Abstract
Simple Summary
Interest in stratification of prognosis for patients with colorectal liver metastases is growing. Numerous clinical prediction models have been developed for this purpose in recent years, either with the aid of traditional statistical methods or by using the aid of artificial intelligence techniques. We herein provide an overview of relevant studies discussing the different types of predictors proven to be of importance and critically assess the variable model development and validation techniques as well as the performance of the reported models.
Abstract
Colorectal liver metastasis (CRLM) is a disease entity that warrants special attention due to its high frequency and potential curability. Identification of “high-risk” patients is increasingly popular for risk stratification and personalization of the management pathway. Traditional regression-based methods have been used to derive prediction models for these patients, and lately, focus has shifted to artificial intelligence-based models, with employment of variable supervised and unsupervised techniques. Multiple endpoints, like overall survival (OS), disease-free survival (DFS) and development or recurrence of postoperative complications have all been used as outcomes in these studies. This review provides an extensive overview of available clinical prediction models focusing on the prognosis of CRLM and highlights the different predictor types incorporated in each model. An overview of the modelling strategies and the outcomes chosen is provided. Specific patient and treatment characteristics included in the models are discussed in detail. Model development and validation methods are presented and critically appraised, and model performance is assessed within a proposed framework.
Keywords: colorectal cancer, liver metastases, machine learning, prognosis, risk assessment
1. Introduction
The prognosis of metastatic colorectal cancer is steadily increasing, and in patients with colorectal liver metastases (CRLMs), resectable disease is a potentially curable entity, with 5-year overall survival (OS) rates between 20 and 58% [1,2]. For potentially resectable disease, administration of neoadjuvant regimens or upfront surgery is still debatable due to limited high-quality evidence, while risk stratification, with quantification of disease burden and selection of “high-risk” patients, is becoming increasingly popular [3]. Apart from surgical resection, multiple treatment modalities, like intra-arterial treatments and local ablative techniques, are becoming available for CRLMs, highlighting the substantial interest in this patient population [2].
Interest in identifying patients with poor prognosis has led to the development of numerous prediction models, and in recent years, artificial intelligence (AI) has been used in model development through a variety of machine learning (ML) techniques, mostly in the form of radiomics-based models [4]. Despite the rise of AI, conventional statistical methods remain the cornerstone of model development, and focus has now shifted in the proper reporting of relevant studies with the release of the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) guidelines [5]. Due to the substantial rise in relevant studies in the last decade, it is imperative to summarize the literature involving all clinical models developed for the prognostication of CRLM patients. Thus, the aim of this review is to provide an overview of the available prognostic models for patients with CRLMs with emphasis on predictors, model development and validation techniques, as well as model performance.
2. Traditional Prediction Models
Clinical prediction models that are still in use in many hepatobiliary units worldwide were developed in the previous decades [6,7,8,9,10,11,12,13,14,15,16,17,18,19]. Details regarding these models can be seen in Supplementary Table S1. The largest single-centre series was published in 1999 by Fong et al. and included 1001 liver resections for CRLM [7]. The classical predictors included in this model, namely nodal status of the primary tumour, disease-free interval from detection of the primary to discovery of the liver metastases < 12 months, number of tumours greater than one, preoperative carcinoembryonic antigen (CEA) level > 200 ng/mL and size of the largest tumour > 5 cm retain their significance in a large percentage of modern CRLM studies. The largest multicentre series was published from France with 1568 patients and included age, T stage, N stage, time interval from primary tumour to metastases, size of largest metastases, number of metastases and clearance (resection margin) as predictors [6]. All studies used traditional Cox regression for model development with univariate screening of predictors and did not report any measures of discrimination or calibration. Two studies utilized bootstrapping as their internal validation technique [6,19], while three studies reported an external validation of their models [9,14,15].
