding motif—appears to reduce C-to-U editing efficiency, particularly under conditions of forced overexpression of Apobec-1 (55). A second site (site II) identified UU residues at positions 6682 to 6683 (Fig. 7) as a binding site for Apobec-1. The latter findings are consistent with findings from earlier studies, specifically the suggestion that Apobec-1 binds to AU-rich regions 3′ of the edited C in apoB RNA and in particular to a region encompassing nt 6678 to 6683 (37). It is possible that the UU residues (site II) identified by circular permutation represent the most exposed nucleotides within the UAUAUU motif spanning nt 6678 to 6683, previously proposed as the Apobec-1 binding site by Navaratnam and coworkers (37, 38). Indeed the model depicted in Fig. 6B positions these UU dinucleotides within a bulge and thus potentially accessible to Apobec-1.
It deserves comment that a different binding site was identified in human apoB RNA by Navaratnam and coworkers, located 5 nt upstream of the edited base (37). The most plausible explanation for the failure to detect this site in our circular permutation assay may be the choice of substrates. While Navaratnam et al. used the human apoB RNA in their studies, which contains the sequence AUAUAU, the present studies used rat apoB RNA as a template. In this species, the corresponding sequence is AUAcgc (sequence divergence in lowercase). The sequence divergence between species at this location suggests that this site may not be absolutely required for Apobec-1-mediated apoB RNA editing. In support of this suggestion, mutational analyses performed within this region (AUAUAU→ugAagU, AUgaAU, or AUAUgc) (mutated bases in lowercase) showed no significant effect on RNA editing (4, 19).
Our predictions for the 105-nt apoB RNA, particularly the region flanking the edited C, taken in conjunction with the predictions of Richardson and colleagues, based upon RNase mapping, thus serve as a foundation for a working model of the minimal editing template. A central feature of both models is a stem-loop structure containing ∼30 nt, with the edited C located within an 8-nt loop (44). These predictions are consistent with earlier findings that a minimum of ∼26 nt of flanking sequence will support C-to-U editing of an apoB RNA template in vitro (1, 3, 19).
Implicit in the proposed structural predictions for apoB RNA is the assumption that Apobec-1 adopts a conformational structure at its active site that will accommodate an RNA substrate of the appropriate size and structure. This issue has been addressed by homology modeling with the crystal structure of E. coli cytidine deaminase, which predicts that a cleft is formed between the two active sites of an Apobec-1 dimer (38). The distance between the two active sites, approximately 21 Å, implies that the RNA must form a higher-order stem-loop structure in which the edited C is exposed (38). The predictions emerging from the present studies with a 105-nt apoB RNA indicate that a tripartite stem-loop structure forms, with a hairpin structure of ∼30 nt containing the edited C. These predictions are consonant with the estimates of Navaratnam and colleagues that the cleft formed at the interface between the two active sites of an Apobec-1 dimer will accommodate a structure with a molecular mass of ∼10 kDa, a value that coincides with the predicted mass of a 30-nt RNA (38). It remains to be determined whether Apobec-1 binds to the RNA target as a monomer or as a dimer and, by extension, whether one or more molecules of apoB RNA are bound per molecule of Apobec-1. Resolution of these issues will require solution of the crystal structure of Apobec-1.
