Abstract
H1 histones bind to linker DNA and nucleosome core particles and facilitate the folding of chromatin into a more compact structure. Mammals contain seven nonallelic subtypes of H1, including testis-specific subtype H1t, which varies considerably in primary sequence from the other H1 subtypes. H1t is found only in pachytene spermatocytes and early, haploid spermatids, constituting as much as 55% of the linker histone associated with chromatin in these cell types. To investigate the role of H1t in spermatogenesis, we disrupted the H1t gene by homologous recombination in mouse embryonic stem cells. Mice homozygous for the mutation and completely lacking H1t protein in their germ cells were fertile and showed no detectable defect in spermatogenesis. Chromatin from H1t-deficient germ cells had a normal ratio of H1 to nucleosomes, indicating that other H1 subtypes are deposited in chromatin in place of H1t and presumably compensate for most or all H1t functions. The results indicate that despite the unique primary structure and regulated synthesis of H1t, it is not essential for proper development of mature, functional sperm.
The histones are a family of basic proteins that are involved in organizing the DNA in the nuclei of eukaryotic cells into a compact structure called chromatin. There are five major classes of histones, the core histones H2A, H2B, H3, and H4 and the linker histone H1. Two molecules of each of the core histones constitute the protein octamer of the nucleosome core particle. H1 histones bind to DNA in the nucleosome core particle and to the linker DNA between nucleosomes. These interactions are thought to facilitate the folding of nucleosomes into the 30-nm chromatin fiber and higher-order chromatin structures (39, 42). Interactions between histones and DNA would be expected to modulate gene activity, and recent evidence clearly shows that both the core histones and H1 can have a profound effect on transcription (reviewed in references 18, 43, and 44).
Among the five classes of histones, the H1 histones exhibit the most diversity. For mice seven H1 subtypes have been described (24, 25), including the “somatic” subtypes H1a through H1e, the replacement subtype H1o, and the testis-specific linker histone H1t. These seven H1 subtypes are also present in humans, and the genomic organization of the genes encoding the H1 subtypes in humans appears to be very similar to that in mice (12, 40).
Within the H1 family of proteins, the testis-specific H1t subtype is unique in that it is the only member exhibiting a truly tissue-specific pattern of expression. Although the other six H1 subtypes display distinct patterns of expression during differentiation and development, they are all expressed in numerous tissues (24–26). On the other hand, the H1t gene is transcribed exclusively in mid- and late-pachytene spermatocytes (10, 13, 16), and H1t protein is found only in pachytene spermatocytes and early, haploid spermatids (11, 17, 29), in which it constitutes up to 55% of the total H1 linker histone in chromatin (29). Moreover, although H1t protein has the tripartite domain organization typical of metazoan linker histones, its primary sequence is highly divergent from that of the other mammalian H1s (7, 10). It has been reported that in vitro H1t binds less tightly than somatic H1 variants to H1-depleted oligonucleosomes, rendering them more sensitive to nucleases (8). It is possible that this property serves to maintain meiotic chromatin in a relatively decondensed state, facilitating meiotic events such as recombination and/or the chromosomal protein transitions that occur during spermatogenesis.
To investigate the role of H1t in mammalian spermatogenesis, we generated a null mutation in the H1t gene by homologous recombination in mouse embryonic stem (ES) cells and then produced mice that transmitted the mutated allele. Surprisingly, mice homozygous for the null mutation were fertile and exhibited no detectable defect in spermatogenesis. The results indicate that H1t is dispensable for normal spermatogenesis. Analysis of chromatin from H1t-deficient germ cells showed that other H1 subtypes are deposited in chromatin in place of H1t and very likely are able to compensate for the lost functions of H1t.
MATERIALS AND METHODS
Disruption of the H1t gene in ES cells and generation of chimeric mice.
Clone WW6 ES cells (21) derived from 129J/Sv mice were cultured according to published procedures (32) except that the medium was supplemented with 1,000 U of leukemia inhibitory factor (GIBCO-BRL) per ml. Twenty-five to 50 μg of NotI-linearized targeting vector was transfected into 2 × 107 to 4 × 107 ES cells by electroporation in phosphate-buffered saline at 400 V and 250 μF using a Bio-Rad Gene Pulser. After incubation at 4°C for 10 min, the cells were distributed to 10 10-cm tissue culture dishes containing G418-resistant mouse embryo fibroblasts inactivated with 4,000 rads of gamma irradiation. The transfected ES cells were cultured at 37°C in a 5% CO2 humidified incubator. After 24 h, 200 μg of active G418 (Geneticin; GIBCO-BRL) per ml was added, and after 48 h, 2 μM ganciclovir (Syntex) was added. The medium was changed with fresh medium containing G418 and ganciclovir every 1 or 2 days. Surviving ES cell clones were picked after 9 to 10 days, transferred to duplicate 48-well plates, and grown in the presence of G418 for 3 days, with a medium change every day. One of the two plates of clones was frozen and the other was used for preparing DNA as described previously (23). Five ES clones containing a modified H1t allele were identified among 200 doubly resistant clones (i.e., resistant to both G418 and ganciclovir) analyzed by Southern blot hybridization. Four clones (T3-1, T3-12, T4-21, and T4-24) were injected into C57BL/6 recipient blastocysts, and the blastocysts were transferred to CD-1 pseudopregnant females to generate chimeric mice as described previously (32). Mouse tail DNA was prepared as described previously (20).