3. Methods
An extensive literature search was performed using the Pubmed database, aiming to identify all studies developing a clinical prediction model using CRLM patients, either with regression-based or ML techniques (supervised and unsupervised). External validation studies of already developed models were excluded. Additionally, reference lists of relevant reviews were also screened for potential studies. The details of the search strategy are presented in the Supplementary methods. The screening process aimed to identify studies focusing on the prognosis of CRLM patients, either short or long term, meaning that studies focusing on the diagnosis of liver metastases were not included in this review. Radiological predictors and studies based on radiomics were also excluded, as these models form a unique category requiring special attention. In order to provide an overview of the performance of published models, studies were included if they at least reported measures of discrimination for their models. A summary of the characteristics of the final studies included in this review is provided in Table 1 and Supplementary Table S2.
Table 1.
Overview of model development studies for the prognosis of patients with colorectal liver metastases.
| First Author (Year) | Data Collection | Model Type | Univariate Screening of Predictors | Outcome(s) | Patients (n) | Internal/External Validation | Missing Data | Risk Groups |
|---|---|---|---|---|---|---|---|---|
| Buisman (2022) [20] | retrospective | Cox regression | no | OS | 4112 | Cross-validation | Multiple imputation | yes (4 groups) |
| Bertsimas (2022) [21] | retrospective | RF, OPT | no | OS and intrahepatic recurrence | 761 | IV: Split sample/external validation cohort | Complete case analysis | no |
| Bao (2021) [22] | retrospective | NGS, Cox and LASSO regression | yes | OS | 144 | External validation (gene signature only) | No information | yes (2 groups) |
| Lam (2023) [23] | retrospective | Cox and LASSO regression | yes | OS and RFS | 572 | Split sample | Multiple imputation | no |
| Reijonen (2023) [24] | retrospective | Cox regression | yes | OS and DFS | 816 | Not performed | The final sum of their risk score points was estimated using the mean of the evaluable predictors | yes (3 groups) |
| Margonis (2018) [25] | retrospective | Cox regression | yes | OS | 502 (development), 747 (validation) | External validation | No information | yes (3 groups) |
| Paredes (2020) [26] | retrospective | Mixed-effects logistic regression | no | Recurrence | 703 (development), 703 (validation) | Split sample, bootstrapping | Multiple imputation | yes (3 groups) |
| Fruhling (2021) [27] | retrospective | Cox regression | yes | OS | 1212 | Cross-validation | Multiple imputation | yes (3 groups) |
| Taghavi (2021) [28] | retrospective | RF | no | Development of metachronous metastases | 70 (development), 21 (validation) | Split sample, cross-validation | Single imputation | no |
| Brudvik (2019) [29] | retrospective | Cox regression | no | OS, RFS | 564 (development), 608 (validation) | External validation | Complete case analysis | no |
| Moaven (2023) [30] | retrospective | GBT and LRB in a leave-one-out cross-validation | no | OS, recurrence | 1004 | Cross-validation, bootstrapping | Variables with more than 20% missing data were eliminated from the model | yes (3 groups) |
| Villard (2022) [31] | retrospective | Cox regression | no | OS | 1013 (development), 391 (validation) | External validation | Multiple imputation | yes (4 groups) |
| Chen (2020) [32] | retrospective | Cox regression | no | RFS | 787 (cohort 1), 162 (cohort 2) | IV: Bootstrapping/temporal validation | Complete case analysis | yes (3 groups) |
| Chen (2022) [33] | retrospective | Cox regression | yes | OS | 1095 | Not performed | Multiple imputation | no |
| Dai (2021) [34] | retrospective | Logistic regression | yes | Early recurrence within 6 months | 150 (development), 52 (validation) | Split sample | Complete case analysis | no |
