Abstract
Background and Aim
Plant disjunctions have fascinated biogeographers and ecologists for a long time. We use tribe Bocageeae (Annonaceae), a predominantly Neotropical plant group distributed across several present-day Neotropical biomes and with an African–American disjunction, to investigate long-distance dispersal mediated by frugivorous animals at both intercontinental and intracontinental scales.
Methods
We reconstructed a species-level phylogeny of tribe Bocageeae with a dataset composed of 116 nuclear markers. We sampled 70 % of Bocageeae species, covering their geographical range and representing all eight genera. We estimated divergence times using BEAST, inferred ancestral range distributions and reconstructed ancestral states for fruit traits related to long-distance dispersal in a Bayesian framework.
Key Results
The ancestral Bocageeae date to the Early Eocene and were inferred to occur in Africa and proto-Amazonia. Their ancestral fruits were large and dehiscent. The first lineage split gave rise to an exclusively Neotropical clade during the Middle Eocene, in proto-Amazonia. Range exchange between the Amazon and the Atlantic Forest occurred at least once during the Miocene, and from Amazonia to Central America and Mexico during the Early Miocene. Transitions in different sets of fruit morphologies were inferred to be related to dispersal events across South American regions/biomes.
Conclusions
In Bocageeae, mammals might have been responsible for long-distance dispersal through the Boreotropics. In the Neotropics, proto-Amazonia is proposed to be the source for dispersal to other tropical American biomes. Long-distance dispersal might have happened via a wide range of dispersal guilds, depending on frugivore radiations, diversity and abundance in particular time periods and places. Hence, inter- and intracontinental dispersal might not rely on a single dispersal syndrome or guild, but more on the availability of frugivorous lineages for seed dispersal.
Keywords: Amazonia, Atlantic Forest, Boreotropics, divergence times, phylogenomics
INTRODUCTION
Traditionally, disjunctions have been explained by vicariance, by extinction in the intervening area of a once widespread taxon or by long-distance dispersal to a new area (Platnick and Nelson, 1978). In plants, long-distance dispersal has been hypothesized to explain the American–African disjunction (sensuThorne, 1973) of the relatively few amphi-Atlantic taxa when compared with the high diversity of both tropical Africa and America (Renner, 2004). Eastern South America and Africa were connected until ~100 million years ago (Ma), when West Gondwana started to break up by the opening of the South Atlantic Ocean (McLoughlin, 2001). Although vicariance has often been invoked to explain the distribution of amphi-Atlantic taxa (e.g. Michalak et al., 2010; Mello-Silva et al., 2011; Luebert et al., 2017), alternative explanations, such as long-distance dispersal, seem to explain the disjunct distributions of younger lineages better (Pennington and Dick, 2004; Beaulieu et al., 2013). Under this scenario, two possible explanations have been proposed. One involves trans-Atlantic dispersal by stepping-stone mechanisms through volcanic lineaments, such as the Walvis Ridge and the Rio Grande rise or the Ceará and Sierra Leone rises (from 100 to 54–36 Ma; Parrish, 1993; Morley, 2000; 2003). Dispersion from Africa to regions of the Northern Hemisphere via geodispersal through the Boreotropical connection (from 52–50 to 34 Ma; Wolfe, 1975) has also been put forward (e.g. Pirie et al., 2006; Smedmark and Anderberg, 2007; Couvreur et al., 2011; Wei et al., 2015).
The general direction of the process that led to amphi-Atlantic distribution in plants might have been mainly from tropical America, i.e. the Neotropics, to tropical Africa (Antonelli et al., 2015; e.g. Malpighiaceae, Davis et al., 2002). This directional trend can be deduced from the phylogenetic history of various tropical lineages when comparing range shifts from and to each tropical region, Africa, America and Asia (Antonelli et al., 2015). A difference in range shifts was observed only for the Neotropics, where range shifts out of the Neotropical region were more frequent than those coming to it during the Cenozoic (Antonelli et al., 2015). However, for some important components of the Neotropical flora, such as Rubiaceae (Antonelli et al., 2009) and Annonaceae (Richardson et al., 2004; Couvreur et al., 2011), a Palaeotropical origin has been hypothesized. In both families, geodispersal from Africa to the Neotropics through the Boreotropical connection (from 52–50 to 34 Ma; Wolfe, 1975) has been invoked (Richardson et al., 2004; Antonelli et al., 2009; Couvreur et al., 2011).
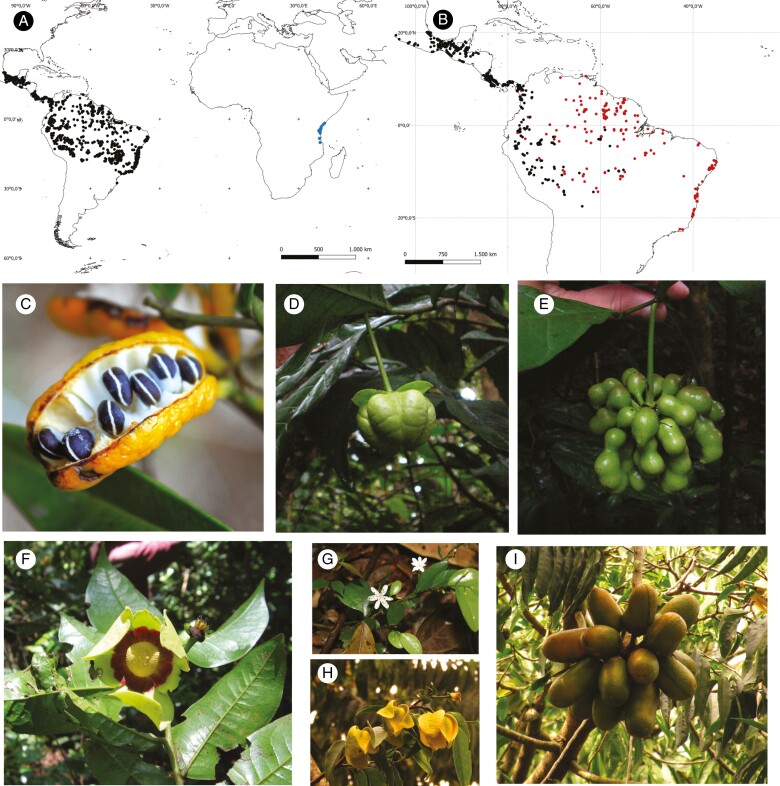
Long-distance dispersal (Jordano, 2016) in a time frame of millions of years is rare (Nathan and Muller-Landau, 2000) and might depend on fruit-eating (frugivory) and seed dispersal by animals of tropical plants. These long-distance dispersal events might have been facilitated primarily by large-bodied and wide-ranging frugivorous megafauna, mammals or strong-flying volant frugivorous birds or bats that could move across large distances and cross barriers (e.g. oceans) (Onstein et al., 2019). In Annonaceae, a major pantropical plant family including ~2500 species of trees, shrubs and lianas (Chatrou et al., 2012) with intercontinental disjunctions (Fig. 1A; Couvreur et al., 2011), fruit traits related to long-distance dispersal and their associated dispersers were investigated with a phylogenetic approach (Onstein et al., 2019). Intercontinental dispersals in Annonaceae were associated with apocarpous fruits with few, large monocarps or with large syncarpous fruits with many seeds, with dull colours (i.e. green or brown; Onstein et al., 2019). These fruit traits are often associated with the mammal dispersal guild (Onstein et al., 2019). The fruit traits in Annonaceae related to the long-distance dispersal events that originated in American–African disjunctions were large fruits on short stipes, with a high probability of being syncarpous and dehiscent (Fig. 1C; Onstein et al., 2019). These features are associated with both mammal and bird trait syndromes (Onstein et al., 2019). However, the fruit trait syndromes related to dispersal within continents have not yet been investigated. Annonaceae, a very diverse plant family in tropical forests (Draper et al., 2021), shows many intracontinental disjunctions, at both the specific level [e.g. Oxandra espintana (Spruce ex Benth.) Baill., Junikka et al. (2016) and Cymbopetalum brasiliense (Vell.) Benth. ex Baill., Fig. 1B; Murray (1993)] and the generic level (e.g. Pseudoxandra, Maas and Westra (2003) and Cymbopetalum, Fig. 1B; Johnson and Murray (1995)).
Fig. 1.
Distribution and morphological diversity of Bocageeae (Annonaceae). (A) Distribution of tribe Bocageeae. Black dots indicate the distribution of the genera Bocageae, Cardiopetalum, Cymbopetalum, Froesiodendron, Hornschuchia, Porcelia and Trigynaea; blue dots indicate the distribution of Mkilua fragrans Verdc. (B) Distribution of Cymbopetalum. Red dots represent the distribution of Cymbopetalum brasiliense (Vell.) Benth. ex Baill.; black dots represent the distribution of the 26 remaining species in the genus. (C) Cardiopetalum calophyllum Schltdl., fruit, dehiscent monocarps. (D, E) Cymbopetalum brasiliense (Vell.) Benth. ex Baill. (D) Flower. (E) Apocarpous fruit. (F) Mkilua fragrans Verdc. Flower. (G) Hornschuchia myrtillus Nee; flowers. (H, I) Porcelia macrocarpa (Warm.) R.E. Fr. (H) Flowers. (I) Apocarpous fruit. Photographs by: Kuhlmann (2018) (C); Tarcísio Leão (D, E); Thomas L. P. Couvreur (F); Renato Mello-Silva (G); and Otávio Marques (H, I).
In the Neotropics, one present-day disjunction pattern is that observed among Amazon–Atlantic Forest taxa, with groups occurring in Amazonian forests in northwestern South America and the Atlantic Forest on the east coast of this continent. This disjunct distribution pattern is separated by a diagonal of open vegetation, which includes the Caatinga, Cerrado and Chaco biomes crossing central South America (Fiaschi and Pirani, 2009).
Within Annonaceae, the tribe Bocageeae presents both disjunction patterns, African–American and Amazon–Atlantic Forest (Johnson and Murray, 1995; Fig. 1A, B). The Bocageeae comprise 66 species distributed among eight genera (Bocagea, Cardiopetalum, Cymbopetalum, Froesiodendron, Hornschuchia, Mkilua, Porcelia and Trigynaea; Fig. 1C–I) (Murray, 1993; Johnson and Murray, 1995; Mello-Silva and Lopes, 2020; Vilela and Lopes, 2022). Most genera are exclusively Neotropical, whereas the monotypic Mkilua is endemic to East Africa (Verdcourt, 1970; Fig. 1A, F). The Neotropical Bocageeae genera are distributed from Mexico to Southern Brazil (Murray, 1993; Johnson and Murray, 1995; Mello-Silva and Lopes, 2020; Fig. 1A). The phylogenetic relationship among Bocageeae genera was first reconstructed by Johnson and Murray (1995) with morphological data. After that, Chatrou et al. (2012) sampled seven species of the tribe, belonging to five genera, whereas Guo et al. (2017) sampled ten species belonging to seven of the eight genera in the tribe; in both those studies, few molecular markers have been used for phylogeny reconstruction. Using the same taxonomic and molecular sampling as Chatrou et al. (2012), Pirie and Doyle (2012) estimated the divergence time for Bocageeae. The tribe is likely to have diverged during the Eocene, with the East African Mkilua fragrans Verdc. as sister to the Neotropical Bocageeae (Chatrou et al., 2012; Pirie and Doyle, 2012; Guo et al., 2017). In turn, Annonaceae is likely to have originated in the African portion of West Gondwana during the Late Cretaceous (Richardson et al., 2004; Couvreur et al., 2011). The American–African disjunction in Annonaceae has been explained by both the trans-Atlantic route and the Boreotropical geodispersal hypothesis (Richardson et al., 2004; Couvreur et al., 2011; Onstein et al., 2019).
Here, we use Bocageeae to investigate long-distance dispersal by frugivorous animals at both intercontinental and intracontinental scales. We hypothesize (H1) that the American–African disjunction resulted from intercontinental dispersal from Africa to America via the Boreotropical route. Furthermore, we hypothesize (H2) that intracontinental dispersal within the Neotropics resulted from proto-Amazonia being the source for dispersal to other biomes and regions (e.g. Atlantic Forest and Central America). Finally, we hypothesize (H3) that inter- and intracontinental dispersals have been facilitated by large-bodied frugivorous mammals, birds or bats. Hence, we predict that ancestral fruit traits related to these dispersal guilds (e.g. large fruits, dehiscent fruits) would be inferred for lineages that shifted between continents or biomes. Towards this goal, we reconstructed the first densely sampled species-level phylogeny of tribe Bocageeae (Annonaceae), using a genomic dataset composed of 116 nuclear molecular markers, including ~70 % of its species (47 of 66 taxa), representing all eight recognized genera and encompassing the distribution of all taxa. In addition, we inferred the ancestral states of fruit traits associated with long-distance dispersal both intercontinentally and within the Neotropics and we estimated ancestral ranges.