Histological analysis of mouse testes.
Mice were sacrificed by cervical dislocation and then perfused with phosphate-buffered saline followed by neutral buffered 10% formalin. The testes were fixed in fresh neutral buffered 10% formalin and embedded in paraffin wax. Tissue sections (5 μm) were stained with hematoxylin and eosin and examined by light microscopy. For electron microscopic analysis the testes were prefixed with 4% glutaraldehyde and postfixed with 1% osmium tetroxide (OsO4). The specimens were stained with uranyl nitrate and lead acetate and examined by electron microscopy.
Epididymal sperm numbers.
Epididymides were removed from 12- to 14-week-old males under sterile conditions and placed in a dish containing 5 ml of Dulbecco modified Eagle medium (DMEM) with 10% fetal calf serum. Sperm were allowed to disperse into the medium for 1 h at 32°C. Sperm mobility and swimming activity were examined by phase-contrast microscopy. Sperm numbers were determined by counting with a hemocytometer.
Preparation and analysis of histones.
Mice were sacrificed by cervical dislocation, and the testes were excised and immediately rinsed with DMEM. Seminiferous epithelial cells were prepared as originally described by Romrell et al. (33) and later modified by Bellve et al. (2, 3). The contents of the excised testes were squeezed gently through a small incision in the tunica albuginea. The decapsulated testes were incubated in 15 ml of DMEM containing 0.5 mg of collagenase per ml for 15 min at 33°C in 5% CO2 in air in a shaking water bath operated at 120 cycles/min. The dispersed seminiferous tubules were isolated after being allowed to sediment in DMEM for 2 to 3 min. The supernatant was decanted and the seminiferous tubules were incubated as described above in 15 ml of DMEM containing 0.5 mg of trypsin per ml and 20 μg of DNase per ml for 10 min. Cell aggregates that remained after trypsin dissociation were sheared gently by repeated pipetting up to 30 times with a Pasteur pipette. After additional shaking for 5 min, 1 ml of serum was added to block trypsin activity, and the cell aggregates were dispersed by pipetting 30 times. The dispersed seminiferous cells were washed twice by centrifugation at 200 × g for 5 min and resuspended in DMEM containing 1 μg of DNase per ml and 0.5% (wt/vol) bovine serum albumin. The final cell pellet was suspended in 10 ml of DMEM containing 0.5% bovine serum albumin and filtered through a Nitex filter cloth (80 mesh), and the cell concentration was adjusted to 107/ml.
Chromatin was prepared from nuclei of isolated germ cells. The cells were placed in sucrose buffer comprised of 0.3 M sucrose, 15 mM NaCl, 10 mM HEPES (pH 7.9), 2 mM EDTA, and 0.5 mM phenylmethylsulfonyl fluoride. The cells were homogenized by 10 strokes of a Teflon-glass dounce homogenizer (B pestle) at 4°C. The homogenate was filtered through Nitex filter cloth (80 mesh) and centrifuged at 1,250 × g for 10 min. The nuclear pellet was resuspended in 2 ml of sucrose buffer containing 0.5% Nonidet P-40 to lyse contaminating erythrocytes and the mixture was centrifuged as described above. The pellet was resuspended in 2 ml of high-salt buffer containing 0.35 M KCl, 10 mM Tris-HCl (pH 7.2), 5 mM MgCl2, and 0.5 mM phenylmethylsulfonyl fluoride, and the released chromatin was sedimented in a bench top microcentrifuge. The chromatin was resuspended in 1 ml of 0.2 N sulfuric acid. The sample was homogenized by using a microcentrifuge tube pestle, incubated on ice overnight, and centrifuged for 30 min at 12,000 rpm in a benchtop microcentrifuge. The pellet was reextracted with acid and centrifuged. The two supernatants were pooled. Histone proteins were precipitated by the addition of 3 volumes of 100% ethanol, centrifuged, and dried under vacuum.
High-performance liquid chromatography (HPLC) analysis was carried out on 50 to 200 μg of histone protein extract dissolved in 1 ml of water containing 0.1% trifluoroacetic acid. The protein mixtures were fractionated on a 0.46- by 25-cm, Vydac 300-Å, 5-mm C18 reverse-phase column as described previously (4, 35, 41), using a Hewlett-Packard 1090 HPLC system. Proteins were eluted with a linear gradient of increasing concentrations of acetonitrile: 0 to 5% for 1 min, 5 to 25% for 10 min, 25 to 30% for 15 min, 30 to 35% for 20 min, 35 to 40% for 20 min, 40 to 43% for 10 min, 43 to 55% for 50 min, 55 to 90% for 5 min, and 90 to 100% for 20 min. The effluent was monitored at 214 nm, and peak areas were determined with a Hewlett-Packard peak integrator program. The percentage of each H1 subtype in the samples was calculated by measuring the ratio of each peak area to the total area of all H1 peaks after normalizing for differences in the number of amino acids in the individual H1 species. The ratio of total H1 to nucleosomes was calculated by dividing the total area of all H1 peaks by one-half the area of the H2b peaks after normalizing for differences in the number of amino acids in H2B and an average-sized H1 molecule.
RESULTS
Generation of ES cells with a disrupted H1t gene.
