Abstract
The SWI-SNF complex has been shown to alter nucleosome conformation in an ATP-dependent manner, leading to increased accessibility of nucleosomal DNA to transcription factors. In this study, we show that the SWI-SNF complex can potentiate the activity of the glucocorticoid receptor (GR) through the N-terminal transactivation domain, τ1, in both yeast and mammalian cells. GR-τ1 can directly interact with purified SWI-SNF complex, and mutations in τ1 that affect the transactivation activity in vivo also directly affect τ1 interaction with SWI-SNF. Furthermore, the SWI-SNF complex can stimulate τ1-driven transcription from chromatin templates in vitro. Taken together, these results support a model in which the GR can directly recruit the SWI-SNF complex to target promoters during glucocorticoid-dependent gene activation. We also provide evidence that the SWI-SNF and SAGA complexes represent independent pathways of τ1-mediated activation but play overlapping roles that are able to compensate for one another under some conditions.
The glucocorticoid receptor (GR) belongs to a large family of ligand-inducible nuclear receptors. When the receptor binds its ligand, associated heat-shock proteins are released, and the receptor can then bind to its cognate DNA response element and either activate or repress transcription of glucocorticoid-regulated genes. The major transactivation domain τ1 (amino acids [aa] 77 to 262 of the human GR), located in the N terminus of the receptor, contains a smaller fragment that represents the minimal core activation domain (τ1c)(aa 187 to 244). Both τ1 and τ1c function efficiently in yeast and have been functionally and structurally characterized (1–3, 12, 13, 15, 21, 26, 29, 30, 46, 47). The current working model suggests that the GR activates transcription by concurrent or sequential recruitment of important target factors to regulated promoters and that the τ1 domain adopts a structural conformation only upon interaction with target factors. Consistent with this, critical hydrophobic residues have been shown to play important roles in both gene activation in vivo (2) and target factor interaction in vitro (3). The τ1c has previously been shown to interact with the TATA binding protein (15), CREB-binding protein (3), and the Ada2 protein (21). Recent studies show that the τ1 can interact with the Ada2-containing histone acetyltransferase (HAT) complex SAGA, but not with the related Ada complex (43). In addition, the Ada-independent NuA4 HAT complex interacts with τ1. Furthermore, both SAGA and NuA4 can stimulate τ1-dependent transcription of chromatin templates in vitro (43).
Current models suggest that gene activation involves both derepression of a repressive chromatin structure within promoters and subsequent activation of transcription, involving recruitment of the transcriptional machinery (35). There is evidence that the GR-τ1 activation domain can participate in both of these steps (29, 30; F. Then Bergh, E. M. Flinn, J. Svaren, A. P. Wright, and W. Hörz, unpublished data). It has been previously shown that GR stimulates the nucleosome-disrupting activity of SWI-SNF complex partially purified either from HeLa cells or from rat liver tissue. The GR-mediated stimulation of SWI-SNF nucleosome disruption depended on the presence of a glucocorticoid response element, suggesting that GR is able to target the nucleosome-disrupting activity of the SWI-SNF complex (35). The SWI-SNF complex, which contains 11 known subunits, was first found in yeast (7), and several yeast SWI-SNF proteins have been shown to enhance GR transactivation activity (49). A mammalian homologue of SWI2-SNF2, hbrm, has previously been shown to potentiate transcriptional activation by GR (32). Furthermore, it has been demonstrated that hormone-dependent activation of the mouse mammary tumor virus (MMTV) promoter by the GR requires the hBRG1 complex, another mammalian SWI-SNF homologue (16). In addition, the progesterone receptor can, together with NF1, synergistically activate the MMTV promoter assembled in minichromosomes, in a process involving ATP-dependent ISWI-containing complexes (14). The SWI-SNF complex has been shown to alter nucleosome conformation in an ATP-dependent manner, which leads to increased accessibility of nucleosomal DNA to transcription factors (10, 25). The in vitro activities of the SWI-SNF complex are consistent with its in vivo functions in altering chromatin structure at promoters and enhancing the binding of transcription factors (6, 17, 48).
An important question regarding SWI-SNF function is how the complex might be targeted to specific promoter regions in chromatin. Recently, there have been reports about targeting directly via transcriptional activators (33, 34, 50). The HAT complexes SAGA and NuA4, which also can be targeted by transcriptional activators (42, 43), alter chromatin structure by acetylation of lysine residues on histones H3 and H4, respectively (19). It has been suggested that the SWI-SNF complex and Gcn5-containing HAT complexes may perform independent but overlapping functions during transcriptional activation (4, 37, 38). In previous studies, we have shown that gene activation mechanisms, in addition to the Ada pathway, are involved in the activity of the τ1c domain (21). In this paper, we address the question of whether the SWI-SNF complex might participate in such a pathway. However, since we have previously shown that the SAGA complex plays an important role specifically for the τ1 activation domain of GR, we first wanted to further characterize the interaction between GR and the SWI-SNF complex and to determine which role τ1 plays in stimulation by SWI-SNF. In this study, we show that the SWI-SNF complex can potentiate the activity of GR through τ1 in both yeast and mammalian cells. GR-τ1 directly interacts with purified yeast SWI-SNF complex. Mutations in τ1 that affected the transactivation activity in vivo also directly affected τ1 interaction with SWI-SNF. Furthermore, the SWI-SNF complex can stimulate Gal4-τ1-driven transcription from chromatin templates in vitro. We show that an SWI-SNF-independent pathway of GR activation exists in yeast, suggesting that Ada-containing HAT complexes and SWI-SNF play parallel roles in mediating τ1c activity. However, overexpression of Ada2 can increase τ1c activity, specifically in yeast cells lacking SWI-SNF, suggesting that Ada-containing HAT complexes and the SWI-SNF complex can have overlapping roles.
MATERIALS AND METHODS
Plasmids and strains.
The plasmids pRS-315-NX (full-length human GR) (28), pRS-315-GRΔτ1 (lacking GR residues 77 to 261) (21), pLGZ-2TAT (GR-responsive lacZ reporter gene) (45), pRS-315-lexA-τ1c, pRS-315-lexA-τ1c mutants, and pLGZ-2lexA (LexA-responsive lacZ reporter gene) (2) have been described previously. pGEX-4T-3, and pGEX-4T-3-τ1 (residues 77 to 262 of the human GR) have also been described previously (15). The plasmids expressing GST τ1 mutants were constructed by inserting the sequence encoding τ1 (residues 77 to 262 of the human GR) with different mutations as a BglII fragment into pGEX-4T-3. The transfection plasmids pCMV-τ1c-GRDBD and pCMV-TK19luc containing the GR response element have been described previously (2). The plasmids pCG-hbrm (4102) and hbrm-NTP-mutant (4137), and C33 and SW13 cells were kindly provided by Moshe Yaniv and have been described previously (32). Yeast strains CY26 (MATα SWI+ ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ1 lys2-801 ade2-101) and CY332 (MATα snf6Δ ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ1 lys2-801 ade2-101) were kindly provided by Craig Peterson. Yeast strains FY1548 (MATa ada2Δ::HIS3 his3Δ200 leu2Δ1 lys2-128δ ura3-52), FY1370 (MATα gcn5Δ::HIS3 his3Δ200 leu2Δ1 ura3-52), FY1550 (MATa ura3-52 lys2-128δ leu2Δ1 his3Δ200 ada2Δ::HIS3 snf2Δ::LEU2), and FY1352 (MATa ura3-52 lys2-173R2 leu2Δ1 his3Δ200 gcn5Δ::HIS3 snf2Δ::LEU2) were a gift from Fred Winston. The plasmid pAda2-HA, described previously (39), was kindly provided by Leonard Guarente.
