Abstract
A secretory defect causes specific and significant transcriptional repression of both ribosomal protein and rRNA genes (K. Mizuta and J. R. Warner, Mol. Cell. Biol. 14:2493–2502, 1994), suggesting the coupling of plasma membrane and ribosome syntheses. In order to elucidate the molecular mechanism of the signaling pathway, we isolated a cold-sensitive mutant with a mutation in a gene termed RRS1 (regulator of ribosome synthesis), which appeared to be defective in the signaling pathway. The rrs1-1 mutation greatly reduced transcriptional repression of both rRNA and ribosomal protein genes that is caused by a secretory defect. RRS1 is a novel, essential gene encoding a nuclear protein of 203 amino acid residues that is conserved in eukaryotes. A conditional rrs1-null mutant was constructed by placing RRS1 under the control of the GAL1 promoter. Rrs1p depletion caused defects in processing of pre-rRNA and assembly of ribosomal subunits.
Balanced synthesis of cellular components is required for normal cell growth. A temperature-sensitive mutation in SLY1, whose gene product is involved in endoplasmic reticulum-to-Golgi trafficking (26), causes the transcriptional repression of both ribosomal protein and rRNA genes in Saccharomyces cerevisiae (20). Further examination using various sec mutants showed that a defect anywhere in the secretory pathway, from a step prior to insertion of the nascent peptide into the endoplasmic reticulum to a step involved in the formation of the plasma membrane, prevents the continued synthesis of the components of the ribosome. Similar results were obtained following treatment of wild-type cells with the secretory inhibitors tunicamycin and brefeldin A (20). Furthermore, many temperature-sensitive mutants in which transcription of ribosomal protein genes is temperature sensitive appear to be defective in the secretory pathway (17). As the membrane is the end product of much of the secretory pathway, these results suggest an important coupling of plasma membrane and ribosome biosynthesis. We proposed the existence of a signal transduction pathway from the plasma membrane to the nucleus. According to this model, a signal generated by the defect in de novo synthesis of membrane should be transmitted to the nucleus and cause specific and significant transcriptional repression of ribosomal genes. It was recently suggested that stress in the plasma membrane is monitored by Pkc1, which initiates a signal transduction pathway that leads to the repression (24). In order to elucidate the molecular mechanism of the signal transduction pathway, we have screened for mutants defective in the response to a secretory defect.
Here we describe the isolation and molecular characterization of RRS1, encoding an essential nuclear protein of 203 amino acids. In the rrs1-1 mutant, a secretory defect fails to cause transcriptional repression of either rRNA or ribosomal protein genes. The mutant gene, rrs1-1, had a single nucleotide difference within codon 114, resulting in a stop codon. The amino acid sequence of Rrs1p is significantly similar to that of a putative protein encoded by human cDNA. In order to analyze functions of Rrs1p, we constructed a conditional rrs1-null mutant. Depletion of Rrs1p affects pre-rRNA processing and assembly of ribosomal subunits, indicating that Rrs1p is required for proper ribosome biogenesis.
MATERIALS AND METHODS
Yeast strains, plasmids, and media.
The yeast strains used in this study are listed in Table 1. Yeast cells were grown in YPD rich medium, YPG, synthetic complete medium containing 2% glucose (SC) or 2% galactose (SCGal), or SC dropout medium, depending on the plasmid markers (10). Yeast transformation was performed by a lithium acetate procedure (9).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| W303 | MATa/α his3-11,15/his3-11,15 ade2-1/ade2-1 ura3-1/ura3-1 leu2-3,112/leu2-3,112 trp1-1/trp1-1 can1-100/can1-100 |
| W303a | MATahis3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 |
| KM003a | MATahis3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 sly1 |
| KM007a | MATa/α his3-11,15/+ ade2-1/ade2-1 ura3-1/ura3-1 leu2-3,112/leu2-3,112 trp1-1/+ can1-100/can1-100 sly1/sly1 |
| KM101 | MATahis3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 sly1 rrs1-1 |
| KM111 | MATahis3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rrs1::LEU2 pRS316-RRS1-HA |
| KM113 | MATahis3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rrs1::LEU2 sly1 pRS316-RRS1-HA |
| KM114 | MATahis3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rrs1::LEU2 sly1 pRS313-rrs1-1-HA |
| KM123 | MATaade2-1 ura3-1 leu2-3,112 can1-100 rrs1-1 |
| KM129 | MATahis3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rrs1::LEU2 pRS313-GAL1-RRS1 |
Derived from KM001 (20).
A library consisting of partial Sau3A fragments of S. cerevisiae genomic DNA inserted into a single-copy yeast vector, YCp50, was generously provided by M. D. Rose (29).
For epitope tagging, an NheI site was introduced by PCR into pRRS1 just after the initiation codon. The DNA cassette encoding three copies of the nine-amino-acid influenza virus hemagglutinin (HA) epitope was obtained by the digestion of pYT11 (33) with NheI and was inserted in frame into the NheI site of pRRS1. The digested DNA fragment including epitope-tagged RRS1 was subcloned into pRS316 (32).
A plasmid containing GAL1 promoter-controlled RRS1 was constructed as follows. pAT-35 (see below) was digested with XbaI and ScaI and the 0.8-kb fragment containing the open reading frame (ORF) of RRS1 was placed under the GAL1-promoter in a multi-copy vector pNV7 (25). The DNA fragment containing the GAL1 promoter, RRS1, and the terminator was cloned into a single-copy vector, pRS313 (32).
Cloning of a mutant allele.
A mutant allele of the chromosomal RRS1 gene was isolated by PCR. Total chromosomal DNAs were isolated from wild-type and rrs1-1 mutant cells. DNA fragments including RRS1 or rrs1-1 were amplified by PCR and cloned into pUC19. PCR was carried out twice, and the DNA sequence was determined by using four independent clones.
Indirect immunofluorescence.
Indirect immunofluorescence microscopy was done by modification of the procedure described by Pringle et al. (27). Cultures of early-log-phase cells growing in SC medium were fixed by addition of formaldehyde to the medium to a final concentration of 3.2%, followed by agitation for 30 min at room temperature. Fixed cells were harvested by centrifugation, washed with KPS buffer (100 mM potassium phosphate buffer, pH 7.5, with 1.2 M sorbitol) and resuspended in KPS. To remove the cell wall, the cell suspension was incubated with 20 μg of Zymolyase 100T per ml and 0.2% β-mercaptoethanol at 37°C for about 10 min. Spheroplasts were washed with KPS and harvested by gentle centrifugation. Resuspended spheroplasts in KPS were spotted onto polylysine-coated multiwell slides and blocked for 5 min with PBS-BSA (0.285% Na2HPO4, 0.02% KH2PO4, 0.8% NaCl, 0.02% KCl [pH 7.5], 1 mg of bovine serum albumin per ml). The cells were incubated with 10 μl of primary antibody (anti-HA.11; Babco) diluted 1:1,000 with PBS-BSA for 1 h at room temperature. After 10 washes with PBS-BSA, the cells were incubated for 1 h with secondary antibody (rhodamine-conjugated goat-anti-mouse immunoglobulin G; Jackson ImmunoResearch Laboratory, Inc.) diluted 1:300. After 10 washes with PBS-BSA and one with PBS, the cells were stained with 1 μg of DAPI (4′,6′-diamidino-2-phenylindole) per ml in mounting medium.
