Abstract
Ligand screening was utilized to isolate a human cDNA that encodes a novel CpG binding protein, human CpG binding protein (hCGBP). This factor contains three cysteine-rich domains, two of which exhibit homology to the plant homeodomain finger domain. A third cysteine-rich domain conforms to the CXXC motif identified in DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. A fragment of hCGBP that contains the CXXC domain binds to an oligonucleotide probe containing a single CpG site, and this complex is disrupted by distinct oligonucleotide competitors that also contain a CpG motif(s). However, hCGBP fails to bind oligonucleotides in which the CpG motif is either mutated or methylated, and it does not bind to single-stranded DNA or RNA probes. Furthermore, the introduction of a CpG dinucleotide into an unrelated oligonucleotide sequence is sufficient to produce a binding site for hCGBP. Native hCGBP is detected as an 88-kDa protein by Western analysis and is ubiquitously expressed. The DNA-binding activity of native hCGBP is apparent in electrophoretic mobility shift assays, and hCGBP trans-activates promoters that contain CpG motifs but not promoters in which the CpG is ablated. These data indicate that hCGBP is a transcriptional activator that recognizes unmethylated CpG dinucleotides, suggesting a role in modulating the expression of genes located within CpG islands.
The human genome contains approximately 45,000 CpG islands, which are discrete clusters of unmethylated CpG dinucleotides (3, 33). More than half of the characterized human gene promoters are associated with these CpG islands, including many housekeeping genes as well as some tissue-specific genes (3, 17, 24, 25, 30). The contribution of CpG islands to the modulation of gene expression has been attributed in part to the presence of multiple binding sites for transcription factors such as Sp1 (11, 34, 44, 52) as well as an open chromatin configuration (4, 53). Despite containing CpG dinucleotides, the target of DNA methyltransferase, most CpG islands remain unmethylated (33). Only a small proportion of CpG islands, such as those associated with genes on the inactive X chromosome and some parentally imprinted genes, are methylated during development (21, 46, 53). It is unclear how CpG islands maintain an unmethylated state despite their open chromatin configuration and free access to DNA methyltransferase. Demethylase (10), Sp1-like cis elements (48), p21WAF1 (5, 13), and histone H1 (56, 57) have been proposed to either remove methyl groups from 5-methylcytosine residues in DNA or protect genomic DNA from methylation.
The cysteine-rich CXXC domain is highly conserved among a small group of proteins, including DNA methyltransferase (9), human trithorax (HRX) (also known as MLL or ALL-1) (19, 27, 32, 45, 55, 58), and methyl-CpG binding domain protein 1 (MBD1/PCM1) (16, 28). The CXXC domain binds zinc and lies within the N-terminal regulatory half of DNA methyltransferase. Removal of the N-terminal domain results in the promiscuous methylation of unmethylated CpG substrates, suggesting that this domain distinguishes between unmethylated and hemimethylated DNA (8). However, the contribution of the CXXC domain to DNA recognition has not been established. The CXXC domain and flanking basic region of HRX binds to salmon sperm DNA and poly(dI-dC), suggesting that the CXXC domain binds DNA without a pronounced sequence specificity (51). The CXXC domain in HRX has also been shown to repress transcription of a reporter gene when expressed as a GAL4 fusion protein (45). Isoforms of MBD1 contain up to three copies of the CXXC domain (23). However, the CXXC domains are not required for binding to oligonucleotides containing CpG motifs, and the function of these domains in MBD1 is not known (16). MBD1 isoforms containing all three CXXC domains suppress both methylated and unmethylated promoters (23).
HRX additionally contains multiple copies of a cysteine-rich domain that conforms to the plant homeodomain (PHD) finger domain, a zinc finger-like structure spanning 50 to 80 amino acids that is characterized by a unique arrangement of histidine and cysteine residues (Cys4-His-Cys3). The function of PHD fingers has not been established, but they have been postulated to mediate protein-protein or protein-DNA interactions (1). This domain has been identified in over 40 proteins, many of which are transcriptional regulators implicated in the modulation of chromatin structure (1).
Here we describe a new member of the family of CpG binding proteins, denoted human CpG binding protein (hCGBP). The deduced amino acid sequence of the full-length hCGBP cDNA reveals two PHD finger-like domains and a CXXC domain. A histidine-tagged hCGBP fusion protein containing the CXXC domain bound to an oligonucleotide containing a single unmethylated CpG dinucleotide but failed to bind to the probe following methylation of the CpG motif. The full-length hCGBP cDNA trans-activated promoters containing CpG motifs when cotransfected into cell lines. Hence, hCGBP is a transcriptional activator that recognizes unmethylated CpG motifs and therefore likely plays a role in the regulation of CpG-rich promoters.
MATERIALS AND METHODS
Oligonucleotides.
The following complementary oligonucleotides were synthesized on an Applied Biosystems model 394 synthesizer (mutated nucleotides are underlined): CpG-neg (bp −30 to −68 of the human gp91phox promoter) (50), 5′-CTATGCTTCTTCTTCCAATGAGGAAATGAAAACAGCAG-3′; CpG-pos (similar to CpG-neg, except that the CCAAT box is mutated to CCGGT) (20), 5′-CTATGCTTCTTCTTCCGGTGAGGAAATGAAAACAGCAG-3′; CGD (similar to CpG-neg, except that it contains two mutations, at bp −55 and −57, that were identified in chronic granulomatous disease patients [40]), 5′-CTATGCTTCTTCTTCCAATGAGGAGAGGAAAACAGCAG-3′; Ets (a high-affinity binding site that was selected from a pool of degenerate oligonucleotides by using partially purified Fli-1 protein [K.S.V., unpublished data]), 5′-GTGGAGACCGGAAGTGGGTGGG-3′; C→T (mutated Ets oligonucleotide), 5′-GTGGAGACTGGAAGTGGGTGGG-3′; G→A (mutated Ets oligonucleotide), 5′-GTGGAGACCAGAAGTGGGTGGG-3′; CG11 (contains 11 CpG motifs), 5′-GATCCGAGCGGTAGCGGTTCGGTACCGGTTTCGAATCCGGGCGGTGCGAATAGACCGGTTGCGGTG-3′; NF-κB (consensus binding site for NF-κB), 5′-GTAGTTGAGGGGACTTTCCCAGGC-3′; NF-κB-CG (NF-κB oligonucleotide mutated to contain a single CpG motif), 5′-GTAGTTGAGCGGACTTTCCCAGGC-3′; and NF-κB-GC (mutated to produce a GpC motif), 5′-GTAGTTGAGGGGCCTTTCCCAGGC-3′. An RNA version of the CpG-pos oligonucleotide (upper strand) was obtained from Dharmacon Research, Inc. (Boulder, Colo.).
Isolation of a novel DNA-binding protein.
A dimethyl sulfoxide-treated HL60 myeloid cell cDNA library was ligand screened by an in situ filter-binding technique (14, 49) as follows. Phages were plated at a density of 5 × 104 per 150-mm-diameter plate. Nitrocellulose filters saturated with 10 mM isopropyl-β-d-thiogalactoside (IPTG) were placed on the plates, which were subsequently incubated for 5 h at 37°C. Duplicate filters were incubated for 10 h at 37°C. The filters were washed for 10 min in a buffer containing 50 mM NaCl, 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1 mM dithiothreitol (DTT), and 0.05% lauryl dimethylamide oxide (LDAO). The filters were then blocked overnight at 4°C in blocking buffer (2.5% dried milk, 25 mM HEPES [pH 8.0], 1 mM DTT, 10% glycerol, 50 mM NaCl, 0.05% LDAO, and 1 mM EGTA) prior to being washed briefly in TNE-50 (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM EGTA, 1 mM DTT). Probes were prepared by phosphorylating double-stranded oligonucleotides, batch ligating them to generate concatenated oligonucleotides (an average of nine copies), and radioactively labeling them by primer extension, using a sequence-specific primer. For ligand screening, filters were incubated in TNE-50 containing concatenated DNA binding site probe at 0.5 × 106 to 1 × 106 cpm/ml and 10 μg of double-stranded herring sperm DNA/ml as a nonspecific competitor. After an incubation of 14 to 16 h at 4°C, the filters were washed three times with cold TNE-50. They were subsequently blotted dry and exposed to X-ray film at −70°C overnight. Primary screenings were performed with concatenated CpG-pos as a probe. Plaques that were positive on duplicate filters were rescreened with both the CpG-pos probe (to confirm DNA-binding activity) and the concatenated CGD oligonucleotide probe (to check for binding specificity). To purify clones that encode sequence-specific DNA-binding factors, four rounds of sequential screening were performed as described above.
Clones of interest were released from the lambda vector by EcoRI digestion and subcloned into pUC19 (New England Biolabs, Beverly, Mass.). The nucleotide sequence of each cDNA was determined by using an Applied Biosystems automated DNA sequencer (DNA sequencing facility, Iowa State University, Ames).
Production of fusion proteins and generation of antiserum.