Transactivation assays in yeast.
Plasmids were transformed into WT and mutant yeast strains by a lithium acetate method (18). Several colonies from each transformation were checked on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates for homogeneity of transactivation activity (5). Representative transformants were grown in minimal medium with 2% galactose to an A600 of 0.2 to 0.3. Extracts were prepared and assayed for β-galactosidase activity as described previously (45). Protein extracts for Western blot analysis were prepared directly in SDS-PAGE loading buffer as described previously (22) and were separated by SDS–18% PAGE. After transfer to nitrocellulose membrane, the samples were incubated for 1 h with polyclonal rabbit anti-LexA antibody. The Western blot was developed by using ECL (Amersham).
Bacterial protein expression and purification.
Plasmids expressing GST, GST- τ1, and GST-τ1 mutants were grown in XL1 cells at 37°C to an A600 of 0.5, followed by induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. Cells were collected by centrifugation, and pellets were resuspended in a 1/20 (vol/vol) culture of phosphate-buffered saline (PBS) and 1 mM phenylmethylsulfonyl fluoride and frozen. Cells were thawed and sonicated. The cellular debris was removed by centrifugation, and 1% Triton-X-100 was added to the supernatant. Glutathione-sepharose beads (Pharmacia) were prewashed in PBS, were added to the supernatant, and were incubated for 2 h at 4°C with constant mixing. The supernatant was removed, and the beads were washed four times with a 10× bead volume of PBS, GST, GST-τ1, and GST-τ1 mutants were eluted from beads with 10 mM imidazole and were dialyzed. Protein concentrations were evaluated by the Bradford assay.
Purification of SWI-SNF complex.
The SWI-SNF complex was purified from yeast (WT strain CY396 or SWI2K798A mutant strain CY397) as described (10) with some minor variations. Elution from Ni2+ agarose was at 300 mM imidazole. The Mono Q column was followed by a heparin sepharose and a DNA cellulose column, with the complex eluting at 340 and 200 mM NaCl, respectively. Purification was monitored by using antibodies to SWI-SNF subunits. The purified SWI-SNF used in these experiments was tested for the presence of SRB-mediator with antibodies against Med4 (data not shown) and against Med2 and Srb2 (33) and found to be devoid of these SRB/Mediator subunits. The fractions were also tested for the presence of SAGA (αTAFII90, αTAFII17, and αTra1) and RSC (remodel the structure of chromatin) components (αRsc6) and found to be devoid of these complexes.
GST pull down.
The SWI-SNF complex was incubated in PDB (150 mM NaCl, 50 mM HEPES [pH 7.5], 10% glycerol, 0.1% Tween-20, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) with the indicated GST-fusion protein for 2 h at 4°C while rotating on a wheel. The supernatant was removed, beads were washed four times in PDB, and equal fractions of both supernatants and beads were used for Western blotting. Proteins were separated by SDS–10% PAGE. After transfer to nitrocellulose membrane, the samples were incubated for 2 h with a polyclonal mouse anti-HA antibody. The Western blot was developed by using ECL (Amersham).
Histone preparation and nucleosome reconstitution.
Core histones and oligonucleosomes were purified from HeLa cells as described (11). Long oligonucleosomes were used in the transcription reactions as competitor nucleosomes. Nucleosomal arrays were reconstituted with core histones by step dilution as described (40).
In vitro transcription.
Transcription experiments were carried out as described previously (24, 40, 41), except that ATP (1 mM final) and MgCl2 (3 mM final) were added to the binding buffer (for remodeling by SWI-SNF). Acetyl coenzyme A and sodium butyrate were not added to any reactions. Fifteen to twenty nanograms of reconstituted G5E4 nucleosomal array (pIC-2085S/G5E4R) or G5E4 DNA was assayed, and 1 to 5 ng of human immunodeficiency virus type 1 (HIV-1) DNA [pHIV(D,N)] was added to each reaction as an internal recovery control. Five hundred micrograms of competitor nucleosomes (long oligonucleosomes) was added to each reaction containing chromatin templates. A 5- to 15-nM final concentration of the Gal4 derivative and 1 μl of purified yeast SWI-SNF (as determined by titration) were added where indicated. For primer extension analysis of the RNA, 25,000–50,000 cpm of 32P-labeled E4 (+86 to +110) and HIV-1 (+50 to +81) primers were used per reaction.
Transient transfections.
C33 or SW13 cells described previously (32) were transiently transfected with plasmid pCMV-τ1c-GRDBD, reporter plasmids pCMV-TK19luc and pCMV-hbrm (4102), or hbrm-NTP-mutant (4137) by using FuGENE 6 transfection reagent (Boehringer Mannheim). Cells were harvested after 30 h, and the levels of luciferase were measured. For Western blotting, C33 cells were transfected with 20 μg of pCMV, pCMV-hbrm, or pCMV-hbrm-NTP-mutant. After 24 h, cells were harvested in lysis buffer (0.5% NP-40, 0.7 M NaCl, 50 mM Tris-HCl [pH 8.0], and protein inhibitors) and were frozen and thawed. Protein concentration was determined with the Bradford protein assay. Twenty micrograms of total protein was separated by SDS–7% PAGE. After transfer to nitrocellulose membrane, the samples were incubated overnight with an affinity-purified polyclonal rabbit anti-hbrm antibody recognizing the amino-terminal end of the protein (32). The Western blot was developed by using ECL (Amersham).
RESULTS
The SWI-SNF complex potentiates the activity of the GR N-terminal transactivation domain in yeast.