| Liu (2021) [35] | retrospective | Cox regression | yes | OS after recurrence | 867 | Bootstrapping | No information | yes (2 groups) |
| Liang (2021) [36] | retrospective | Cox regression | yes | Post-recurrence survival | 251 (development), 125 (validation) | Split sample, bootstrapping | Complete case analysis | yes (3 groups) |
| Wu (2021) [37] | retrospective | Cox regression | yes | Recurrence, PFS | 229 (development), 128 (validation) | Temporal validation | Complete case analysis | yes (3 groups) |
| Sasaki (2022) [38] | prospective | Cox regression | yes | OS | 1205 (development), 1307 + 1058 (validation) | External validation | No information | yes (3 groups) |
| Huiskens (2019) [39] | retrospective | Logistic regression | yes | 90-day mortality (after stage 2) | 486 | Not performed | Complete case analysis | yes (3 groups) |
| Bai (2022) [40] | retrospective | Cox regression | yes | OS and RFS | 341 (development), 325 (validation) | External validation | Complete case analysis | yes (3 groups) |
| Fang (2022) [41] | retrospective | Cox regression | yes | OS | 237 | Not performed | Complete case analysis | yes (3 groups) |
| Qin (2022) [42] | prospective | Cox regression | yes | ihPFS | 314 | Not performed | No information | yes (3 groups) |
| Kawaguchi (2021) [43] | prospective | Cox regression | yes | OS | 810 (development), 673 (validation) | External validation | Complete case analysis | no |
| Zhang (2023) [44] | retrospective | Cox and LASSO regression | yes | OS | 415 (development), 207 (validation) | IV: Split sample/External validation cohort | No information | yes (2 groups) |
| Chen (2021) [45] | retrospective | Logistic and Cox regression | yes | Postoperative complications, PFS, OS | 380 | Not performed | Complete case analysis | yes (3 groups) |
| Jin (2022) [46] | retrospective | Cox regression | yes | CSS | 881 (development), 169 (validation) | IV: Split sample/External validation cohort | Complete case analysis | yes (2 groups) |
| Zhai (2022) [47] | retrospective | Cox regression | yes | Liver RFS | 147 | Not performed | Complete case analysis | yes (3 groups) |
| Liu (2021) [48] | retrospective | Cox regression | yes | PFS | 532 (development), 237 (validation) | External validation | No information | yes (2 groups) |
| Moro (2020) [49] | retrospective | CART analysis | no | OS | 1123 | Bootstrapping | Multiple imputation | yes (4 groups) |
| Chen (2021) [50] | retrospective | Logistic and Cox regression | yes | Complications, PFS, OS | 169 | Not performed | Complete case analysis | yes (3 groups) |
| Yao (2021) [51] | retrospective | Logistic and Cox regression | yes | Presence of LN metastases, PFS | 241 | Not performed | Complete case analysis | no |
| Kazi (2023) [52] | retrospective | Logistic and Cox regression | yes | Serious complications | 92 | Bootstrapping | No information | yes (4 groups) |
| Meng (2021) [53] | retrospective | Cox regression | yes | OS | 174 (development), 60 (validation) | Split sample | Complete case analysis | yes (2 groups) |
| Imai (2016) [54] | prospective | Cox regression | yes | OS | 439 | Not performed | No information | yes (4 groups) |
| Chen (2022) [55] | retrospective | Logistic regression | yes | Early recurrence (<11 months) | 144 (development), 40 (validation) | Another cohort from the same hospital | Complete case analysis | no |
| Cheng (2022) [56] | retrospective | Cox regression | yes | CSS | 1314 (development), 560 (validation) | Split sample | Complete case analysis | yes (2 groups) |
| Kulik (2018) [57] | retrospective | Logistic regression | yes | OS | 965 | Not performed | Complete case analysis | no |
| Bai (2021) [58] | retrospective | Cox regression | yes | OS | 490 | Not performed | Complete case analysis | yes (7 and 6 groups) |
| Wang (2021) [59] | retrospective | Cox and LASSO regression | no | OS | 113 (development), 114 (validation), 168 (external validation) | IV: Split sample/external validation cohort | Complete case analysis | yes (2 groups) |
| Xu (2021) [60] | retrospective | Logistic regression | yes | Major pathologic response to chemotherapy | 241 (development), 241 (validation) | Split sample | Complete case analysis | yes (2 groups) |