During the course of identifying a consensus high-affinity binding site for Apobec-1 through analysis of multiple candidate AU-rich targets, it was simultaneously discovered that Apobec-1 demonstrated high-affinity binding of other targets, including several RNAs that contain sequence motifs known to be involved in regulating mRNA degradation (reviewed in references 13 and 48). The best characterized of such motifs is the AU-rich element (ARE), which varies in size and location but generally contains multiple copies of a pentanucleotide stretch, AUUUA, coupled with a context of abundant U or AU residues (13). A number of RNA binding proteins have been identified that exhibit affinity for and interact with AREs (reviewed in references 9 and 13), and their biological properties and functional characteristics have been the focus of considerable investigation. Many of these proteins contain highly conserved RNA binding domains that place them within the RRM (RNA recognition motif) superfamily while containing differences within the hinge domain that presumably permit conformational flexibility (9, 13). These features are of interest, since Apobec-1 contains neither a recognizable RNA recognition domain nor a region similar to the hinge domain found in these proteins. Homology modeling and mutagenesis of Apobec-1 indicate that the RNA binding domain of Apobec-1 is located in the amino-terminal half of the protein and most plausibly involves the crevice created between the two monomers (38). RNA binding has been demonstrated to include some of the zinc-coordinating residues (H63 and C93) that occupy the active site of the enzyme (34, 37), as well as F66 and F87, which are positioned in the cleft and distant from the active site. While the ARE may function as a destabilizing element, this property is conferred in a highly individualized context, requiring a combination of structural features and sequence motifs (45). Additionally, the binding affinity of ARE-binding proteins may vary directly or inversely with the observed stability of the target RNA (13, 48). For example, overexpression of AUF1–hnRNP-D has been demonstrated to mediate the rapid degradation of c-myc mRNA (33), while overexpression of HuR leads to stabilization of ARE-containing mRNAs (20, 42). The present demonstration that overexpression of Apobec-1 leads to stabilization of c-myc mRNA is thus consistent with the preceding observations demonstrating high-affinity binding to one of the AREs present in the 3′ UTR of this mRNA (18). To clarify the functional properties of Apobec-1 that may mediate this observation, two mutants were utilized in transfection experiments. An H61R mutant was selected, since it lacks RNA binding and C-to-U RNA-editing activity and is also defective in cytidine deaminase activity (34, 37), while the F66,87L mutant, which is also defective in RNA binding and C-to-U editing (37), retains full enzymatic activity on a monomeric substrate (data not shown). The data demonstrate conclusively that RNA binding activity of Apobec-1 is required for interaction with the c-myc ARE, since both the H61R and the F66,87L mutants are without effect on c-myc mRNA turnover (Fig. 8). The possibility was considered that the C located within the loop of the c-myc ARE might itself be a target for deamination, in view of the high-affinity binding in vitro and the observed effects of overexpression of Apobec-1 on its stability. However, no evidence was found for C-to-U editing of any cytidine nucleotide within the ARE following primer extension analysis of c-myc RNA extracted after overexpression of Apobec-1 in F442A cells (data not shown).
A further point for consideration is that c-myc mRNA contains multiple destabilizing elements that are located both in the protein coding region and the 3′ UTR (45). Further studies will be required to determine whether Apobec-1 binds to any or all of these elements in vitro. Nevertheless, it is tempting to speculate whether the gain-of-function phenotype described with the forced overexpression of Apobec-1 in transgenic mice and in rabbit liver (52), which produces dysplasia and carcinoma and is associated with aberrant editing of other target RNAs, including NAT1 (52, 53, 55), may also result in alterations in mRNA turnover. By corollary, it is important to delineate the role, if any, of Apobec-1 in regulating the stability of c-myc mRNA in vivo. Along these lines, it bears emphasizing that although Apobec-1 expression is limited to the human gastrointestinal tract, Apobec-1 mRNA overexpression has been observed in colonic tumors and colon tumors metastatic to the liver (32). Whether Apobec-1 functions in mediating mRNA stability in humans in locations distant from the small intestine remains to be tested. These and other questions concerning the interaction of Apobec-1 with target RNAs will be the focus of future reports.
ACKNOWLEDGMENTS
We appreciate the assistance of T. Lindsten (University of Chicago) in providing the AU-rich plasmids. We are grateful to all members of the Davidson laboratory for helpful discussions.
This work was supported by NIH grant HL 38180 to N.O.D.