Ligand screening resulted in isolation of a 720-bp cDNA encoding a novel DNA-binding activity (hCGBP). To obtain bacterially expressed hCGBP, the 720-bp cDNA fragment was subcloned into both the glutathione S-transferase (GST) fusion plasmid pGEX-5X-1 (Amersham Pharmacia Biotech, Piscataway, N.J.) and the histidine-tagged plasmid pET32a (Novagen, Madison, Wis.). The GST fusion plasmid was introduced into Escherichia coli DH5α, while the histidine-tagged construct was introduced into E. coli BL21(DE3). In addition, a truncated version of histidine-tagged hCGBP (546 bp) was produced by removal of 174 bp from the 3′ end of the original hCGBP cDNA fragment by digestion with BamHI. Cells were grown at 37°C to an optical density at 600 nm of 1.0. IPTG was added to 0.1 mM, and cells were incubated for an additional 4 to 6 h. The cells were harvested and resuspended in 50 μl of ice-cold phosphate-buffered saline per ml of culture. GST-hCGBP fusion protein was affinity purified, using glutathione-Sepharose and reduced glutathione in accordance with the protocol provided by Amersham Pharmacia Biotech. The histidine-tagged hCGBP fusion protein was affinity purified by the His-Trap purification protocol (Amersham Pharmacia Biotech).
Chickens and New Zealand White rabbits were immunized with the 720-bp GST-hCGBP fusion protein by Covance, Inc. (Denver, Pa.) and HRP, Inc. (Denver, Pa.), respectively. Sodium dodecyl sulfate (SDS)-denatured histidine-tagged hCGBP fusion protein was used in Western blot analysis to determine the serum antibody titer. Western blot analysis was performed following electrophoresis of fusion protein on SDS-Tris-glycine–4 to 12% polyacrylamide gels (Novex, San Diego, Calif.). Proteins were transferred to nitrocellulose membranes (MSI, Westborough, Mass.), immunoblotted with a 1:3,000 dilution of rabbit hCGBP polyclonal antibody, and detected by enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech) in accordance with the manufacturer's instructions.
In vitro DNA-binding assays.
Crude nuclear extract was isolated from a 100-liter culture of the human chronic myelogenous leukemia cell line K562 grown at the Cell Culture Center (Minneapolis, Minn.), using the method of Dignam et al. (18). Crude nuclear extract was adsorbed to a heparin-Sepharose column in buffer D (20 mM HEPES [pH 7.9], 20% glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, and 5 μg of aprotinin/ml) (39). Proteins were eluted with a gradient of increasing KCl concentration in buffer D. The peak hCGBP fractions (eluting at 0.24 to 0.35 M KCl) were pooled and then concentrated with a Centriprep 50 concentrator (Amicon, Beverly, Mass.).
Oligonucleotide probes were radiolabeled by T4 polynucleotide kinase, using [γ-32P]ATP (Amersham Pharmacia Biotech), and then annealed with an equimolar amount of complementary-strand oligonucleotide. Methylation of the oligonucleotide probes was accomplished by incubating them with SssI methylase and S-adenosylmethionine as recommended by the manufacturer (New England Biolabs). Radiolabeled probes were resolved by 10% native polyacrylamide gel electrophoresis and eluted by the crush-and-soak method (47).
Electrophoretic mobility shift assays (EMSAs) were performed as described previously (50) with slight modifications. Briefly, 0.05 to 0.5 μg of histidine-tagged hCGBP fusion protein (720-bp cDNA) or 3 μg of heparin-fractionated nuclear extract derived from K562 cells was incubated with 0.25 μg of herring sperm DNA or 2 μg of poly(dA-dT), respectively, on ice for 15 min in a 40-μl reaction volume prior to the addition of competitor oligonucleotides. After an additional 30-min incubation on ice, 104 cpm of oligonucleotide probe was added to the reaction and the mixture was incubated for 30 min on ice. Chicken antiserum raised against hCGBP was added in some experiments after the probe incubation, and samples were incubated on ice for an additional 1 h. Immunodepletion of antiserum was performed by incubating the antiserum with an affinity resin prepared by incubating histidine-tagged hCGBP protein with His-Trap beads (Amersham Pharmacia Biotech). The affinity resin and bound immunoglobulins were pelleted by centrifugation, and the supernatant was used in EMSAs. EMSA samples were loaded onto 0.5× Tris-borate-EDTA–3.5 to 5% nondenaturing polyacrylamide gels, and electrophoresis was performed at 230 V and 4°C for 2.5 h.
DNase I footprinting assays were performed essentially as described previously (50), except that poly(dA-dT) was used as a nonspecific competitor and samples were incubated with DNase I for 90 s. A G+A sequence ladder was generated by incubating the probe with acidic loading dye (98% formamide, 20 mM Tris acetate, pH 6.0) at 95°C for 2 h as previously described (54).
Southern and Northern blot analyses.
Genomic DNA was isolated from the human cell line K562 as previously described (37). Twenty micrograms of genomic DNA was digested with the appropriate restriction enzyme, fractionated by agarose gel electrophoresis, and transferred to a nitrocellulose membrane as described elsewhere (12). The blot was hybridized in a solution containing 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 10× Denhardt's solution, 2% SDS, 50% formamide, and 100 μg of denatured herring testis DNA/ml and probed with randomly primed 720-bp hCGBP cDNA. The blots were subsequently washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.05% SDS for 30 min at room temperature and then with 0.1× SSC–0.1% SDS for 40 min at 50°C, and autoradiography was performed.
Human tissue mRNA blots were purchased from Clontech (Palo Alto, Calif.). Northern blots were probed with the 720-bp hCGBP cDNA fragment or actin cDNA (to determine RNA loading and integrity) for 18 h at 42°C. The blots were hybridized and washed as described above for Southern analysis.
Construction of plasmids.
The 720-bp hCGBP cDNA clone was used as a probe to screen an HL60 λgt11 library, and a 2.2-kb cDNA clone was obtained. A database search revealed an expressed sequence tag (EST) clone (AA325016) that extended further upstream. Together, the 2.2-kb cDNA clone and the EST clone constitute a full-length hCGBP cDNA, and the nucleotide sequence of both strands was determined by the dideoxy chain termination method with an Amplicycle sequencing kit (Perkin-Elmer, Branchburg, N.J.) and α-33P-labeled deoxynucleoside triphosphates in accordance with the manufacturer's protocol. A full-length hCGBP open reading frame (ORF) was constructed by ligating the 5′ end of the EST clone to the 3′ end of the 2.2-kb cDNA clone (at a StuI site) and subcloning the resulting construct into Bluescript vector (Stratagene, La Jolla, Calif.) digested with EcoRI. Expression constructs were generated by transferring this hCGBP cDNA clone into XhoI- and XbaI-digested pcDNA3.1(+) (Invitrogen, Carlsbad, Calif.). The cytomegalovirus (CMV)-luciferase reporter construct was generated by subcloning the CMV promoter-enhancer into HindIII- and BamHI-digested luciferase reporter gene vector pXP2 (41). The CMV–β-galactosidase (β-gal) reporter construct was a generous gift of Yu Chung Yang (Indiana University, Indianapolis). Dimer CpG-pos–luciferase and dimer CpG-neg–luciferase reporter constructs were generated by subcloning two copies of the CpG-pos or CpG-neg oligonucleotide into a minimal TATA box-pXP2 luciferase vector (the TATA vector was a generous gift of Ellis Neufeld, Harvard University School of Medicine, Boston, MA) digested with HindIII and BamHI. Plasmids were purified by using a Maxiprep kit (Promega, Inc., Madison, Wis.) followed by ultracentrifugation in a cesium chloride gradient and were then transfected into human erythroleukemia (HEL) cells by electroporation.
Cell culture and transfection.
The human chronic myelogenous leukemia cell line K562, human erythroleukemia cell line HEL, and the human cervical carcinoma epithelial cell line HeLa were obtained from the American Type Culture Collection (Manassas, Va.). HeLa cells were cultured in Dulbecco's modified Eagle's medium and K562 and HEL cells were cultured in RMPI 1640 medium at 37°C and 5% CO2. Both media were supplemented with 10% fetal bovine serum (Sigma Chemical Co., St. Louis, Mo.), 50 Units of penicillin/ml, 50 μg of streptomycin/ml, and 0.2 mM glutamine (GIBCO-BRL, Gaithersburg, Md.).
Cotransfection assays for HEL cells were performed by resuspending 107 cells in 350 μl of culture medium and electroporating them in the presence of 25 μg of plasmid DNA (5 μg of reporter plasmid and 20 μg of expression vector) at 960 μF and 220 V in 4-mm-path-length cuvettes with a Bio-Rad Gene Pulser. Electroporated cells were transferred to 100-mm-diameter tissue culture dishes, each containing 12 ml of prewarmed medium, which were subsequently incubated at 37°C in an atmosphere of 5% CO2. After incubation for 15 h, the cells were washed with phosphate-buffered saline. Cell pellets were resuspended in either 150 μl of lysis buffer (Promega, Inc.), for luciferase and β-gal assays, or nuclear extraction buffer, for preparation of a mini-nuclear extract as described by Andrews and Faller (2). Transfection of K562 cells was performed as described above except that 50 μg of expression vector was used. For HeLa cell transfections, 6 × 106 cells were suspended in 570 μl of culture medium and electroporated in the presence of 25 to 30 μg of plasmid DNA (10 μg of TATA box-luciferase reporter plasmid or 5 μg of CMV-luciferase reporter plasmid, and 20 μg of expression vector) at 960 μF and 250 V. After a 15-h incubation, the cells were harvested and total cellular protein was isolated as described above. Multiple preparations of each plasmid construct were examined in cotransfection assays, and multiple experiments were performed with each plasmid preparation.