| Sasaki (2018) [61] | retrospective | A priori selection of predictors and interactions | no | OS | 604 (development) | External validation | No information | yes (3 groups) |
| Wada (2022) [62] | retrospective | Cox and LASSO regression | no | Recurrence | 169 (development), 151 (validation) | External validation | No information | yes (2 groups) |
| Kim (2020) [63] | retrospective | Cox regression | yes | Recurrence | 197 (development), 98 (validation) | Split sample | No information | yes (2 groups) |
| Dupre (2019) [64] | prospective | Cox regression | yes | OS | 364 (development), 219 (validation) | External validation | No information | yes (2 groups) |
| Qi (2023) [65] | retrospective | Automated tissue classification and quantification of CRLM SOFs derived from histology images with deep learning and Cox regression | yes | OS | 433 (development), 403 (validation) | External validation | Complete case analysis | yes (SOF scoring system 2 groups, SOF-CRS 3 groups) |
| Wu (2021) [66] | retrospective | Cox regression | yes | PFS | 158 | Not performed | Complete case analysis | yes (3 groups) |
| Dasari (2023) [67] | retrospective | Cox and LASSO regression | yes | OS | 927 (development), 309 (validation) | Split sample | Complete case analysis | yes (5 groups) |
| Liu (2023) [68] | retrospective | Cox and LASSO regression | yes | OS | 295 (development), 295 (validation) | Split sample | Complete case analysis | yes (2 groups) |
| Amygdalos (2023) [69] | retrospective | GBT with the Top6 selected predictors | no | OS | 389 (development), 98 (validation) | Split sample | Complete case analysis | yes (2 groups) |
| Chen (2023) [70] | retrospective | Cox regression | yes | OS | 85 | Not performed | Complete case analysis | yes (3 groups) |
| Wu (2018) [71] | retrospective | Cox regression | yes | OS and CSS | 4825 (development), 4790 (validation) | Split sample | Complete case analysis | no |
| Deng (2023) [72] | retrospective | Logistic regression | yes | Early recurrence (<13 months) | 323 (development), 71 (validation) | External validation | Complete case analysis | no |
| Berardi (2023) [73] | prospective | Logistic regression | yes | Treatment failure (recurrence or death within 12 months) | 535 (development), 248 (validation) | Split sample | No information | yes (2 groups) |
| Liu (2019) [74] | retrospective | Cox regression | yes | DFS | 447 (development), 117 (validation) | External validation | No information | yes (3 groups) |
| Welsh (2008) [75] | prospective | Logistic regression | yes | R1 resection margin | 911 | Bootstrapping | Single (median) imputation | no |
| Famularo (2023) [76] | prospective | Survival RF to estimate the best possible treatment, then CART was used to develop a decision tree | no | OS | 448 | Cross-validation | Multiple imputation | yes (7 groups) |
| He (2023) [77] | retrospective | Logistic regression | yes | Benefit from upfront surgery (survival > 15 months) | 572 (development), 242 (validation) | Split sample | Complete case analysis | no |
| Kattan (2008) [78] | retrospective | Cox regression | yes | DSS | 1477 | Bootstrapping | No information | no |
| Wensink (2023) [79] | retrospective | Cox regression | no | Early extrahepatic recurrence (at 6 and 12 months) | 1077 | Bootstrapping and internal–external cross-validation | Multiple imputation | yes (4 groups) |
| Fendler (2015) [80] | retrospective | Cox regression | yes | OS | 100 (development), 25 (validation) | IV: Bootstrapping/external validation cohort | No information | no |
| Marfa (2016) [81] | prospective | CART analysis | no | OS | 57 (development), 28 (validation) | Split sample | No information | yes (2 groups) |
| Jiang (2023) [82] | retrospective | Cox regression | yes | OSS and CSS | 225 (development), 180 (validation) | External validation | Complete case analysis | no |
| Endo (2023) [83] | retrospective | OPT analysis | no | OS and RFS | 679(development), 679 (validation) | Split sample | Multiple imputation | yes (multiple nodes) |