Western blot analysis.
Western blotting followed standard techniques, and signals were visualized by enhanced chemiluminescence (Amersham).
Northern blot and [methyl-3H]methionine pulse-chase analysis.
Northern blot analysis was carried out using 1.5% agarose gels for mRNAs and 1.2% agarose gels for steady-state level of rRNAs in formaldehyde as described previously (4, 20). The oligonucleotides used for analyzing mature rRNAs and pre-rRNAs were as follows; probes a, b, and c correspond to oligonucleotides B, C and D, respectively, as described by Zanchin and Goldfarb (41), probes d and e correspond to oligonucleotides a and g, respectively, as described by Bergès et al. (1).
[methyl-3H]methionine pulse-chase analysis was carried out using strains grown in SC lacking methionine (SC−Met). Each culture was pulsed with [methyl-3H]methionine and was chased with nonradioactive methionine (500 μg/ml). Samples were taken by pouring cultures onto crushed sterile ice. Total RNA was prepared and analyzed on agarose gels.
Polyribosome analysis.
Yeast cells were grown in 100 ml of medium to mid-log phase and harvested immediately following the addition of cycloheximide (100 μg/ml). The pellet was washed twice with CH buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 30 mM MgCl2, 50 μg of cycloheximide per ml, 200 μg of heparin per ml) and suspended in 0.3 ml of CH buffer. After glass-bead lysis of yeast cells, aliquots of supernatant corresponding to 10 A260 units were overlaid on top of 12 ml of a 10 to 40% (wt/wt) sucrose gradient made in 50 mM Tris-HCl (pH 7.6)–12 mM magnesium acetate–50 mM ammonium acetate–1 mM dithiothreitol and centrifuged for 2 h at 40,000 rpm at 4°C in a Beckman SW40 Ti rotor.
RESULTS
Isolation of yeast mutants unable to respond to the secretory defect.
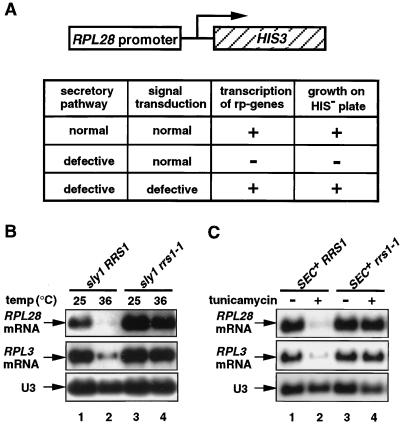
In order to obtain genes which are involved in the signaling pathway, we screened for mutants in which a secretory defect did not cause the transcriptional repression of ribosomal protein genes. The strategy is shown in Fig. 1A. A plasmid was generated in which the HIS3 gene was driven by the promoter of the ribosomal protein gene RPL28 (according to new nomenclature in reference 18; formerly called CYH2). This plasmid was transformed into a strain with the genotype his3− sly1ts. These cells are phenotypically His+ at 25°C, but at 31°C, just below the nonpermissive temperature for sly1, they grow extremely slowly on SC−His plates containing 3-aminotriazole because the HIS3 transcription is repressed. The cells were treated with ethyl methanesulfonate to a survival frequency of 25% and grown on SC−His plates containing 3-aminotriazole at 31°C. A mutant defective in the signaling pathway was expected to grow much faster than the parental strain. Of an estimated 8 × 104 cells plated, 24 colonies that grew rapidly were picked. Each mutant was tested for its ability to repress transcription of ribosomal protein genes RPL28 and RPL3 (formerly TCM1) in response to a secretory defect; five mutant strains were selected. In this study, one of them, termed rrs1-1 (regulator of ribosome synthesis), was chosen for further analysis. In rrs1-1 cells, the transcription of ribosomal protein genes is not significantly repressed in response to a secretory defect which is caused by transfer of cells to the restrictive temperature in the background of sly1 (Fig. 1B) or by addition of a secretory inhibitor, tunicamycin (Fig. 1C). The rrs1-1 mutation does not suppress the temperature sensitivity of sly1 (data not shown), indicating that rrs1-1 is not a suppressor of the secretory defect of the sly1 mutation. Genetic analysis showed that the mutation is recessive; when the mutant cells were crossed to parental sly1 cells of the opposite mating type, RPL28 was significantly repressed at the restrictive temperature (data not shown). On sporulation and dissection of asci, the phenotype for the defect in the response segregated 2:2, indicating that this phenotype is due to a single mutation in a nuclear gene.
FIG. 1.
Isolation of rrs1-1 mutant. (A) The strategy used to isolate yeast mutants defective in the signaling in response to a secretory defect. The reporter gene shown in the diagram consists of the coding sequence of the yeast HIS3 gene fused to the promoter of RPL28. Haploid KM003 (sly1 his3) cells carrying this reporter construct cannot grow at 31°C on SC−His plates containing 5 mM 3-aminotriazole. Mutant cells that do not respond to a secretory defect are expected to grow. (B and C) Northern analysis of the rrs1-1 mutant with the secretory defect. (B) Yeast KM003 (sly1 RRS1) and KM101 (sly1 rrs1-1) cells were grown in YPD medium to log phase (optical density at 600 nm = 0.4 to 0.6) at 25°C. Half of the culture was shifted to 36°C, and after 90 min, the cells were harvested. (C) Yeast strains W303a (SEC+ RRS1) and KM123 (SEC+ rrs1-1) were grown to log phase (optical density at 600 nm = 0.4) at 25°C. Half of the culture was treated with tunicamycin at a final concentration of 1 μg/ml for 4 h at 25°C, and the cells were harvested. Total RNA was prepared and separated by gel electrophoresis. Northern blot analysis was carried out using 32P-labeled DNA probes specific for RPL28 and RPL3. SnoRNA U3 was used as a marker to check equal loading. Ten micrograms of total RNA was loaded in each lane.
The rrs1-1 mutation also affects the repression of rDNA transcription in response to a secretory defect.