In vitro transcription and translation.
The cDNA for hCGBP (in Bluescript vector) was transcribed and translated in vitro, using a rabbit reticulocyte lysate assay and 1 μg of the plasmid DNA, in accordance with the manufacturer's instructions (Promega, Inc.). Five microliters of each reaction mixture was resolved by SDS–5 to 12% polyacrylamide gel electrophoresis, and the gels were fixed, soaked in Amplify enhancer (Amersham Pharmacia Biotech), dried, and then exposed to X-ray film overnight.
Nucleotide sequence extension number.
The hCGNP cDNA sequence has been deposited in the GenBank database (accession no. AF149758).
RESULTS
Isolation of a novel DNA-binding factor.
We previously demonstrated the binding of a hematopoiesis-associated factor (HAF-1) to the bp −68 to bp −30 bp region of the gp91phox promoter (20), which is active in mature myeloid cells. During the course of efforts to further characterize HAF-1, we ligand screened an HL60 λgt11 cDNA library with a concatenated version of the bp −68 to bp −30 gp91phox gene promoter. The concatenated oligonucleotide (CpG-pos) carries a mutation of an inverted CCAAT box found in the gp91phox promoter (changed to CCGGT) to eliminate interaction with CCAAT-box-binding factors. Importantly, this mutation also introduces a CpG motif into the oligonucleotide.
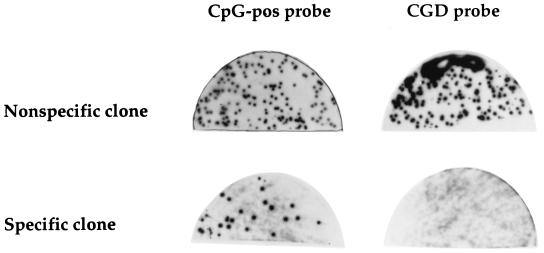
Ligand screening of 2 × 106 plaques yielded nine novel sequence-specific DNA-binding clones that bound to the CpG-pos probe but not to a mutated version of the probe (CGD) containing two mutations identified in chronic granulomatous disease patients and lacking a CpG motif (Fig. 1). The nine novel clones contain a novel 720-bp cDNA insert (see below), which is the subject of this report.
FIG. 1.
Isolation by ligand screening of cDNA clones that encode DNA-binding factors. Phages derived from an HL60 λgt11 expression library were induced to express fusion proteins and incubated with CpG-pos (20) or CGD oligonucleotide probe (40) as described in Materials and Methods. Shown are a representative phage clone that encodes a non-sequence-specific DNA-binding activity (top) and a representative phage clone that exhibits sequence-specific DNA-binding activity (bottom).
The novel DNA-binding protein requires a CpG dinucleotide for binding.
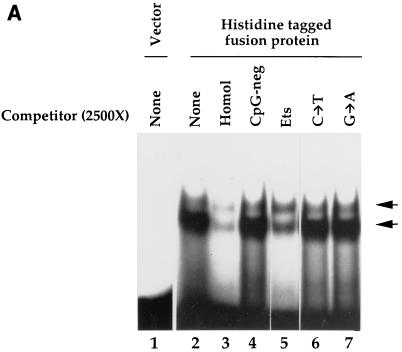
To further examine the DNA-binding properties of the 720-bp cDNA clone, a histidine-tagged fusion protein was isolated and EMSA was performed with the CpG-pos oligonucleotide probe. No DNA-protein complex was detected with the affinity-purified histidine tag (189 amino acids) alone (Fig. 2A, lane 1). The histidine-tagged fusion protein bound to the CpG-pos probe and formed two complexes (lane 2). The predominant, faster-migrating EMSA complex was efficiently disrupted by a molar excess of unlabeled CpG-pos oligonucleotide (lane 3) but not by the wild-type bp −68 to bp −30 gp91phox promoter oligonucleotide (CpG-neg) that lacks the CpG motif (lane 4). Hence, this factor does not interact with the wild-type gp91phox promoter element. A consensus binding site for the Ets family of DNA-binding factors, which also contains a CpG motif, efficiently disrupted the EMSA complex (lane 5). However, mutation of either nucleotide of the CpG motif within the Ets oligonucleotide competitor abolished binding affinity (lanes 6 and 7). In a separate experiment using more fusion protein and larger molar excess of unlabeled competitor, the slower-migrating complex was also disrupted by CpG-pos and Ets oligonucleotides but not by oligonucleotides lacking the CpG motif (data not shown). We conclude that the novel DNA-binding factor requires a CpG motif for efficient binding and hereafter refer to this factor as human CpG binding protein (hCGBP).
FIG. 2.
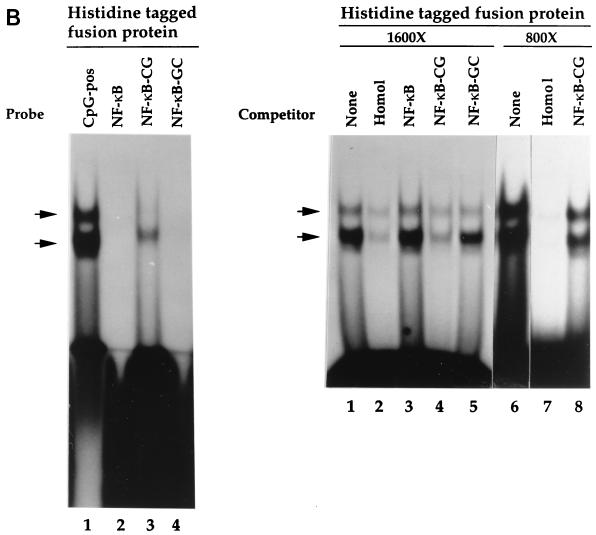
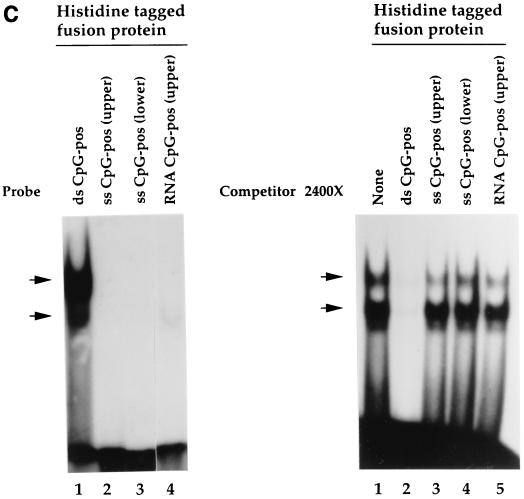
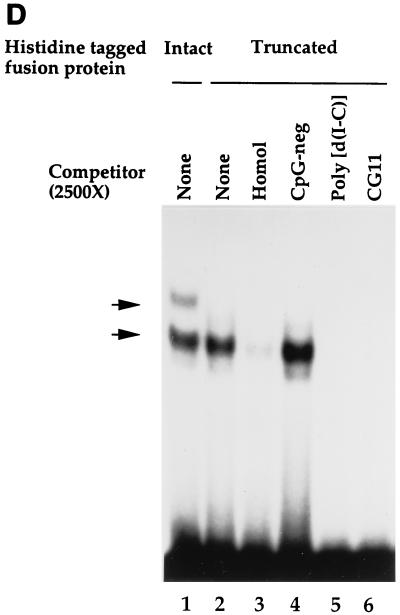
The histidine-tagged fusion protein exhibits sequence-specific DNA-binding activity. (A) Binding specificity of the histidine-tagged fusion protein. EMSA was performed as described in Materials and Methods, using purified histidine-tagged fusion protein (720-bp cDNA fragment) and the CpG-pos probe. Competitor oligonucleotides were added as indicated. Lane 1, vector alone; lanes 2 to 7, addition of histidine-tagged fusion protein (lane 2, no competitor; lane 3, homologous competitor; lane 4, CpG-neg competitor [40]; lane 5, Ets oligonucleotide competitor (which contains a CpG motif); lane 6, mutated Ets oligonucleotide competitor in which the cytosine of the CpG motif is mutated to a thymine [C→T]; lane 7, same as lane 6, except the guanine of the CpG motif is mutated to adenine [G→A]). (B) Introduction of a CpG motif into an unrelated oligonucleotide is sufficient to produce a binding site for the histidine-tagged fusion protein. EMSA was performed as described for panel A. (Left panel) Lane 1, CpG-pos probe; lane 2, NF-κB probe; lane 3, NF-κB–CG probe; lane 4, NF-κB–GC probe. (Right panel) Competitor oligonucleotides (1,600- or 800-fold molar excess) were added to samples containing the CpG-pos probe. Lanes 1 and 6, no competitor; lanes 2 and 7, homologous competitor; lane 3, NF-κB competitor; lanes 4 and 8, NF-κB–CG competitor; lane 5, NF-κB–GC competitor. (C) The histidine-tagged fusion protein fails to interact with single-stranded nucleic acids. EMSA was performed as described for panel A. (Left panel) Lane 1, double-stranded CpG-pos probe; lane 2, single-stranded CpG-pos probe (upper strand); lane 3, single-stranded CpG-pos probe (lower strand); lane 4, RNA CpG-pos probe (upper strand). (Right panel) EMSA was performed with the CpG-pos probe and the following oligonucleotide competitors: lane 1, no competitor; lane 2, homologous competitor; lane 3, single-stranded CpG-pos (upper strand); lane 4, single-stranded CpG-pos (lower strand); and lane 5, RNA CpG-pos (upper strand). (D) A truncated histidine-tagged fusion protein retains DNA-binding activity. EMSA was performed with the CpG-pos probe, the truncated histidine-tagged fusion protein encoded by the 546-bp hCGBP cDNA fragment, and a 2,500-fold molar excess of double-stranded competitor where indicated. Lane 1, peptide encoded by the 720-bp cDNA; lanes 2 to 6, peptide encoded by the 546-bp cDNA [lane 2, no competitor; lane 3, homologous competitor; lane 4, CpG-neg competitor; lane 5, competition with poly(dI-dC); lane 6, CG11 competitor, which contains 11 CpG motifs]. Arrows indicate positions of retarded EMSA complexes.