MATERIALS AND METHODS
Taxon sampling
We sampled all eight genera and 47 of 66 species of tribe Bocageeae of the subfamily Annonoideae: Bocagea A. St.-Hil. (two of four species), Cardiopetalum Schltdl. (two of three species), Cymbopetalum Benth. (17 of 27 species), Froesiodendron R.E.Fr. (two of three species), Hornschuchia Nees (10 of 12 species), Mkilua Verdc. (one of one species), Porcelia Ruiz and Pav. (seven of seven species) and Trigynaea Schltdl. (six of nine species; Table 1; Fig. 1C–I; Murray, 1993; Johnson and Murray, 1995; Lobão, 2017; Mello-Silva and Lopes, 2020; Vilela and Lopes, 2022). Five species were sequenced previously (Couvreur et al., 2019); the remaining 42 species were newly sequenced for this study.
Table 1.
Taxa sampling from the ingroup, tribe Bocageeae (Annonaceae: Annonoideae) and outgroups. Vouchers with collector and associated collector number and herbaria according to Thiers (updated continuously).
| Species | Country | Voucher | GenBank accession |
|---|---|---|---|
| Bocagea asymmetrica Mello-Silva & J.C. Lopes | Brazil | Mello-Silva, R. 3142 (SPF) | PRJNA508895 |
| Bocagea longepedunculata Mart. | Brazil | Demuner, V. 4181 (U) | SUB11858406 |
| Cardiopetalum calophyllum Schltdl. | Brazil | Silva, A.S.L. 6 (U) | SUB12129452 |
| Cardiopetalum surinamense R.E. Fr. | French Guyana | Mori, S.A. 25325 (NY) | SUB11858406 |
| Cymbopetalum aequale N.A. Murray | Ecuador | Couvreur, T.L.P. 1352 (P) | SUB11463464 |
| Cymbopetalum baillonii R.E. Fr. | Mexico | Liebmann, F.M. 29 (P) | PRJNA508895 |
| Cymbopetalum brasiliense (Vell.) Benth. ex Baill. | Brazil | Lopes, J.C. 20 (SPF) | PRJNA508895 |
| Cymbopetalum coriaceum N.A. Murray | Ecuador | Couvreur, T.L.P. 1390 (P) | PRJNA508895 |
| Cymbopetalum costaricense (Donn. Sm.) Saff. | Costa Rica | Chatrou, L. W. 58 (U) | PRJNA508895 |
| Cymbopetalum fosteri N.A. Murray | Peru | Gentry, A 26851 (U) | SUB11861053 |
| Cymbopetalum gracile R.E. Fr. | Mexico | Hinton, B. 11679 (P) | PRJNA508895 |
| Cymbopetalum hintonii Lundell | Mexico | Murray, N.A. 1457 (U) | SUB11463464 |
| Cymbopetalum lanugipetalum Schery | Panama | Murray, NA 1492 (U) | SUB11861053 |
| Cymbopetalum longipes Benth. ex Diels | Peru | Pirie, M. D. 26 (U) | PRJNA508895 |
| Cymbopetalum mayanum Lundell | Belize | Davidse, G. 35791 (MO) | PRJNA508895 |
| Cymbopetalum mirabile R.E. Fr. | Guatemala | Contreras, E. 9134 (P) | SUB11861053 |
| Cymbopetalum oppositiflorum Aristeg. ex N.A. Murray | Colombia | Fuchs, H.P. 22208 (U) | PRJNA508895 |
| Cymbopetalum sanchezii N.A. Murray | Colombia | Maas, P.J.M. 10485 (WAG) | PRJNA508895 |
| Cymbopetalum schunkei N.A. Murray | Peru | Schunke Vigo, J 6977 (U) | SUB11858406 |
| Cymbopetalum stenophyllum Donn. Sm. | Mexico | Murray, NA 1407 (U) | SUB11861053 |
| Cymbopetalum torulosum G.E. Schatz | Costa Rica | Chatrou, L.W. 56 (U) | PRJNA508895 |
| Froesiodendron amazonicum R.E. Fr. | Brazil | Maas, P.J.M. P 12734 (U) | SUB11861053 |
| Froesiodendron urceocalyx N.A. Murray | Peru | Prance, G.T. 17155 (U) | SUB12129452 |
| Hornschuchia alba (A. St.-Hil.) R.E. Fr. | Brazil | Maas, P.J.M. 8818 (U) | PRJNA508895 |
| Hornschuchia bryotrophe Nees | Brazil | Mello-Silva, R. 3141 (SPF) | PRJNA508895 |
| Hornschuchia cauliflora Maas & Setten | Brazil | dos Santos, T.S. 2193 (NY) | SUB11861053 |
| Hornschuchia citriodora D.M. Johnson | Brazil | Lopes, J.C. 110 (SPF) | PRJNA508895 |
| Hornschuchia leptandra D.M. Johnson | Brazil | Hage, J.L. 1447 (U) | SUB11463464 |
| Hornschuchia lianarum D.M. Johnson | Brazil | Mello-Silva, R. 3140 (SPF) | PRJNA508895 |
| Hornschuchia mediterranea Mello-Silva & D.M. Johnson | Brazil | Mello-Silva, R. 3138 (SPF) | PRJNA508895 |
| Hornschuchia myrtillus Nees | Brazil | Lopes, J.C. 122 (SPF) | PRJNA508895 |
| Hornschuchia polyantha Maas | Brazil | Mello-Silva, R. 3132 (SPF) | PRJNA508895 |
| Hornschuchia santosii D.M. Johnson | Brazil | Mello-Silva, R. 3134 (SPF) | PRJNA508895 |
| Mkilua fragrans Verdc. | Tanzania | Couvreur, T.L.P. 25 (WAG) | PRJNA508895 |
| Porcelia macrocarpa (Warm.) R.E. Fr. | Brazil | Glaziou, A. s.n. (P) | PRJNA508895 |
| Porcelia magnifructa (Schery) R.E. Fr. | Costa Rica | Maas, P.J.M. 9489 (U) | PRJNA508895 |
| Porcelia mediocris N.A. Murray | Ecuador | Neill, D.A. 6916 (U) | SUB11463464 |
| Porcelia nitidifolia Ruiz & Pav. | Peru | Ruiz, H. s.n. (P) | PRJNA508895 |
| Porcelia ponderosa (Rusby) Rusby | Brazil | Maas, P.J.M. 9264 (U) | SUB12129452 |
| Porcelia steinbachii (Diels) R.E. Fr. | Bolivia | Chatrou, L. W. 341 (U) | PRJNA508895 |
| Porcelia venezuelensis Pittier | Venezuela | Stergios, B. 20976 (U) | PRJNA508895 |
| Trigynaea axilliflora D.M. Johnson & N.A. Murray | Brazil | Maas, P.J.M. 8817 (U) | SUB11463464 |
| Trigynaea caudata (R.E. Fr.) R.E. Fr. | Guyana | Raes, N. 44 (U) | PRJNA508895 |
| Trigynaea cinnamomea D.M. Johnson & N.A. Murray | Peru | Chatrou, L.W. 222 (L) | PRJNA508895 |
| Trigynaea duckei (R.E. Fr.) R.E. Fr. | Ecuador | Couvreur, T.L.P. 1323 (P) | SUB11463464 |
| Trigynaea oblongifolia Schltdl. | Brazil | Farney, C. 2478 (U) | SUB11463464 |
| Trigynaea triplinervis D.M. Johnson & N.A. Murray | Ecuador | Pérez Castañeda, A.J. 10586 (QCA) | PRJNA508895 |
| Outgroup | |||
| Annona coriacea Mart. | Brazil | Lobão, A.Q. 606 (RB) | SUB11858406 |
| Duguetia antioquensis León & Maas | Colombia | Maas, P.J.M. 10477 (L) | PRJNA508895 |
| Guatteria alticola Scharf & Maas | Guyana | Clarke, H.D. 9247 (U) | SUB11861053 |
| Monodora hastipetala Couvreur | Tanzania | Couvreur, T.L.P. 44 (WAG) | PRJNA508895 |
| Sanrafaelia ruffonammari Verdc. | Tanzania | Dagallier, L-P. M.J. 31 (MPU) | PRJNA508895 |
| Uvaria acuminata Oliv. | Kenya | Robertson, S.A. 7557 (WAG) | SUB11463464 |
| Xylopia aethiopica (Dunal) A. Rich. | Gabon | Couvreur, T.L.P. 543 (MPU) | PRJNA508895 |
| Anaxagorea phaeocarpa Mart. | Bolivia | Chatrou, L.W. 295 (U) | SUB11463464 |
| Ambavia gerrardii (Baill.) Le Thomas | Madagascar | Gautier, L. 5380 (P) | SUB11861053 |
| Drepananthus ramuliflorus Maingay ex Hook. f. & Thomson | Malaysia | Sauquet, H. 167 (P) | PRJNA508895 |
| Meiocarpidium oliverianum (Baill.) D.M. Johnson & N.A. Murray | Gabon | Couvreur, T.L.P. 920 (WAG) | PRJNA508895 |
| Tetrameranthus globuliferus Westra | Ecuador | Guevarra, J.-E. 5891 (QCA) | SUB11858406 |
| Annickia ambigua (Robyns & Ghesq.) Setten & Maas | Congo | Couvreur, T.L.P. 788 (WAG) | PRJNA508895 |
| Cremastosperma monospermum (Rusby) R.E. Fr. | Bolivia | Pirie, M.D. 4 (U) | SUB11463464 |
| Ephedranthus boliviensis Chatrou & Pirie | Bolivia | Chatrou, L. W. 301 (U) | PRJNA508895 |
| Oxandra riedeliana R.E. Fr. | Ecuador | Pérez Castañeda, A.J. 10501 (QCA) | SUB11463464 |
| Desmopsis lanceolata Lundell | Mexico | Ortiz Rodriguez, A.E. 1322 (MEXU) | SUB11861053 |
| Meiogyne tiebaghiensis (Däniker) Heusden | New Caledonia | Grignon 148 (P) | PRJNA508895 |
| Polyalthia dolichopoda (Merr.) I.M. Turner | Malaysia | Onstein, R.E. 50 (P) | PRJNA508895 |
| Greenwayodendron gabonicum (Pellegr. ex Le Thomas) Lissambou & Couvreur | Gabon | Wieringa, J.J. 8417 (WAG) | PRJNA508895 |
| Sirdavidia solannona Couvreur & Sauquet | Gabon | Couvreur, T.L.P. 1127 (P) | PRJNA508895 |
| Eupomatia laurina R. Br. | Oceania | Sauquet, H. 165(P) | PRJNA508895 |
For fossil calibration and divergence time estimation within Bocageeae, we included 22 outgroup species, one from the sister family Eupomatiaceae, one from subfamily Anaxagoreoideae, four Ambavioideae, seven Annonoideae from other tribes besides Bocageeae, and nine Malmeoideae, representing the main lineages of the Annonaceae phylogeny (Chatrou et al., 2012; Guo et al., 2017; Table 1).
DNA extraction and sequencing
DNA was extracted from silica-dried or herbarium-dried leaves, as described by Couvreur et al. (2019). Illumina libraries were constructed following Couvreur et al. (2019) based on the original protocol of Rohland and Reich (2012). Hybridization of targeted regions was undertaken using the baiting kit for the Annonaceae, which targets a total of 469 exons (Couvreur et al., 2019). Libraries were sequenced on a NovaSeq PE150 platform. Pair-end reads of 150 bp were generated (Novogene, Cambridge, UK) with ~18 pmol of the capture-amplified DNA libraries deposited on the flow cell.
Bioinformatics, contig assembly and multi-sequence alignment
To demultiplex the raw sequences, the demultadapt script (https://github.com/Maillol/demultadapt) was used with a zero-mistmatch threshold following the protocol described by Couvreur et al. (2019). The pipeline HybPiper (v.1.2) (Johnson et al., 2016) was used under the default settings to map the sequences onto a reference and assemble the genes (Couvreur et al., 2019); if contigs were slightly overlapping, they were combined into ‘supercontigs’ that contained both target and off-target sequence data (Johnson et al., 2016). After assembly, the supercontigs were aligned using MAFFT (v.7.305) (Katoh and Standley, 2013) with the ‘-auto’ option. The alignments were cleaned with GBLOCKS (v.0.91b) (Castresana, 2000) using the default parameters and all allowed gap positions (‘-b5=a’). HybPiper flags potential paralogues when multiple contigs map well to a single reference sequence (Johnson et al., 2016). Flagged paralogues were excluded from subsequent phylogenetic analyses, and the remaining dataset (assumed now to be only single-copy loci) was used for phylogenetic analyses.