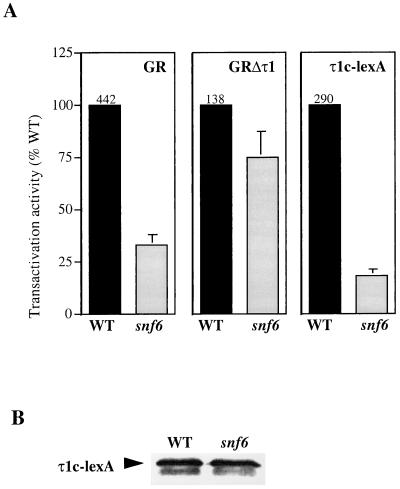
Yamamoto's group has previously shown that several SWI-SNF proteins are important for GR function in yeast (49). We wished to investigate which specific role τ1, as the major N-terminal transactivation domain, would play during SWI-SNF-stimulated transcription. Therefore, we measured the transactivation activity of intact GR and various GR derivatives in a yeast strain defective in the snf6 gene. Plasmids expressing intact GR and GR lacking the N-terminal transactivation activity, GRΔτ1, were transformed together with a GR-responsive lacZ reporter plasmid into a yeast strain which contained a deletion in the snf6 gene and into an otherwise isogenic wild-type (WT) strain. Strains expressing GR and GRΔτ1 were grown in the presence of hormone (10 μM triamcinolone acetonide), exponentially growing cells were harvested, and β-galactosidase levels were measured. Figure 1A shows that the transactivation activity for GR is substantially reduced in the snf6 mutant strain compared to WT. The activity of the GRΔτ1 protein is reduced to a lesser extent in the snf6 mutant yeast strain, indicating a profound role of the Snf6 protein in the activity of the τ1 domain. To pursue this issue further, we expressed a protein representing the functional core of the τ1 domain (τ1c), fused to the DNA-binding domain of the LexA protein. As shown in Fig. 1A, the expression of β-galactosidase from a LexA-dependent lacZ reporter gene was reduced in the snf6 mutant strain, to an even greater extent than that seen with full-length GR. The Western blot shown in Fig. 1B shows that the expression level of the τ1c-LexA fusion protein was similar in the WT and the snf6 mutant strain, indicating that the reduced activity of the τ1c domain in the snf6 strain was not due to a lower expression level.
FIG. 1.
The SWI-SNF complex is important for the GR τ1 transactivation activity. (A) Transactivation activities of the GR, GRΔτ1, and τ1c proteins were obtained from a WT and a snf6 mutant strain by measuring β-galactosidase activity (nanomoles of O-nitrophenyl-β-d-galactopyranoside per milligram of protein per minute). Strains expressing GR and GRΔτ1 were grown in the presence of 10 μM triamcinolone acetonide. The graphs show mean values from five independent experiments, where the GR derivative activities measured in the snf6 strain are relative to their activities in the WT strain (actual WT values are shown above the columns). (B) Western blot showing expression levels of the τ1c-LexA protein in the WT and the snf6 mutant strain.
The τ1 domain interacts directly with the yeast SWI-SNF complex.
While an interaction of the GR DNA-binding domain with SWI3 protein has been detected previously in yeast cell extract (49), it is still unclear whether GR activation domains interact directly with the SWI-SNF complex. We therefore tested the τ1 activation domain in a “pull down” assay with purified yeast SWI-SNF complex. The τ1 domain was expressed in Escherichia coli as a fusion protein with glutathione S-transferase (GST) and was purified. The fusion proteins were coupled to glutathione-agarose beads and then incubated with purified SWI-SNF complex. When interacting with the GST fusion proteins, SWI-SNF complex was pelleted with the glutathione-agarose beads. The presence of SWI-SNF complex in the pellet and supernatant fractions was determined by Western blotting with an antibody against the hemagglutinin (HA)-tagged SWI2-subunit. The results (Fig. 2) show that the GST protein alone does not interact with the SWI-SNF complex since HA-SWI2 protein is only found in the supernatant fraction. However, the GST-τ1 protein precipitated a majority of the SWI-SNF complex.
FIG. 2.
The SWI-SNF complex interacts with τ1, the N-terminal transactivation domain of GR. GST pull down assays were performed with either GST-τ1 or GST alone bound to gluthathione-sepharose beads and the SWI-SNF complex. Supernatants (S) and beads (B) were subjected to Western blotting, and SWI-SNF complex was detected by using an antibody against the HA-tagged SWI2 subunit.
Binding of τ1 mutants to the yeast SWI-SNF complex in vitro correlates with their transactivation activity in vivo.
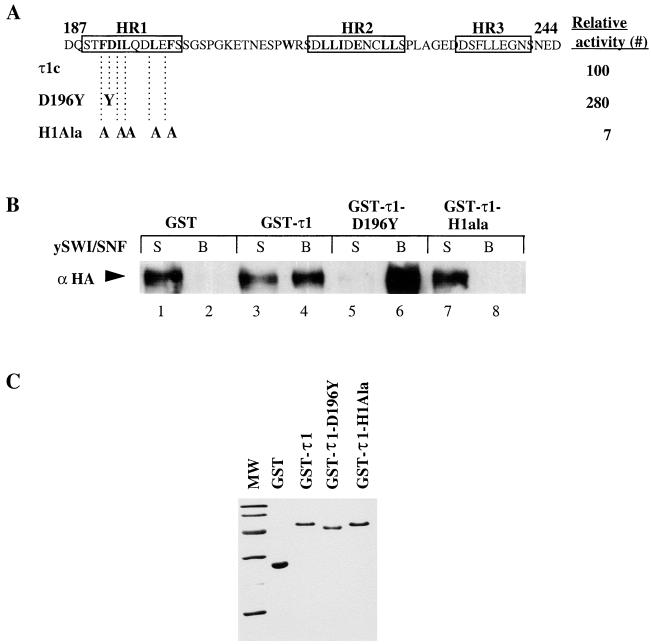
To see whether mutations that affect the activity of the τ1 domain would have an effect on binding of τ1 to the SWI-SNF complex, we selected two τ1 mutants with different aa substitutions. The mutant D196Y is two to three times more active than unmutated τ1c in transactivation assays in vivo, while the mutant H1ala is less than 10% as active as WT τ1c (Fig. 3A). As shown in Fig. 3B, the high-activity mutant D196Y bound to the SWI-SNF complex at least three times more strongly than WT τ1. However, there is very little binding of the SWI-SNF complex to the low-activity mutant H1ala. The amounts of each τ1 mutant protein used in a pull down assay is visualized by a Coomassie-blue-stained sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 3C). Clearly, there is a good correlation between the ability of these mutant τ1 proteins to bind to the SWI-SNF complex and their activities in vivo.
FIG. 3.
Binding of τ1 mutant proteins to the SWI-SNF complex. (A) Schematic representation showing the amino acid substitutions in the τ1 mutants used. Boxes indicate the locations of putative helical regions I, II, and III. Mean relative β-galactosidase activity (#) of τ1-core-LexA fusion proteins are shown as percentage of WT level (taken from reference 2). (B) Immunoblot showing coprecipitation of purified GST-τ1 mutant proteins with the SWI-SNF complex, which was detected by using an antibody raised against the HA-tagged SWI2 subunit. (C) Coomassie blue staining of GST-τ1 mutant proteins used in a pull down assay with the SWI-SNF complex.