As a secretory defect causes transcriptional repression of DNA encoding rRNA (rDNA) as well as ribosomal protein genes (20), we examined whether the rrs1-1 mutation had any effect on the repression of rDNA. We can monitor the synthesis and processing of rRNA by [methyl-3H]methionine pulse-chase analysis, since newly synthesized precursor rRNA is methylated immediately (30, 37). As shown in Fig. 2, a secretory defect by shifting sly1 cells to the restrictive temperature leads to strong repression of rDNA transcription (lane 3), and newly synthesized precursor rRNA is processed very slowly (lane 4), consistent with previous data (20). On the other hand, in rrs1-1 mutant cells, a secretory defect does not cause repression of rDNA transcription (Fig. 2, lanes 7 and 8), indicating that the rrs1-1 mutation also affects the signaling pathway from a secretory defect to the repression of rDNA transcription. When the cells were pulse-labeled with [3H]uracil, its incorporation into the trichloroacetic acid-insoluble fraction was also strongly reduced under the conditions in which the secretory pathway was blocked, indicating that a secretory defect causes repression of rDNA transcription rather than inhibition of pre-rRNA methylation (data not shown).
FIG. 2.
The rrs1-1 mutation also affects the transcriptional repression of rDNA in response to a secretory defect. Strains KM003 (sly1 RRS1) and KM101 (sly1 rrs1-1) were grown to log phase (optical density at 600 nm = 1.0) in SC−Met medium at 25°C. Aliquots from each culture were shifted to 37°C for 90 min. Each culture was pulsed with [methyl-3H]methionine (60 μCi/ml) for 5 min and chased with nonradioactive methionine (500 μg/ml). Samples were taken and chilled by pouring onto crushed sterile ice at the time of addition of cold methionine (t = 0) and after a chase time of 10 min. Total RNA was prepared, and 20 μg of each sample was analyzed by electrophoresis. The gel was divided into two pieces. The upper gel was soaked in En3Hance (NEN), dried, and exposed to a film for 3 days (top panel) or for 10 days (middle panel). The lower gel was blotted and probed for U3 (bottom panel).
RRS1 is required for the repression of ribosomal protein genes in response to a secretory defect but not for the heat shock response.
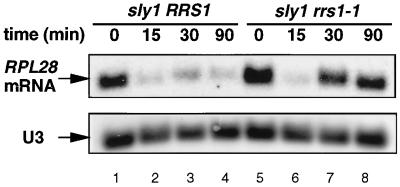
Mild heat shock causes the temporary repression of transcription of ribosomal protein genes (7, 11). To determine whether this repression acts in the same way as that due to a failure in the secretory pathway, we examined the effect of the rrs1-1 mutation on the repression through heat shock at 36°C. As shown in Fig. 3, a ribosomal protein gene, RPL28, was significantly repressed 15 min after the temperature shift-up in both the RRS1 and the rrs1-1 strains, indicating that the rrs1-1 mutation had no effect on the heat shock response. In the background of sly1, the RRS1 strain did not recover its RPL28 mRNA level in response to a secretory defect, whereas rrs1-1 recovered due to a defect in the secretory response, consistent with the data in Fig. 1B.
FIG. 3.
The rrs1-1 mutation affects the secretory defect response but not the heat shock response. Yeast KM003 (sly1 RRS1) and KM101 (sly1 rrs1-1) cells were grown to log phase (optical density at 600 nm = 0.5 to 0.8) in YPD medium at 25°C, and then the culture was shifted to 36°C. Aliquots were taken at the times indicated, and total RNA was prepared. Northern blot analysis was carried out using 32P-labeled DNA probe specific for RPL28. U3 was used as a marker to check equal loading. Five micrograms of total RNA was loaded in each lane.
Cloning of the RRS1 gene.
rrs1-1 mutant cells exhibited a prolonged generation time (about 7 h) at 18°C when cultured in YPD liquid medium. At higher temperatures, the growth rate was not so severely reduced; the generation time of rrs1-1 cells was 40% longer than that of wild-type cells at 30°C and 20% longer at 37°C. On a YPD plate, the rrs1-1 cells ceased to proliferate at 18°C. The cold sensitivity for growth segregated 2:2 in three sets of tetrads from RRS1/rrs1-1 diploid cells, and the cold sensitivity was linked to the phenotype for the defect in the signaling detected in Northern blot analysis (data not shown). Therefore, the wild-type RRS1 gene was cloned by complementation of the cold sensitivity of rrs1-1 cells. rrs1-1 cells were transformed with a library of yeast genomic DNA constructed in a URA3+ centromere-based vector, YCp50 (29). Of 2 × 104 Ura+ transformants, five could grow at 18°C on a YPD plate. Restriction maps of the plasmid DNAs recovered from these transformants revealed that each plasmid has an identical 18-kb insert. We determined a partial DNA sequence of the plasmid. A database search revealed that the insert carries six complete ORFs and two partial ORFs. To identify the region of the 18-kb DNA insert required for complementation, six subclones were constructed and their complementing activities for the cold-sensitivity of rrs1-1 were checked. The complementing activity was fully recovered in the plasmid, termed pAT-35, containing the 1.6-kb EcoRI-SacI fragment. Northern analysis revealed that the plasmid complements the defect in the signaling of rrs1-1 (data not shown). To confirm that pAT-35 contains the wild-type version of the rrs1-1 mutant gene, pAT-35 was subcloned into YIp5 and a fragment from the resultant plasmid was integrated into the rrs1-1 strain. After the transformant was crossed with a wild-type haploid, dissection of 17 tetrads yielded 17 sets of four spores, all of which could grow at 18°C (data not shown). These results indicate that pAT-35 contains RRS1.
Sequences of RRS1.