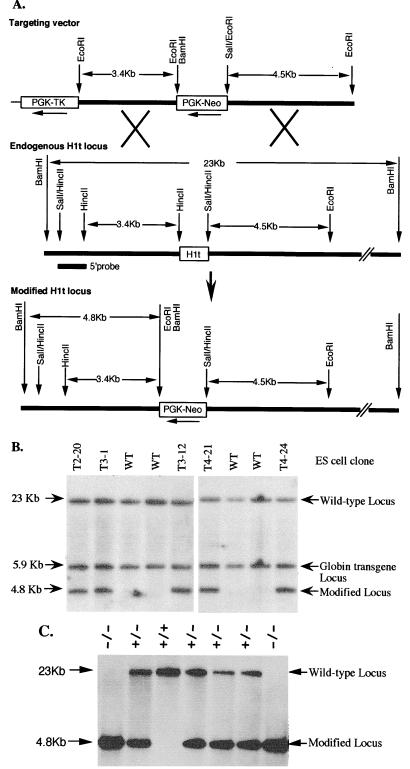
The mouse H1t gene encodes a protein of 207 amino acids. It does not contain introns (10). H1t genomic clones were isolated from a mouse strain 129J/Sv genomic library. To generate an H1t-targeting vector (Fig. 1A), a 3.4-kb HincII fragment, the 3′ end of which lies 285 bp upstream of the H1t start codon, was inserted into the EcoRI site of the pPNT vector (37). pPNT contains a PGK-Neo cassette that allows positive selection for transfectants with G418 and a PGK-TK cassette that allows enrichment of transfectants which have undergone homologous recombination and have lost the PGK-TK cassette by selection with ganciclovir (5). Next, a 4.5-kb SalI/EcoRI fragment beginning 110 bp downstream of the H1t stop codon was subcloned into the vector. Homologous recombination between the targeting vector and the endogenous H1t locus results in the generation of a modified H1t allele in which the entire H1t coding region together with 285 bp of upstream sequence and 110 bp of downstream sequence is replaced by the PGK-Neo cassette (Fig. 1A). The targeting vector was linearized by NotI digestion and electroporated into WW6 ES cells (21), and cells resistant to G418 and ganciclovir were selected. The double selection resulted in a 3.6-fold enrichment of G418-resistant colonies. DNA from 200 doubly resistant clones was pooled (two clones per pool), digested with BamHI, and analyzed by Southern blot hybridization with a 5′ probe lying outside of the targeting construct (Fig. 1A). Five pools gave a 4.8-kb BamHI hybridizing fragment expected from the modified allele (Fig. 1A), and on further analysis only one member of each pool showed the 4.8-kb BamHI fragment (Fig. 1B).
FIG. 1.
Targeted disruption of the H1t gene in mouse ES cells and mice. (A) Homologous recombination strategy in ES cells. The H1t targeting vector (top) was constructed by inserting a 3.4-kb HincII fragment lying 285 bp upstream of the H1t start codon into the pPNT vector (37) and then inserting a 4.5-kb blunt-ended SalI/EcoRI fragment beginning 110 bp downstream of the stop codon. A homologous recombination event (X's) between the targeting vector and the endogenous H1t locus results in production of a modified H1t locus in which a 1.0-kb fragment, including the entire H1t coding sequence along with 285 bp of 5′ noncoding sequence and 110 bp of 3′ noncoding sequence, is removed. A 1.1-kb HincII fragment (5′ probe) lying outside of the targeting construct was used as a probe for Southern blotting analysis of DNA from ES cells and mice. (B) Identification of ES cell clones containing the modified H1t allele. ES cell DNA (10 μg) was digested with BamHI and blot hybridized with the 5′ probe shown in panel A. The expected positions of the hybridizing fragments from the unmodified (wild-type [WT]) and modified H1t loci and their respective sizes are indicated. The common 5.9-kb hybridizing band (globin transgene locus) is due to hybridization between contaminating plasmid sequences in the 5′ probe and multiple copies of a globin transgene present in the parental WW6 ES cells (21). (C) Genotype analysis of offspring from parents heterozygous for the modified H1t allele. Ten micrograms of tail DNA from offspring was digested with BamHI and blot hybridized with the 5′ probe shown in panel A. Other details are as described for panel B. The deduced genotype of each mouse is indicated above each lane.
Generation of H1t null mice.
Two ES cell clones (T3-12 and T4-24) containing the modified H1t allele were injected into C57BL/6 blastocysts to generate chimeric mice. Both cell lines gave rise to chimeras ranging in chimerism from 40 to 99% as judged by coat color. Male chimeras from both ES cell lines produced agouti progeny, indicating that both lines could contribute to the germ line. Of 31 agouti offspring analyzed by Southern blot hybridization, 15 were heterozygous for the modified H1t alleles, whereas 16 had only wild-type H1t alleles. Mice heterozygous for the modified H1t allele were interbred, and tail DNA from the progeny was analyzed by Southern blot hybridization. Examples of hybridization to DNA from F2 progeny are shown in Fig. 1C. Of 118 F2 animals examined in this way, 27 (23%) carried only wild-type alleles, 54 (46%) had one copy of the modified allele, and 37 (31%) had two copies of the modified allele. The ratio of the three classes of mice is consistent with Mendelian transmission of the two alleles. H1t homozygous mice were normal in weight and size and they were indistinguishable from heterozygous and wild-type littermates. These results indicate that the presence of the modified H1t allele does not impair normal development.