Additional studies were performed to assess whether a CpG motif is sufficient to provide a binding site for the novel fusion protein. A single-base-pair mutation that creates a CpG motif was introduced into a consensus binding site for NF-κB. The wild-type NF-κB oligonucleotide failed to serve as a binding site for the fusion protein, as it failed to produce a retarded complex in EMSA when used as a probe (Fig. 2B, left panel, lane 2) and also failed to disrupt the complex formed with the CpG-pos probe when used as a competitor (Fig. 2B, right panel, lane 3). Importantly, introduction of a CpG moiety into this sequence produced a significant binding site for the hCGBP fusion protein (Fig. 2B, left panel, lane 3, and right panel, lane 4). Introduction of a GpC motif into the NF-κB oligonucleotide failed to create a binding site for the fusion protein (Fig. 2B, left panel, lane 4, and right panel, lane 5). Competition analysis indicated that the affinity of the fusion protein for the NF-κB-CG oligonucleotide is lower than that for the CpG-pos sequence (Fig. 2B, right panel, lanes 6 and 8). Hence, sequence flanking the CpG motif also appears to contribute to the binding specificity of hCGBP.
EMSA was performed to further characterize the binding specificity of the histidine-tagged hCGBP fusion protein. hCGBP failed to bind to probes corresponding to either strand of the CpG-pos sequence (Fig. 2C, left panel, lanes 2 and 3), and these single-stranded oligonucleotides failed to disrupt the EMSA complex formed with the double-stranded CpG-pos probe when used as competitors (Fig. 2C, right panel, lanes 3 and 4). Analysis of the RNA equivalent of the upper strand of the CpG-pos sequence detected a very weak binding affinity (Fig. 2C, left panel, lane 4, and right panel, lane 5). We concluded that hCGBP requires double-stranded DNA for effective binding.
A truncated version of the hCGBP fusion protein was generated to further delineate the DNA-binding domain. The 3′-most 174 bp were removed from the 720-bp construct, leaving 546 bp of hCGBP cDNA. This truncated fusion protein bound efficiently to the CpG-pos probe and produced a single EMSA complex whose mobility was similar to that of the faster-migrating complex produced by the original hCGBP fusion protein (Fig. 2D, compare lanes 1 and 2). The truncated hCGBP peptide also exhibited sequence-specific DNA-binding activity and was disrupted by homologous competition (lane 3) but not by the CpG-neg oligonucleotide (lane 4). This EMSA complex was also efficiently disrupted by the addition of poly(dI-dC) (lane 5) or an oligonucleotide (CG11) that contains 11 CpG motifs (lane 6). EMSA complexes produced by the longer hCGBP fragment were also disrupted by poly(dI-dC) and CG11 (data not shown). We concluded that the DNA-binding domain of hCGBP resides within a 546-bp fragment of cDNA.
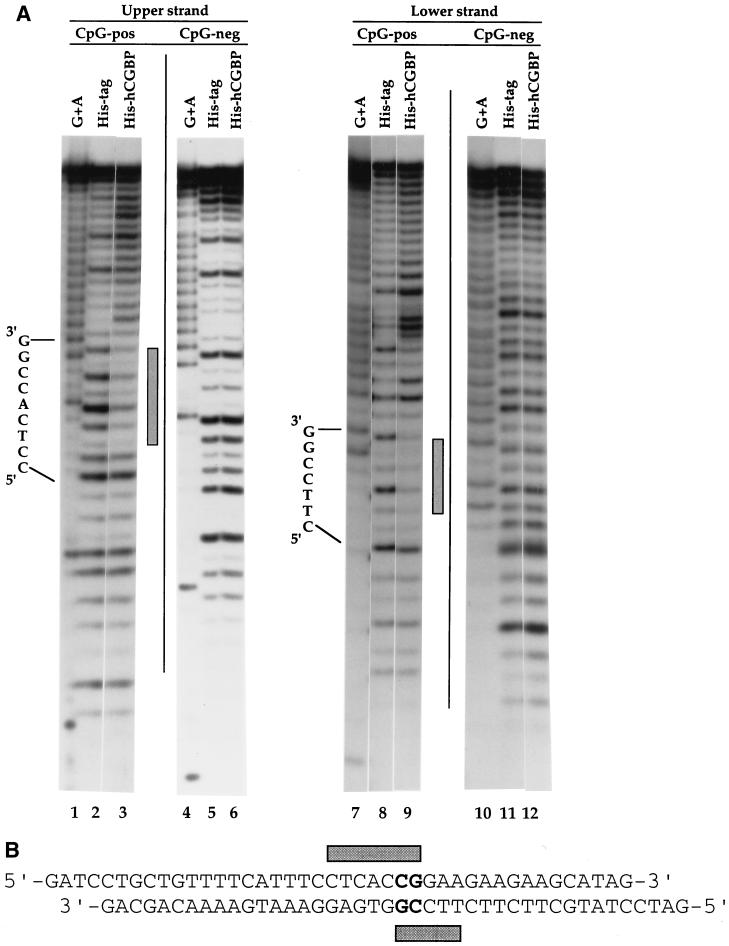
DNase I footprinting was performed to further characterize the DNA-binding properties of the histidine-tagged hCGBP. A protected region of 5 to 7 bp was detected when the CpG-pos sequence was used as a probe (Fig. 3). Interestingly, the footprint was staggered on the two strands of the binding site, with only a central CpG motif in common. No footprint was detected when the CpG-neg sequence, which is similar to CpG-pos but lacks the CpG moiety, was utilized as a probe.
FIG. 3.
DNase I footprinting analysis of the histidine-tagged fusion protein binding site. (A) DNase I footprinting was performed as described in Materials and Methods, using the CpG-pos or CpG-neg oligonucleotide as a probe and 5 μg of either the histidine-tagged hCGBP fusion protein (720-bp cDNA fragment) or the histidine tag (189 aa) alone. Sequence of the region containing the CpG motif is indicated. Shaded bars denote footprinted regions. (B) Schematic representation of the hCGBP footprint produced on the CpG-pos sequence.
Cloning of a full-length hCGBP cDNA.
The 720-bp hCGBP cDNA was used to screen an HL60 λgt11 library to obtain a full-length cDNA clone. The nucleotide sequence of the longest recovered cDNA (2.2 kb) overlaps that of the original 720-bp cDNA clone. A database search revealed a human EST clone (AA325016) whose sequence overlaps that of the 5′ end of the 2.2 kb cDNA clone. An hCGBP cDNA clone of 2.45 kb that includes a complete ORF was constructed by ligating the 5′ end of the Est cDNA clone to the 3′ end of the 2.2-kb λgt11 cDNA clone (GenBank accession no. AF149758). Conceptual translation of the hCGBP cDNA sequence revealed an ORF extending 656 amino acids (aa) downstream from a putative initiation methionine (Fig. 4A). The original 720-bp cDNA clone isolated by ligand screening encodes amino acids 106 to 345, while the truncated version of hCGBP that retained DNA-binding activity encodes amino acids 106 to 287. In addition, a highly basic region (65% basic residues) rich in lysine and arginine residues is found at amino acid positions 321 to 360. Importantly, the 720-bp hCGBP fragment originally isolated by ligand screening contains only a portion of this basic region, and the 546-bp hCGBP fragment that retained DNA-binding activity completely lacks the basic region. Hence, the basic domain is not necessary for the DNA-binding activity of hCGBP. The full-length cDNA also contains 237 bp of 5′ untranslated sequence and 247 bp of 3′ untranslated sequence (data not shown). The ORF does not extend further upstream, as multiple stop codons are present upstream of the initiation codon (data not shown). The predicted polypeptide has a mass of 76 kDa and is basic (pI = 8.66).
FIG. 4.
Deduced amino acid sequence of the novel DNA-binding protein hCGBP. Underlined amino acid residues 27 to 73, 164 to 208, and 485 to 591 identify three cysteine-rich domains. The lysine- and arginine-rich basic region (aa 321 to 360) is indicated by a dashed underline. Heavily underlined residues (aa 430 to 471) denote a predicted coiled-coil domain. Horizontal arrows denote the region encoded by the 720-bp hCGBP cDNA clone recovered from ligand screening. The vertical arrow denotes the 3′ end of the truncated hCGBP cDNA clone that retains DNA-binding activity. This sequence has been deposited in the GenBank database (accession no. AF149758). (B) Diagram of hCGBP showing the relative positions of the identified protein domains.