The SWI-SNF complex stimulates Gal4-τ1-driven transcription in vitro.
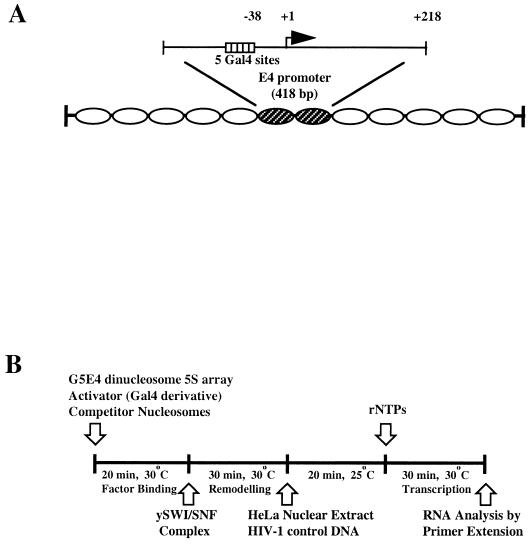
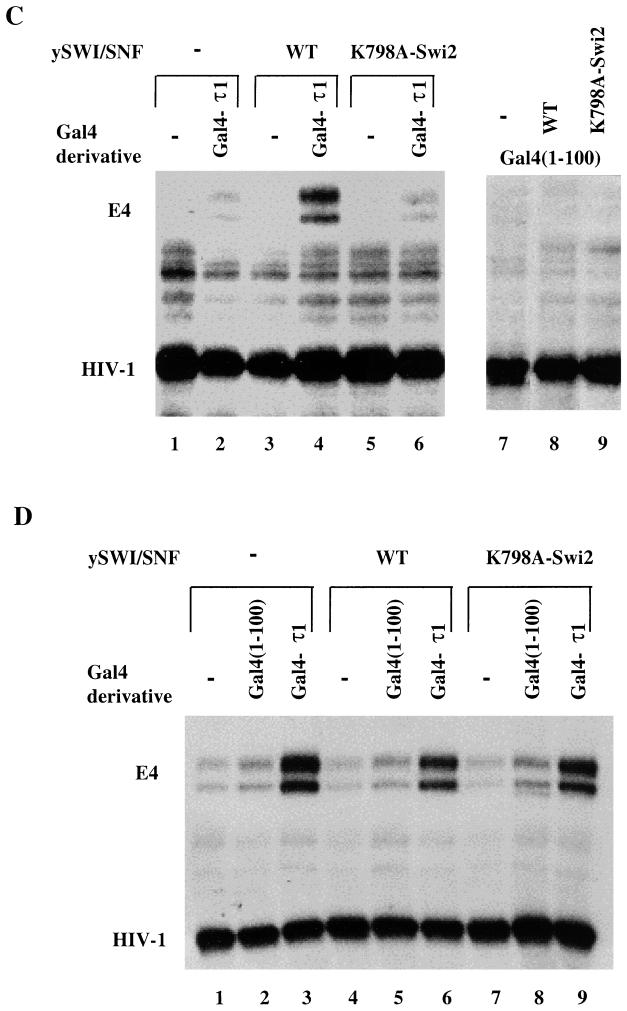
Previous reports have indicated that the SWI-SNF complex can enhance the activity of nuclear receptors, but it has not previously been shown that this involves recruitment of SWI-SNF to target promoters by DNA-bound receptor proteins. To determine whether SWI-SNF could specifically enhance τ1 activation potential in an in vitro chromatin-reconstituted system, we used a nucleosomal array template containing five Gal4-binding sites upstream of a minimal adenovirus E4 promoter (Fig. 4A). The nucleosomal templates were incubated with Gal4(1-100) or Gal4(1-100)-τ1 and SWI-SNF complex followed by transcription analysis. Transcription reactions were performed in the presence of a 30-fold excess of competitor nucleosomes in order to increase the demand for SWI-SNF recruitment to the nucleosome array (Fig. 4B). Under these conditions, the SWI-SNF complex needed to be targeted to the promoter by the τ1 domain in order to specifically stimulate transcription. Figure 4C shows that, with competitor nucleosomes present, the SWI-SNF complex alone could not stimulate transcription (lane 3). In addition, Gal4(1-100) alone did not stimulate transcription in the absence or presence of SWI-SNF (lanes 7 and 8). By contrast, Gal4-τ1-driven transcription from the nucleosome array was enhanced greater than fivefold in the presence of the SWI-SNF complex (compare lanes 2 and 4).
FIG. 4.
The SWI-SNF complex stimulates Gal4-τ1-driven transcription from chromatin templates. (A) A diagram showing the nucleosomal 5S-G5E4 array template. (B) A schematic representation of the in vitro transcription assay indicating the order in which the reagents were added. (C) The nucleosomal array template was transcribed following activator binding in the presence or absence of WT SWI-SNF complex or ATPase-deficient SWI-SNF complex (K798A-Swi2) as indicated. The HIV-1 DNA template was used as an internal recovery control. The transcripts marked E4 are subjected to regulation by the added Gal4 and Gal4-τ1 proteins. (D) Transcription of naked DNA template in the presence or absence of Gal4 or Gal4-τ1 and WT or ATPase-deficient (K798A-Swi2) SWI-SNF complex. The transcription conditions were the same as described for panel C, except for the replacement of the nucleosome array template with naked DNA and the absence of competitor nucleosomes.
To investigate whether the SWI-SNF complex would mediate Gal4-τ1 activity through its chromatin-remodeling activities, we tested an ATPase-deficient SWI-SNF complex (K798A-Swi2) in the transcription assay. As shown in Fig. 4C, this ATPase-deficient complex could not significantly stimulate transcription alone (lane 5) or in the presence of Gal4 (lane 9) or Gal4-τ1 (lane 6). The τ1 activity was only increased 1.4-fold by the mutant SWI-SNF complex (compare lane 2 to lane 6). To further examine whether the function of the SWI-SNF complex would be independent of its activity on nucleosomes, we also tested if SWI-SNF could further potentiate Gal4-τ1-driven transcription from a naked DNA template. However, neither the WT SWI-SNF complex nor the ATPase-deficient SWI-SNF complex (K798A-Swi2) could increase Gal4-τ1 activity (Fig. 4D, compare lane 3 to lanes 6 and 9) or affect the low levels of transcription observed in the lanes with Gal4 (Fig. 4D, compare lane 2 to lanes 5 and 8). Taken together, these results clearly indicate that recruitment of SWI-SNF and subsequent chromatin remodeling is an important mechanism by which the GR can activate gene expression.
The human homologue of SWI2 enhances transactivation of GR-τ1c in mammalian cells.