| Rees (2008) [84] | prospective | Cox regression | yes | CSS | 929 | Bootstrapping | Single (median) imputation | yes (5 groups) |
| Zakaria (2007) [85] | retrospective | Cox regression | yes | DFS, recurrence | 662 | Not performed | Complete case analysis | yes (3 groups) |
| Tan (2008) [86] | retrospective | Cox regression | yes | OS | 296 | Not performed | Multiple imputation | yes (3 groups) |
| Hill (2012) [87] | retrospective | Cox regression | yes | Survival following resection for recurrence | 280 | Bootstrapping | No information | yes (3 groups) |
| Takeda (2021) [88] | retrospective | Cox regression | yes | OS | 341 (development), 309 (validation) | External validation | Complete case analysis | yes (4 groups) |
| Wang (2017) [89] | retrospective | Cox regression | yes | OS | 300 | Not performed | No information | yes (4 groups) |
| Spelt (2013) [90] | retrospective | ANN and Cox regression | yes | OS | 241 | Cross-validation | Multiple imputation | no |
OS: overall survival, RF: random forest, OPT: optimal policy tree, IV: internal validation, NGS: next-generation sequencing, LASSO: least absolute shrinkage and selection operator, RFS: recurrence-free survival, DFS: disease-free survival, GBT: gradient-boosted trees, LRB: logistic regression with bootstrapping, ihPFS: intrahepatic progression-free survival, CART: classification and regression tree, LN: lymph node, CSS: cancer-specific survival, CRLM: colorectal liver metastasis, SOFs: spatial organization features, CRS: clinical risk score, ANN: artificial neural network.
4. Outcome Types
A variety of short- and long-term outcomes related to the prognosis of CRLM patients have been used in the literature. The outcomes chosen for each model can be seen in Table 1.
4.1. Postoperative Complications/Mortality
Prediction of postoperative complications was the goal of three studies, all of which used the Clavien–Dindo classification for categorization of the grade of complications [45,50,52,91]. One study focused on 90-day mortality following Associating Liver Partition and Portal vein Ligation for Staged hepatectomy (ALPPS) [39]. All studies developed their models using regression-based techniques, reported moderate discriminative performances and fair or good calibration.
4.2. Survival
Most studies (n = 59, 83.1%) focused on patient survival, either as OS, disease-specific survival (DSS), disease-free survival (DFS), cancer-specific survival (CSS), progression-free survival (PFS) or intrahepatic progression-free survival (ihPFS) (Table 1). Regarding long-term survival, two regression-based models for 10-year OS were proposed [20,57]. Over 20 significant predictors were identified in these two studies, including patient-, disease- and treatment-related variables (Supplementary Table S3). Post-recurrence survival was studied in three models [35,36,87]. Unique predictors incorporated in these models are pattern of recurrence (liver only, lung only, extrahepatic), time from hepatectomy to recurrence and treatment of the recurrent disease [36,87].
4.3. Recurrence
Ten studies developed prognostic models for prediction of recurrence [21,26,30,34,55,62,63,72,79,85]. Regression-based methods were used in seven studies [26,34,55,63,72,79,85], and the remaining three studies used ML techniques, including the least absolute shrinkage and selection operator (LASSO) [62], gradient-boosted trees (GBT) [30] and random forest (RF) with a globally optimal decision tree (OPT) analysis [21]. The latter was employed to identify the ideal margin width that minimizes the probability of intrahepatic recurrence within 5 years, and margins between 9 and 11 mm were proposed according to the diameter of the largest CRLM, the primary tumour nodal status and the primary tumour site [21]. Four studies attempted to predict early recurrence, defined either as recurrence within 6 months using a previously established definition [34,79] or by performing an additional analysis that suggested cut-offs at 11 or 13 months [55,72].