Phylogenetic reconstruction
We generated two matrices: (1) a ‘complete dataset’ with all nuclear markers selected and used for phylogenetic inferences; and (2) a reduced ‘molecular dating dataset’ containing a selection of 30 molecular markers. Owing to computational time constraints, a subset of 30 loci instead of the complete dataset was used for phylogenetic dating. The molecular dating dataset was generated using the SortaDate pipeline (Smith et al., 2018) to select the most clock-like 30 loci from the complete dataset based on the root-to-tip variance filtering from SortaDate. We analysed the data using a concatenated approach and a coalescent approach.
In the concatenated approach, all loci were concatenated into a single data matrix, and we undertook a maximum likelihood (ML) phylogenetic analysis using RAxML (v.8.2.9) (Stamatakis, 2014). We selected the GTR+GAMMA substitution model (Abadi et al., 2019) without the proportion of invariant sites, to avoid overparametrization (Yang, 2006). Node support was assessed by fast-bootstrap using 100 non-parametric bootstrap pseudo-replicates. Bootstrap (BS) values of ≥80 were considered as strong support.
For the coalescent approach, individual gene trees were inferred using RAxML (v.8.2.9) (Stamatakis, 2014). In the coalescent approach, we used the gene trees to infer the relationships under the multi-species coalescent model (MSCM) as implemented by ASTRAL III (v.5.6.3) (Zhang et al., 2018), referred to as the MSC tree. For the MSC tree local posterior probability (LPP; Sayyari and Mirarab, 2016), support values were also generated for each node.
Divergence time estimation
We used the molecular dating dataset to estimate divergence time in BEAST (v.2.5.2) (Bouckaert et al., 2019). To reach convergence in BEAST within a reasonable time frame (limit of a month), we constrained the starting tree topology in BEAST to the one inferred under ML from the complete dataset. As a starting tree, a chronogram obtained through the penalized likelihood (PL) approach (Sanderson, 2002), as implemented in treePL (Smith and O’Meara, 2012), was used. An uncorrelated lognormal relaxed molecular clock model and a birth–death tree prior model were chosen (Sarver et al., 2019). The substitution model was GTR+Γ (Yang, 2006; Abadi et al., 2019). Two independent searches ran for 500 million Markov chain Monte Carlo (MCMC) generations each, sampling every 1000th generation. The first 20 % of runs were discarded as burn‐in. The output was analysed using Tracer (v.1.7.1) (Rambaut et al., 2018) to check for convergence and an effective sample size (ESS) of >200. LogCombiner (v.2.5.2) (Bouckaert et al., 2019) was used to combine converging runs into a single chain (with a burn-in of 20 % for each analysis). We retrieved the maximum clade credibility (MCC) tree, in addition to the mean age and 95 % highest posterior density (HPD) with TreeAnnotator (v.2.5.2) (Bouckaert et al., 2019). Two calibration points provided by fossils were used (see next paragraph). For the MSC tree, we inferred branch lengths with RAxML (v.8.2.9) (Stamatakis, 2014) and estimated time divergence by PL in treePL (Smith and O’Meara, 2012).
Two fossils were used for calibration: (1) Endressinia (Mohr and Bernardes-de-Oliveira, 2004) from the Late Aptian of Brazil (113–126 Ma); and (2) Futabanthus (Takahashi et al., 2008) from the Early Coniacian of Japan (86.3–89.8 Ma). Endressinia provides a minimum age estimate for the crown node of Magnoliineae (Magnoliaceae, Degeneriaceae, Himantandraceae, Eupomatiaceae and Annonaceae; Doyle and Endress, 2010). The stem node in our phylogeny was calibrated using a uniform prior distribution with minimum age of 112 Ma (maximum age of 150 Ma, Massoni et al., 2015) in the BEAST analysis and as the maximum age for the PL analysis. Futabanthus is associated with the crown node of Annonaceae (Takahashi et al., 2008; Pirie and Doyle, 2012). Therefore, the crown node of Annonaceae was set to a minimum age of 89 Ma in a uniform prior distribution (maximum age of 149 Ma, Massoni et al., 2015) in the BEAST analysis and as the minimum age in the PL analysis.
Ancestral range estimation
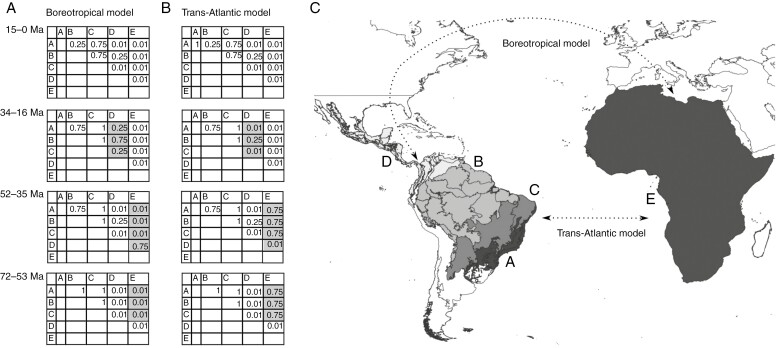
To address our hypotheses (H1 and H2), we estimated ancestral ranges with a maximum likelihood approach for both topologies, the ML and the MSC trees. Species occurrence data for the species sampled were obtained from GBIF (2022) and cleaned using the R package ‘CoordinateCleaner’ (Zizka et al., 2019), keeping only preserved specimens and removing those without coordinates and with imprecise coordinates, records older than 1945. The distribution of each species of Bocageeae was assessed using QGIS v.3.24 (QGIS Development Team, 2009). Specimens collected outside the distribution known from the literature were considered outliers and excluded from the analyses (Verdcourt, 1970; Murray, 1993; Johnson and Murray, 1995; Mello-Silva and Lopes, 2020; Vilela and Lopes, 2022). Owing to the low sampling of the Annonoideae outgroups, we coded them as being present in ‘Africa’ independently of the real distribution, based on the ancestral range estimation of Annonoideae by Onstein et al. (2019). For this reason, we coded the whole African continent, without Madagascar, as a single area, to encompass the distribution of Mkilua fragrans, in Eastern Africa, in addition to the outgroups (Fig. 1A). The biogeographical regions proposed by Morrone et al., (2022) were used as a guide to define the biogeographical areas; taxa sampled were coded into five discrete areas: A, Atlantic Forest; B, Amazon; C, Caatinga, Cerrado and Chaco; D, North-Central America; and E, Africa (Fig. 2; see Supplementary Data Table S1.1). We used the R package BioGeoBEARS (Matzke, 2013) to estimate ancestral ranges for tribe Bocageeae taxa. BioGeoBEARS tests diversification-independent biogeographical models and estimates ancestral ranges based on ML (Matzke, 2013). We tested the biogeographical models implemented in BioGeoBEARS, DEC (Ree, 2005; Ree and Smith, 2008), DIVALIKE (Ronquist, 1997; Matzke, 2013) and BAYAREALIKE (Landis et al., 2013; Matzke, 2013).
Fig. 2.
Delimitation of five areas assigned to the species of Bocageeae and dispersal matrix for two alternative biogeographical models tested in this study. (A) Dispersal matrices for the Boreotropical hypothesis. (B) Dispersal matrices for the trans-Atlantic hypothesis. Probabilities of dispersal: 0.01 is none or low; 0.25 is medium–low; 0.5 is medium; 0.75 is medium–high; and 1 is high. Ma = mega annum. (C) Biogeographical units: A, Atlantic Forest; B, Amazon; C, Caatinga, Cerrado and Chaco; D, North-Central America; E, Africa.
To address the American–African disjunction (H1), using the biogeographical model with the lowest Akaike Information Criterion (AIC) score, we tested three models: the Boreotropical and the trans-Atlantic models, and a null model unconstrained, without time stratification. For each scenario, we prepared a dispersal transition matrix divided into four major time slices favouring dispersal or not under these hypotheses: 72–53 Ma, trans-Atlantic direct connections (Parrish, 1993; Morley, 2000, 2003); 52–35 Ma, the existence of the Boreotropical flora (Wolfe, 1975); 34–16 Ma, connections between North and South America up to the origin of the Cerrado (Iturralde-Vinent and MacPhee, 1999); 15–0 Ma, the origin of the Cerrado in Central Brazil (Azevedo et al., 2020; Fig. 2). We considered a range expansion probability of >70 %. The maximum range size was set to two, the maximum range observed in the data. Model selection was done using the Akaike information criterion (AIC) (Akaike, 1974) and the AIC corrected for small data (Sugiura, 1978).
Ancestral fruit morphology reconstruction
To address our hypothesis H3, i.e. that inter- and intracontinental dispersals have been facilitated by dispersal by large-bodied frugivorous mammals, birds or bats, we inferred the ancestral states of fruit traits using BayesTraits v.4.0 (Pagel and Meade, 2006), which allows the inclusion of polymorphic characters with missing data. We performed the analysis using the tree with time divergence estimation generated by BEAST. The fruit traits related to the mammal trait and bird/bat trait syndromes were selected based on the study by Onstein et al. (2019). The dispersal syndroms are associated to certain traits depending on the animal group: mammals: (1) few, large, apocarpous monocarps with few large seeds, or large syncarpous fruits with many seeds and fruits with dull colour, i.e. green, brown; birds (2 and 3) are associated with: (2) many monocarps, brightly coloured, i.e. yellow, red, purple, black, orange; the monocarps are small with long stipes (fruit stalk), and with few and small seeds, and (3) dehiscent fruits with small seeds; bats (4): (4) dull-coloured, cauliflorous fruits (Onstein et al., 2019). In total, eight characters related to fruit morphology of Bocageeae were coded based on the literature (Table 2; Verdcourt, 1970; Murray, 1993; Johnson and Murray, 1995; Mello-Silva and Lopes, 2020; Vilela and Lopes, 2022; see Supplementary Data Table S1.2). For Trigynaea, all the species were coded as having dehiscent fruits; however, this information was documented only for Trigynaea caudata and Trigynaea duckei (Johnson and Murray, 1995). For the outgroup, we used the results from a broader analysis and coded the Annonoideae species as its inferred ancestral state for each fruit trait, except for character 6, seed length, in which we removed the outgroup from the analysis (Onstein et al., 2019). We followed the same approach described by Onstein et al. (2019).
Table 2.
List of fruit traits related to long-distance dispersal and their character state coding used for the ancestral state inference for Bocageeae (Annonaceae).