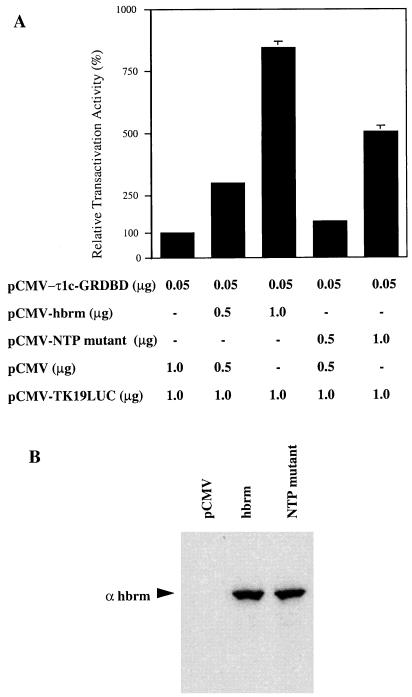
It has previously been reported that hbrm can potentiate GR activity in mammalian cells (32). We wanted to test whether hbrm might play a functional role specifically for τ1 during GR-mediated transcriptional activation. Figure 5 shows the results of experiments in which plasmids expressing the τ1c-GRDBD and hbrm were cotransfected with a GR-responsive luciferase reporter gene into C33 cells. It has previously been shown that C33 cells do not express endogenous hbrm, and most probably only low amounts of GR (32). The level of transactivation by the τ1c was enhanced eightfold at the highest tested level of cotransfected hbrm expression plasmid (Fig. 5A). Transfection of the reporter plasmid or the hbrm plasmid alone did not result in any significant activation (data not shown).
FIG. 5.
hbrm potentiates GR τ1 activity in mammalian C33 cells. (A) Transactivation activity of the τ1c after cotransfection with expression plasmids τ1c-GRDBD and hbrm or hbrm-NTP mutant. The activity of the luciferase reporter gene in each condition is expressed relative to the activity of the τ1c in the absence of cotransfected plasmid encoding hbrm. The graphs represent mean values obtained from three independent transfections. (B) Western blot with affinity-purified hbrm antibodies. Extracts were prepared from C33 cells transfected with 20 μg of pCMV, pCMV-hbrm, or pCMV-NTP mutant.
It has been previously shown that the cooperativity of hbrm with full-length GR was potentiated by the putative ATPase activity of hbrm (32). To determine how much the τ1c contributes to this cooperativity, we also used an hbrm-nucleoside triphosphate (NTP)-binding mutant with reduced ATPase activity due to a mutation at aa 749 in the hbrm protein. With this mutant, cotransfection with τ1c resulted in a weaker increase in activity (fivefold) compared to intact hbrm (eightfold) (Fig. 5A). To determine whether hbrm and the NTP mutant were present at similar levels, Western blotting was performed on the total extract from C33 cells transfected with pCMV, hbrm, or the NTP mutant, and a polyclonal antibody prepared against the first 178 aa of hbrm was used. As shown in Fig. 5B, hbrm and the NTP mutant seem to be expressed equally well in the C33 cells. These data suggest that the ATPase activity of hbrm, which is required for its nucleosome disruption activity, contributes but is not essential for activation by GR. The detected interactions of GR with SWI-SNF (above) and of SWI-SNF with RNA polymerase holoenzyme (44) are consistent with a potential coactivator function of the complex. Our results show that the human SWI-SNF pathway appears to play an important role in the activity of the τ1 domain during GR-mediated gene activation in mammalian cells, involving the putative ATPase activity of hbrm.
Additional pathways can mediate τ1 activation independently of SWI-SNF.
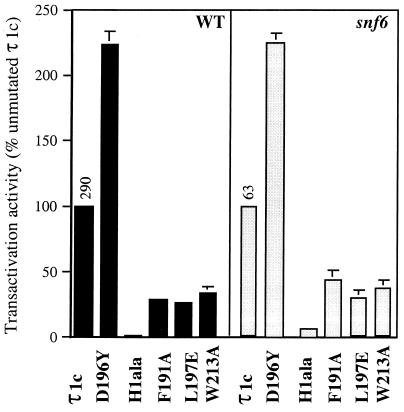
We have shown that the SWI-SNF complex can potentiate τ1-mediated transactivation both in vivo and in vitro. The residual τ1 activity in snf6 strains as well as previously published evidence suggests that τ1 activity can also be mediated by other coactivator pathways (3, 15, 21, 43), at least some of which are affected by a set of τ1 mutations that also affected interaction with the SWI-SNF pathway. To further characterize the nature of the pathways functioning in SWI-SNF-defective strains, we measured the activity of a range of τ1c mutants with reduced or increased activity in a snf6 mutant yeast strain and an otherwise isogenic WT strain. Figure 6 shows the relative activities of proteins with mutations in different segments of τ1c. All of the τ1c-mutants show similar deficiencies in activity in both the WT and the snf6 mutant strain. Furthermore, the effects of τ1 mutants on τ1 activity in snf6 mutant strains parallels their ability to interact with HAT complexes (43). It is therefore possible that at least one of the residual activation mechanisms in snf6 strains is due to the activity of HAT complexes.
FIG. 6.
Transactivation activities of τ1c mutants in the WT and a snf6 mutant strain. The graphs show mean values from five independent experiments, where the β-galactosidase activity (nanomoles of O-nitrophenyl-β-d-galactopyranoside per milligram of protein per minute) was measured. The activities of the τ1c mutants are relative to unmutated τ1c in the WT and the snf6 mutant strain, respectively (actual values for unmutated τ1c are shown above the columns).
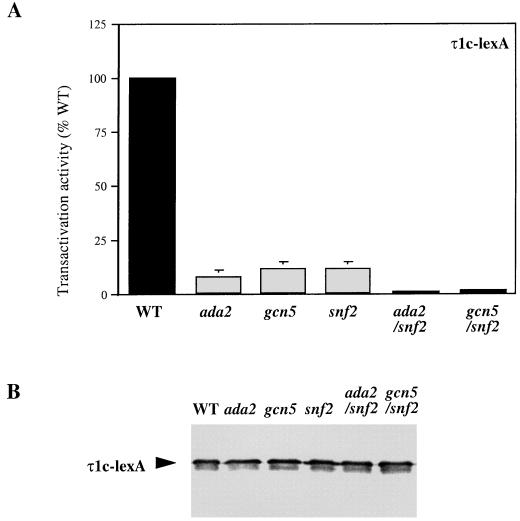
The activity of GR-τ1c is further decreased in yeast strains with deletions in both the SWI-SNF and Ada genes.