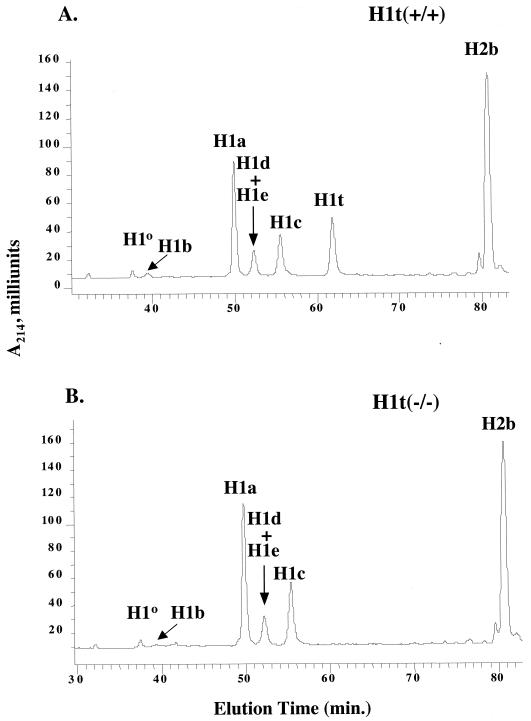
Since the modified H1t allele has a deletion of the entire H1t coding region, it is expected that H1t−/− mice would be completely lacking in H1t protein. To confirm that homozygous mutant animals do not produce H1t protein, total histone extracts of germ cell chromatin were analyzed by reverse-phase HPLC. Previous work (4, 27, 28, 35, 41) showed that this method resolves the five somatic H1 histones and H1o. The HPLC chromatogram of germ cell histones from wild-type animals contains a peak eluting at approximately 62 min (Fig. 2A) which is absent from histone extracts from other tissues (41). We isolated this peak from the HPLC chromatogram and showed that it migrated in sodium dodecyl sulfate-containing and acid-urea-polyacrylamide gels like the testis-specific H1 histone described previously (34) (data not shown). This peak is also absent in the chromatogram of germ cell histone extracts from H1t−/− animals (Fig. 2B). To prove that this peak was H1t, we collected the material eluting in this position and subjected it to time-of-flight mass spectrometry. The mass spectrograph showed a single component with a molecular mass of 21,610 Da (data not shown), consistent with the molecular mass of 21,626 Da for the H1t protein predicted from the H1t gene sequence. Since this peak is missing from the chromatogram of germ cells extracts from H1t−/− mice, we conclude that the modified H1t allele is indeed a null mutation.
FIG. 2.
Reverse-phase HPLC analysis of histones from wild-type and homozygous H1t mutant mice. Approximately 200 μg of total histone extract of chromatin from purified germ cells of a 3-month-old wild-type mouse (A) and a homozygous H1t mutant littermate (B) were fractionated by reverse-phase HPLC as described in Materials and Methods. The identity of the histone subtype(s) in each peak is indicated (see text and reference 41).
Characterization of H1t-deficient mice.
H1t is synthesized in mid- and late-pachytene spermatocytes, and H1t protein is detectable only in these cells and early haploid spermatids. To determine whether H1t is required for male fertility, H1t−/− males were bred with both wild-type and H1t−/− females. Litter sizes from these matings were indistinguishable from those of wild-type matings, and the progeny all appeared normal. However, mice can be fertile with less than 10% the normal number of mature sperm (6). Therefore, it was important to determine the number of mature sperm in H1t−/− mice and to examine the testes for any abnormalities. Testis weights were determined for 3-month-old wild-type, heterozygous, and homozygous mutant littermates. No significant differences were detected among the three classes of mice (Table 1). The testis weights of F3 H1t null mutant mice generated from H1t homozygous mutant mouse matings also did not show any statistically significant difference from those of normal mice (Table 1). To determine whether the number of mature sperm produced by H1t mutant mice was similar to that of normal mice, the sperm were isolated from the cauda epididymis and counted in a hemocytometer. No significant differences in sperm number were detected among the three classes of F2 animals and homozygous F3 mutant mice (Table 1). Furthermore, no significant differences were found among these mice in the motility of their sperm as determined by phase-contrast microscopy. Examination of hematoxylin-eosin-stained paraffin-embedded sections of testes from 3-month-old wild-type and H1t−/− mice did not reveal any abnormal histological features in the mutant mice (data not shown). Like normal testes, the testes of H1t−/− mice contained closely packed seminiferous tubules and limited interstitial space. Moreover, the diameter of the seminiferous tubules, the thickness of the seminiferous epithelium, and the size of the lumen appeared normal in H1t−/− mice. The seminiferous epithelium of H1t−/− testes contained, in addition to Sertoli cells and spermatogonia, multiple layers of spermatocytes, including pachytene spermatocytes, round spermatids, and condensed spermatids; the lumen contained mature sperm. Therefore, no abnormalities in the process of spermatogenesis could be detected in the testes of H1t−/− mice by these light microscopic examinations.
TABLE 1.
Sperm number and testis weights of wild-type and H1t mutant micea
| Generation and genotype | F2 +/+ | F2 +/− | F2 −/− | F3 −/− |
|---|---|---|---|---|
| Testis wt (mg) | 80 ± 9.5 | 91.2 ± 11.0 | 84.5 ± 13.6 | 109 ± 10.8 |
| Sperm no. (107) | 2.84 ± 1.52 | 3.0 ± 1.75 | 2.51 ± 0.46 | 3.14 ± 0.6 |
Values are means ± standard deviations of individual determinations made for four F2 +/+, six F2 +/−, six F2 −/−, and four F3 −/− 3-month-old mice. F2 are progeny of F1 parents heterozygous for the modified H1t allele. F3 are progeny of F2 parents homozygous for the modified H1t allele. Sperm numbers are the numbers of sperm obtained from two epididymides.