| 1. Fruit length (in centimetres): continuous |
| 2. Number of monocarps: 1–4 (0); 5–19 (1); 20–50 (2) |
| 3. Fruit colour: dull: green, brown (0); bright: yellow, red, purple, black, orange (1) |
| 4. Dehiscence: absent (0); present (1) |
| 5. Stipe length (in millimetres): continuous |
| 6. Seed length (in millimetres): continuous |
| 7. Number of seeds per monocarp: 1–10 (0); 11–28 (1) |
| 8. Cauliflory: absent (0); present (1) |
RESULTS
Bioinformatics and phylogenetic reconstruction
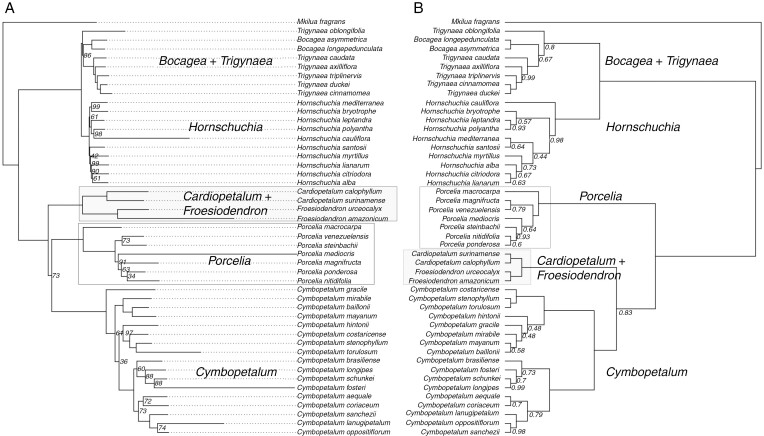
We recovered all 469 exons present in the Annonaceae bait kit (Couvreur et al., 2019). Of these, 273 markers had 75 % of their length reconstructed in 75 % of the samples; 188 of these loci were flagged as paralogues and removed from downstream analyses. The complete dataset contained 116 markers and was 180 885 bp long. After selecting the 30 genetic markers by clock-likeness, the molecular dating dataset was 41 988 bp long. The concatenated and coalescent approaches retrieved mostly the same relationships (Fig. 3; Table 3). Tribe Bocageeae was recovered as monophyletic with strong support (BS = 100; LPP = 1). Mkilua fragrans, the East African taxon, appeared as sister to the remaining Neotropical genera (Bocagea, Cardiopetalum, Cymbopetalum, Froesiodendron, Hornschuchia, Porcelia and Trigynaea; hereafter, the Neotropical clade). The Neotropical clade was recovered as monophyletic with strong support (BS = 100; LPP = 1), with two main clades. One included Bocagea, Trigynaea and Hornschuchia, strongly supported (BS = 100; LPP = 1), and the other clade included Cardiopetalum, Cymbopetalum, Froesiodendron and Porcelia, reconstructed as monophyletic with strong support (BS = 100; LPP = 1). In the clade formed by Bocagea, Trigynaea and Hornschuchia, the two sampled species of Bocagea were nested within the genus Trigynaea (BS = 100; LPP = 0.8). Bocagea and Trigynaea appeared as sister to Hornschuchia, which was recovered as monophyletic with strong support (BS = 100; LPP = 1). In the clade including Cardiopetalum, Cymbopetalum, Froesiodendron and Porcelia, all genera were recovered as monophyletic with strong support (BS = 100; LPP = 1). However, one incongruence was noted between the concatenated and the coalescent approach. In the ML analysis, Froesiodendron and Cardiopetalum formed a clade with strong support (BS = 100), while Cymbopetalum was recovered as sister to Porcelia with moderate support (BS = 73), with the latter appearing as sister to Froesiodendron and Cardiopetalum (Fig. 3A; Table 3). In the MSCM analysis, Cymbopetalum was recovered as sister to the clade formed by Froesiodendron and Cardiopetalum with moderate support (LPP = 0.83), with these three genera sister to Porcelia with high support (LPP = 1) (Fig. 3B).
Fig. 3.
Phylogenetic relationships recovered for tribe Bocageeae (Annonaceae). Values close to the nodes are branch support. (A) Maximum likelihood tree inferred by RAxML with the complete dataset; branch support with bootstrap = 100 was omitted. (B) Tree inferred by the multi-species coalescent model implemented in ASTRAL with the gene trees; branch support with local posterior probability = 1 was omitted.
Table 3.
Ages of crown nodes for clades and genera of Bocageeae (Annonaceae). Divergence times [in millions of years ago (Ma)] are presented; numbers in parentheses are the 95 % highest posterior density. Node supports are shown for specific nodes, bootstrap (BS) and local posterior probability (LPP). Asterisk (*) indicates a different phylogenetic relationship. Ancestral range reconstructions with relative probabilities > 0.05 for Bocageeae obtained from BioGeoBEARS with the DEC with time stratification model are also presented. Area codes: A, Atlantic Forest; B, Amazon; C, Caatinga, Cerrado and Chaco; D, North-Central America; and E, Africa.
| Lineage | BEAST | PL | BS | LLP | Ancestral range |
|---|---|---|---|
| Bocageeae | 52.5 (35.5–74.5) | 34.6 | 100 | 1 | AE (0.25), BE (0.73) |
| Neotropical clade | 46.5 (29.5–64.5) | 32.3 | 100 | 1 | A (0.13), B (0.6), AB (0.22) |
| Bocagea, Hornschuchia, Trigynaea | 16.5 (9–23.9) | 9.9 | 100 | 1 | A (0.26), AB (0.73) |
| Hornschuchia | 11.3 (6.7–16) | 7.9 | 100 | 1 | A (0.99) |
| Bocagea, Trigynaea | 14.5 (8.2–21) | 9.3 | 100 | 0.8 | A (0.26), AB (0.73) |
| Cardiopetalum, Cymbopetalum, Froesiodendron, Porcelia | 38.2 (25.7–52.2) | 28.4 | 100 | 1 | B (0.64), AB (0.2), BD (0.14) |
| Porcelia, Cymbopetalum | 35.6 (24.2–49.5) | 27.8 | 73 | * | B (0.57), D (0.1), AB (0.22), BD (0.09) |
| Porcelia | 21.9 (12–31.4) | 21.1 | 100 | 1 | AB (0.83), AD (0.12) |
| Cymbopetalum | 21.8 (13.5–30.9) | 16.4 | 100 | 1 | D (0.18), BD (0.81) |
| Cardiopetalum, Froesiodendron | 26.2 (15.2–39.3) | 21.6 | 100 | 1 | B (0.94), BC (0.05) |
| Froesiodendron | 14.5 (5–24) | 15.7 | 100 | 1 | B (0.99) |
| Cardiopetalum | 15.7 (6.9–26.1) | 12.6 | 100 | 1 | B (0.77), BC (0.22) |
Divergence time estimation
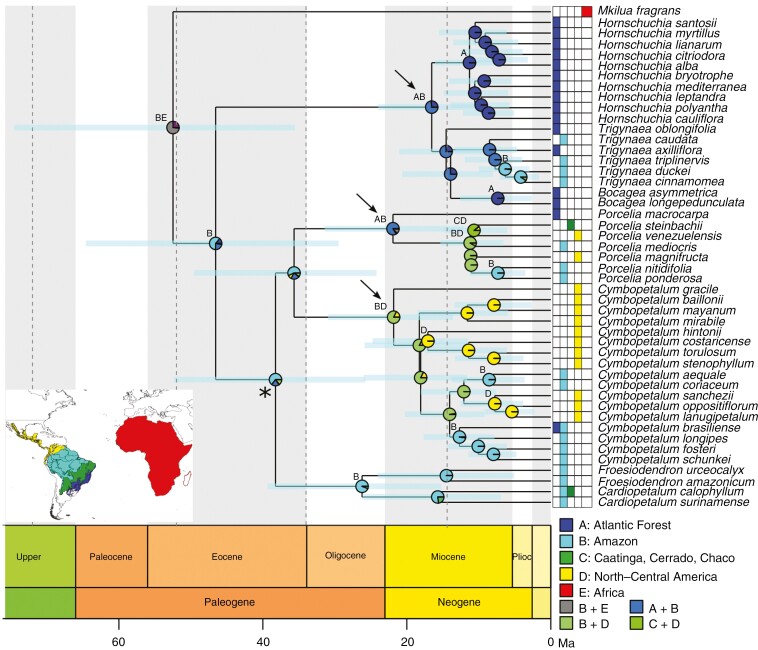
The age estimations under PL were more recent than those obtained by BEAST; nevertheless, they fall within the 95 % HPD (Table 3; Fig. 4; Supplementary Data Table S1.3). For this reason, we present the results from BEAST in the ML topology here. The most recent common ancestor (MRCA) of Bocageeae diverged during the Early Eocene (mean age, 95 % HPD: 52.5, 35.5–74.5 Ma). The estimated crown age of the Neotropical clade dates to the Middle Eocene (mean age, 95 % HPD: 46.5, 29.5–64.5 Ma). In the Neotropical clade, the two main clades [(1) Hornschuchia and Bocagea plus Trigynaea and (2) Cardiopetalum, Cymbopetalum, Froesiodendron and Porcelia] diverged during the Middle Miocene, at 16.5 (mean age, 95 % HPD: 9–23.9) Ma, and the Late Eocene, at 38.2 (mean age, 95 % HPD: 25.7–52.2) Ma, respectively. The estimated crown age for the clade formed by Bocagea and Trigynaea dates to the Middle Miocene (mean age, 95 % HPD: 14.5, 8.2–21 Ma). The MRCA of Hornschuchia diverged during the Late Miocene (mean age, 95 % HPD: 11.3, 6.7–16 Ma). The estimated crown age for the clade formed by Porcelia and Cymbopetalum dates to the Late Eocene (mean age, 95 % HPD: 35.6, 24.2–49.5 Ma), with the divergence of the MRCA of Porcelia (mean age, 95 % HPD: 21.9, 12–31.4 Ma) and Cymbopetalum (21.8, 13.5–30.9 Ma) both dating to the Early Miocene. The divergence time of the crown node from the group formed by Cardiopetalum and Froesiodendron was dated to the Late Oligocene (mean age, 95 % HPD: 26.2, 15.2–39.3 Ma), with the divergence of the MRCA of Cardiopetalum (mean age, 95 % HPD: 15.7, 6.9–26.1 Ma) and Froesiodendron (mean age, 95 % HPD: 14.5, 5–24 Ma) both dating to the Middle Miocene (Table 3; Fig. 4; Supplementary Data Table S1.3).
Fig. 4.
Phylogenetic relationships, divergence times and ancestral range estimates of tribe Bocageeae (Annonaceae). Phylogenetic relationships by maximum likelihood, complete dataset, divergence time estimation by BEAST, molecular dating dataset, trimmed tree showing only the ingroup. All nodes for relationships of genera are highly supported, except for the node indicated by an asterisk, which is moderately supported. Light blue bars on nodes represent the 95 % highest posterior density. Pie charts at nodes depict the probabilities of different ancestral areas, and letters indicate the most likely ancestral areas obtained by the DEC Boreotropical model; letters are omitted when the ancestral range is the same as the node below. Arrows close to the nodes indicate range expansion. Time slices: 72–53, 52–35, 34–16 and 15–0 Ma. Biogeographical units: A, Atlantic Forest; B, Amazon; C, Caatinga, Cerrado and Chaco; D, North-Central America; E, Africa.
Ancestral range estimation
We obtained 7576 species occurrence records for species of Bocageeae. After data cleaning, 3393 georeferenced records remained (Fig. 1A, B). The biogeographical model with the lowest AIC/c* score selected was the DEC model l, the Boreotropical hypothesis, corroborating our first hypothesis (H1) for explaining the American–African disjunction (Fig. 2A), which has different dispersal transition probabilities for specific time slices when compared with the trans-Atlantic route (Fig. 2B; Supplementary Data Tables S1.4 and S1.5). Given that the ML and MSC tree topologies present some conflicts, we describe both results (Fig. 4; Supplementary Data Fig. S2.1). In the ML topology, an area including the Amazon and Africa represented the most likely ancestral range for the MRCA of Bocageeae (P = 0.73; Table 3; Fig. 4). For the MSC topology, the Atlantic Forest and Africa were the most likely ancestral range (P = 0.61; Supplementary Data Table S1.3; Fig. S2.1). In the ML topology, the most likely distribution for the MRCA of the Neotropical Bocageeae was Amazonia (P = 0.6; Fig. 4), whereas for the MSC topology the Atlantic Forest was the most likely ancestral range (P = 0.52; Supplementary Data Fig. S2.1). The first split within this clade gave rise to two sub-clades: (1) Bocagea, Hornschuchia and Trigynaea; and (2) Cardiopetalum, Cymbopetalum, Froesiodendron and Porcelia. The ancestral range of the latter clade was the Amazon (P = 0.64) in the ML topology (Fig. 4) and an area including the Atlantic Forest and Amazon (P = 0.67) in the MSC topology (Supplementary Data Fig. S2.1). The Atlantic Forest was the ancestral range for Porcelia in the ML topology (P = 0.83), and an area including the Atlantic Forest and Amazon was the most likely ancestral range in the MSC topology (P = 0.97). A range expansion to Central America occurred in Cymbopetalum, whose ancestral range was the Amazon and North-Central America in both topologies (ML P = 0.81, MSC P = 0.99). For the lineage including Bocagea, Hornschuchia and Trigynaea, the most likely ancestral range was the Atlantic Forest and the Amazon in the ML topology (P = 0.73; Table 3; Fig. 4) and the Atlantic Forest in the MSC topology (P = 0.63; Supplementary Data Table S1.3; Fig. S2.1).