We wished to investigate how τ1-mediated activation via the SWI-SNF complex interacts with other activation pathways, in particular the Ada pathway investigated previously (21, 43). Thus, the transactivation activity of the τ1c-LexA was measured in yeast strains defective in both SWI-SNF and Ada genes. A plasmid expressing τ1c-LexA was transformed together with a LexA-responsive lacZ reporter plasmid into yeast mutant strains ada2, gcn5, snf2, snf2-ada2, and gcn5-snf2 and into an otherwise isogenic WT strain, exponentially growing cells were harvested, and β-galactosidase levels were measured. The activity of τ1c is reduced in the ada2 (6%), gcn5 (10%), and snf2 (10%) strains compared to its activity in the WT strain (100%) (Fig. 7A). However, the remaining activity of τ1c is further decreased in the snf2-ada2 (1.5%) and gcn5-snf2 (2.1%) strains compared to the single-mutation strains. The τ1c protein is expressed at the same level in all of the mutant yeast strains, as shown by the Western blot in Fig. 7B. These results indicate that the SWI-SNF and Ada pathways both contribute to τ1-mediated activation in complementary ways and in that sense they represent alternative pathways.
FIG. 7.
GR τ1c transactivation activity is further decreased in yeast strains lacking both SWI-SNF and Ada proteins. (A) Transactivation activity of the τ1c protein was obtained from WT or ada2, gcn5, snf2, ada2-snf2, or gcn5-snf2 mutant strains by measuring β-galactosidase activity (nanomoles of O-nitrophenyl-β-d-galactopyranoside per milligram of protein per minute). The graphs show mean values from three independent experiments, where the τ1c activity measured in all the mutant strains are relative to its activity in the WT strain. (B) Western blot showing expression levels of the τ1c-LexA protein in the WT and the mutant strains.
Overlapping function for SWI-SNF and Ada proteins in GR activation.
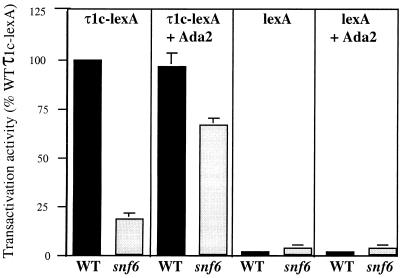
Recent reports have suggested that the SWI-SNF complex and HAT complexes function in overlapping pathways when mediating the function of some yeast activators (4, 37, 38). Since both the Ada-containing SAGA complex and the SWI-SNF complex have been shown to interact with and potentiate GR-τ1 activity in vivo and in vitro, we wanted to further investigate whether the complexes could have overlapping functions when mediating τ1c activity. We transformed yeast cells defective in SWI-SNF (snf6) and an otherwise isogenic WT strain with plasmids expressing τ1c-LexA and a LexA-dependent lacZ reporter gene and overexpressing Ada2. Figure 8 shows that the β-galactosidase levels measured in the WT strain are similar when τ1c-LexA is expressed alone or together with overexpressed Ada2. Interestingly though, there is a threefold increase in activation measured in the snf6 strain when τ1c-LexA is expressed together with overexpressed Ada2, compared to τ1c-LexA alone in the snf6 strain. The activities measured in the WT and the snf6 mutant strain transformed with LexA are similar and very low with or without overexpressed Ada2. These results clearly indicate that overexpression of Ada2 can compensate in part for the deficiency in snf6. Thus, while the SWI-SNF and Ada proteins perform distinct functions, they appear to have the ability to play overlapping roles in mediating GR-τ1c transactivation activity.
FIG. 8.
Overexpression of the Ada2 protein can increase GR-τ1 activity in snf6 mutant yeast cells by compensating for the absent Snf6 protein. Transactivation activities of the τ1c-LexA and LexA proteins in the absence or presence of a plasmid expressing Ada2 in a WT and a snf6 mutant strain. The graph shows mean values from five independent experiments, where the β-galactosidase activity (nanomoles of O-nitrophenyl-β-d-galactopyranoside per milligram of protein per minute) was measured.
DISCUSSION
The SWI-SNF complex potentiates the activity of the GR-τ1 transactivation domain in yeast and mammalian cells.
Previous studies from Yamamoto's group have shown that deletion of swi genes in yeast cells reduces the transactivation capacity of full-length GR (49). The SWI1, SWI2, and SWI3 proteins were found to be essential for GR function in yeast, while snf5 and snf6 mutations resulted in an approximately fourfold decrease in GR activity. Yamamoto's group also showed that a GR derivative lacking the steroid-binding domain was dependent on the SWI-SNF proteins for activity (49). Since we wished to investigate which role τ1, as the major N-terminal transactivation domain, specifically would play during SWI-SNF-stimulated transcription, we compared the activity of τ1c to that of full-length GR in snf6 mutant yeast cells. We also saw a four-fold decrease in the activity of full-length GR in snf6 mutant cells as compared to that in WT yeast cells. The activity for a GR derivative lacking τ1, GR-Δτ1, was reduced to a much lesser extent in snf6 as compared to WT cells, indicating that SWI-SNF stimulates transcription by GR predominantly through the τ1 domain. This was further supported by measuring the activity of τ1c-LexA in snf6 and WT cells, since the reduction of activity was even stronger than for full-length GR. However, it is possible that other activation domains in GR, like AF-2, may use the SWI proteins for transactivation in vivo. Such interaction has been observed in the yeast two-hybrid system for the estrogen receptor α-LBD, which can interact with the human homologues of the SWI2 protein in a ligand-dependent manner (23).
Previous results from Yaniv's group (32) show that hbrm is important for GR transactivation in mammalian cells involving the ATPase activity of hbrm. The hbrm-stimulated increase in τ1 activation that we observed was almost identical to the increase in GR activation reported by Yaniv's group (32). When we transfected an hbrm-NTP-binding mutant with reduced ATPase activity together with τ1c, we saw the same decrease in activity as reported for full-length GR. The transfection experiments were repeated with another hbrm-defective mammalian cell line (SW13), from which we obtained very similar results (data not shown). Taken together, the yeast and mammalian cell data suggest that the τ1c region of the GR is important for SWI-SNF-mediated potentiation of GR function.
The τ1 domain interacts directly with the yeast SWI-SNF complex, and binding of τ1 mutants to the yeast SWI-SNF complex in vitro correlates with their transactivation activities in vivo.
It has previously been shown that GR-DBD can precipitate the SWI3 protein from yeast cell extract, and this interaction required SWI1 and SWI2, perhaps indicating that a complex of SWI proteins interacts with the receptor (49). Here we show that GR-τ1 can bind directly to purified yeast SWI-SNF complex. We also tested GST-GRLBD in a pull down assay with SWI-SNF complex, but in our assay the GRLBD did not precipitate any SWI-SNF complex in the presence or absence of ligand (data not shown). To date, there have been several reported correlations between the transcriptional activity of activators and their binding to target factors (8, 20, 31, 39). We previously showed that the binding of τ1 mutants to the HAT complex SAGA in vitro corresponded to the activity of the mutants in vivo and in vitro (43). Here we show that a high-activity mutant, D196Y, interacts more strongly with the SWI-SNF complex, while a low-activity mutant, H1ala, interacts less efficiently. The relationship between the activity of the mutants and their binding to the SWI-SNF complex further supports our model that the τ1 mutations in question affect the ability of τ1 to fold into a structured form that is competent to interact with target proteins. Interestingly, previous results have shown that the D196Y and H1ala mutants were affected for interaction with the SAGA complex in the same way as shown here for the SWI-SNF complex, indicating that these complexes share a binding determinate on GR-τ1.