5. Specific Patient and Treatment Characteristics
5.1. Simultaneous Resections
Six recent Asian studies focused on patients undergoing simultaneous resections of the primary and metastatic tumours [37,51,52,53,72,82]. Outcomes studied included OS [53,82], CSS [82], PFS [37,51], recurrence [37,72], serious postoperative complications [52] and presence of lymph node (LN) metastases [51]. The latter was unique as an outcome of choice since a positive primary LN status is a strong predictor of poor outcomes in CRLM patients in many studies. All models were based on regression-based techniques, and notably, two of the studies performed a decision curve analysis (DCA) [72,82]. In the study by Deng et al., clinical utility was found in only a narrow range of risk thresholds [72], while Jiang et al. demonstrated superior net benefit of their model compared to AJCC stage [82].
5.2. Upfront Surgery versus Neoadjuvant Chemotherapy
Prediction models studying which patients will benefit from upfront surgery (UPS) instead of neoadjuvant treatment were developed in three recent studies [73,76,77]. Famularo et al. employed survival random forest (RF) to estimate the best possible treatment (BPT) for each patient [76]. Following this step, a classification and regression tree (CART) analysis was used to develop a decision tree, showing the possibility of being assigned to UPS or to a neoadjuvant regimen according to five predictors (planned R1 vascular resection, number of intrahepatic metastases, colon tumour localization, CEA and sex) [76]. He et al. used a cohort of 814 patients from the Surveillance, Epidemiology, and End Results (SEER) database to develop a logistic regression-based model using benefit from UPS (defined as survival >15 months) as their outcome [77]. The authors presented a nomogram in which a lower N stage, lower histological grade, negative CEA, chemotherapy following primary resection and absence of lung metastases were associated with a higher possibility of benefit from UPS [77]. Conversely, a study with 783 UPS patients from the US developed a nomogram for prediction of treatment failure following primary resection, defined as recurrence or death within 12 months [73]. Predictors included in the final model were primary location, interval from primary to CRLM, LN positive primary, T stage and number and size of CRLMs. Notably, continuous predictors were not dichotomized in this study before entering the nomogram [73].
5.3. Systemic Therapies
In the debatable field of systemic therapies for CRLMs, benefit from and response to chemotherapy were studied in recent papers. ML with an OPT analysis was employed in a multinational cohort of 1358 patients to identify which patients would benefit from adjuvant chemotherapy through a higher OS or RFS [83]. In a logistic regression model from China, disease-free interval < 12 months, tumour size, tumour number and RAS status were shown to predict major pathologic response to chemotherapy, defined as less than 50% remnant viable cells [60]. Tumour response to chemotherapy, in terms of non-progressive disease, was also an independent predictor of ihPFS in patients with unresectable disease receiving radiofrequency ablation (RFA) [66]. Two Chinese studies presented prediction models for CRLM patients receiving neoadjuvant chemotherapy followed by resection [74,89]. In this high-burden patient subgroup, an increase in tumour diameter during first-line chemotherapy was entered in the nomogram predicting DFS by Liu et al. [74], while in the model presented by Wang et al., named the tumour biology score, KRAS mutation, Fong score > 2 and poor preoperative chemotherapy response were decisive for 5-year OS [89].