Ancestral fruit morphology analyses
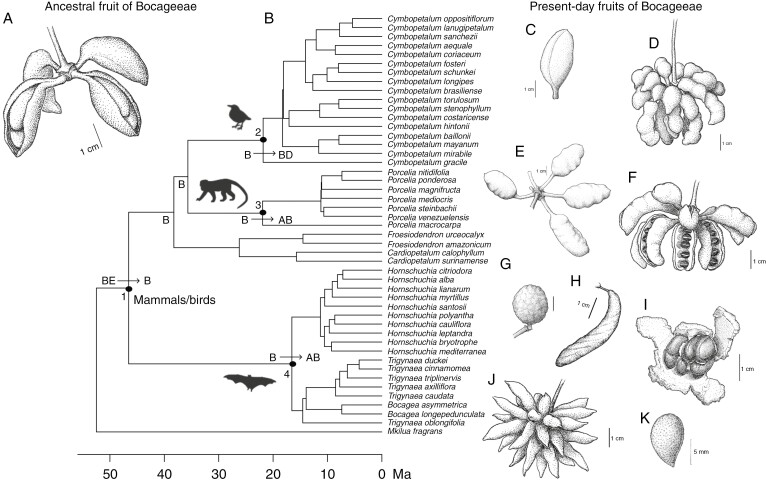
The inferred ancestral fruit of the tribe Bocageeae was characterized as being large (~3 cm), dehiscent and with few monocarps (one to four), with bright colours (e.g. yellow, red, purple, black, orange), short stipes (~5 mm long) and few (one to ten), large (33.3-mm-long) seeds; cauliflory was ambiguously reconstructed (Fig. 5A). The reconstructed ancestral fruits for Bocagea (Fig. 5K), Hornschuchia (Fig. 5G, H) and Trigynaea (Fig. 5I) were cauliflorous, with medium to large (~2.5 cm), dehiscent fruits and few monocarps (1–4), with dull colours (e.g. green, brown), short stipes (~2.5 mm long) and a few (one to ten) medium-sized (14.6 mm long) seeds. The ancestral fruit of Porcelia (Figs 1I and 5E) had very large (~6 cm), indehiscent fruits and few monocarps (1–4), with dull colours and medium stipe lengths (~12 mm long), many (11–28) medium to large (24.2 mm long) seeds, and was not cauliflorous. The inferred ancestral fruit of Cymbopetalum (Figs 1E and 5D) had large (~4 cm), dehiscent fruits and many monocarps (5–19), with bright colours and medium stipe lengths (~10.7 mm long), with few (one to ten), medium-sized (15.3 mm long) seeds, and was not cauliflorous (Table 4; Fig. 5; Supplementary Data Figs S2.2−S2.9). All these traits are related to dispersal guilds by large-bodied frugivorous mammals, birds or bats, according to our third hypothesis (H3).
Fig. 5.
Phylogenetic relationships and reconstructed ancestral fruit of tribe Bocageeae (Annonaceae) and present-day fruits. (A) Ancestral fruit of Bocageeae; line drawing based on the results of BayesTraits. (B) Phylogenetic relationships by maximum likelihood, complete dataset, divergence time estimation by BEAST, molecular dating dataset, trimmed tree showing only the ingroup. Numbers refer to ancestral fruit reconstruction by BayesTraits and inferred trait syndromes: 1 = mammal bird traits syndrome; 2 = bird trait syndrome; 3 = mammal trait syndrome; 4 = bat trait syndrome. Letters refer to ancestral range reconstruction under the DEC Boreotropical model, and arrows indicate range expansion: AB = Atlantic Forest + Amazon; B = Amazon; BD = Amazon + North-Central America; BE = Amazon + Africa. (C–K) Present-day fruits of Bocageeae. (C) Froesiodendron amazonicum R.E. Fr. (D) Cymbopetalum brasiliense (Vell.) Benth. ex Baill. (E) Porcelia macrocarpa (Warm.) R.E. Fr. (F) Cardiopetalum calophyllum Schltdl. (G) Hornschuchia bryotrophe Nees. (H) Hornschuchia santosii D.M. Johnson. (I) Trigynaea spp. (J) Mkilua fragrans Verdc. (K) Bocagea asymmetrica Mello-Silva & J.C. Lopes. Vouchers: R.L. Fróes 20861 (NY) (C); E.M. Saddi 780 (RB) (D); N.M. Ivanauskas (SPF 123557) (E); C. Figueiredo 548 (NY) (F); Vilela and Lopes, 2022 (G, H); B.A.S. Pereira 3672 (IBGE) (I); W1991GR01649 (U) (J); and Mello-Silva and Lopes, 2020 (K). Herbarium acronyms follow Thiers (updated continuously). Line drawing by Laura Montserrat.
Table 4.
Summary of the results of Bayesian ancestral state analyses for fruit traits related to long-distance dispersal for the crown node of Bocageeae and the crown node of the Neotropical clade; probabilities of <30 % were omitted; measurements are median values.
| Fruit trait | Bocageeae | Bocagea, Hornschuchia and Trigynaea | Porcelia | Cymbopetalum |
|---|---|---|---|---|
| 1. Monocarp length (cm) | 3.1 | 2.5 | 6 | 4 |
| 2. Number of monocarps | 81.3 % (state 0: 1–4) | 99.98 % (state 0: 1–4) | 99.47 % (state 0: 1–4) | 98.95 % (state 1: 5–19) |
| 3. Fruit colour | 86.52 % (state 1: bright) | 99.73 % (state 0: dull) | 96.62 % (state 0: dull) | 98.58 % (state 1: bright) |
| 4. Dehiscence | 95.62 % (state 1: present) | 99.89 % (state 1: present) | 99.36 % (state 0: absent) | 99.79 % (state 1: present) |
| 5. Stipe length (mm) | 4.9 | 2.46 | 12 | 10.74 |
| 6. Seed length (mm) | 33.3 | 14.63 | 24.19 | 15.32 |
| 7. Number of seeds per monocarp | 99.68 % (state 0: 1–10) | 99.99 % (state 0: 1–10) | 96.89 % (state 1: 11–28) | 99.92 % (state 0: 1–10) |
| 8. Cauliflory | 51.37 % (state 0: absent); 48.63 % (state 1: present) | 85.65 % (state 1: present) | 78.43 % (state 0: absent) | 89.6 % (state 0: absent) |
DISCUSSION
African–American disjunction
To gain further insights into the origin and evolution of the Neotropical Annonaceae, in this study we focused on the tribe Bocageeae, a clade centred in the Neotropics but disjunctly distributed between tropical east Africa and the Americas.
Mkilua fragrans, the only African taxon, was recovered as sister to all the Neotropical genera in Bocageeae, which is also supported by other studies (Johnson and Murray, 1995; Chatrou et al., 2012; Guo et al., 2017). In Africa, a once-continuous rainforest probably extended from west to east Africa during the Eocene (Morley, 2000; Couvreur et al., 2021). After that, and linked to different palaeo-geoclimatic events, this proto-rainforest contracted and possibly re-expanded throughout the Cenozoic, isolating west/central from east African forest blocks. The Mkilua lineage originated during the early Eocene (Fig. 4) and was probably affected in several ways by the different extinction events known to have occurred during the rest of the Cenozoic (Jacobs et al., 2010; Couvreur et al., 2021). Indeed, diversification studies of African Annonaceae (Brée et al., 2020; Dagallier et al., 2023) detected signs of extinction during the middle Miocene Climate Transition (MCT), a period with overall drier conditions and the expansion of open habitats. Along the Mkilua evolutionary branch, the MCT probably led to a complete extinction of the west/central representatives, leaving a single surviving lineage in East Africa. The MCT was also suggested as the main cause of the origin of the monotypic Tanzanian endemic genus Mwasumbia (Brée et al., 2020), although this lineage still has a single central African lineage surviving today (the monotypic genus Sirdavidia).
Bocageeae was estimated to have diverged during the Early Eocene (52.5 Ma; 95 % HPD: 35.5–74.5 Ma); this result is also supported by previous studies that included fewer samples of the tribe (Pirie and Doyle, 2012). Our analyses estimated Africa and Amazonia, or Africa and the Atlantic Forest, as the ancestral range of Bocageeae (Fig. 4; Supplementary Data Fig. S2.1). This supports two scenarios: migration from Africa to South America, or vice versa. In general, tropical America has been suggested as a source of lineages through time (Antonelli et al., 2015), favouring the hypothesis of a Neotropical origin for Bocageeae. However, biogeographical analyses (Doyle and Le Thomas, 1997; Richardson et al., 2004; Scharaschkin and Doyle, 2005; Onstein et al., 2019), palynological data (Le Thomas, 1980) and the fossil record (Chesters, 1955) suggest that the Annonaceae are likely to have originated in the African portion of West Gondwana. Africa has been inferred as the ancestral location for several groups of Annonaceae (Erkens et al., 2009; Surveswaran et al., 2010; Couvreur et al., 2011; Zhou et al., 2012; Stull et al., 2017). Given the African origin of Annonoideae, in which Bocageeae is the sister to all other tribes (Couvreur et al., 2011; Onstein et al., 2019), it is most likely that the MRCA of the Neotropical Bocageeae dispersed into the Neotropics before the Middle Eocene (Fig. 4; Table 3).
The Boreotropical flora was a northern tropical rainforest distributed throughout the Northern Hemisphere (Wolfe, 1975). From the Middle Palaeocene (59 Ma) to the Early Eocene (52 Ma), climatic changes led to increases in global temperatures, reaching a maximum during the Early Eocene climatic optimum, 52–50 Ma (Zachos et al., 2001). These ideal climatic conditions for megathermal plants (Morley, 2000) led to the development of a Boreotropical flora that lasted until the onset of northern glaciations at the Eocene–Oligocene boundary (~34 Ma) (Wolfe, 1975; Zachos et al., 2001). Land connections between western Europe and North America are also supported by geological evidence from the Late Palaeocene to the Late Eocene (Tiffney, 1985) and by migration of tropical plants eastwards and westwards.
Given that the Boreotropical flora was restricted to the Northern Hemisphere, other migration pathways need to be invoked to explain the distribution of the Neotropical clade. Three exchange pathways between North and South America have existed at different times, namely: (1) the Proto-Greater Antilles pathway (Iturralde-Vinent and MacPhee, 1999), between the Middle Eocene (~50 Ma) to Late Eocene (34 Ma); (2) GAARlandia, the Greater Antilles and Aves ridge pathway (Iturralde-Vinent and MacPhee, 1999) during the Eocene–Oligocene boundary (35–33 Ma); and (3) the Central American land bridge pathway (Bermingham and Martin, 1998), during the Late Miocene (~7–4 Ma).
However, the ancestral range for Bocageeae was an area including Africa and Amazonia or the Atlantic Forest, excluding Central America and tropical North America, other possibilities should be considered (Fig. 4). The African–American disjunctions have also been explained by direct trans-Atlantic dispersion by stepping-stone mechanisms through volcanic lineaments (Morley, 2000, 2003). Trans-Atlantic dispersals might have happened through the Northern portions, through Ceará and Sierra Leone ridges (O’Connor and Duncan, 1990; Morley, 2003), or through the Southern portions of the Atlantic Ocean, through the Walvis Ridge and the Rio Grande Rise. These geological formations possibly played a role in the trans-Atlantic dispersal from the Late Cretaceous (~100 Ma) to the Eocene (54–36 Ma) (Morley, 2003).
Given that both the Boreotropical and trans-Atlantic dispersal routes should be considered while addressing our first hypothesis (H1), we compared the likelihood of different models explaining the amphi-Atlantic distribution in Bocageeae (Fig. 2). The most likely route between Africa and South America might have been by geodispersal through the Boreotropical flora (Fig. 4). Moreover, the fossil record (Chandler, 1964; Manchester, 1994) has documented the presence of Annonaceae in the Northern Hemisphere when the Boreotropical flora dominated. Besides Annonaceae, there are also other tropical plant groups with amphi-Atlantic taxa in which the Boreotropical hypothesis has also been invoked, such as Malpighiaceae (Davis et al., 2002), Arecaceae (Bacon et al., 2012; Baker and Couvreur, 2013), Athyriaceae (Wei et al., 2015), Sapotaceae (Smedmark and Anderberg, 2007) and Rubiaceae (Antonelli et al., 2009).
Frugivores might have facilitated this million-year-old long-distance dispersal between Africa and South America during the Eocene, our third hypothesis (H3) (Fig. 5). The inferred ancestral fruit of the Neotropical clade was large, with few monocarps, and brightly coloured on a short stipe, dehiscent, with few but large seeds per monocarp (Table 4; Fig. 5). The reconstructed ancestral fruit of Bocageeae suggests that both the mammal trait syndrome, with large fruits and seeds and few monocarps, and the bird trait syndrome, with dehiscent, brightly coloured fruits, might have been possible. In the Boreotropical region, specifically, where Europe is currently located, the diversity of large mammals and herbivore browsers, which feed on leaves, fruits and soft shoots, increased after the thermal maximum during the Middle Eocene (Hooker, 2000). However, the avifauna in the Northern Hemisphere during the time when the Boreotropical flora existed had a high diversity of forms with no or limited flight capabilities and large bodies (Martin, 2010). Besides that, the radiation of several lineages of modern groups of extant frugivorous birds took place during the Eocene; however, there is no substantial evidence that they were fruit eaters by this time, except for trogons (Eriksson, 2016). The ancestral fruit of Bocageeae, despite presenting both mammal and bird trait syndromes, was most likely to be dispersed by the existent frugivorous mammalian fauna in the Northern Hemisphere. These results are in accordance with the general trend for all angiosperms, with mammals being the main frugivorous dispersers in the early phase of the angiosperm–frugivore interaction (Eriksson, 2016).