REFERENCES
- 1.Anant S, MacGinnitie A J, Davidson N O. apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, is a novel RNA-binding protein. J Biol Chem. 1995;270:14762–14767. [PubMed] [Google Scholar]
- 2.Anant S, MacGinnitie A J, Davidson N O. The binding of apobec-1 to mammalian apo B RNA is stabilized by the presence of complementation factors which are required for posttranscriptional editing. Nucleic Acids Symp Ser. 1995;33:99–102. [PubMed] [Google Scholar]
- 3.Backus J W, Schock D, Smith H C. Only cytidines 5′ of the apolipoprotein B mRNA mooring sequence are edited. Biochim Biophys Acta. 1994;1219:1–14. doi: 10.1016/0167-4781(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 4.Backus J W, Smith H C. Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res. 1992;20:6007–6014. doi: 10.1093/nar/20.22.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya S, Navaratnam N, Morrison J R, Scott J, Taylor W R. Cytosine nucleoside/nucleotide deaminases and apolipoprotein B mRNA editing. Trends Biochem Sci. 1994;19:105–106. doi: 10.1016/0968-0004(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 6.Bohjanen P R, Petryniak B, June C H, Thompson C B, Lindsten T. AU RNA-binding factors differ in their binding specificities and affinities. J Biol Chem. 1992;267:6302–6309. [PubMed] [Google Scholar]
- 7.Bohjanen P R, Petryniak B, June C H, Thompson C B, Lindsten T. An inducible cytoplasmic factor (AU-B) binds selectively to AUUUA multimers in the 3′ untranslated region of lymphokine mRNA. Mol Cell Biol. 1991;11:3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bostrom K, Lauer S J, Poksay K S, Garcia Z, Taylor J M, Innerarity T L. Apolipoprotein B48 RNA editing in chimeric apolipoprotein EB mRNA. J Biol Chem. 1989;264:15701–15708. [PubMed] [Google Scholar]
- 9.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 10.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carey J, Cameron V, de Haseth P L, Uhlenbeck O C. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983;22:2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- 12.Chan L, Chang B H, Nakamuta M, Li W H, Smith L C. Apobec-1 and apolipoprotein B mRNA editing. Biochim Biophys Acta. 1997;1345:11–26. doi: 10.1016/s0005-2760(96)00156-7. [DOI] [PubMed] [Google Scholar]
- 13.Chen C Y, Shyu A B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 14.Chen S H, Habib G, Yang C Y, Gu Z W, Lee B R, Weng S A, Silberman S R, Cai S J, Deslypere J P, Rosseneu M, Gotto A M, Li W H, Chan L. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987;238:363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- 15.Chen S H, Li X X, Liao W S, Wu J H, Chan L. RNA editing of apolipoprotein B mRNA. Sequence specificity determined by in vitro coupled transcription editing. J Biol Chem. 1990;265:6811–6816. [PubMed] [Google Scholar]
- 16.Davidson N O, Anant S, MacGinnitie A J. Apolipoprotein B messenger RNA editing: insights into the molecular regulation of post-transcriptional cytidine deamination. Curr Opin Lipidol. 1995;6:70–74. doi: 10.1097/00041433-199504000-00002. [DOI] [PubMed] [Google Scholar]
- 17.Davies M S, Wallis S C, Driscoll D M, Wynne J K, Williams G W, Powell L M, Scott J. Sequence requirements for apolipoprotein B RNA editing in transfected rat hepatoma cells. J Biol Chem. 1989;264:13395–13398. [PubMed] [Google Scholar]
- 18.DeMaria C T, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 19.Driscoll D M, Lakhe-Reddy S, Oleksa L M, Martinez D. Induction of RNA editing at heterologous sites by sequences in apolipoprotein B mRNA. Mol Cell Biol. 1993;13:7288–7294. doi: 10.1128/mcb.13.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Funahashi T, Giannoni F, DePaoli A M, Skarosi S F, Davidson N O. Tissue-specific, developmental and nutritional regulation of the gene encoding the catalytic subunit of the rat apolipoprotein B mRNA editing enzyme: functional role in the modulation of apoB mRNA editing. J Lipid Res. 1995;36:414–428. [PubMed] [Google Scholar]