During mammalian spermiogenesis, the linker and core histones are replaced by transition proteins. This change is correlated with the appearance of specific morphological features in the spermatid nuclei, including the development of smooth, condensed chromatin fibers (31). To further investigate possible effects of loss of H1t on the process of spermatogenesis, the testes from wild-type and H1t homozygous mutant littermates were examined by electron microscopy. The results from the ultrastructural examination supported the conclusions based on light microscopic studies indicating development of normal sperm in H1t homozygous mutant mice (data not shown). The major characteristic features of pachytene spermatocytes, including patchy condensed chromatin, short profiles of synaptonemal complexes, mitochondria with dilated intracrista vesicles, and extensive endoplasmic reticulum, were found in testes of H1t−/− mice. Major typical features of round spermatids, including well-developed acrosomal caps, scattered profiles of endoplasmic reticulum, and peripherally located, condensed mitochondria, were found in mutant testes. Electron-dense, fully condensed chromatin was also seen in spermatozoa of the mutants. Thus, all of the major morphological transitions that occur in the nuclei of developing male germ cells during the period when H1t accumulates and then is replaced by transition proteins appear to occur in H1t−/− mice.
Compensation for loss of H1t by other H1 subtypes.
The absence of any detectable abnormality in spermatogenesis in H1t−/− mice suggests that other H1 subtypes may compensate for the absence of H1t. To investigate this possibility, we compared the H1 subtype composition and the stoichiometry of linker histones and nucleosomes in germ cell chromatin of H1t−/− mice and wild-type littermates. Since H1t constitutes nearly 30% of the H1 histone in germ cell chromatin (34) (Table 2), any change in either of these parameters should be readily detectable in samples from H1t-deficient testes.
TABLE 2.
H1 subtype composition of testis chromatin from wild-type and H1t−/− micea
| Genotype | % of indicated subtype
|
Total H1/nucleosome ratiod | |||||
|---|---|---|---|---|---|---|---|
| H1o | H1b | H1a | H1d + H1e | H1c | H1t | ||
| Total H1b | |||||||
| +/+ | 1.4 ± 0.7 | 1.2 ± 0.7 | 38.6 ± 2.4 | 10.8 ± 1.1 | 20.1 ± 1.6 | 27.7 ± 2.3 | 0.67 ± 0.08 |
| −/− | 2.2 ± 0.3 | 0.4 ± 0.1 | 51.2 ± 4.2 | 14.0 ± 2.2 | 31.8 ± 2.9 | 0 | 0.68 ± 0.08 |
| H1a–H1e, H1oc | |||||||
| +/+ | 2.0 ± 0.9 | 1.5 ± 0.9 | 53.4 ± 2.1 | 15.0 ± 1.8 | 28.2 ± 1.3 | ||
| −/− | 2.2 ± 0.3 | 0.4 ± 0.1 | 51.2 ± 4.2 | 14.0 ± 2.2 | 31.8 ± 2.9 | ||
Data were from HPLC analyses like that in Fig. 2. Values are means ± standard deviations of individual determinations made for three 3-month-old littermates of each of the indicated genotypes.
Determined by the ratio of the A214 of the indicated H1 peak to the total A214 of all H1 peaks. The percentages were adjusted to account for the differences in the number of peptide bonds in each H1 subtype.
Determined by the ratio of the A214 of the indicated H1 peak to the total A214 of the H1a to H1e and H1o peaks.
Determined by the ratio of the total A214 of all H1 peaks to half of the total A214 of the H2b peaks. The A214 values of the H1 and H2b peaks were adjusted to account for the differences in the number of peptide bonds in each H1 subtype and H2b.
Total chromatin-associated histones were extracted from germ cell chromatin with sulfuric acid and fractionated by reverse-phase HPLC (Fig. 2). This method resolves the H1 histones in germ cell chromatin into six peaks; H1d and H1e migrate as a single peak and therefore their amounts cannot be estimated separately unless the peak containing them is collected and subjected to mass spectrometry (41). The method also separates the H1 histones from the nucleosomal core histones, allowing an estimate of the linker histone-to-nucleosome ratio by measuring the total amount of all of the H1 subtypes relative to a nucleosomal histone, such as H2B.
Quantitative measurements from analyses like that shown in Fig. 2, carried out on the germ cell chromatin from three H1t−/− mice and three wild-type littermates, showed that the H1/nucleosome ratio in chromatin from both types of animals is the same (Table 2). The observed values (0.67 to 0.68) were close to that in liver chromatin (0.71) obtained by the same method (35), which in turn is in good agreement with previous measurements on liver chromatin obtained by other methods (1). These results indicate that other H1 subtypes can compensate for the deficiency of H1t in homozygous mutant animals to maintain the stoichiometry between H1 linker histones and nucleosomes in germ cell chromatin.