Amazonia–Atlantic Forest disjunction and diversification in the Neotropics
This is the first study to include all genera of Bocageeae, one from Africa and seven from the Neotropics, a taxon sampling of 70 % and a broad molecular sampling. The internal phylogenetic relationships agree, in part, with previous studies: the Neotropical Bocageeae include two main clades: (1) Bocagea, Hornschuchia and Trigynaea; and (2) Cardiopetalum, Cymbopetalum, Froesiodendron and Porcelia (Johnson and Murray, 1995; Chatrou et al., 2012; Guo et al., 2017). However, the internal relationships among the genera in these clades have presented dissimilar results compared with other works and with two different analyses in this study. We recovered Bocagea plus Trigynaea as sister to Hornchuchia (Fig. 3; Guo et al., 2017); however, based on morphological data alone, Hornschuchia was recovered as sister to Bocagea and Trigynaea as sister to the latter two (Johnson and Murray, 1995). For the clade including Cardiopetalum, Cymbopetalum, Froesiodendron and Porcelia, the relationships among these genera conflicted between the concatenated and coalescent approaches (Fig. 3), both of which were different from the results recovered by morphological data (Johnson and Murray, 1995). We inferred the ancestral range for both the ML and the MSC topologies.
Neotropical Bocageeae were estimated to diverge in proto-Amazonia during the Middle Eocene (Fig. 4; Supplementary Data Fig. S2.1). In the Neotropics, the Amazon has been hypothesized as the primary source of lineages for all other biomes in the region (Antonelli et al., 2018), which is in accordance with our second hypothesis (H2). Similar to Africa, rainforests were widespread throughout South America by the Middle Eocene (Morley 2000; Jaramillo et al. 2010). Fossil data suggest that the rainforests of South America were established during the Early Palaeogene, at ~64 Ma (Burnham and Graham, 1999; Morley, 2000). Between the Early Eocene climatic optimum (52–50 Ma) and the Eocene–Oligocene boundary (~34 Ma), the South American climate was warmer and more humid than today (Zachos et al., 2001), favouring extensive rainforest vegetation (Burnham and Graham, 1999; Morley, 2000; Fine and Ree, 2006; Wing et al., 2009; Jaramillo and Cárdenas, 2013; Dick and Pennington, 2019; Carvalho et al., 2021). The subsequent establishment of a dry diagonal, the eastern South American Dry Diagonal, composed of the Chaco (Argentina and Paraguay), the Cerrado (central Brazil) and the Caatinga (northeastern Brazil; Luebert, 2021), fragmented the once-continuous South American rainforest into the Atlantic Forest to the east and Amazonia to the northwest. The savanna vegetation is estimated to have originated in the Middle to Late Miocene (Herbert et al., 2016; Henrot et al., 2017; Azevedo et al., 2020).
Our analysis indicates at least one dispersal event between Amazonia and the Atlantic Forest (H3; Fig. 4; Supplementary Data Fig. S2.1). Following the results from the ML topology (Fig. 4), the first dispersal between the two tropical forests occurred in the ancestor of the genus Porcelia during the Early Miocene (21.9, 12–31.4 Ma). The ancestral fruit was reconstructed as being very large, with few indehiscent monocarps, with dull colours and medium stipes and medium to large seeds (Figs 1I and 5E; Table 4), suggesting dispersal mediated by mammals (Onstein et al., 2019). This range exchange took place when the rainforest in South America was still continuous. In the extant species of Porcelia, monkeys and pacas and other rodents have been reported to feed on the fruits (Murray, 1993), but these animals might be seed predators. Birds were also reported ‘attacking’ the fruits (Warming, 1873).
The second dispersal event occurred in the ancestor of Hornschuchia and Bocagea plus Trigynaea during the Middle Miocene (16.5, 9–23.9 Ma; Figs 4 and 5; Tables 3 and 4). Thus, this second dispersal event happened across an already established dry barrier of open vegetation during the Middle Miocene (Fig. 4). Although the Cerrado might already have been established by that time, the two rainforest domains, the Amazon and the Atlantic Forest, might not yet have been isolated completely from each other (Por, 1992; Batalha-Filho et al., 2013). The oldest forest corridor thought to have linked western Amazonia and the southern Atlantic Forest passed through the southern portion of the eastern South American Dry Diagonal (nowadays, the Chaco) from the Middle to Late Miocene (Por, 1992; Batalha-Filho et al., 2013; Cheng et al., 2013; Ledo and Colli, 2017). The timing of the inferred range expansions of the MRCA of Bocagea, Hornschuchia and Trigynaea is consistent with biotic exchanges through this southern forest corridor, across present-day Chaco (Fig. 4; Table 3). The reconstructed ancestral fruit of Bocagea, Hornschuchia and Trigynaea (Table 4; Fig. 5G–I, K) was probably cauliflorous, with medium to large, dehiscent fruits and few monocarps, with dull colours, short stipes and few, medium-sized seeds, suggesting bat dispersal (Onstein et al., 2019). However, the main groups of Neotropical frugivorous bats, from the family Phyllostomideae, diverged by the Middle to Late Miocene, after the divergence of the MRCA of Bocagea, Hornschuchia and Trigynaea, in addition to other plant lineages with fruits dispersed by bats, such as Piper, Ficus and Solanum (Sánchez and Giannini, 2018). Thus, it is possible that the ancestral fruit might also have been dispersed by other frugivorous animals.
A range expansion from the Amazon back to Central America and Mexico is inferred to have occurred in the ancestors of Cymbopetalum (Fig. 4; Supplementary Data Fig. S2.1). In fact, most species of South American Cymbopetalum occur in western Amazonia, with two species occurring in Chocó, Cymbopetalum lanugipetalum Schery and Cymbopetalum oppositiflorum Aristeg. ex N.A. Murray (Murray, 1993; Fig. 1B). These species from Chocó plus Cymbopetalum sanchezii N.A. Murray from Colombia are sister to two western Amazonian species, Cymbopetalum aequale N.A. Murray and Cymbopetalum coriaceum N.A. Murray (Fig. 4). The time of divergence between the Chocó and western Amazonian clades, ~12 Ma, suggests that it might be attributable to vicariance as a result of Andean orogeny (Hoorn et al., 2010), as in the Annonaceae genera Cremastosperma and Mosannona (Pirie et al., 2018). Cymbopetalum is inferred to have had the ancestral states of large, dehiscent fruits, many brightly coloured monocarps with medium stipe lengths and few medium-sized seeds (Figs 1E and 5D; Table 4). This combination of traits suggests that its range expansion in the Neotropics, beginning in the Early Miocene (21.8, 13.5–30.9 Ma; Fig. 4), was mediated by birds (Onstein et al., 2019). Dispersal was documented in an extant Mexican species, Cymbopetalum baillonii, which exhibits the dispersal-related traits common to species of the genus: the aggregate of asynchronously ripening red- to rose-coloured monocarps dehisce outwards such that the black seeds with orange-red arils are displayed against the cream-coloured endocarp; the seed is the diaspore, and the aril provides a food reward (Murray, 1993). A wide variety of bird species were observed taking seeds, including 26 resident species and five migrants that nest in North America (summarized by Murray 1993); most residents were passerines. Passerines, which include many frugivorous species, diversified in the Southern Hemisphere by the Early Eocene and colonized the Northern Hemisphere during the Oligocene (Eriksson, 2016), coinciding with the hypothesized divergence time of the MRCA of Cymbopetalum (Fig. 4).
The biogeographical history of tribe Bocageeae illustrates past connections between the South American and African floras, indicating that mammals were likely to be responsible for long-distance dispersal between Africa and Amazonia through the Boreotropical flora. In the Neotropics, proto-Amazonia was the source for other Neotropical biomes. Range exchange between the Amazon and the Atlantic Forest occurred at least once during the Miocene. From Amazonia, a colonization to Central America and Mexico occurred during the Early Miocene. Long-distance dispersal might have resulted from a range of dispersal guilds, depending on the frugivore radiations, diversity and abundance at different times and places. Hence, inter- and intracontinental dispersal might not rely on a single dispersal syndrome or guild, but more on the availability of frugivore lineages for seed dispersal.
SUPPLEMENTARY DATA
Supplementary data are available at Annals of Botany online and consist of the following.
Table S1.1: species geographical range for Bocageeae (Annonaceae). Area codes: A, Atlantic Forest; B, Amazon; C, eastern South American Dry Diagonal; D, North-Central America; and E, Africa. Table S1.2: matrix of taxa/morphological character states for fruit traits related to long-distance dispersal for Bocageeae (Annonaceae) and outgroups. Table S1.3: ages of crown nodes for clades and genera of Bocageeae (Annonaceae) multi-species coalescent (MSC) topology. Divergence times [in millions of years ago (Ma)] by penalized likelihood. Ancestral range reconstructions with relative probabilities > 0.05 for Bocageeae obtained from BioGeoBEARS with the DEC with the time stratification model are also presented. Area codes: A, Atlantic Forest; B, Amazon; C, Caatinga, Cerrado and Chaco; D, North-Central America; and E, Africa. Table S1.4: summary of the statistics comparing the models implemented in BioGeoBEARS on the maximum likelihood tree topology, with time divergence estimation by BEAST. Table S1.5: summary of the statistics comparing the DEC with three biogeographical models: model 0, unconstrained; model 1, Boreotropical hypothesis; and model 2, trans-Atlantic route. Figure S2.1: Phylogenetic relationships, divergence times and ancestral range estimates of tribe Bocageeae (Annonaceae). Phylogenetic relationships by multi-species coalescent model, complete dataset, divergence time estimation by penalized likelihood. All nodes for relationships of genera are highly supported, except for the node indicated by an asterisk, which is moderately supported. Pie charts at nodes depict the probabilities of different ancestral areas, and letters indicate the most likely ancestral areas obtained by DEC Boreotropical model. Time slices: 53–35, 34–16 and 15–0 Ma. Biogeographical units: A, Atlantic Forest; B, Amazon; C, Caatinga, Cerrado and Chaco; D, North-Central America; and E, Africa. Figure S2.2: Phylogenetic relationships and ancestral state inference for fruit length (in centimetres), in tribe Bocageeae (Annonaceae). Phylogenetic relationships by maximum likelihood, matrix with 116 molecular markers, divergence time estimation by BEAST, matrix with 30 molecular markers, trimmed tree showing only the ingroup and its sister group. Pie charts at nodes depict the probabilities of different ancestral areas obtained by BayesTraits. Figure S2.3: ancestral state inference for number of monocarps [1–4 (0), 5–19 (1) and 20–50 (2)] in tribe Bocageeae (Annonaceae). Figure S2.4: ancestral state inference for fruit colour [dull (0), bright (1)] in tribe Bocageeae (Annonaceae). Figure S2.5: ancestral state inference for dehiscence [absent (0), present (1)] in tribe Bocageeae (Annonaceae). Figure S2.6: ancestral state inference for stipe length (in millimetres) in tribe Bocageeae (Annonaceae). Figure S2.7: ancestral state inference for seed length (in millimetres) in tribe Bocageeae (Annonaceae). Figure S2.8: ancestral state inference for number of seeds per monocarp [1–10 (0), 11–28 (1)] in tribe Bocageeae (Annonaceae). Figure S2.9: ancestral state inference for cauliflory [absent (0), present (1)] in tribe Bocageeae (Annonaceae).
ACKNOWLEDGEMENTS
We thank Gisele Alves, Andressa Cabral and Laura Montserrat for assistance with the preparation of figures; José Rubens Pirani for insights into the project; Annelise Frazão and the curators of the herbaria (L, MEXU, MO, MPU, NY, OWU, P, QCA, SPF, RB, U and WAG) for assistance with herbarium sampling; Otávio Marques, Tarcísio Leão and Marcelo Kuhlmann for providing us with photographs; and Isabel Sanmartín, Lars Chatrou and two anonymous reviewers for suggestions on the manuscript. We acknowledge the ISO 9001-certified IRD i-Trop HPC (South Green Platform) at IRD Montpellier for providing HPC resources that have contributed to the research results reported within this paper. D.M.J. and N.M. acknowledge the support of the Jason Swallen Herbarium (OWU) of Ohio Wesleyan University. This work is dedicated to Renato Mello-Silva (in memoriam).