- 22.Gott J M, Pan T, LeCuyer K A, Uhlenbeck O C. Using circular permutation analysis to redefine the R17 coat protein binding site. Biochemistry. 1993;32:13399–13404. doi: 10.1021/bi00212a004. [DOI] [PubMed] [Google Scholar]
- 23.Greeve J, Lellek H, Rautenberg P, Greten H. Inhibition of the apolipoprotein B mRNA editing enzyme-complex by hnRNP C1 protein and 40S hnRNP complexes. Biol Chem. 1998;379:1063–1073. doi: 10.1515/bchm.1998.379.8-9.1063. [DOI] [PubMed] [Google Scholar]
- 24.Hadjiagapiou C, Giannoni F, Funahashi T, Skarosi S F, Davidson N O. Molecular cloning of a human small intestinal apolipoprotein B mRNA editing protein. Nucleic Acids Res. 1994;22:1874–1879. doi: 10.1093/nar/22.10.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris S G, Sabio I, Mayer E, Steinberg M F, Backus J W, Sparks J D, Sparks C E, Smith H C. Extract-specific heterogeneity in high-order complexes containing apolipoprotein B mRNA editing activity and RNA-binding proteins. J Biol Chem. 1993;268:7382–7392. [PubMed] [Google Scholar]
- 26.Hersberger M, Innerarity T L. Two efficiency elements flanking the editing site of cytidine 6666 in the apolipoprotein B mRNA support mooring-dependent editing. J Biol Chem. 1998;273:9435–9442. doi: 10.1074/jbc.273.16.9435. [DOI] [PubMed] [Google Scholar]
- 27.Jones T R, Cole M D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol Cell Biol. 1987;7:4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laird-Offringa I A, Elfferich P, Knaken H J, de Ruiter J, van der Eb A J. Analysis of polyadenylation site usage of the c-myc oncogene. Nucleic Acids Res. 1989;17:6499–6514. doi: 10.1093/nar/17.16.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau P P, Chen S H, Wang J C, Chan L. A 40 kilodalton rat liver nuclear protein binds specifically to apolipoprotein B mRNA around the RNA editing site. Nucleic Acids Res. 1990;18:5817–5821. doi: 10.1093/nar/18.19.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau P P, Zhu H J, Baldini A, Charnsangavej C, Chan L. Dimeric structure of a human apolipoprotein B mRNA editing protein and cloning and chromosomal localization of its gene. Proc Natl Acad Sci USA. 1994;91:8522–8526. doi: 10.1073/pnas.91.18.8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau P P, Zhu H J, Nakamuta M, Chan L. Cloning of an Apobec-1-binding protein that also interacts with apolipoprotein B mRNA and evidence for its involvement in RNA editing. J Biol Chem. 1997;272:1452–1455. doi: 10.1074/jbc.272.3.1452. [DOI] [PubMed] [Google Scholar]
- 32.Lee R M, Hirano K I, Anant S, Baunoch D, Davidson N O. An alternatively spliced form of apobec-1 messenger RNA is overexpressed in human colon cancer. Gastroenterology. 1998;115:1096–1103. doi: 10.1016/s0016-5085(98)70080-0. [DOI] [PubMed] [Google Scholar]
- 33.Loflin P, Chen C Y, Shyu A B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacGinnitie A J, Anant S, Davidson N O. Mutagenesis of apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, reveals distinct domains that mediate cytosine nucleoside deaminase, RNA binding, and RNA editing activity. J Biol Chem. 1995;270:14768–14775. [PubMed] [Google Scholar]
- 35.Mehta A, Banerjee S, Driscoll D M. Apobec-1 interacts with a 65-kDa complementing protein to edit apolipoprotein B mRNA in vitro. J Biol Chem. 1996;271:28294–28299. doi: 10.1074/jbc.271.45.28294. [DOI] [PubMed] [Google Scholar]
- 36.Mehta A, Driscoll D M. A sequence-specific RNA-binding protein complements apobec-1 to edit apolipoprotein B mRNA. Mol Cell Biol. 1998;18:4426–4432. doi: 10.1128/mcb.18.8.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Navaratnam N, Bhattacharya S, Fujino T, Patel D, Jarmuz A L, Scott J. Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell. 1995;81:187–195. doi: 10.1016/0092-8674(95)90328-3. [DOI] [PubMed] [Google Scholar]