5.4. Special Treatment Modalities (RFA, MWA, HAIP, SIRT, ALPPS)
Apart from the already mentioned study by Wu et al., in which patients undergoing RFA were enrolled [66], multiple treatment modalities, now available for CRLM patients, have been studied in recent model development papers. Qin et al. focused on patients who underwent US-guided percutaneous microwave ablation (MWA) in a selected cohort of 314 patients with a number of CRLM less than nine and size of CRLMs < 5 cm [42]. A model for ihPFS (at 1, 2 and 3 years) was developed with five predictors: maximal size of CRLM, number of CRLMs, ablative margin, primary tumour lymph node status and chemotherapy, with areas under the curve (AUCs) between 0.695 and 0.782 and fair calibration [42]. Regarding hepatic arterial infusion pump chemotherapy (HAIP chemotherapy), it has only been reported as a predictor in a Dutch model developed for 10-year OS in a cohort of patients after curative resection and/or ablation of CRLM [20]. Fendler et al. focused on patients undergoing selective internal radiation therapy (SIRT) through hepatic arterial delivery of Yttrium-90 [90Y] microspheres [80]. A nomogram incorporating four predictors, namely no prior liver surgery, CEA, transaminase toxicity ≥2.5× ULN and CRLM size ≥10 cm, demonstrated an AUC of 0.81 in the training cohort and 0.83 in an external validation cohort [80]. Lastly, in a multinational study of 486 patients receiving ALPPS as therapy for CRLM, two different models were developed using 90-day mortality after stage 2 as their target outcome [39]. The two models, using predictors available before the 1st and 2nd stage, respectively, had a moderate discrimination (AUCs: 0.70–0.72) and good calibration with the Hosmer–Lemeshow (HL) test [39].
6. Predictor Types
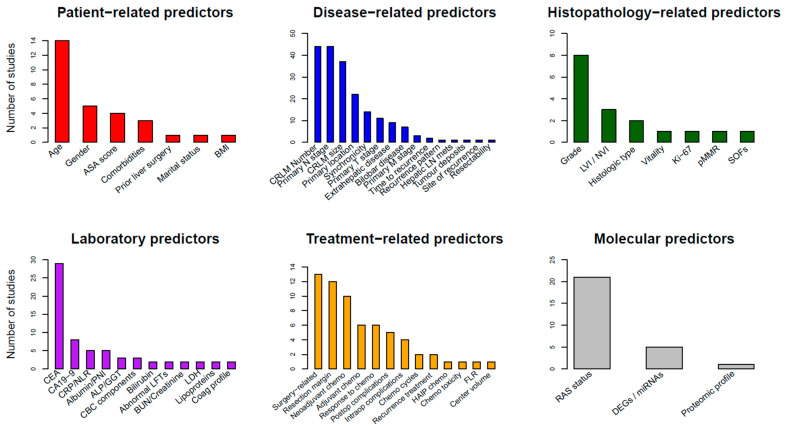
The different predictors included in the models are seen in detail in Supplementary Table S3 and are depicted graphically in Figure 1.
Figure 1.
Frequency of predictors included in studies developing a clinical prediction model for prognosis of patients with colorectal liver metastases, stratified by predictor type. ASA: American Society of Anesthesiologists, BMI: body mass index, CRLM: colorectal liver metastasis, LN: lymph node, LVI: lymphovascular invasion, NVI: neurovascular invasion, pMMR: mismatch repair proficiency, SOFs: spatial organization features, CEA: carcinoembryonic antigen, CA19-9: carbohydrate antigen 19-9, PNI: prognostic nutrition index, ALP: alkaline phosphatase, GGT: gamma-glutamyl transferase, CBC: complete blood count, LFTs: liver function tests, BUN: blood urea nitrogen, LDH: lactate dehydrogenase, HAIP: hepatic arterial infusion pump, FLR: future liver remnant, RAS: rat sarcoma virus, DEGs: differentially expressed genes.
6.1. Patient-Related Predictors
Few patient-related predictors are included in prediction models for patients with CRLMs. The most commonly used are patient age (14 studies), gender (5 studies), American Society of Anesthesiologists (ASA) score and comorbidities (each in 3 studies). Comorbidities were defined using the Charlson comorbidity score in two of three studies [23,83]. Body mass index (BMI), marital status and no prior liver surgery were also reported as variables (each in one study).