To determine whether specific H1 subtypes selectively compensate for the loss of H1t, the relative amounts of each H1 subtype were calculated from the above-mentioned analyses. The results showed that each H1 subtype increased proportionally to compensate for the loss of H1t (Table 2). This is most easily seen by calculating the amount of each H1 subtype as a percentage of all H1s other than H1t (percent H1a to H1e and H1o in Table 2). This type of analysis shows that the relative proportion of each H1 subtype among the non-H1t subtypes is the same in H1t-deficient and wild-type animals. Thus, each H1 subtype contributes proportionally, according to its representation in germ cells, to maintain a constant H1-to-nucleosome stoichiometry in the absence of H1t.
DISCUSSION
The present work was undertaken to investigate the role of the testis-specific linker histone H1t in mammalian spermatogenesis. We generated a null mutation in the H1t gene by using homologous recombination to remove the entire H1t coding region and portions of the upstream and downstream DNA sequence in one copy of the gene in mouse ES cells. The modified ES cells were then used to produce mice that transmitted the mutant allele. Mice homozygous for the mutation were completely lacking H1t protein in germ cell chromatin. Nevertheless, these mice did not exhibit any detectable abnormality in spermatogenesis. Homozygous mutant males were fertile and had normal testis weights, numbers of mature sperm, sperm motility, and testis histology. Moreover, their pachytene spermatocytes and round spermatids, stages in which H1t is very abundant, were found to have normal ultrastructural features when examined by electron microscopy. By all these criteria the process of spermatogenesis does not appear to be affected by the absence of H1t.
The absence of any detectable abnormality in H1t-deficient testis is unexpected because both the regulation of H1t gene expression and the primary sequence of the H1t protein are different from those of other H1 linker histone subtypes. H1t is the only H1 subtype which is expressed in a truly tissue-specific manner. H1t mRNA is transcribed exclusively in mid- and late-pachytene spermatocytes (10, 13, 16), and H1t protein is present only in these cells and early, haploid spermatids (11, 17, 29). In contrast, although expression of the six other H1 genes is differentially regulated, they are all expressed in numerous tissues (24–26). Furthermore, whereas expression of the five somatic H1 genes (H1a to H1e) is generally coordinately regulated with DNA synthesis, transcription of the H1t gene occurring in meiotic prophase appears to be independent of DNA replication.
Likewise, the H1t protein is highly divergent in primary sequence from the other H1s. Although the H1t histone consists of a three-domain structure like other H1s, the mouse H1t amino acid sequence shares only 50% identity with its closest relative among the mouse H1s. Homologs of H1t have been identified in many mammals (7, 10, 14, 34), and germ cell-specific linker histones have been described for other phyla as well (19, 22, 36). Germ cell-specific core histone genes and proteins have also been described for H2a, H2b, H3 (15, 30, 38). Thus, during evolution mammals and other organisms have acquired the capacity to express specific nucleosomal and linker histones in their male germ cells. However, the role of such testis-specific chromatin components in the processes that occur during spermatogenesis remains unknown. In mammals, male germ cell chromatin undergoes two major transitions in protein composition; during spermiogenesis all of the histones are replaced by small lysine- and arginine-rich proteins called transition proteins, which are themselves replaced by the protamines (31). It may be that the testis-specific histones, including H1t, facilitate the first transition. It has also been suggested that H1t in pachytene spermatocytes may facilitate meiotic recombination (31). Either of these processes might be expected to require decondensed chromatin structures, and indeed chromatin reconstituted in vitro with H1t has been reported to have a more open structure than that reconstituted with other H1 subtypes (8). We have not made direct measurements of the rates of chromosomal protein transitions or recombination in germ cells of H1t−/− mice, and therefore it is still possible that the efficiency of these processes is affected by the absence of H1t.
There are now many reports of gene inactivation experiments with mice in which a phenotype is not observed. In some cases the general function of the protein may not be known, and it also may be difficult to prove that the gene targeting resulted in a null mutation. Neither of these uncertainties applies to this report. In nearly all instances in which a phenotype was not observed, it is inferred that another protein with similar function was able to compensate for the deficiency, but in most cases this conclusion has been difficult to prove. However, in the work described here we showed that the stoichiometry of total linker histones to nucleosomes is not reduced in H1t−/− germ cells. This result indicates that the other H1 subtypes must be deposited in sites in chromatin that are normally occupied by H1t. Because H1t normally constitutes nearly 28% of the linker histone in germ cells, an inability of the other H1s to enter such sites would have been readily detected as a reduction in the H1/nucleosome ratio. Thus, it is most likely that other H1 subtypes are able to compensate for all of the functions of H1t in germ cells, although it remains possible that they do so with a reduced efficiency.
Earlier gene inactivation studies from our laboratory showed that the H1o linker histone, another highly divergent H1 subtype, is not absolutely required for normal mouse development (35). H1o accumulates in terminally differentiated cells from many lineages, at about the time when the cells cease dividing (9). Here too our analysis of H1o−/− mice indicated that other H1 subtypes are able to compensate for the loss of H1o in cells in which this linker histone is very abundant. Thus, at least under the conditions in which laboratory mice are maintained, the functions of the two most highly divergent H1s appear to be redundant with those of other members of the family. Nevertheless, the mice described in this report should prove to be especially valuable for future work aimed at studying the in vivo properties of H1-depleted chromatin. As shown in Table 2, 50% of the H1 in germ cells of H1t−/− mice is of a single subtype, H1a. Thus, by combining the H1t null allele with a similarly inactivated allele of H1a, it will be possible to create a much larger linker histone deficiency in male germ cells. Because normal germ cells are not required for normal development and survival of the mice, the doubly mutated germ cells should be useful for studying the general role of H1 linker histones in development and differentiation in a specific cell type.