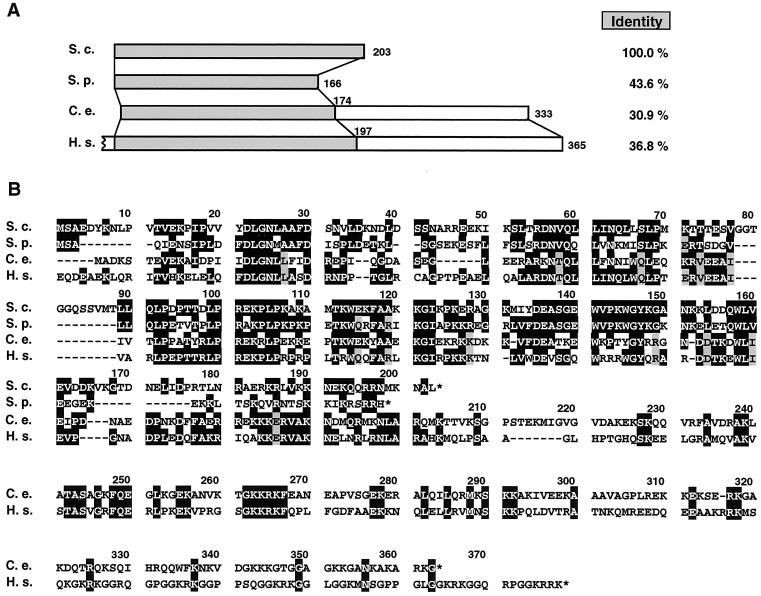
We determined the entire nucleotide sequence of the insert in pAT-35. RRS1 corresponds to ORF YOR294w, identified in the yeast genome project. RRS1 encodes a novel protein consisting of 203 amino acids with a predicted molecular mass of 23 kDa and an isoelectric point of 9.9. Although no conserved motifs were detected by using Prosite, a coiled-coil structure was predicted from amino acid 34 to 63 by using CoilScan (GCG). A database search revealed that DNA sequences from Homo sapiens, Caenorhabditis elegans, and Schizosaccharomyces pombe can encode proteins with significant similarity to Rrs1p (Fig. 4) with 36.8, 30.9, and 43.6% identities, respectively. The amino acid sequences of the putative human and C. elegans homologs are longer than that of Rrs1p at C-terminal domain, where their sequence similarity (identity, 26.7%) is lower.
FIG. 4.
Alignment of Rrs1p and related sequences from other species. The amino acid sequence of Rrs1p was predicted by nucleotide sequence that was determined using plasmid pAT-35 by dideoxy chain termination method with a Pharmacia DNA sequencer. The sequence was aligned with the predicted sequences from a database; Schizosaccharomyces pombe SPBC29A3.16 (S.p.) (DDBJ/EMBL/GenBank accession number AL022299), C. elegans cosmid C15H11 (C.e.) (39), and H. sapiens KIAA0112 (H.s.) (23). (A) Schematic alignment of the open reading frames. Amino acid residue numbers are marked. The initial codon of the H. sapiens sequence has not been determined yet and amino acids are numbered from the first methionine of the ORF. Identities to Rrs1p across the highlighted regions are listed on the right. (B) Alignment in single-letter code. Deletions needed for the alignment are indicated by dashes. Identical amino acids are boxed in black. When there are two sets of identical amino acids, one set is boxed in black and the other is shaded in gray.
rrs1-1 produces a truncated protein.
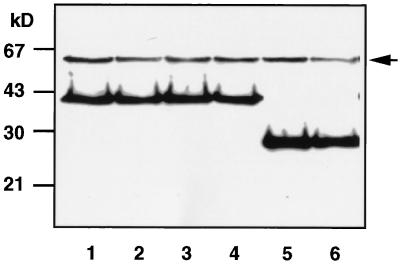
In order to determine the mutation point(s), the rrs1-1 gene was amplified by PCR using chromosomal DNA prepared from the mutant cells. Sequence analysis showed that rrs1-1 had only one nucleotide difference (a G-to-A transition), within codon 114, resulting in a stop codon. In order to compare the expression and molecular size between wild-type and mutant Rrs1p, three tandem copies of a sequence encoding an epitope from the influenza virus HA protein was inserted just after the initial codon of RRS1 or rrs1-1. Each of the fused genes was subcloned into a CEN plasmid. The plasmid containing RRS1-HA could complement both the lethality of the rrs1-null mutation and the cold sensitivity of the rrs1-1 mutation. The plasmid containing rrs1-1-HA could complement the lethality of the rrs1-null mutation and resulted in both cold sensitivity for growth and a defect in the signaling in response to a secretory defect (data not shown). These results indicate that the constructions are biologically functional. As shown in Fig. 5, the HA-tagged proteins, designated Rrs1-HA and rrs1-1-HA, were detected as 40-kDa and 26-kDa bands on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, considerably larger than the predicted molecular masses of the proteins. The levels of Rrs1-HA and rrs1-1-HA do not change in response to a secretory defect. It is noteworthy that the truncated protein appears to have stability similar to that of the wild-type protein.
FIG. 5.
Western blot analysis of Rrs1-HA and rrs1-1-HA. Haploid SEC+ cells expressing RRS1-HA (KM111; lanes 1 and 2) and sly1 cells expressing RRS1-HA (KM113; lanes 3 and 4) or rrs1-1-HA (KM114; lanes 5 and 6) were grown in SC medium to log phase at 25°C (lanes 1, 3, and 5). Half of the culture was shifted to 36°C for 90 min (lanes 2, 4, and 6). Crude extracts were denatured in SDS sample buffer and heated at 95°C for 5 min. Equal amounts of protein (44 μg) were subjected to SDS-PAGE and Western blotting using anti-HA. An arrow indicates nonspecific bands. The positions of size markers are shown on the left.
The RRS1 gene product is an essential nuclear protein.
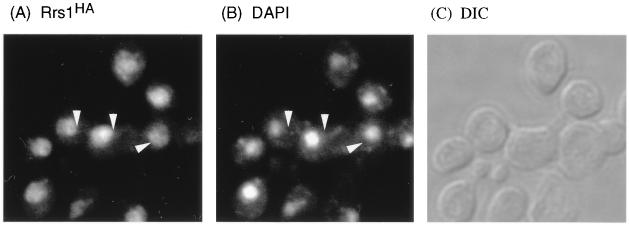
An rrs1-null allele was created by replacing the XbaI-BstEII fragment with the LEU2 gene. The DNA fragment containing the rrs1::LEU2 disruption was used to transform diploid W303 and KM007 (sly1/sly1). Southern blot analysis of genomic DNA isolated from the transformants both of W303 and KM007 demonstrated that the resultant diploid cells carried one intact RRS1 gene and one disrupted by insertion of LEU2 (data not shown). Thirteen tetrads from the transformant of W303 and nine tetrads from the transformant of KM007 were dissected. All tetrads yielded only two viable spores (data not shown), both of which were Leu−. Microscopic inspection of the other spores showed that they stopped dividing after several divisions (data not shown), indicating that RRS1 is essential for vegetative growth. Indirect immunofluorescent microscopy revealed that Rrs1-HA was localized in the nucleus at 25°C (Fig. 6). The staining region of Rrs1-HA was somewhat larger than that of DAPI, suggesting that Rrs1-HA was localized in both the nucleoplasm and the nucleolus.
FIG. 6.