A database search revealed that the hCGBP cDNA encodes a novel protein. Cysteine-rich domains were identified at the N terminus (aa 27 to 73), central region (aa 164 to 208), and C terminus (aa 485 to 591) of the protein (Fig. 4). Sequence analysis with the ExPASy computer sequence analysis program PAIRCOIL (6) identified a putative coiled-coil domain residing at aa 430 to 471 of hCGBP (Fig. 4). The coiled-coil motif has been found in several DNA-binding proteins, such as leucine zipper factors, and facilitates homo- or heterodimerization (29, 42, 43). The relative positions of these hCGBP domains are illustrated schematically in Fig. 4B.
The amino- and carboxyl-terminal cysteine-rich domains of hCGBP exhibit similarity to the PHD finger (1). The PHD finger is a zinc finger-like motif defined by a unique arrangement of cysteine and histidine residues (Cys4-His-Cys3). PHD fingers have been identified in over 40 proteins, including Drosophila polycomb and trithorax, which have been implicated in chromatin-mediated transcriptional regulation (1). The PHD1 domain of hCGBP exhibits similarity to domains found in heterologous proteins in humans, Schizosaccharomyces pombe, Drosophila melanogaster, and Caenorhabditis elegans (Fig. 5A). The PHD2 domain of hCGBP is imperfect in that it does not conform to the consensus arrangement of cysteine and histidine residues. This sequence exhibits similarity to domains found in proteins identified in humans, D. melanogaster, and C. elegans. Importantly, PHD2 exhibits significant similarity to PHD domains found in HRX and Drosophila trithorax (Fig. 5B).
FIG. 5.
Similarity of the cysteine-rich regions of hCGBP (Hu-CGBP) to conserved motifs. (A) Alignment of hCGBP PHD1 domain (aa 27 to 73) and sequences with highest degrees of similarity. Ye_EST1 and Ye_EST2 (EST clones AL031523 and AL031852, respectively), S. pombe putative transcriptional regulatory proteins of PHD finger family; Ce_EST1, C. elegans EST clone Z81515; Hu_EST1, human EST clone AF044076; p33ING1, candidate tumor suppressor; Hu_EST2, human EST clone AL031852; Hu_RBB2, human retinoblastoma binding protein 2; Dr_PCL_B, Drosophila polycomb-like protein. (B) Alignment of hCGBP PHD2 domain (aa 485 to 591) and sequences with highest degrees of similarity: Dr_EST, putative Drosophila homologue of hCGBP (EST clone AI404379); Hu_EST3, human EST clone AL009172; Ce_EST2, C. elegans EST clone Z82268; Hu_HRX, human trithorax protein (27, 55); and Dr_TRX, Drosophila trithorax protein (36). (C) Alignment of hCGBP CXXC domain (aa 164 to 208) and sequences with highest levels of similarity: MBD1 (MBD1a, aa 174 to 218; MBD1b, aa 223 to 265; MBD1c, aa 336 to 379) (23), HRX (27, 32, 55), and DNMT (DNA methyltransferase) (7). (D) Alignment of hCGBP (aa 374 to 547) with putative Drosophila homologue (Dr_EST; EST clone AI404379) (aa 19 to 190). In all panels, identical amino acids are boxed.
The central cysteine-rich domain of hCGBP is located within the DNA-binding domain (Fig. 4). This domain exhibits high degree of homology to the zinc-binding CXXC domain which is conserved in DNA methyltransferase (9) and HRX (MLL/ALL-1) (19, 27, 32) and is found in three copies within methyl-CpG binding domain protein 1 (MBD1/PCM1) (16, 28) (Fig. 5C). The hCGBP CXXC domain exhibits 50% identity to that of DNA methyltransferase and HRX and 40, 38, and 64% identity to the three CXXC domains of MBD1.
A database search detected a 570-bp Drosophila EST clone (AI404379) that exhibits 49% sequence identity and 69% sequence similarity to hCGBP over 172 aa, including a portion of the PHD2 domain (Fig. 5B and D). This similarity has a smallest-sum probability of 5.4−49, suggesting that this Drosophila EST clone is likely a homologue of hCGBP. However, this conclusion is tentative because the extent of homology beyond the available Drosophila EST sequence is unknown.
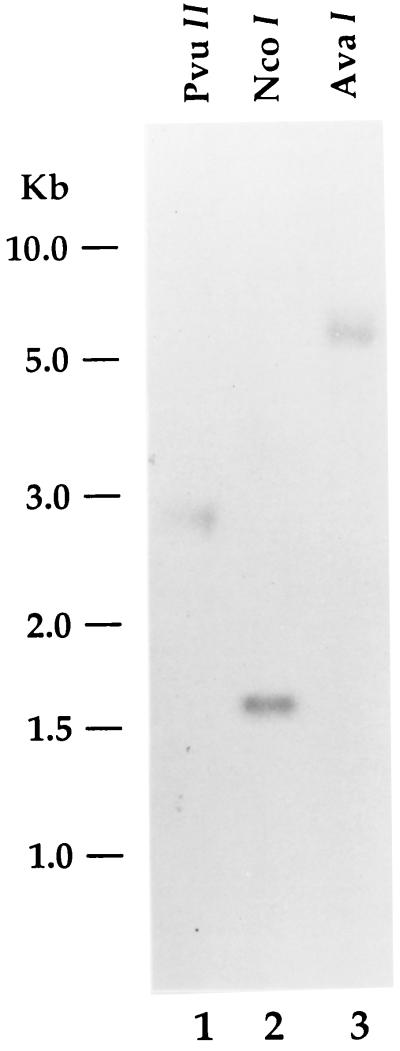
Southern blot analysis was performed with the 720-bp hCGBP cDNA fragment that was recovered by ligand screening as a probe (Fig. 6). Digestion of human genomic DNA with several different restriction enzymes generated a single dominant band, suggesting that hCGBP is encoded by a unique gene.
FIG. 6.
Southern blot analysis of the hCGBP gene. Human genomic DNA was isolated and analyzed as described in Materials and Methods. Samples were digested with PvuII, NcoI, or AvaI as indicated and probed with the 720-bp hCGBP cDNA fragment. Positions of molecular size markers are indicated on left.
Analysis of native hCGBP.
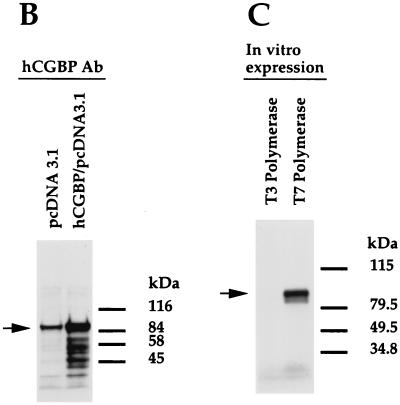
Additional studies were performed to examine the behavior of native hCGBP. Antiserum raised against the GST-hCGBP fusion protein effectively disrupted the EMSA complexes produced by the histidine-tagged hCGBP fusion protein, while preimmune chicken serum had no effect (Fig. 7A). Western blot analysis of a nuclear extract derived from K562 cells transfected with an expression vector containing the full-length hCGBP cDNA detected an abundant band of 88 kDa, slightly greater than hCGBP's predicted mass of 76 kDa (Fig. 7B). A less-intense band of the same size was detected in an extract of cells transfected with empty expression vector. This band presumably corresponds to endogenous hCGBP, because a band of similar size and intensity was apparent for untransfected cells (data not shown). Preimmune rabbit serum failed to detect this protein (data not shown). A band of similar size was produced from the full-length hCGBP cDNA following in vitro transcription-translation (Fig. 7C), and it was also recognized by the hCGBP antiserum (data not shown). These results confirm that the hCGBP cDNA assembled from two cDNA clones encodes an authentic full-length hCGBP protein.
FIG. 7.
Assessment of hCGBP antiserum and expression construct. (A) Chicken antiserum raised against the GST-hCGBP fusion protein (720-bp cDNA fragment) disrupts the histidine-tagged hCGBP EMSA complexes. EMSA was performed as described in Materials and Methods, using the CpG-pos probe and purified histidine-tagged hCGBP fusion protein. One microliter of hCGBP or preimmune serum was added to each of the indicated samples. Arrows indicate positions of retarded EMSA complexes. (B) Rabbit hCGBP antiserum detects an 88-kDa protein in K562 nuclear extract that is overexpressed upon transfection with the full-length hCGBP cDNA expression vector. Transfections and Western blot analysis were performed as described in Materials and Methods. pcDNA3.1 denotes the parental expression vector. The arrow indicates the 88-kDa hCGBP protein. Positions of protein standards are shown on the right. (C) The full-length hCGBP cDNA produces an 88-kDa protein following in vitro transcription-translation. In vitro expression and electrophoresis of hCGBP were performed as described in Materials and Methods, using either the T3 (antisense) or T7 (sense) promoter. The arrow denotes the 88-kDa hCGBP protein. Protein standards are shown on the right.