ACKNOWLEDGMENTS
We thank the Albert Einstein College of Medicine Cancer Center Gene Targeting Facility, the Laboratory of Macromolecular Analysis, and the Analytical Imaging Facility. We thank Paula Cohen and Stuart Moss for instruction and advice in several aspects of testis and germ cell analysis, Yuhong Fan for helpful discussion, and Hui Xu for technical assistance.
This work was supported by National Institutes of Health grant CA 79057.
REFERENCES
- 1.Bates D L, Thomas J O. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 1981;9:5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellve A R, Cavicchia J C, Millette C F, O'Brien D A, Bhatnagar Y M, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellve A R, Millette C F, Bhatnagar Y M, O'Brien D A. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem. 1977;25:480–494. doi: 10.1177/25.7.893996. [DOI] [PubMed] [Google Scholar]
- 4.Brown D T, Sittman D B. Identification through overexpression and tagging of the variant type of the mouse H1e and H1c genes. J Biol Chem. 1993;268:713–718. [PubMed] [Google Scholar]
- 5.Capecchi M R. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 6.Cohen P E, Chisholm O, Arceci R J, Stanley E R, Pollard J W. Absence of colony-stimulating factor-1 in osteopetrotic (csfmop/ csfmop) mice results in male fertility defects. Biol Reprod. 1996;55:310–317. doi: 10.1095/biolreprod55.2.310. [DOI] [PubMed] [Google Scholar]
- 7.Cole K D, Kandala J C, Kistler W S. Isolation of the gene for the testis-specific H1 histone variant H1t. J Biol Chem. 1986;261:7178–7183. [PubMed] [Google Scholar]
- 8.De Lucia F, Faraone-Mennella M R, D'Erme M, Quesada P, Caiafa P, Farina B. Histone-induced condensation of rat testis chromatin: testis-specific H1t versus somatic H1 variants. Biochem Biophys Res Commun. 1994;198:32–39. doi: 10.1006/bbrc.1994.1005. [DOI] [PubMed] [Google Scholar]
- 9.Doenecke D, Alonso A. Organization and expression of the developmentally regulated H1(o) histone gene in vertebrates. Int J Dev Biol. 1996;40:395–401. [PubMed] [Google Scholar]
- 10.Drabent B, Bode C, Doenecke D. Structure and expression of the mouse testicular H1 histone gene (H1t) Biochim Biophys Acta. 1993;1216:311–313. doi: 10.1016/0167-4781(93)90162-7. [DOI] [PubMed] [Google Scholar]
- 11.Drabent B, Bode C, Miosge N, Herken R, Doenecke D. Expression of the mouse histone gene H1t begins at premeiotic stages of spermatogenesis. Cell Tissue Res. 1998;291:127–132. doi: 10.1007/s004410050986. [DOI] [PubMed] [Google Scholar]
- 12.Drabent B, Franke K, Bode C, Kosciessa U, Bouterfa H, Hameister H, Doenecke D. Isolation of two murine H1 histone genes and chromosomal mapping of the H1 gene complement. Mamm Genome. 1995;6:505–511. doi: 10.1007/BF00356166. [DOI] [PubMed] [Google Scholar]
- 13.Drabent B, Kardalinou E, Bode C, Doenecke D. Association of histone H4 genes with the mammalian testis-specific H1t histone gene. DNA Cell Biol. 1995;14:591–597. doi: 10.1089/dna.1995.14.591. [DOI] [PubMed] [Google Scholar]
- 14.Drabent B, Kardalinou E, Doenecke D. Structure and expression of the human gene encoding testicular H1 histone (H1t) Gene. 1991;103:263–268. doi: 10.1016/0378-1119(91)90284-i. [DOI] [PubMed] [Google Scholar]
- 15.Franklin S G, Zweidler A. Non-allelic variants of histones 2a, 2b, and 3 in mammals. Nature. 1977;266:273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- 16.Grimes S R, Wolfe S A, Koppel D A. Tissue-specific binding of testis nuclear proteins to a sequence element within the promoter of the testis-specific histone H1t gene. Arch Biochem Biophys. 1992;296:402–409. doi: 10.1016/0003-9861(92)90590-s. [DOI] [PubMed] [Google Scholar]
- 17.Grimes S R., Jr Nuclear proteins in spermatogenesis. Comp Biochem Physiol B. 1986;83:495–500. doi: 10.1016/0305-0491(86)90285-3. [DOI] [PubMed] [Google Scholar]
- 18.Hartzog G A, Winston F. Nucleosomes and transcription: recent lessons from genetics. Curr Opin Genet Dev. 1997;7:192–198. doi: 10.1016/s0959-437x(97)80128-1. [DOI] [PubMed] [Google Scholar]
- 19.Hnilica L S, Johnson A W. Fractionation and analysis of nuclear proteins in sea urchin embryos. Exp Cell Res. 1970;63:261–270. doi: 10.1016/0014-4827(70)90212-0. [DOI] [PubMed] [Google Scholar]
- 20.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 21.Ioffe E, Liu Y, Bhaumik M, Poirier F, Factor S M, Stanley P. WW6: an embryonic stem cell line with an inert genetic marker that can be traced in chimeras. Proc Natl Acad Sci USA. 1995;92:7357–7361. doi: 10.1073/pnas.92.16.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaye J S, McMaster-Kaye R. Histones of spermatogenous cells in the house cricket. Chromosoma. 1974;46:397–419. doi: 10.1007/BF00331629. [DOI] [PubMed] [Google Scholar]