Intracellular localization of Rrs1-HA detected by indirect immunofluorescence. Haploid cells expressing RRS1-HA (KM111) were grown in SC medium at 25°C to early log phase (optical density at 600 nm = 0.1 to 0.15) and stained with anti-HA antibodies (A), DAPI (B), and differential-interference-contrast (DIC) (C). Arrowheads are put at the same positions in panels A and B.
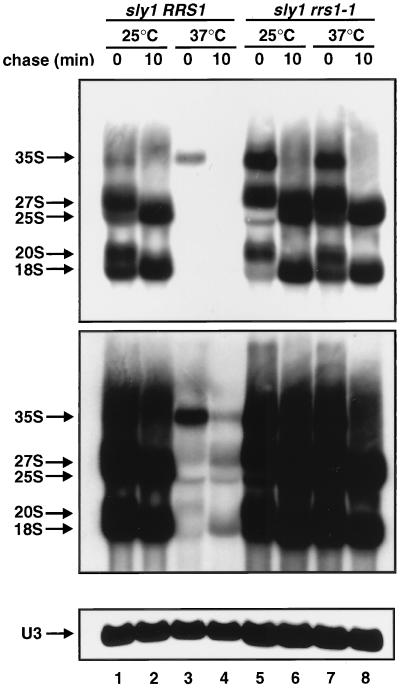
rrs1-1 is defective in rRNA processing and ribosome assembly.
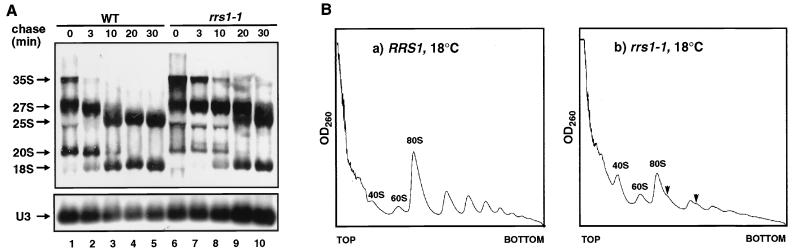
Because some Rrs1 protein appears to be localized in the nucleolus, Rrs1p is expected to have a role in ribosome biogenesis. Furthermore, as shown in Fig. 2, maturation of 25S rRNA in rrs1-1 mutant cells (lanes 5 and 6) appears to be slower than in wild-type cells (lanes 1 and 2) at a permissive temperature. Thus, we performed [methyl-3H]methionine pulse-chase and polyribosome analyses with wild-type and rrs1-1 cells at 18°C. rrs1-1 cells cannot proliferate at 18°C on a YPD plate, but they continue to grow at 18°C in liquid culture at an extremely low rate. As shown in Fig. 7A, in rrs1-1 cells, the rate of processing of pre-rRNA was low and less mature rRNA was produced than in wild-type cells. The polyribosome profile showed that in rrs1-1 cells, 80S ribosomes and polyribosomes were decreased, 40S subunits were accumulated, and half-mer polyribosomes appeared (Fig. 7B), suggesting that 60S subunit production is compromised more than 40S production.
FIG. 7.
rrs1-1 mutation causes defect in rRNA processing and ribosome assembly. (A) Pulse-chase analysis of rRNA synthesis. W303a (RRS1, wild type [WT]) and KM123 (rrs1-1) were cultured in SC−Met medium at 18°C overnight. Each culture was pulsed with [methyl-3H]methionine (10 μCi/ml) for 3 min and chased with nonradioactive methionine (500 μg/ml). Samples were taken and chilled by pouring onto crushed sterile ice at the time of addition of cold methionine (t = 0) and after the indicated chase times. Total RNA was prepared and 20 μg of each sample was analyzed by electrophoresis and blotted to a Nytran membrane. The membrane was sprayed with En3Hance (NEN) and exposed to a film for 2 days. The lower gel was blotted and probed for U3. (B) Polyribosome profiles from W303a (a) and KM123 (b). The cells were cultured in YPD at 18°C overnight. Free ribosomal subunits and polyribosomes in cell extracts were separated by sucrose density gradient centrifugation. Arrows indicate half-mer polyribosomes.
RRS1 is required for ribosome biogenesis.
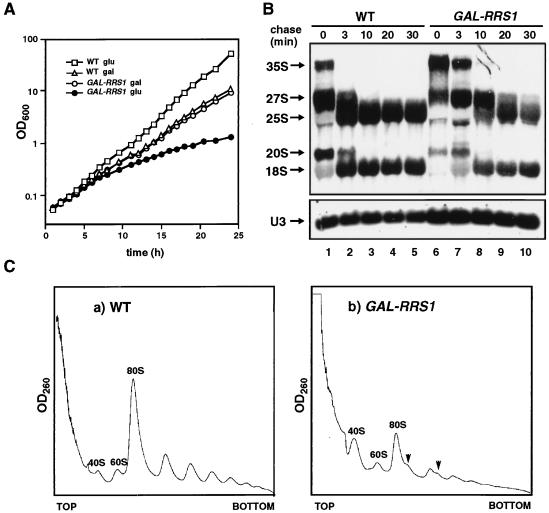
In order to achieve conditional expression of RRS1, we constructed a strain in which the chromosomal RRS1 gene was disrupted and a plasmid containing a GAL1-promoter driven RRS1 was introduced. This strain, KM129, could grow on a YPG (galactose) plate but not on a YPD (glucose) plate (data not shown). In a liquid galactose medium, KM129 cells grew as fast as wild-type cells. After the shift to a glucose medium, the growth of KM129 cells slowed gradually (Fig. 8A). We performed [methyl-3H]methionine pulse-chase analysis to investigate pre-rRNA synthesis, its processing, and the stability of the rRNAs. In Rrs1p-depleted cells, the rate of processing of pre-rRNA was low and less 25S rRNA was produced than in wild-type cells (Fig. 8B). After a 30-min chase, amounts of 25S rRNA appeared to be somewhat decreased compared to the level at 20-min chase, suggesting that newly formed 25S rRNA is unstable in Rrs1p-depleted cells. This experiment was done in duplicate, and similar results were obtained. Accumulation of 18S rRNA in Rrs1p-depleted cells was also slower than that in wild-type cells, but the defect appeared to be less severe than for 25S rRNA. The polyribosome profile in Rrs1p-depleted cells showed a decrease in the levels of 80S ribosomes and polyribosome, accumulation of 40S subunits, and the appearance of half-mer polyribosomes (Fig. 8C). These results suggest that Rrs1p depletion affects 25S rRNA and 60S subunit production much more than 18S rRNA and 40S subunit production.
FIG. 8.