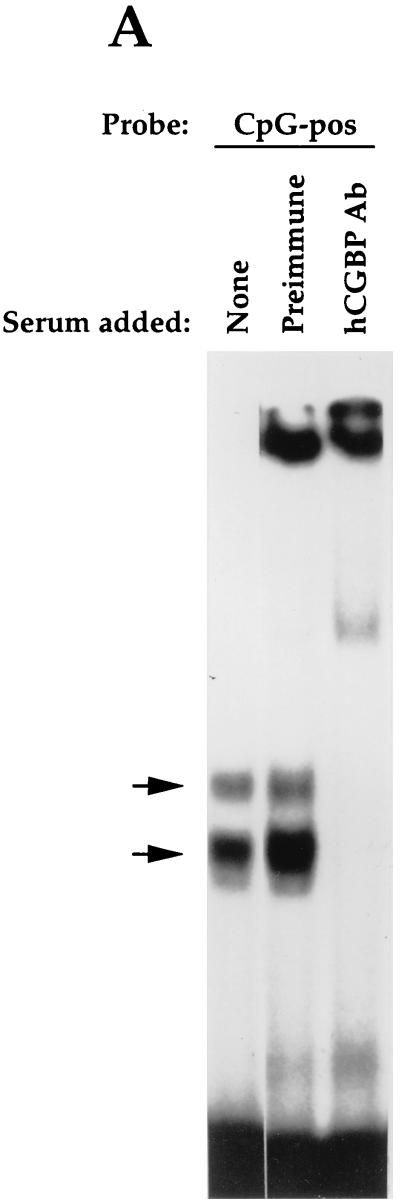
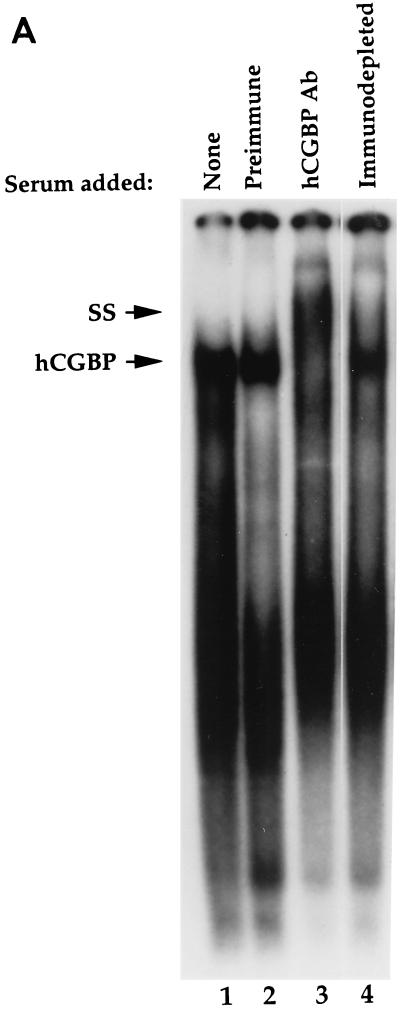
The DNA-binding behavior of native hCGBP was assessed by EMSA. A low-mobility EMSA complex was revealed following incubation of a heparin-fractionated nuclear extract derived from K562 cells with a probe containing a string of 11 CpG motifs (CG11) (Fig. 8A, lane 1). Addition of hCGBP antiserum supershifted the putative hCGBP EMSA complex (lane 3), while preimmune chicken serum had no effect (lane 2). Furthermore, supershifting of the hCGBP complex was abolished following immunodepletion of the antiserum with the histidine-tagged hCGBP fusion protein (lane 4).
FIG. 8.
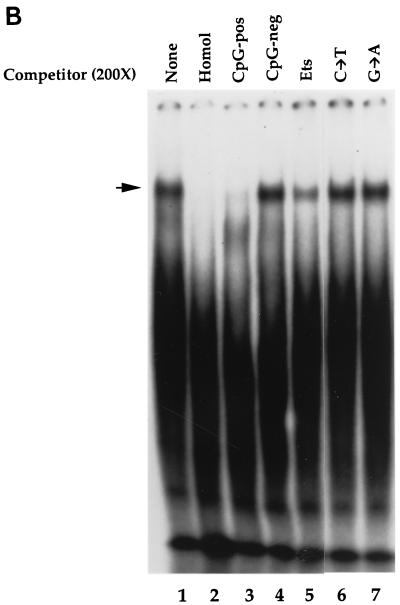
Detection of the endogenous hCGBP DNA-binding activity. (A) Detection of endogenous DNA-binding activity due to hCGBP epitopes. EMSA was performed as described in Materials and Methods, using the CG11 probe and a heparin-fractionated nuclear extract derived from K562 cells. Chicken antiserum raised against the hCGBP fusion protein (720-bp cDNA fragment) (hCGBP Ab), preimmune serum, or immunodepleted antiserum was added where indicated. The arrows indicate the positions of hCGBP and supershifted (SS) complexes. (B) Analysis of the DNA-binding specificity of native hCGBP. EMSA was performed as described above, with the addition of a 200-fold molar excess of the indicated oligonucleotide competitors (as described for Fig. 2). The arrow indicates the position of the hCGBP complex. Homol, homologous competitor.
Native hCGBP exhibits a binding specificity consistent with that observed for the histidine-tagged hCGBP DNA-binding domain (Fig. 8B). The native hCGBP EMSA complex (lane 1) was disrupted by homologous (CG11) competition (lane 2) and by the CpG-pos oligonucleotide (lane 3) but not by the CpG-neg competitor (lane 4). The complex was also significantly disrupted by the Ets oligonucleotide competitor that contains a CpG dinucleotide (lane 5). However, similar to the histidine-tagged hCGBP fusion protein, the competition efficiency of the Ets binding site oligonucleotide for native hCGBP was dramatically lowered by mutation of either nucleotide of the CpG motif (lanes 6 and 7).
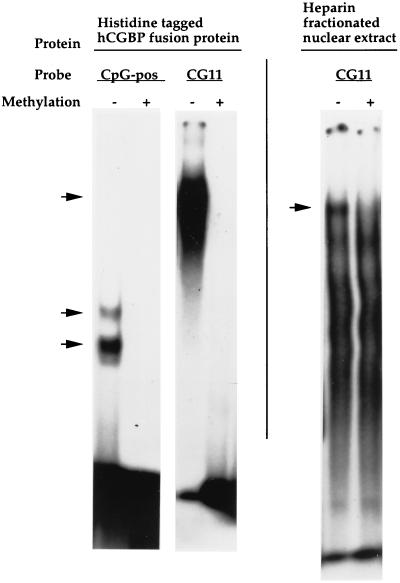
Because CpG is the substrate for DNA methyltransferase, we examined whether hCGBP binds to methylated DNA. These studies were performed with the CpG-pos or CG11 probe. Methylated or unmethylated probes were incubated with the histidine-tagged hCGBP fusion protein or a heparin-fractionated nuclear extract derived from K562 cells to examine native hCGBP. DNA methylation abolished the binding affinity of both the histidine-tagged hCGBP fragment and the native hCGBP for these probes (Fig. 9).
FIG. 9.
hCGBP fails to bind to methylated CpG motifs. EMSA was performed as described in Materials and Methods, using either the CpG-pos or CG11 oligonucleotide as the probe. Purified histidine-tagged hCGBP fusion protein (720-bp cDNA fragment) (0.5 μg) or 3 μg of heparin-fractionated nuclear extract derived from K562 cells was added where indicated. Probes were methylated by SssI methylase as described in Materials and Methods. The arrows indicate the positions of retarded EMSA complexes.
Ubiquitous expression of hCGBP mRNA.
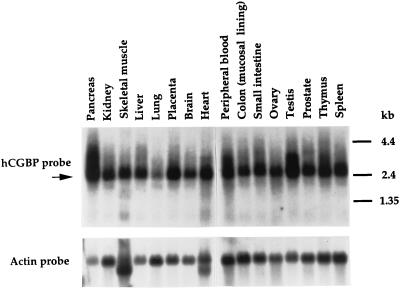
Northern blot analysis was performed with the 720-bp hCGBP cDNA fragment as a probe to determine the distribution of hCGBP expression. hCGBP mRNA is expressed predominantly as a 2.6-kb transcript in a wide variety of human tissues (Fig. 10). hCGBP is highly expressed in the pancreas, placenta, heart, testis, and spleen tissue, while the lowest level of expression is in the lungs. Transcripts of larger size were detected in tissue of the pancreas, peripheral blood, and testis, possibly indicating alternative splicing events. Consistent with this distribution, hCGBP cDNA clones were identified as both human and mouse Est clones derived from macrophages, neuronal precursors, liver stem cells, and tissue of the heart, testis, brain, lung, uterus, mammary gland, pineal gland, cerebellum, myotube, lymph node, and embryon (data not shown). These results indicate that hCGBP is ubiquitously expressed.
FIG. 10.
hCGBP is ubiquitously expressed. Northern blots were hybridized with a radiolabeled 720-bp hCGBP cDNA probe (top panel) as described in Materials and Methods. The lower panel shows hybridization with human actin cDNA as a control for RNA loading and integrity. The arrow indicates the 2.6-kb transcript corresponding to hCGBP. The positions of molecular size markers are indicated on the right.
hCGBP trans-activates promoters containing CpG motifs.