Contributor Information
Jenifer C Lopes, Departamento de Botânica, Instituto de Biociências, Universidade de São Paulo, São Paulo, Brazil; Department of Organismic and Evolutionary Biology, Harvard University Herbaria, 22 Divinity Avenue, Cambridge, MA, USA.
Luiz Henrique M Fonseca, Department of Biology, Ghent University, Ghent, Belgium.
David M Johnson, Department of Biological Sciences, Ohio Wesleyan University, Delaware, OH, USA.
Federico Luebert, Departamento de Ciencias Ambientales y Recursos Naturales Renovables Universidad de Chile, Santiago, Chile; Departmento de Silvicultura y Conservación de la Naturaleza, Universidad de Chile, Santiago, Chile.
Nancy Murray, Department of Biological Sciences, Ohio Wesleyan University, Delaware, OH, USA.
Francis J Nge, IRD, UMR DIADE, Université de Montpellier, Montpellier, France.
Carlos Rodrigues-Vaz, IRD, UMR DIADE, Université de Montpellier, Montpellier, France; Institut de Systématique, Evolution, Biodiversité (ISYEB), Muséum National d’Histoire Naturelle-CNRS-SU-EPHE-UA, Paris, France.
Vincent Soulé, IRD, UMR DIADE, Université de Montpellier, Montpellier, France.
Renske E Onstein, Naturalis Biodiversity Center, Leiden, The Netherlands; German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, Leipzig, Germany.
Lúcia G Lohmann, Departamento de Botânica, Instituto de Biociências, Universidade de São Paulo, São Paulo, Brazil; University and Jepson Herbaria, and Department of Integrative Biology, University of California, Berkeley, CA, USA.
Thomas L P Couvreur, IRD, UMR DIADE, Université de Montpellier, Montpellier, France; Naturalis Biodiversity Center, Leiden, The Netherlands.
FUNDING
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), a postdoctoral fellowship to J.C.L. [2018/11272-5; 2022/08659-0] and a research grant to L.G.L. [2018/23899-2]. Additional funds were provided by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) through a Pq-1B grant to L.G.L. [210871/2017-4], by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme [grant agreement no. 865787] to T.L.P.C.
AUTHOR CONTRIBUTIONS
J.C.L. and Renato Mello-Silva (in memoriam) designed the study with the assistance of L.G.L., T.C., F.L. and F.J.N. C.R.V. and V.S. sampled the species and did the sequencing. J.C.L. conducted the analyses, with the assistance of L.H.M.F., F.L., F.J.N. and R.O. D.M.J. and N.M. provided samples and contributed with taxonomic expertise. J.C.L. led the writing, with the assistance of all authors.
CONFLICT OF INTERESTS
The authors declare no competing interests, financial or otherwise.
DATA AVAILABILITY
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference numbers SUB11858406, SUB11861053, SUB12129452, SUB11463464 and PRJNA508.
LITERATURE CITED
- Abadi S, Azouri D, Pupko T, Mayrose I.. 2019. Model selection may not be a mandatory step for phylogeny reconstruction. Nature Communications 10: 934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike H. 1974. A new look at the statistical model identification. IEEE Transactions on Automatic Control 19: 716–723. [Google Scholar]
- Antonelli A, Nylander JAA, Persson C, Sanmartin I.. 2009. Tracing the impact of the Andean uplift on Neotropical plant evolution. Proceedings of the National Academy of Sciences of the United States of America 106: 9749–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonelli A, Zizka A, Silvestro D, Scharn R, Miñana-Cascales B, Bacon CD.. 2015. An engine for global plant diversity: highest evolutionary turnover and emigration in the American tropics. Frontiers in Genetics 6: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonelli A, Zizka A, Carvalho FA, et al. 2018. Amazonia is the primary source of Neotropical biodiversity. Proceedings of the National Academy of Sciences of the United States of America 115: 6034–6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo JAR, Collevatti RG, Jaramillo CA, et al. 2020. On the young savannas in the land of ancient forests. In: Rull V, Carnaval A, eds. Neotropical diversification: patterns and processes. Berlin: Springer, 271–297. [Google Scholar]
- Bacon CD, Baker WJ, Simmons MP.. 2012. Miocene dispersal drives Island radiations in the palm tribe Trachycarpeae (Arecaceae). Systematic Biology 61: 426–442. [DOI] [PubMed] [Google Scholar]
- Baker WJ, Couvreur TLP.. 2013. Global biogeography and diversification of palms sheds light on the evolution of tropical lineages. I. Historical biogeography. Journal of Biogeography 40: 274–285. [Google Scholar]
- Batalha-Filho H, Fjeldsa J, Fabre PH, Miyaki CY.. 2013. Connections between the Atlantic and the Amazonian forest avifaunas represent distinct historical events. Journal of Ornithology 154: 41–50. [Google Scholar]
- Beaulieu JM, Tank DC, Donoghue MJ.. 2013. A Southern Hemisphere origin for campanulid angiosperms, with traces of the break-up of Gondwana. BMC Evolutionary Biology 13: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham E, Martin AP.. 1998. Comparative mtDNA phylogeography of neotropical freshwater fishes: testing shared history to infer the evolutionary landscape of lower Central America. Molecular Ecology 7: 499–517. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Vaughan TG, Barido-Sottani J, et al. 2019. BEAST 25: an advanced software platform for Bayesian evolutionary analysis. PLoS Computational Biology 15: e1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brée B, Helmstetter AJ, Bethune K, Ghogue J-P, Sonké B, Couvreur TLP.. 2020. Diversification of African rainforest restricted clades: Piptostigmateae and Annickieae (Annonaceae). Diversity 12: 227. [Google Scholar]
- Burnham RJ, Graham A.. 1999. The history of neotropical vegetation: new developments and status. Annals of the Missouri Botanical Garden 86: 546–589. [Google Scholar]
- Carvalho MR, Jaramillo C, de la Parra F, et al. 2021. Extinction at the end-Cretaceous and the origin of modern Neotropical rainforests. Science 372: 63–68. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Chandler MEJ. 1964. The lower Tertiary floras of southern England. 4. A summary and survey of findings in the light of recent botanical observations. London: British Museum of Natural History. [Google Scholar]
- Chatrou LW, Pirie MD, Erkens RHJ, et al. 2012. A new subfamilial and tribal classification of the pantropical flowering plant family Annonaceae informed by molecular phylogenetics. Botanical Journal of the Linnean Society 169: 5–40. [Google Scholar]
- Cheng H, Sinha A, Cruz FW, et al. 2013. Climate change patterns in Amazonia and biodiversity. Nature Communications 4: 1411. [DOI] [PubMed] [Google Scholar]
- Chesters KIM. 1955. Some plant remains from the upper Cretaceous and Tertiary of west Africa. Annals and Magazine of Natural History 12: 489–504. [Google Scholar]
- Couvreur TLP, Pirie MD, Chatrou LW, et al. 2011. Early evolutionary history of the flowering plant family Annonaceae: steady diversification and boreotropical geodispersal. Journal of Biogeography 38: 664–680. [Google Scholar]
- Couvreur TLP, Helmstetter AJ, Koenen EJM, et al. 2019. Phylogenomics of the major tropical plant family Annonaceae using targeted enrichment of nuclear genes. Frontiers in Plant Science 9: 1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur TLP, Dauby G, Blach‐Overgaard A, et al. 2021. Tectonics, climate and the diversification of the tropical African terrestrial flora and fauna. Biological Reviews 96: 16–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagallier L-PMJ, Condamine FL, Couvreur TLP.. 2023. Sequential diversification with Miocene extinction and Pliocene speciation linked to mountain uplift explains the diversity of the African rain forest clade Monodoreae (Annonaceae). Annals of Botany 133: 677–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CC, Bell CD, Mathews S, Donoghue MJ.. 2002. Laurasian migration explains Gondwanan disjunctions: evidence from Malpighiaceae. Proceedings of the National Academy of Sciences of the United States of America 99: 6833–6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick CW, Pennington RT.. 2019. History and geography of neotropical tree diversity. Annual Review of Ecology, Evolution, and Systematics 50: 279–301. [Google Scholar]
- Doyle JA, Endress PK.. 2010. Integrating Early Cretaceous fossils into the phylogeny of living angiosperms: Magnoliidae and eudicots. Journal of Systematics and Evolution 48: 1–35. [Google Scholar]
- Doyle JA, Le Thomas A.. 1997. Phylogeny and geographic history of Annonaceae. Géographie Physique et Quaternaire 51: 353–361. [Google Scholar]
- Draper FC, Costa FRC, Arellano G, et al. 2021. Amazon tree dominance across forest strata. Nature Ecology & Evolution 5: 757–767. [DOI] [PubMed] [Google Scholar]
- Eriksson O. 2016. Evolution of angiosperm seed disperser mutualisms: the timing of origins and their consequences for coevolutionary interactions between angiosperms and frugivores. Biological Reviews of the Cambridge Philosophical Society 91: 168–186. [DOI] [PubMed] [Google Scholar]
- Erkens RHJ, Maas JW, Couvreur TLP.. 2009. From Africa via Europe to South America: migrational route of a species-rich genus of Neotropical lowland rain forest trees (Guatteria, Annonaceae). Journal of Biogeography 36: 2338–2352. [Google Scholar]
- Fiaschi P, Pirani JR.. 2009. Review of plant biogeographic studies in Brazil. Journal of Systematics and Evolution 47: 477–496. [Google Scholar]
- Fine PVA, Ree RH.. 2006. Evidence for a time‐integrated species‐area effect on the latitudinal gradient in tree diversity. The American Naturalist 168: 796–804. [DOI] [PubMed] [Google Scholar]
- GBIF.org. 2022. GBIF Occurrence Download. 10.15468/dl.df8yw2 (16 December 2022, date last accessed). [DOI]
- Guo X, Tang CC, Thomas DC, Couvreur TLP, Saunders RMK.. 2017. A mega-phylogeny of the Annonaceae: taxonomic placement of five enigmatic genera and support for a new tribe, Phoenicantheae. Scientific Reports 7: 7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrot AJ, Utescher T, Erdei B, et al. 2017. Middle Miocene climate and vegetation models and their validation with proxy data. Palaeogeography, Palaeoclimatology, Palaeoecology 467: 95–119. [Google Scholar]
- Herbert TD, Lawrence KT, Tzanova A, Peterson LC, Caballero-Gill R, Kelly CS.. 2016. Late Miocene global cooling and the rise of modern ecosystems. Nature Geoscience 9: 843–847. [Google Scholar]
- Hooker JJ. 2000. Ecological response of mammals to global warming in the late Paleocene and early Eocene. GFF 122: 77–79. [Google Scholar]
- Hoorn C, Wesselingh FP, ter Steege H, et al. 2010. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science 330: 927–931. [DOI] [PubMed] [Google Scholar]
- Iturralde-Vinent MA, MacPhee RDE.. 1999. Paleogeography of the Caribbean region: implications for Cenozoic biogeography. Bulletin of the American Museum of Natural History 238: 1–95. [Google Scholar]
- Jacobs BF, Pan AP, Scotese CR. 2010. A review of the Cenozoic vegetation history of Africa. In: Werdelin L, Sanders J, eds. Cenozoic mammals of Africa. Berkeley: University of California Press, 57–72. [Google Scholar]
- Jaramillo C, Cárdenas A.. 2013. Global warming and neotropical rainforests: a historical perspective. Annual Review of Earth and Planetary Sciences 41: 741–766. [Google Scholar]
- Jaramillo C, Hoorn MC, Silva S, , et al. 2010. The origin of the modern Amazon rainforest: implications from the palynological and paleobotanical record. In: Hoorn MC, Wesselingh FP, eds. Amazonia, landscape and species evolution. Oxford: Blackwell, 317–334. [Google Scholar]
- Johnson DM, Murray NA.. 1995. Synopsis of the tribe Bocageeae (Annonacecae), with revisions of Cardiopetalum, Froesiodendron, Trigynaea, Bocagea, and Hornschuchia. Brittonia 47: 248–319. [Google Scholar]
- Johnson MG, Gardner EM, Liu Y, et al. 2016. HybPiper: extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Applications in Plant Sciences 4: 1600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordano P. 2016. What is long-distance dispersal? And a taxonomy of dispersal events. Journal of Ecology 105: 75–84. [Google Scholar]
- Junikka L, Maas PJM, Maas-van de Kamer H, Westra LYT.. 2016. Revision of Oxandra (Annonaceae). Blumea - Biodiversity, Evolution and Biogeography of Plants 61: 215–266. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann M. 2018. Frutos e Sementes do Cerrado: espécies atrativas para fauna. Brasília: Ipsis gráfica e editora. [Google Scholar]