Depletion of Rrs1p causes growth arrest and defect in ribosome biogenesis and in rRNA processing. (A) Growth curves of W303a (RRS1, wild type [WT]) and KM129 (GAL-RRS1) cultured at 25°C in YPGal and shifted to YPD. The change in optical density at 600 nm was monitored after the shift. The cell cultures were diluted to keep the optical density lower than 1.0, and the values were calculated as changes from initial values. (B) Pulse-chase analysis of rRNA synthesis. W303a and KM129 (GAL-RRS1) were cultured in SCGal−Met and shifted to SC−Met for 12 h. Each culture was pulsed with [methyl-3H]methionine (10 μCi/ml) for 3 min and chased with nonradioactive methionine (500 μg/ml). Samples were taken and chilled by pouring onto crushed sterile ice at the time of addition of cold methionine lite(t = 0) and after the indicated chase times. Total RNA was prepared and 20 μg of each sample was analyzed by electrophoresis and blotted to a Nytran membrane. The membrane was sprayed with En3Hance (NEN) and exposed to a film for 2 days. The lower gel was blotted and probed for U3. (C) Polyribosome profiles from W303a (a) and KM129 (GAL-RRS1) (b). The cells were cultured in YPGal and shifted to YPD for 12 h. Free ribosomal subunits and polyribosomes in cell extracts were separated by sucrose density gradient centrifugation. Arrows indicate half-mer polyribosomes.
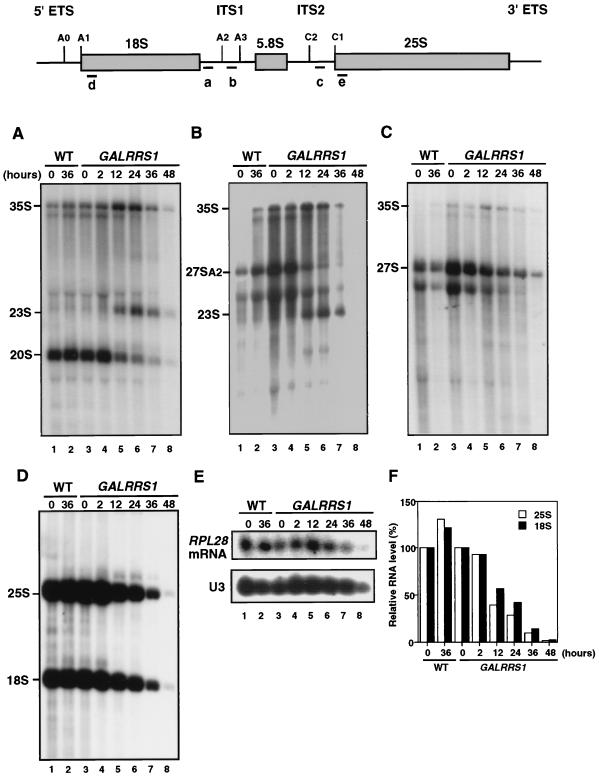
Steady-state levels of pre-rRNAs in GAL-RRS1 cells were analyzed after the shift to glucose medium for up to 48 h. Following transfer of the GAL-RRS1 strain to glucose medium, the 35S pre-rRNA accumulated slightly up to 24 h (Fig. 9A to C), consistent with the results of pulse-chase labeling. The reduced levels of both 27SA2 (Fig. 9B) and 20S (Fig. 9A) suggest that cleavage at A2 is retarded in accordance with the depletion of Rrs1p. In Rrs1p-depleted cells, 23S RNA appears via an aberrant pathway in which the 35S pre-rRNA is not processed at sites A0, A1, and A2 and is instead cleaved at site A3 in the internal transcribed spacer 1 (Fig. 9A and B). The steady-state level of the 25S rRNA decreased during the time course of the experiments and levels of both the 20S pre-rRNA and the mature 18S rRNA were also reduced (Fig. 9A and D). The level of 25S rRNA declined more rapidly than that of 18S rRNA; after 12 h of growth in glucose medium, the amount of 25S rRNA was reduced to 39% and the amount of 18S rRNA was 57% (Fig. 9F). On the other hand, the level of ribosomal protein L28 mRNA was not significantly decreased up to 24 h (Fig. 9E), indicating that Rrs1p is not required for transcription of ribosomal protein genes.
FIG. 9.
Northern analysis for steady-state level of pre-rRNAs. The structure of the pre-rRNA and locations of oligonucleotide hybridization probes are shown on the top. W303a (RRS1, wild type [WT]) and KM129 (GAL-RRS1), cultured at 25°C in YPGal, were shifted to YPD and cultured for up to 48 h. Total RNA equivalent to cells at an optical density at 600 nm of 0.5 was used for each sample. Northern blot analysis was carried out using 32P-labeled DNA probes: probe a (A), probe b (B), probe c (C), probe d for 18S and probe e for 25S (D), and probe for RPL28 and probe for U3 (E). (F) RNA levels were quantified using BAS-1000 (Fuji Photo Film Co.), and the ratio of the radioactivity value at each time point to that at time zero is shown.
DISCUSSION
Because of the huge amount of ribosome biosynthesis in growing cells, its regulation is essential for the economy of the cells. A functional secretory pathway is required for the normal synthesis of ribosomes, suggesting a coupling between plasma membrane and ribosome syntheses (20). In order to elucidate this regulatory mechanism, we isolated the rrs1-1 mutant in which a secretory defect (sly1ts) failed to repress transcription of ribosomal protein genes. RRS1 is an essential gene whose function has not been previously described. Comparison of the Rrs1p sequence with database sequences showed that the protein is conserved throughout eukaryotes, but no information about its function is available. The amino acid sequence of Rrs1p has no readily recognizable protein motifs.
It is unlikely that rrs1-1 suppresses the defect in the secretory pathway of sly1, because rrs1-1 does not suppress temperature sensitivity for growth (data not shown) and the rrs1-1 mutation results in derepression of ribosomal protein genes when the secretory pathway is blocked by using tunicamycin as well as by shifting sly1 cells to the restrictive temperature (Fig. 1). Western blotting analysis of functional epitope-tagged Rrs1-HA and rrs1-1-HA indicates that rrs1-1-HA has stability similar to that of the wild-type version and that a secretory defect does not cause a significant change in their level, suggesting that the effect of rrs1-1 mutation on the response to a secretory defect is not due to a change of rrs1-1 concentration in the cell.