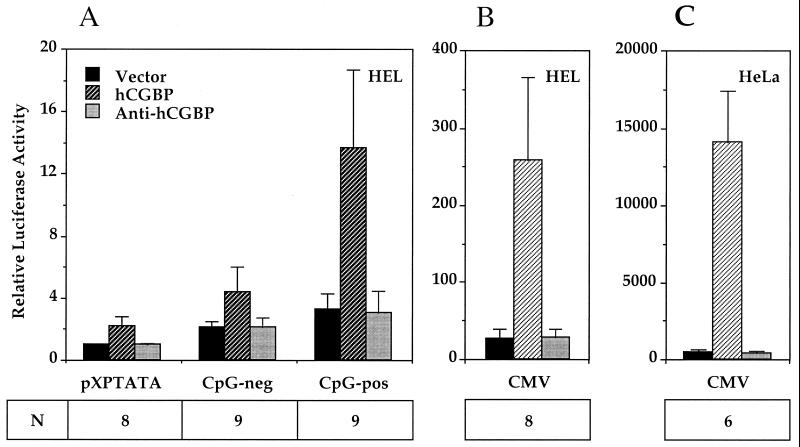
Cotransfection experiments were performed to assess whether hCGBP is a transcriptional regulator. The hCGBP expression vector was cotransfected into HEL or HeLa cells along with putative promoter targets linked to a luciferase reporter gene. Dimers of either the CpG-pos or CpG-neg oligonucleotide were introduced upstream of a minimal TATA-box promoter linked to the luciferase reporter gene. hCGBP bound efficiently to the CpG-pos sequence, which contains a CpG motif, but failed to interact with the CpG-neg oligonucleotide sequence, which lacks a CpG motif (Fig. 2 and 8). Fold activation was calculated following subtraction of the background level of luciferase expression generated by each reporter construct upon cotransfection with the empty expression vector. Comparison of the CpG-pos and CpG-neg reporter plasmids reveals a 4.5-fold trans-activation produced by hCGBP via the CpG motif within the CpG-pos oligonucleotide (P < 0.001) (Fig. 11A). The background expression observed with the minimal TATA-box promoter is presumably due to a cryptic hCGBP binding site(s) in the vector backbone. Inspection of the nucleotide sequence surrounding the TATA box revealed several CpG motifs (data not shown). Introduction of the antisense hCGBP expression vector had no effect on reporter gene expression. We concluded that hCGBP trans-activates transcription via binding to CpG motifs.
FIG. 11.
hCGBP trans-activates promoters containing CpG motifs. HEL or HeLa cells were cotransfected with CMV promoter-luciferase (CMV), CpG-pos dimer TATA promoter-luciferase, CpG-neg dimer TATA promoter-luciferase, or TATA promoter-luciferase (pXPTATA) reporter plasmids and expression vectors containing sense (hCGBP) or antisense (anti-hCGBP) hCGBP or vector alone (vector) as described in Materials and Methods. Luciferase activity is presented relative to that of the pXPTATA-luciferase vector. Data (means ± standard deviations) presented are from multiple experiments performed in duplicate, using four independent plasmid preparations. N denotes the number of independent experiments performed.
During preliminary experiments it was noted that expression of an internal control vector (CMV–β-gal) was consistently higher in cells receiving the hCGBP expression vector. Inspection of the CMV promoter revealed in excess of 30 CpG motifs; hence, we hypothesized that hCGBP trans-activates the CMV promoter via these elements. EMSA experiments demonstrated that the histidine-tagged hCGBP fusion protein bound each of two analyzed CMV promoter fragments that contain 12 or 13 CpG motifs (data not shown). Further cotransfection experiments were performed with HEL or HeLa cells, the CMV-luciferase reporter plasmid, and hCGBP expression vectors. CMV promoter activity is induced 9.4-fold in HEL cells by hCGBP and 29-fold in HeLa cells (Fig. 11B and C). Antisense hCGBP had no effect on reporter gene expression.
DISCUSSION
In this article we report the cloning of hCGBP, a novel mammalian CpG-binding protein. This factor is widely expressed, binds specifically to unmethylated CpG motifs, and trans-activates promoters that contain CpG motifs. The affinity of hCGBP for unmethylated CpG motifs, which is characteristic of a typical CpG island, is consistent with hCGBP functioning as an activator of genes residing within CpG islands. This is also consistent with the ubiquitous pattern of hCGBP expression. Identification of natural target genes of hCGBP will be of great interest.
We hypothesize that the CXXC domain of hCGBP that is conserved in DNA methyltransferase, MBD1, and HRX contributes to the observed DNA-binding activity. A fragment of hCGBP which contains the CXXC domain (aa 106 to 345) binds to distinct oligonucleotides that contain CpG motifs. Mutation of either nucleotide within the CpG motif abolished hCGBP's affinity for target binding sites. Like HRX, hCGBP contains a stretch of basic residues (aa 321 to 360) adjacent to the CXXC domain. However, a shorter hCGBP fragment (aa 106 to 287) lacks the basic domain but maintains strong DNA-binding activity, indicating that the flanking basic region is not required for DNA-binding activity. The fragment of hCGBP (aa 106 to 345) that contains a portion of the basic region formed an additional, lower-mobility EMSA complex, suggesting that the basic domain adjacent to the CXXC domain may facilitate dimerization. Consistent with the DNA-binding behavior of the CXXC domain of HRX (51), hCGBP EMSA complexes are efficiently disrupted by the addition of poly(dI-dC). Additional studies, utilizing truncated and mutated hCGBP fragments, are required to directly assess the contribution of the CXXC domain to the observed DNA-binding activity.
The hCGBP sequence also contains two PHD finger-like domains, which have been identified in more than 40 proteins, many of which are implicated in chromatin-mediated transcriptional control (1). The function of the PHD domain remains unclear, but its significance in vivo has been established by studying PHD-containing proteins implicated in human disease. For example, mutation of a PHD domain within the ATRX gene results in X-linked α-thalassemia/mental retardation syndrome (ATR-X) (26), and mutations that affect PHD domains within the AIRE gene cause autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED) (38). The HRX gene has several PHD domains and is involved in chromosomal translocations associated with acute leukemia (27), while in Drosophila the removal of a PHD finger from the trithorax gene is lethal (36).
The function of the PHD fingers in hCGBP remains to be determined. Importantly, fragments of hCGBP that lack both PHD domains exhibit a DNA-binding specificity similar to that shown by native hCGBP. It is intriguing that hCGBP contains several domains implicated in protein-protein interactions, such as the PHD finger domains and coiled-coil domain (42). Interestingly, in EMSA, native hCGBP exhibits an unusually low mobility for an 88-kDa protein. The hCGBP complex migrates to a position above that of the heterotrimeric CCAAT-box-binding factor CP1 (data not shown), which has a mass of approximately 120 kDa (35). Although it is difficult to draw conclusions based on mobility in native gels, this observation suggests that other proteins may be present in the hCGBP EMSA complex.
Interestingly, three genomic clones (GenBank accession no. AJ132338, AJ132339, and AJ236590) derived from human chromosome 18 contain both hCGBP and MBD1 sequences. This relationship was also recently noted by Cross et al. (15). Examination of these sequences indicated that the genes encoding hCGBP and MBD1 are located within approximately 800 bp of each other in the human genome. Examination of a 700-bp genomic fragment that surrounds the 5′ flanking region of the gene encoding hCGBP revealed in excess of 60 CpG motifs, and a 500-bp region upstream of the MBD1 gene was found to contain in excess of 50 CpG motifs (data not shown). Hence, the genes encoding hCGBP and MBD1 both reside within CpG islands.
The relationship between the genes encoding hCGBP and MBD1 is intriguing. These tightly linked genes each contain CXXC domains. MBD1 isoforms bind to methylated or unmethylated CpG motifs and function as transcriptional repressors (16, 23), while hCGBP binds to unmethylated CpG motifs and functions as a transcriptional activator. It will be of interest to determine if these two tightly linked activities are coordinately regulated by common control elements. In this regard, it is interesting that the gene encoding hCGBP resides within a CpG island, suggesting that perhaps this gene is autoregulated.
Although a number of proteins that bind to methylated or hemimethylated CpG motifs have been described (7, 16, 28, 31), hCGBP is to our knowledge the first identified factor that exhibits a specific affinity for unmethylated CpG motifs. Fisscher et al. (22) described a CpG-binding protein (CGBP-1), derived from tobacco nuclear extract, that binds with higher affinity to oligonucleotides containing increasing numbers of CpG dinucleotides, a feature similar to that of hCGBP. However, CGBP-1 has not been cloned; hence, the relationship between CGBP-1 and hCGBP remains to be determined. The cloning of hCGBP will permit further dissection of the role of this CpG-binding transcription factor in the regulation of genes located within CpG islands.
ACKNOWLEDGMENTS
We are grateful to Robert Hromas for providing the HL60 λgt11 cDNA library and to Nancy Andrews and Harinder Singh for helpful discussions regarding protein purification and ligand screening, respectively. We also acknowledge the Cell Culture Center and the National Center for Research Resources for providing large-scale tissue culture services at a subsidized rate.