- 38.Navaratnam N, Fujino T, Bayliss J, Jarmuz A, How A, Richardson N, Somasekaram A, Bhattacharya S, Carter C, Scott J. Escherichia coli cytidine deaminase provides a molecular model for ApoB RNA editing and a mechanism for RNA substrate recognition. J Mol Biol. 1998;275:695–714. doi: 10.1006/jmbi.1997.1506. [DOI] [PubMed] [Google Scholar]
- 39.Navaratnam N, Shah R, Patel D, Fay V, Scott J. Apolipoprotein B mRNA editing is associated with UV crosslinking of proteins to the editing site. Proc Natl Acad Sci USA. 1993;90:222–226. doi: 10.1073/pnas.90.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan T, Gutell R R, Uhlenbeck O C. Folding of circularly permuted transfer RNAs. Science. 1991;254:1361–1364. doi: 10.1126/science.1720569. [DOI] [PubMed] [Google Scholar]
- 41.Pan T, Uhlenbeck O C. Circularly permuted DNA, RNA and proteins—a review. Gene. 1993;125:111–114. doi: 10.1016/0378-1119(93)90317-v. [DOI] [PubMed] [Google Scholar]
- 42.Peng S S, Chen C Y, Xu N, Shyu A B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powell L M, Wallis S C, Pease R J, Edwards Y H, Knott T J, Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 44.Richardson N, Navaratnam N, Scott J. Secondary structure for the apolipoprotein B mRNA editing site. AU-binding proteins interact with a stem loop. J Biol Chem. 1998;273:31707–31717. doi: 10.1074/jbc.273.48.31707. [DOI] [PubMed] [Google Scholar]
- 45.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott J. A place in the world for RNA editing. Cell. 1995;81:833–836. doi: 10.1016/0092-8674(95)90002-0. [DOI] [PubMed] [Google Scholar]
- 47.Shah R R, Knott T J, Legros J E, Navaratnam N, Greeve J C, Scott J. Sequence requirements for the editing of apolipoprotein B mRNA. J Biol Chem. 1991;266:16301–16304. [PubMed] [Google Scholar]
- 48.Siomi H, Dreyfuss G. RNA-binding proteins as regulators of gene expression. Curr Opin Genet Dev. 1997;7:345–353. doi: 10.1016/s0959-437x(97)80148-7. [DOI] [PubMed] [Google Scholar]
- 49.Teng B, Burant C F, Davidson N O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 50.Teng B, Davidson N O. Evolution of intestinal apolipoprotein B mRNA editing. Chicken apolipoprotein B mRNA is not edited, but chicken enterocytes contain in vitro editing enhancement factor(s) J Biol Chem. 1992;267:21265–21272. [PubMed] [Google Scholar]
- 51.Teng B B, Ochsner S, Zhang Q, Soman K V, Lau P P, Chan L. Mutational analysis of apolipoprotein B mRNA editing enzyme (APOBEC1). Structure-function relationships of RNA editing and dimerization. J Lipid Res. 1999;40:623–635. [PubMed] [Google Scholar]
- 52.Yamanaka S, Balestra M E, Ferrell L D, Fan J, Arnold K S, Taylor S, Taylor J M, Innerarity T L. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci USA. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamanaka S, Poksay K S, Arnold K S, Innerarity T L. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 1997;11:321–333. doi: 10.1101/gad.11.3.321. [DOI] [PubMed] [Google Scholar]
- 54.Yamanaka S, Poksay K S, Balestra M E, Zeng G Q, Innerarity T L. Cloning and mutagenesis of the rabbit ApoB mRNA editing protein. A zinc motif is essential for catalytic activity, and noncatalytic auxiliary factor(s) of the editing complex are widely distributed. J Biol Chem. 1994;269:21725–21734. [PubMed] [Google Scholar]
- 55.Yamanaka S, Poksay K S, Driscoll D M, Innerarity T L. Hyperediting of multiple cytidines of apolipoprotein B mRNA by APOBEC-1 requires auxiliary protein(s) but not a mooring sequence motif. J Biol Chem. 1996;271:11506–11510. doi: 10.1074/jbc.271.19.11506. [DOI] [PubMed] [Google Scholar]
- 56.Young S G. Recent progress in understanding apolipoprotein B. Circulation. 1990;82:1574–1594. doi: 10.1161/01.cir.82.5.1574. [DOI] [PubMed] [Google Scholar]