Rrs1-HA appears to be localized throughout the nucleus, including the nucleolus (Fig. 6). The nucleolus is the site of rRNA synthesis, rRNA processing, and ribosomal subunit assembly (reviewed in reference 38). Ribosomal proteins newly synthesized in the cytoplasm need to be transported into the nucleolus and assembled with rRNAs. The assembled preribosomal subunits are then transported into the cytoplasm. During ribosome assembly, pre-rRNAs are modified and cleaved to mature rRNAs. Pre-rRNA processing and assembly of ribosomal subunits are tightly linked to each other because many ribosomal proteins associate with the pre-rRNA at early steps prior to cleavage. For example, mutants of the 60S subunit protein L11 (according to a new nomenclature in reference 18; formerly called L16) exhibit a shortage of mature 25S rRNA and accumulation of rRNA precursors (22). Many trans-acting factors have been found to be involved in pre-rRNA processing and ribosomal subunit assembly steps (reviewed in reference 34). Some of them have an enzymatic activity such as RNA cleavage, methylation, or pseudouridine formation (2, 12, 13, 19, 31). Others may have a role in maintaining the structure of the complex containing pre-rRNA(s) and many ribosomal proteins. Depletion of such factors may cause structural changes of the complex and prevent further processing of pre-rRNA. In Rrs1p-depleted cells, pre-rRNA processing is retarded at the A0, A1, and A2 sites, and consequently, less 25S and 18S mature rRNA is produced. Instead, aberrant cleavage produces 23S rRNA, as was shown in other mutants (5, 14, 16, 41). In [methyl-3H]methionine-pulse chase analysis of rrs1-1 and GAL-RRS1 (Fig. 7A and 8B), the 20S band appears to be a doublet. The upper band seems to be 21S produced by aberrant cleavage (35). Steady-state levels of 18S and 25S rRNAs decline along with depletion of Rrs1p. These results suggest that Rrs1p has a role in maintaining the structure of preribosomal subunit particles rather than a role as a specific enzyme in a step of pre-rRNA processing.
Interestingly, the rrs1-1 mutation strongly diminished transcriptional repression of both ribosomal protein genes and rDNA in response to a secretory defect (Fig. 1 and 2). These results suggest that Rrs1p may have an important role in transcriptional regulation of both rRNA and ribosomal protein genes. However, neither the rrs1-1 mutation nor the depletion of Rrs1p causes a significant defect in transcription of ribosomal protein genes under the conditions in which the secretory pathway is normal. Rrs1p may have a role in maintenance of balanced transcription between the components of the ribosome. Depletion of Rrs1p using a conditional rrs1-null mutation caused defects in pre-rRNA processing and ribosomal subunit assembly; slow processing of pre-rRNA, reduced production of 25S rRNA, decrease in levels of 80S ribosomes and polyribosomes, accumulation of 40S ribosomal subunits, and the appearance of half-mer polyribosomes which contain 43S initiation complexes stalled at the AUG start codon. The result indicates that Rrs1p depletion strongly affects maturation of 25S rRNA and 60S subunit assembly rather than that of 18S rRNA and 40S subunit assembly. [methyl-3H]methionine pulse-chase analysis and the polyribosome pattern from the rrs1-1 mutant cells cultured at 18°C also suggest its defect in 25S rRNA production and 60S subunit assembly (Fig. 7B). These results suggest that rrs1-1 or Rrs1p depletion affects primarily 25S rRNA maturation and 60S subunit assembly. The decline of the 18S rRNA level in Rrs1p-depleted cells may be a secondary effect. The cold-sensitive phenotype of the rrs1-1 mutant may be explained by its defect in assembly of ribosomal subunits; many yeast mutants that had defects in ribosomal subunit assembly were isolated by screening from a collection of cold-sensitive strains (28).
Transcription of ribosomal protein genes is temporarily repressed by heat shock (7, 11). The rrs1-1 allele has little effect on the temporary repression of transcription brought about by mild heat shock, in contrast to its effect on the secretory response (Fig. 3). This suggests that RRS1 is not required for the temporary repression of ribosomal protein genes after temperature shift-up. This also indicates that the mechanism of the secretory response is different from that of the heat shock response, consistent with the results obtained by using a rap1-17 mutant (21).
The great majority of ribosomal protein genes are driven by a Rap1-binding site(s) (15, 40). A few ribosomal protein genes, including RPL3 (encoding ribosomal protein L3), have no site for Rap1 but have a single Abf1-binding site instead (3, 6, 8). Yet the transcription of all the ribosomal protein genes appears to be coordinated under several experimental conditions. A secretory defect also causes the coordinated repression of ribosomal protein genes (20). We showed that the repression of both RPL28 (Rap1 driven) and RPL3 (Abf1 driven) was diminished by the C-terminally truncated rap1-17 mutation (21). This suggests that Rap1 is essential for the repression in response to a secretory defect but Rap1-binding sites are not necessary as cis-acting element for the repression. In this paper, we have demonstrated that the rrs1-1 mutation affects the repression of both RPL28 and RPL3 (Fig. 1 and 2), suggesting that a common mechanism regulates the transcription of both types of genes.
The mechanism of transcriptional repression of both rRNA and ribosomal protein genes in response to a secretory defect remains to be elucidated. Our data suggest that Rrs1p is involved in this mechanism, although there are several alternative possibilities. One possibility is that the slow growth of rrs1-1 may result in insensitivity to a secretory defect. However, other mutants which exhibit slow growth still respond to a defect in the secretory pathway (data not shown). The second possibility is that a feedback effect of a defect in ribosome synthesis of rrs1-1 mutation can overcome the effect caused by a secretory defect. We cannot eliminate this possibility completely, but the rrs1-1 allele has little effect on the temporary repression of transcription of ribosomal protein genes brought about by mild heat shock (Fig. 3). This suggests that the rrs1-1 cells still have a potential to lead the repression of ribosomal protein genes after temperature shift-up. Thus, we prefer an alternative possibility: the signal from a secretory defect might be transduced through the ribosome assembly machinery, including Rrs1p. A two-hybrid screen using RRS1 as a bait has suggested that Rrs1p may interact with Rpl11 (Y. Matsui et al., unpublished data). As Rpl11 is a ribosomal protein localized on the surface of the large subunit (36), Rrs1p might associate with the ribosome through interaction with Rpl11. The possible association of Rrs1p with the ribosome is now under investigation. It is possible that ribosomal assembly machinery that involves an interaction of Rrs1p with Rpl11 might be important for the regulation. According to this model, the mechanism of transcriptional repression in response to a secretory defect is tightly linked to the normal regulatory mechanism that maintains ribosome synthesis. The C-terminal region of Rrs1p that is deleted in rrs1-1 might be responsible for the signaling. In this case, transcriptional repression of ribosomal protein genes may follow the repression of rRNA genes and the C-terminal region of Rrs1p may be required for the signaling.