- 23.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lennox R W, Cohen L H. The alterations in H1 histone complement during mouse spermatogenesis and their significance for H1 subtype function. Dev Biol. 1984;103:80–84. doi: 10.1016/0012-1606(84)90009-5. [DOI] [PubMed] [Google Scholar]
- 25.Lennox R W, Cohen L H. The histone H1 complements of dividing and nondividing cells of the mouse. J Biol Chem. 1983;258:262–268. [PubMed] [Google Scholar]
- 26.Lennox R W, Oshima R G, Cohen L H. The H1 histones and their interphase phosphorylated states in differentiated and undifferentiated cell lines derived from murine teratocarcinomas. J Biol Chem. 1982;257:5183–5189. [PubMed] [Google Scholar]
- 27.Lindner H, Helliger W, Dirschlmayer A, Talasz H, Wurm M, Sarg B, Jaquemar M, Puschendorf B. Separation of phosphorylated histone H1 variants by high-performance capillary electrophoresis. J Chromatogr. 1992;608:211–216. doi: 10.1016/0021-9673(92)87126-s. [DOI] [PubMed] [Google Scholar]
- 28.Lindner H, Helliger W, Puschendorf B. Separation of rat tissue histone H1 subtypes by reverse-phase h.p.l.c. Identification and assignment to a standard H1 nomenclature. Biochem J. 1990;269:359–363. doi: 10.1042/bj2690359. . (Erratum, 271:842.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meistrich M L, Bucci L R, Trostle-Weige P K, Brock W A. Histone variants in rat spermatogonia and primary spermatocytes. Dev Biol. 1985;112:230–240. doi: 10.1016/0012-1606(85)90137-x. [DOI] [PubMed] [Google Scholar]
- 30.Moss S B, Orth J M. Localization of a spermatid-specific histone 2B protein in mouse spermiogenic cells. Biol Reprod. 1993;48:1047–1056. doi: 10.1095/biolreprod48.5.1047. [DOI] [PubMed] [Google Scholar]
- 31.Oko R J, Jando V, Wagner C L, Kistler W S, Hermo L S. Chromatin reorganization in rat spermatids during the disappearance of testis-specific histone, H1t, and the appearance of transition proteins TP1 and TP2. Biol Reprod. 1996;54:1141–1157. doi: 10.1095/biolreprod54.5.1141. [DOI] [PubMed] [Google Scholar]
- 32.Robertson E J. Teratocarcinomas and embryonic stem cells. Oxford, England: IRL; 1987. [Google Scholar]
- 33.Romrell L J, Bellve A R, Fawcett D W. Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev Biol. 1976;49:119–131. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- 34.Seyedin S M, Cole R D, Kistler W S. H1 histones from mammalian testes. The widespread occurrence of H1t. Exp Cell Res. 1981;136:399–405. doi: 10.1016/0014-4827(81)90019-7. [DOI] [PubMed] [Google Scholar]
- 35.Sirotkin A M, Edelmann W, Cheng G, Klein-Szanto A, Kucherlapati R, Skoultchi A I. Mice develop normally without the H1(0) linker histone. Proc Natl Acad Sci USA. 1995;92:6434–6438. doi: 10.1073/pnas.92.14.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subirana J A. Nuclear proteins from a somatic and a germinal tissue of the echinoderm Holothuria tubulosa. Exp Cell Res. 1970;63:253–260. doi: 10.1016/0014-4827(70)90211-9. [DOI] [PubMed] [Google Scholar]
- 37.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 38.Unni E, Zhang Y, Kangasniemi M, Saperstein W, Moss S B, Meistrich M L. Stage-specific distribution of the spermatid-specific histone 2B in the rat testis. Biol Reprod. 1995;53:820–826. doi: 10.1095/biolreprod53.4.820. [DOI] [PubMed] [Google Scholar]
- 39.Van Holde K E. Chromatin. New York, N.Y: Springer-Verlag; 1988. [Google Scholar]
- 40.Wang Z F, Krasikov T, Frey M R, Wang J, Matera A G, Marzluff W F. Characterization of the mouse histone gene cluster on chromosome 13: 45 histone genes in three patches spread over 1Mb. Genome Res. 1996;6:688–701. doi: 10.1101/gr.6.8.688. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z F, Sirotkin A M, Buchold G M, Skoultchi A I, Marzluff W F. The mouse histone H1 genes: gene organization and differential regulation. J Mol Biol. 1997;271:124–138. doi: 10.1006/jmbi.1997.1166. [DOI] [PubMed] [Google Scholar]
- 42.Wolffe A P. Chromatin—structure and function. New York, N.Y: Academic Press; 1998. [Google Scholar]
- 43.Wolffe A P. Histone H1. Int J Biochem Cell Biol. 1997;29:1463–1466. doi: 10.1016/s1357-2725(97)00026-5. [DOI] [PubMed] [Google Scholar]
- 44.Wolffe A P, Khochbin S, Dimitrov S. What do linker histones do in chromatin? Bioessays. 1997;19:249–255. doi: 10.1002/bies.950190311. [DOI] [PubMed] [Google Scholar]