- Landis M, Matzke NJ, Moore BR, Huelsenbeck JP.. 2013. Bayesian analysis of biogeography when the number of areas is large. Systematic Biology 62: 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledo RMD, Colli GR.. 2017. The historical connections between the Amazon and the Atlantic Forest revisited. Journal of Biogeography 44: 2551–2563. [Google Scholar]
- Le Thomas A. 1980. Ultrastructural characters of the pollen grains of African Annonaceae and their significance for the phylogeny of primitive Angiosperms. Pollen Spores 22: 267–342. [Google Scholar]
- Lobão AQ. 2017. A new species of Trigynaea (Annonaceae) endemic to the Atlantic Forest of Brazil. Phytotaxa 309: 193–196. [Google Scholar]
- Luebert F. 2021. The two South American dry diagonals. Frontiers of Biogeography 13: e51267. [Google Scholar]
- Luebert F, Couvreur TLP, Gottschling M, Hilger HH, Miller JS, Weigend M.. 2017. Historical biogeography of Boraginales: West Gondwanan vicariance followed by long-distance dispersal? Journal of Biogeography 44: 158–169. [Google Scholar]
- Maas PJM, Westra LYT.. 2003. Revision of the Neotropical genus Pseudoxandra (Annonaceae). Blumea - Biodiversity, Evolution and Biogeography of Plants 48: 201–259. [Google Scholar]
- Manchester SR. 1994. Fruits and seeds of the Middle Eocene nut beds flora, Clarno Formation, Oregon. Palaeontographica Americana 58: 1–205. [Google Scholar]
- Martin LD. 2010. Paleogene avifauna of the Holarctic. Vertebrata Palasiatica 48: 367–374. [Google Scholar]
- Massoni J, Couvreur TLP, Sauquet H.. 2015. Five major shifts of diversification through the long evolutionary history of Magnoliidae (angiosperms). BMC Evolutionary Biology 15: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke NJ. 2013. Probabilistic historical biogeography: new models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Frontiers of Biogeography 5: 242–248. [Google Scholar]
- McLoughlin S. 2001. The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Australian Journal of Botany 49: 271–300. [Google Scholar]
- Mello-Silva R, Lopes JC.. 2020. The Brazilian Atlantic Forest genus Bocagea (Annonaceae) revisited, with two new species. Phytotaxa 475: 279–288. [Google Scholar]
- Mello-Silva R, Santos DYAC, Salatino MLF, et al. 2011. Five vicarious genera from Gondwana: the Velloziaceae as shown by molecules and morphology. Annals of Botany 108: 87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak I, Zhang LB, Renner SS.. 2010. Trans-Atlantic, trans-Pacific and trans-Indian Ocean dispersal in the small Gondwanan Laurales family Hernandiaceae. Journal of Biogeography 37: 1214–1226. [Google Scholar]
- Mohr BAR, Bernardes-de-Oliveira MEC.. 2004. Endressinia brasiliana, a magnolialean angiosperm from the Lower Cretaceous Crato Formation (Brazil). International Journal of Plant Sciences 165: 1121–1133. [Google Scholar]
- Morley RJ. 2000. Origins and evolution of tropical rain forests. Chichester: John Wiley & Sons. [Google Scholar]
- Morley RJ. 2003. Interplate dispersal routes for megathermal angiosperms. Perspectives in Plant Ecology, Evolution and Systematics 6: 5–20. [Google Scholar]
- Morrone JJ, Escalante T, Rodríguez-Tapia G, Carmona A, Arana M, Gómez-Mercado JD.. 2022. Biogeographic regionalization of the Neotropical region: new map and shapefile. Anais da Academia Brasileira de Ciências 94: e2021116. [DOI] [PubMed] [Google Scholar]
- Murray NA. 1993. Revision of Cymbopetalum and Porcelia (Annonaceae). Systematic Botany Monographs 40: 1–121. [Google Scholar]
- Nathan R, Muller-Landau HC.. 2000. Spatial patterns of seed dispersal, their determinants and consequences for recruitment. Trends in Ecology & Evolution 15: 278–285. [DOI] [PubMed] [Google Scholar]
- O’Connor JM, Duncan RA.. 1990. Evolution of the Walvis Ridge-Rio Grande Rise hot spot system: implications for African and South American plate motions over plumes. Journal of Geophysical Research 95: 17475–17502. [Google Scholar]
- Onstein RE, Kissling WD, Chatrou LW, Couvreur TLP, Morlon H, Sauquet H.. 2019. Which frugivory-related traits facilitated historical long-distance dispersal in the custard apple family (Annonaceae)? Journal of Biogeography 46: 1874–1888. [Google Scholar]
- Pagel M, Meade A.. 2006. Bayesian analysis of correlated evolution of discrete characters by reversible jump Markov chain Monte Carlo. The American Naturalist 167: 808–825. [DOI] [PubMed] [Google Scholar]
- Parrish JT. 1993. The palaeogeography of the opening South Atlantic. In: George W, Lavocat R, eds. The Africa–South America connection. Oxford: Clarendon Press, 8–27. [Google Scholar]
- Pennington RT, Dick CW.. 2004. The role of immigrants in the assembly of the South American rainforest tree flora. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences 359: 1611–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirie MD, Doyle JA.. 2012. Dating clades with fossils and molecules: the case of Annonaceae. Botanical Journal of the Linnean Society 169: 84–116. [Google Scholar]
- Pirie MD, Chatrou LW, Mols JB, Erkens RHJ, Oosterhof J.. 2006. ‘Andean-centred’ genera in the short-branch clade of Annonaceae: testing biogeographic hypotheses using phylogeny reconstruction and molecular dating. Journal of Biogeography 33: 31–46. [Google Scholar]
- Pirie MD, Maas PJM, Wilschut RA, Melchers-Sharrott H, Chatrou LW.. 2018. Parallel diversifications of Cremastosperma and Mosannona (Annonaceae), tropical rainforest trees tracking Neogene upheaval of South America. Royal Society Open Science 5: 171561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platnick, NI, Nelson G.. 1978. A method of analysis for historical biogeography. Systematic Zoology 27: 1–16. [Google Scholar]
- Por FD. 1992. Sooretama: the Atlantic rain forest of Brazil. The Hague: SPB Academic. [Google Scholar]
- QGIS Development Team 2009. QGIS Geographic Information System. Open Source. https://qgis.org/pt_BR/site/ (16 December 2022, date last accessed). [Google Scholar]
- Rambaut A., Drummond AJ, Xie D, Baele G, Suchard MA.. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Systematic Biology 67: 901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ree RH. 2005. Detecting the historical signature of key innovations using stochastic models of character evolution and cladogenesis. Evolution 59: 257–265. [PubMed] [Google Scholar]
- Ree RH, Smith SA.. 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology 57: 4–14. [DOI] [PubMed] [Google Scholar]
- Renner S. 2004. Plant dispersal across the tropical Atlantic by wind and sea currents. International Journal of Plant Sciences 165: S23–S33. [Google Scholar]
- Richardson JE, Chatrou LW, Mols JB, Erkens RHJ, Pirie MD.. 2004. Historical biogeography of two cosmopolitan families of flowering plants: Annonaceae and Rhamnaceae. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences 359: 1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Reich D.. 2012. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Research 22: 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F. 1997. Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Systematic Biology 46: 195–203. [Google Scholar]
- Sánchez MS, Giannini NP.. 2018. Trophic structure of frugivorous bats in the Neotropics: emergent patterns in evolutionary history. Mammal Review 48: 90–107. [Google Scholar]
- Sanderson MJ. 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Molecular Biology and Evolution 19: 101–109. [DOI] [PubMed] [Google Scholar]
- Sarver BAJ, Pennell MW, Brown JW, et al. 2019. The choice of tree prior and molecular clock does not substantially affect phylogenetic inferences of diversification rates. PeerJ 7: e6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayyari E, Mirarab S.. 2016. Fast coalescent-based computation of local branch support from quartet frequencies. Molecular Biology and Evolution 33: 1654–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharaschkin T, Doyle JA.. 2005. Phylogeny and historical biogeography of Anaxagorea (Annonaceae) using morphology and non-coding chloroplast sequence data. Systematic Botany 30: 712–735. [Google Scholar]
- Smedmark JEE, Anderberg AA.. 2007. Boreotropical migration explains hybridization between geographically distant lineages in the pantropical clade Sideroxyleae (Sapotaceae). American Journal of Botany 94: 1491–1505. [DOI] [PubMed] [Google Scholar]
- Smith, SA, O’Meara BC.. 2012. treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28: 2689–2690. [DOI] [PubMed] [Google Scholar]
- Smith SA, Brown JW, Walker JF.. 2018. So many genes, so little time: a practical approach to divergence-time estimation in the genomic era. PLoS One 13: e0197433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull GW, Johnson DM, Murray NA, Couvreur TLP, Reeger JE, Roy CM.. 2017. Plastid and seed morphology data support a revised infrageneric classification and an African origin of the Pantropical genus Xylopia (Annonaceae). Systematic Botany 42: 211–225. [Google Scholar]
- Sugiura N. 1978. Further analysis of the data by Akaike’s information criterion and the finite corrections. Communications in Statistics - Theory and Methods 7: 13–26. [Google Scholar]
- Surveswaran S, Wang RJ, Su YCF, Saunders RMK.. 2010. Generic delimitation and historical biogeography in the early-divergent ‘ambavioid’ lineage of Annonaceae: Cananga, Cyathocalyx and Drepananthus. Taxon 59: 1721–1734. [Google Scholar]
- Takahashi M, Friis EM, Uesugi K, Suzuki Y, Crane PR.. 2008. Floral evidence of Annonaceae from the Late Cretaceous of Japan. International Journal of Plant Sciences 169: 908–917. [Google Scholar]
- Thiers BM. (updated continuously). Index Herbariorum.https://sweetgum.nybg.org/science/ih/. (16 December 2022, date last accessed).
- Thorne, RF. 1973. Floristic relationships between tropical Africa and tropical America. In Meggers BJ, Ayensu ES, Duckworth WD, eds. Tropical forest ecosystems in Africa and South America: a comparative review. Washington: Smithsonian Institution Scholarly Press, 27–47. [Google Scholar]
- Tiffney BH. 1985. The Eocene North Atlantic land bridge: its importance in Tertiary and modern phytogeography of the Northern Hemisphere. Journal of the Arnold Arboretum 66: 243–273. [Google Scholar]
- Verdcourt B. 1970. A new genus of Annonaceae from the east African coastal forests. Kew Bulletin 24: 449–453. [Google Scholar]
- Vilela L, Lopes JC.. 2022. Hornschuchia (Annonaceae), an endemic and threatened genus from the Brazilian Atlantic Forest. European Journal of Taxonomy 828: 75–108. [Google Scholar]
- Warming E. 1874 [‘1873’]. Symbolae ad floram Brasiliae centralis cognoscendam, particula XVI. Videnskabelige meddelelser fra den Naturhistoriske Forening i Kjöbenhavn 1873: 142–143. [Google Scholar]
- Wei R, Xiang Q, Schneider H, et al. 2015. Eurasian origin, boreotropical migration and transoceanic dispersal in the pantropical fern genus Diplazium (Athyriaceae). Journal of Biogeography 42: 1809–1819. [Google Scholar]
- Wing SL, Herrera F, Jaramillo CA, Gomez-Navarro C, Wilf P, Labandeira CC.. 2009. Late Paleocene fossils from the Cerrejon formation, Colombia, are the earliest record of Neotropical rainforest. Proceedings of the National Academy of Sciences of the United States of America 106: 18627–18632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe JA. 1975. Some aspects of plant geography of the Northern Hemisphere during the Late Cretaceous and Tertiary. Annals of the Missouri Botanical Garden 62: 264–279. [Google Scholar]
- Yang Z. 2006. Computational molecular evolution. Oxford: Oxford University Press. [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K.. 2001. Trends, rhythms and aberrations in global climate 65 Ma to present. Science 292: 686–693. [DOI] [PubMed] [Google Scholar]
- Zhang C, Rabiee M, Sayyari E, Mirarab S.. 2018. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Su YCF, Thomas DC, Saunders RMK.. 2012. ‘Out-of-Africa’ dispersal of tropical floras during the Miocene climatic optimum: evidence from Uvaria (Annonaceae). Journal of Biogeography 39: 322–335. [Google Scholar]
- Zizka A, Silvestro D, Andermann T, et al. 2019. CoordinateCleaner: standardized cleaning of occurrence records from biological collection databases. Methods in Ecology and Evolution 10: 744–751. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference numbers SUB11858406, SUB11861053, SUB12129452, SUB11463464 and PRJNA508.