ACKNOWLEDGMENTS
We thank E. Tsuchiya, D. Hirata, and A. Wada for valuable discussion, M. D. Rose for yeast genomic DNA library, K. Matsumoto and Y. Ohya for a plasmid, H. Yoshida and Y. Maki for help in polyribosome analysis, and N. Tanaka for help in the database search. We also thank Y. Matsui and A. Toh-e for communicating unpublished data. We are particularly grateful to J. R. Warner for critical reading of the manuscript.
This research was supported by grants from the Ministry of Education, Science and Culture of Japan and Special Coordination Funds for Promoting Science and Technology of the Science and Technology Agency of the Japanese government.
REFERENCES
- 1.Bergès T, Petfalski E, Tollervey D, Hurt E C. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu S, Archer R H, Zengel J M, Lindahl L. The RNA of Rnase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorsman J C, Doorenbosch M M, Maurer C T C, de Winde J H, Mager W H, Planta R J, Grivell L A. An ARS/silencer binding factor also activates two ribosomal protein genes in yeast. Nucleic Acids Res. 1989;17:4917–4923. doi: 10.1093/nar/17.13.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eng F J, Warner J R. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991;65:797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- 5.Gautier T, Bergès T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamil K G, Nam H G, Fried H M. Constitutive transcription of yeast ribosomal protein gene RPL3 is promoted by uncommon cis- and trans-acting elements. Mol Cell Biol. 1988;8:4328–4341. doi: 10.1128/mcb.8.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herruer M H, Mager W H, Raué H A, Vreken P, Wilms E, Planta R J. Mild temperature shock affects transcription of yeast ribosomal protein genes as well as the stability of their mRNAs. Nucleic Acids Res. 1988;16:7917–7929. doi: 10.1093/nar/16.16.7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herruer M H, Mager W H, Doorenbosch T M, Wessels P L M, Wassenaar T M, Planta R J. The extended promoter of the gene encoding ribosomal protein S33 in yeast consists of multiple protein binding elements. Nucleic Acids Res. 1989;17:7427–7439. doi: 10.1093/nar/17.18.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1994. [Google Scholar]
- 11.Kim C H, Warner J R. Mild temperature shock alters the transcription of a discrete class of Saccharomyces cerevisiae genes. Mol Cell Biol. 1983;3:457–465. doi: 10.1128/mcb.3.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiss-Laszlo Z, Henry Y, Bachellerie J, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 13.Lafontaine D L J, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lafontaine D L J, Preiss T, Tollervey D. Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol Cell Biol. 1998;18:2360–2370. doi: 10.1128/mcb.18.4.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lascaris R F, Mager W H, Planta R. DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics. 1999;15:267–277. doi: 10.1093/bioinformatics/15.4.267. [DOI] [PubMed] [Google Scholar]
- 16.Lee S J, Baserga S J. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol Cell Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Warner J R. Mutation of the Rab6 homologue of Saccharomyces cerevisiae, YPT6, inhibits both early Golgi function and ribosome biosynthesis. J Biol Chem. 1996;271:16813–16819. doi: 10.1074/jbc.271.28.16813. [DOI] [PubMed] [Google Scholar]
- 18.Mager W H, Planta R J, Ballesta J-P G, Lee J C, Mizuta K, Suzuki K, Warner J R, Woolford J. A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 20.Mizuta K, Warner J R. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol Cell Biol. 1994;14:2493–2502. doi: 10.1128/mcb.14.4.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuta K, Tsujii R, Warner J R, Nishiyama M. The C-terminal silencing domain of Rap1p is essential for the repression of ribosomal protein genes in response to a defect in the secretory pathway. Nucleic Acids Res. 1998;26:1063–1069. doi: 10.1093/nar/26.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moritz M, Pulaski B A, Woolford J L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991;11:5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagase T, Miyajima N, Tanaka A, Sazuka T, Seki N, Sato S, Tabata S, Ishikawa K-I, Kawarabayasi Y, Kotani H, Nomura N. Prediction of the coding sequences of unidentified human genes. III. The coding sequences of 40 new genes (KIAA0081–KIAA0120) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1995;2:37–43. doi: 10.1093/dnares/2.1.37. [DOI] [PubMed] [Google Scholar]
- 24.Nierras C R, Warner J R. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J Biol Chem. 1999;274:13235–13241. doi: 10.1074/jbc.274.19.13235. [DOI] [PubMed] [Google Scholar]
- 25.Ninomiya-Tsuji J, Nomoto S, Yasuda H, Reed S I, Mutsumoto K. Cloning of a human cDNA encoding a CDC2-related kinase by complementation of a budding yeast cdc28 mutation. Proc Natl Acad Sci USA. 1991;88:9006–9010. doi: 10.1073/pnas.88.20.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ossig R, Dascher C, Trepte H-H, Schmitt H D, Gallwitz D. The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol Cell Biol. 1991;11:2980–2993. doi: 10.1128/mcb.11.6.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 28.Ripmaster T L, Vaughn G P, Woolford J L., Jr DRS1 to DRS7, novel genes required for ribosome assembly and function in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7901–7912. doi: 10.1128/mcb.13.12.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 30.Rotenberg M O, Woolford J L., Jr Tripartite upstream promoter element essential for expression of Saccharomyces cerevisiae ribosomal protein genes. Mol Cell Biol. 1986;6:674–687. doi: 10.1128/mcb.6.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt M E, Clayton D A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sikorski R, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takita Y, Ohya Y, Anraku Y. The CLS2 gene encodes a protein with multiple membrane-spanning domains that is important Ca2+ tolerance in yeast. Mol Gen Genet. 1995;246:269–281. doi: 10.1007/BF00288599. [DOI] [PubMed] [Google Scholar]
- 34.Tollervey D. trans-acting factors in ribosome synthesis. Exp Cell Res. 1996;229:226–232. doi: 10.1006/excr.1996.0364. [DOI] [PubMed] [Google Scholar]
- 35.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsay Y-F, Shankweiler G, Lake J, Woolford J L., Jr Localization of Saccharomyces cerevisiae ribosomal protein L16 on the surface of 60S ribosomal subunits by immunoelectron microscopy. J Biol Chem. 1994;269:7579–7586. [PubMed] [Google Scholar]
- 37.Udem S A, Warner J R. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J Mol Biol. 1972;65:227–242. doi: 10.1016/0022-2836(72)90279-3. [DOI] [PubMed] [Google Scholar]
- 38.Warner J R. The nucleolus and ribosome formation. Curr Opin Cell Biol. 1990;2:521–527. doi: 10.1016/0955-0674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 39.Wilson R, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 40.Woolford J L, Jr, Warner J R. The ribosome and its synthesis. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 587–626. [Google Scholar]
- 41.Zanchin N I T, Goldfarb D S. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol Cell Biol. 1999;19:1518–1525. doi: 10.1128/mcb.19.2.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]