This work was supported by Public Health Service grant CA58997 from the National Cancer Institute, awarded to D.G.S.; an Arthritis Foundation postdoctoral fellowship, awarded to K.S.V., an American Heart Association postdoctoral fellowship, awarded to D.L.C.; and a GAANN fellowship from the Department of Education, awarded to B.M.J.
REFERENCES
- 1.Aasland R, Gibson T J, Stewart F. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 2.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antequera F, Macleod D, Bird A. Specific protection of methylated CpGs in mammalian nuclei. Cell. 1989;58:509–517. doi: 10.1016/0092-8674(89)90431-5. [DOI] [PubMed] [Google Scholar]
- 5.Baylin S B. Tying it all together: epigenetics, genetics, cell cycle, and cancer. Science. 1997;277:1948–1949. doi: 10.1126/science.277.5334.1948. [DOI] [PubMed] [Google Scholar]
- 6.Berger B, Wilson D, Wolf E, Tonchev T, Milla M, Kim P S. Predicting coiled coils by use of pairwise correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bestor T H. Cloning of a mammalian DNA methyltransferase. Gene. 1988;74:9–12. doi: 10.1016/0378-1119(88)90238-7. [DOI] [PubMed] [Google Scholar]
- 8.Bestor T H. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bestor T H, Verdine G L. DNA methyltransferases. Curr Opin Cell Biol. 1994;6:380–389. doi: 10.1016/0955-0674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya S K, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 11.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 12.Brown T. Analysis of DNA sequences by blotting and hybridization. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1993. pp. 2.9.1–2.9.15. [Google Scholar]
- 13.Chuang L S, Ian H I, Koh T W, Ng H H, Xu G, Li B F. Human DNA-(cytosine-5)methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 14.Cowell I G, Hurst H C. Cloning transcription factors from a cDNA expression library. In: Latchman D S, editor. Transcription factors: a practical approach. Oxford, United Kingdom: IRL Press; 1993. pp. 105–124. [Google Scholar]
- 15.Cross S H, Clark V H, Bird A P. Isolation of CpG islands from large genomic clones. Nucleic Acids Res. 1999;27:2099–2107. doi: 10.1093/nar/27.10.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cross S H, Meehan R R, Nan X S, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat Genet. 1997;16:256–279. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 17.Delgado S, Gomez M, Bird A, Antequera F. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 1998;17:2426–2435. doi: 10.1093/emboj/17.8.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domer P H, Fakharzadeh S S, Chen C S, Jockel J, Johansen L, Silverman G A, Kersey J H, Korsmeyer S J. Acute mixed-lineage leukemia t(4;11)(q21;q23) generates an MLL-AF4 fusion product. Proc Natl Acad Sci USA. 1993;90:7884–7888. doi: 10.1073/pnas.90.16.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eklund E A, Skalnik D G. Characterization of a gp91-phox promoter element that is required for interferon gamma-induced transcription. J Biol Chem. 1995;270:8267–8273. doi: 10.1074/jbc.270.14.8267. [DOI] [PubMed] [Google Scholar]
- 21.Ferguson-Smith A C, Sasaki H, Cattanach B M, Surani M A. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature. 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- 22.Fisscher U, Weisbeek P, Smeekens S. A tobacco nuclear protein that preferentially binds to unmethylated CpG-rich DNA. Eur J Biochem. 1996;235:585–592. doi: 10.1111/j.1432-1033.1996.00585.x. [DOI] [PubMed] [Google Scholar]
- 23.Fujita N, Takebayashi S-I, Okumura K, Kudo S, Chiba T, Saya H, Nakao M. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol. 1999;19:6415–6426. doi: 10.1128/mcb.19.9.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 25.Gardiner-Garden M, Frommer M. Transcripts and CpG islands associated with the pro-opiomelanocortin gene and other neurally expressed genes. J Mol Endocrinol. 1994;12:365–382. doi: 10.1677/jme.0.0120365. [DOI] [PubMed] [Google Scholar]
- 26.Gibbons R J, Bachoo S, Picketts D J, Aftimos S, Asenbauer B, Bergoffen J, Berry S A, Dahl N, Fryer A, Keppler K, Kurosawa K, Levin M L, Masuno M, Neri G, Pierpont M E, Slaney S F, Higgs D R. Mutations in transcriptional regulator of ATRX establish the functional significance of a PHD-like domain. Nat Genet. 1997;17:146–148. doi: 10.1038/ng1097-146. [DOI] [PubMed] [Google Scholar]
- 27.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 28.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landschulz W H, Johnson P F, McKnight S L. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989;243:1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- 30.Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 31.Lewis J D, Meehan R R, Henzel W J, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 32.Ma Q, Alder H, Nelson K K, Chatterjee D, Gu Y, Nakamura T, Canaani E, Croce C M, Siracusa L D, Buchberg A M. Analysis of the murine ALL-1 gene reveals conserved domains with human ALL-1 and identifies a motif shared with DNA methyltransferase. Proc Natl Acad Sci USA. 1993;90:6350–6354. doi: 10.1073/pnas.90.13.6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macleod D, Ali R R, Bird A. An alternative promoter in the mouse major histocompatibility complex class II I-Aβ gene: implications for the origin of CpG islands. Mol Cell Biol. 1998;18:4433–4443. doi: 10.1128/mcb.18.8.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macleod D, Charlton J, Mullins J, Bird A P. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 35.Maity S N, Sinha S, Ruteshouser E C, de Crombrugghe C B. Three different polypeptides are necessary for DNA binding of the mammalian heteromeric CCAAT binding factor. J Biol Chem. 1992;267:16574–16580. [PubMed] [Google Scholar]
- 36.Mazo A M, Huang D H, Mozer B A, Dawid I B. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci USA. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller S A, Dykes D D, Polesky H F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagamine K, Peterson P, Scott H S, Kudoh J, Minoshima S, Heino M, Krohn K J, Lalioti M D, Mullis P E, Antonarakis S E, Kawasaki K, Asakawa S, Ito F, Shimizu N. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–398. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 39.Neufeld E J, Skalnik D G, Lievens P M-J, Orkin S H. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat Genet. 1992;1:50–55. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- 40.Newburger P E, Skalnik D G, Hopkins P J, Eklund E A, Curnutte J T. Mutations in the promoter region of the gene for gp91-phox in X-linked chronic granulomatous disease with decreased expression of cytochrome b558. J Clin Investig. 1994;94:1205–1211. doi: 10.1172/JCI117437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nordeen S K. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques. 1988;6:454–458. [PubMed] [Google Scholar]
- 42.O'Shea E K, Klemm J D, Kim P S, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 43.O'Shea E K, Ruthkowski R, Kim P S. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science. 1989;243:538–542. doi: 10.1126/science.2503872. [DOI] [PubMed] [Google Scholar]
- 44.Pfeifer G P, Tanguay R L, Steigerwald S D, Riggs A D. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 1990;4:1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- 45.Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce C M, Canaani E. Domains with transcriptional regulatory activity within the ALL-1 and AF4 proteins involved in acute leukemia. Proc Natl Acad Sci USA. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riggs A D, Pfeifer G P. X-chromosome inactivation and cell memory. Trends Genet. 1992;8:169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 48.Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi B-Z, Cedar H. DNA methylation represses transcription in vivo. Nat Genet. 1999;22:203–206. doi: 10.1038/9727. [DOI] [PubMed] [Google Scholar]
- 49.Singh H, LeBowitz J H, Baldwin A S, Jr, Sharp P A. Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell. 1988;52:415–423. doi: 10.1016/s0092-8674(88)80034-5. [DOI] [PubMed] [Google Scholar]
- 50.Skalnik D G, Strauss E C, Orkin S H. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J Biol Chem. 1991;266:16736–16744. [PubMed] [Google Scholar]
- 51.Slany R K, Lavau C, Cleary M L. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol Cell Biol. 1998;18:122–129. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Somma M P, Pisano C, Lavia P. The housekeeping promoter from the mouse CpG island HTF9 contains multiple protein-binding elements that are functionally redundant. Nucleic Acids Res. 1991;19:2817–2824. doi: 10.1093/nar/19.11.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tazi J, Bird A. Alternative chromatin structure at CpG islands. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- 54.Teerawatanasuk N, Skalnik D G, Carr L G. CCAAT displacement protein (CDP/Cut) binds a negative regulatory element in the human tryptophan hydroxylase gene. J Neurochem. 1999;72:29–39. doi: 10.1046/j.1471-4159.1999.0720029.x. [DOI] [PubMed] [Google Scholar]
- 55.Tkachuk D C, Kohler S, Cleary M L. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 56.Zardo G, Caiafa P. The unmethylated state of CpG islands in mouse fibroblasts depends on the poly(ADP-ribosyl)ation process. J Biol Chem. 1998;273:16517–16520. doi: 10.1074/jbc.273.26.16517. [DOI] [PubMed] [Google Scholar]
- 57.Zardo G, D'Erme M, Reale A, Strom R, Perilli M, Caiafa P. Does poly(ADP-ribosyl)ation regulate the DNA methylation pattern? Biochemistry. 1997;36:7937–7943. doi: 10.1021/bi970241s. [DOI] [PubMed] [Google Scholar]
- 58.Zeleznik-Le N J, Harden A M, Rowley J. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcription repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci USA. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]