Gal4-τ1-driven transcription in vitro is stimulated by the SWI-SNF complex.
Since we found that GR-τ1 could interact directly with purified SWI-SNF complex, we wanted to know whether this interaction could be important for the τ1 transactivation activity. Using an in vitro system with reconstituted chromatin templates, we have shown that SWI-SNF needs to be targeted by the τ1 activation domain in order to stimulate transcription in this system. In the absence of activator, or in the presence of the DNA-binding domain of Gal4 alone, the SWI-SNF complex did not stimulate transcription. The SWI-SNF complex could not, however, increase Gal4-τ1 activity from a naked DNA template. In addition, an ATPase-deficient SWI-SNF complex could not stimulate transcription in the absence or presence of Gal4-τ1. These observations strongly suggest that the yeast SWI-SNF complex can mediate Gal4-τ1 activity through its chromatin-remodeling activities. Although the capacity of the GR to recruit remodeling activity to DNA has been shown previously (35), it has not previously been demonstrated in the literature that GR-dependent recruitment of SWI-SNF can lead to transcriptional activation.
Ada-containing HAT complexes and the SWI-SNF complex represent independent but complementary functions that can have overlapping roles in mediating GR-τ1c activity.
We have previously shown that the chromatin-modifying complex SAGA is important for GR-τ1 transactivation activity (43). However, by using yeast strains with deletions in the ada2, ada3, or gcn5 genes, we found that the Ada proteins alone were not solely responsible for τ1 activity and that other pathways mediating τ1 activity seemed to be independent of the Ada pathway (21). Since we did not know whether the Ada-independent pathway would be mediated by the SWI-SNF proteins alone or if additional unknown proteins would be involved, we measured the τ1c activity in yeast strains with deletions in both the ada and the swi-snf genes. The activity of τ1c is further decreased in yeast strains with deletions in both the snf2 and ada genes, compared to a single deletion in these genes. This clearly indicates that the SAGA complex and the SWI-SNF complex are two major pathways that can mediate τ1c transactivation activity.
It has previously been suggested that while HAT complexes and the SWI-SNF complex have separate activities, they might perform overlapping functions during transcriptional activation by yeast activators (4, 37). Since both the SAGA complex and the SWI-SNF complex have been shown to interact with and potentiate GR-τ1 activity, we tested whether components of these two complexes would have the capability of compensating for each other when mediating τ1 transactivation activity. By overexpressing Ada2 in yeast cells reduced in SWI-SNF (snf6), we found a threefold increase in activation in the snf6 strain when τ1c and Ada2 were expressed together, compared to τ1c alone in the snf6 strain. In the WT strain, the activity for τ1c was similar in the presence or absence of overexpressed Ada2. Thus, our results suggest that SWI-SNF and Ada proteins play distinct but overlapping roles in stimulating GR-τ1c activity. It is possible that overexpression of the Ada histone acetylase activity can substitute for chromatin remodeling under these conditions, but the exact mechanisms involved need to be further investigated. It is also not clear whether the SWI-SNF and the Ada protein function together or at different stages in τ1-mediated transcription. Both the SWI-SNF complex and the SAGA complex can be recruited by GR-τ1 in order to remodel chromatin, but in vivo there is most probably an ordered pathway of this recruitment, as has been suggested for the SWI5 activator (9, 27).
ACKNOWLEDGMENTS
We thank Moshe Yaniv and Christian Muchardt for generously providing C33 and SW13 cells, hbrm-plasmids, and hbrm antibodies and Anki Östlund-Farrants for advice on the use of the antibodies in a Western blot. We thank Craig Peterson for the snf6 yeast strain, Fred Winston for the ada2, gcn5, snf2, ada2/snf2, and gcn5/snf2 yeast strains, and Leonard Guarente for the yeast Ada2 plasmid. We also thank Elisabeth Flinn, Jane Thompson, and Patrick Grant for valuable discussions.
J.L.W. is an Associate Investigator of the Howard Hughes Medical Institute. This work was supported by grants from the National Institute of General Medical Sciences awarded to J.L.W., the Swedish Natural Sciences Research Council awarded to A.P.H.W., the Swedish Medical Research Council awarded to J.-Å.G. (13×-2819) and A.E.W. (K98-03RM-12413), and the Erik and Edith Fernströms foundation awarded to A.E.W.
REFERENCES
- 1.Almlöf T, Wright A P H, Gustafsson J-Å. Role of acidic and phosphorylated residues in gene activation by the glucocorticoid receptor. J Biol Chem. 1995;270:17535–17540. doi: 10.1074/jbc.270.29.17535. [DOI] [PubMed] [Google Scholar]
- 2.Almlöf T, Gustafsson J-Å, Wright A P H. Role of hydrophobic amino acid clusters in the transactivation activity of the human glucocorticoid receptor. Mol Cell Biol. 1997;17:934–945. doi: 10.1128/mcb.17.2.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almlöf T, Wallberg A E, Gustafsson J-Å, Wright A P H. Role of important hydrophobic amino acids in the interaction between the glucocorticoid receptor τ1-core activation domain and target factors. Biochemistry. 1998;37:9586–9594. doi: 10.1021/bi973029x. [DOI] [PubMed] [Google Scholar]
- 4.Biggar S R, Crabtree G R. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohen S P, Yamamoto K R. Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc Natl Acad Sci USA. 1993;90:11424–11428. doi: 10.1073/pnas.90.23.11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns L G, Peterson C L. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns B R, Kim Y J, Sayre M H, Laurent B C, Kornberg R D. A multisubunit complex containing the Swi1/Adr6, Swi2/Snf2, Swi3, Snf5, and Snf6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang Y C, Komarnitsky P, Chase D, Denis C L. ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J Biol Chem. 1996;271:32359–32365. doi: 10.1074/jbc.271.50.32359. [DOI] [PubMed] [Google Scholar]
- 9.Cosma M-P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodelling factors to a cell cycle and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 10.Côte J, Quinn J, Workman J L, Peterson C L. Stimulation of Gal4 derivative binding to nucleosomal DNA by the yeast Swi/Snf complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 11.Côte J, Utley R T, Workman J L. Basic analysis of transcription factor binding to nucleosomes. Methods Mol Genet. 1995;6:108–129. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- 12.Dahlman-Wright K, Almlöf T, McEwan I J, Gustafsson J-Å, Wright A P H. Delination of a small region within the major transactivation domain of the human glucocorticoid receptor that mediates transactivation of gene expression. Proc Natl Acad Sci USA. 1994;91:1619–1623. doi: 10.1073/pnas.91.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlman-Wright K, Baumann H, McEwan I J, Almlöf T, Wright A P H, Gustafsson J-Å, Härd T. Structural characterization of a minimal functional transactivation domain from the human glucocorticoid receptor. Proc Natl Acad Sci USA. 1995;92:1699–1703. doi: 10.1073/pnas.92.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Crose L, Koop R, Venditti P, Westphal H M, Nightingale K P, Corona D F V, Becker P B, Beato M. Two-step synergism between the progesterone receptor and the DNA-binding domain of nuclear factor 1 on MMTV minichromosomes. Mol Cell. 1999;4:45–54. doi: 10.1016/s1097-2765(00)80186-0. [DOI] [PubMed] [Google Scholar]
- 15.Ford J, McEwan I J, Wright A P H, Gustafsson J-Å. Involvement of the TFIID protein complex in gene activation by the N-terminal transactivation domain of the glucocorticoid receptor in vitro. Mol Endocrinol. 1997;11:1467–1475. doi: 10.1210/mend.11.10.9995. [DOI] [PubMed] [Google Scholar]
- 16.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 17.Gavin I M, Simpson R T. Interplay of yeast global transcriptional regulators Ssn6p-Tup1p and Swi-Snf and their effect on chromatin structure. EMBO J. 1997;16:6263–6271. doi: 10.1093/emboj/16.20.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gietz R D, Schiestl R H. Applications of high efficiency lithium acetate transformation of intact yeast cells using single stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 19.Grant P A, Duggan L, Côte J, Roberts S M, Brownell J E, Candau R, Ohba R, Owenhughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 20.Hagemeier C, Cook A, Kouzarides T. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res. 1993;21:4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henriksson A, Almlöf T, Ford J, McEwan I J, Gustafsson J-Å, Wright A P H. Role of the Ada adaptor complex in gene activation by the glucocorticoid receptor. Mol Cell Biol. 1997;17:3065–3073. doi: 10.1128/mcb.17.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvath A, Riezman H. Rapid protein extraction from Saccharomyces cerevisiae. Yeast. 1994;10:1305–1310. doi: 10.1002/yea.320101007. [DOI] [PubMed] [Google Scholar]
- 23.Ichinose H, Garnier J-M, Chambon P, Losson R. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene. 1997;188:95–100. doi: 10.1016/s0378-1119(96)00785-8. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda K, Steger D, Eberharter A, Workman J L. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol Cell Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 26.Iniguez-Lluhi J A, Lou D Y, Yamamoto K R. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J Biol Chem. 1997;272:4149–4156. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- 27.Krebs J E, Kuo M-H, Allis C D, Peterson C L. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lind U, Carlstedt-Duke J, Gustafsson J-Å, Wright A P H. Identification of single amino acid substitutions of Cys-736 that affect the steroid-binding affinity and specificity of the glucocorticoid receptor using phenotypic screening in yeast. Mol Endocrinol. 1996;10:1358–1370. doi: 10.1210/mend.10.11.8923462. [DOI] [PubMed] [Google Scholar]
- 29.McEwan I J, Wright A P, Dahlman-Wright K, Carlstedt-Duke J, Gustafsson J-Å. Direct interaction of the tau 1 transactivation domain of the human glucocorticoid receptor with the basal transcription machinery. Mol Cell Biol. 1993;13:399–407. doi: 10.1128/mcb.13.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McEwan I J, Almlöf T, Wikström A-C, Dahlman-Wright K, Wright A P, Gustafsson J-Å. The glucocorticoid receptor functions at multiple steps during transcription initiation by RNA polymerase II. J Biol Chem. 1994;269:25629–25636. [PubMed] [Google Scholar]
- 31.Melcher K, Johnston S A. GAL4 interacts with TATA-binding protein and coactivators. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Natarajan K, Jackson B M, Zhou H, Winston F, Hinnebusch A G. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/Mediator. Mol Cell. 1999;4:657–664. doi: 10.1016/s1097-2765(00)80217-8. [DOI] [PubMed] [Google Scholar]
- 34.Neely K E, Hassan A H, Wallberg A E, Steger D J, Cairns B R, Wright A P H, Workman J L. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 35.Östlund-Farrants A-K, Blomquist P, Kwon H, Wrange Ö. Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol Cell Biol. 1997;17:895–905. doi: 10.1128/mcb.17.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paranjape S M, Kamakaka R T, Kadonaga J T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 37.Pollard K J, Peterson C L. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/Mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silverman N, Agapite J, Guarente L. Yeast Ada2 protein binds to the VP16 protein activation domain and activates transcription. Proc Natl Acad Sci USA. 1994;91:11665–11668. doi: 10.1073/pnas.91.24.11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steger D J, Eberharter A, John S, Grant P A, Workman J L. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc Natl Acad Sci USA. 1998;95:12924–12929. doi: 10.1073/pnas.95.22.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steger, D. J., and J. L. Workman. Transcriptional analysis of purified histone acetyltransferase complexes. Methods, in press. [DOI] [PubMed]
- 42.Utley R T, Ikeda K, Grant P A, Côte J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators target histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 43.Wallberg A E, Neely K E, Gustafsson J-Å, Workman J L, Wright A P H, Grant P A. Histone acetyltransferase complexes can mediate transcriptional activation by the major glucocorticoid receptor activation domain. Mol Cell Biol. 1999;19:5952–5959. doi: 10.1128/mcb.19.9.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodelling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 45.Wright A P H, Carlstedt-Duke J, Gustafsson J-Å. Ligand-specific transactivation of gene expression by a derivative of the human glucocorticoid receptor expressed in yeast. J Biol Chem. 1990;265:14763–14769. [PubMed] [Google Scholar]
- 46.Wright A P H, McEwan I J, Dahlman-Wright K, Gustafsson J-Å. High level expression of the major transactivation domain of the human glucocorticoid receptor in yeast cells inhibits endogenous gene expression and cell growth. Mol Endocrinol. 1991;5:1366–1372. doi: 10.1210/mend-5-10-1366. [DOI] [PubMed] [Google Scholar]
- 47.Wright A P H, Gustafsson J-Å. Mechanism of synergistic transcriptional transactivation by the human glucocorticoid receptor. Proc Natl Acad Sci USA. 1991;88:8283–8287. doi: 10.1073/pnas.88.19.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu L, Winston F. Evidence that Snf-Swi controls chromatin structure over both the TATA and UAS regions of the SUC2 promoter in Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4230–4234. doi: 10.1093/nar/25.21.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 50.Yudkovsky N, Logie C, Hahn S, Peterson C L. Recruitment of the SWI/SNF chromatin complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]