6.2. Laboratory Biomarkers
A wide variety of laboratory biomarkers have been reported as predictors. The most widely used were tumour markers, namely CEA and carbohydrate antigen 19-9 (CA19-9). CEA was included in 29 (40.8%) models, while CA19-9 was reported in 8 studies. Inflammatory markers, like C-reactive protein (CRP) and neutrophil-to-lymphocyte ratio (NLR), were used in 7% of the models, and nutritional predictors, like albumin or the Prognostic Nutrition Index (PNI), defined as serum albumin (g/L) + 5 × total lymphocyte count (109/L), were also used in 7% of the models. Abnormal liver function tests (LFTs, two studies), bilirubin (two studies) and gamma-glutamyl transferase/alkaline phosphatase (GGT/ALP, three studies) were also documented to be related to poor outcomes.
6.3. Disease-Related Predictors
Predictors related to CRLMs or the primary disease were consistently reported in the majority of the prediction models. Number (n = 44, 62.0%) and size (n = 37, 52.1%) of the liver metastases were amongst the most essential predictors, along with N stage (n = 44, 62.0%) and location (n = 22, 31.0%) of the colorectal primary. Other important variables include synchronicity (interval between primary and metastatic tumour diagnosis, 14 studies), primary T stage (11 studies), concurrent extrahepatic disease (9 studies) and bilobar distribution of the metastases (7 studies).
6.4. Histopathological Predictors
Variables included in pathology reports were also of importance in several studies. Tumour grade of differentiation was the most widely used (eight studies, 11.3%), followed by lymphovascular invasion (LVI), neurovascular invasion (NVI) biliary invasion (three studies) and histopathological growth pattern (desmoplastic or not) or histologic type (two studies). Other reported predictors included vitality (percentage in pathology report), Ki-67, pMMR (mismatch repair proficiency) and SOFs (spatial organization features from histology), each in one model. The latter were identified using deep learning techniques through fully automated tissue classification and quantification of SOFs that were correlated with poorer outcomes [65].
6.5. Treatment-Related Predictors
Over 20 different treatment-specific predictors were included in the reported models. The most significant were surgery-related predictors (13 studies, 18.3%), like major resection (either defined as resection of greater than three or four liver segments), non-anatomical resection, bilateral resection, one- or two-stage hepatectomy and planned R1 vascular resection. Other important variables include resection margin in 12 studies (16.9%), neoadjuvant (10 studies) or adjuvant chemotherapy (6 studies), response to chemotherapy or tumour regression grade (6 studies) and postoperative complications (5 studies). Resection margin was modelled differently in the included studies, most commonly as positive or negative, dichotomized (with cut-offs at 1 or 5 mm) [24,71], trichotomized [42,67] or kept in a continuous scale in mm [57].
6.6. RAS Status and Molecular Predictors
Molecular predictors are gaining interest and are becoming part of clinical prediction models due to the increasing availability of sequencing techniques. The most important predictor for CRLM patients is the mutant rat sarcoma virus (RAS) oncogene, which is part of almost 30% of modern models (n = 21, 29.6%). Regarding other molecular biomarkers, Marfa et al. used the CART analysis to construct a proteomic signature in a series of 85 patients, which differentiates mild from severe cases (based on predicted OS) according to the four most significant protein peaks [81]. Another emerging predictor is differentially expressed genes (DEGs), or differentially expressed exosomal miRNAs, which has already been used in five studies [22,44,59,62,68]. After selection of candidate genes, LASSO regression, which applies a penalty to candidate predictors and eliminates some variables from the final model, was applied in all studies to construct the final gene panel. All five studies combined the DEGs with clinical predictors and reported merged scores [22,44,59,62,68].
7. Development and Validation Techniques
The characteristics of the included studies are summarized in Table 1. Data were collected prospectively in 10 studies (14.1%). The majority used regression-based techniques (53 studies, 74.6%), while 10 studies (14.1%) employed a mix of regression-based and ML methods. Seven models (9.9%) were developed with ML or deep learning techniques. Most studies (n = 54, 76.1%) performed a univariate screening of candidate predictors, and selected variables were entered into multivariate models. Continuous predictors were mostly dichotomized (52 studies, 73.2%) and were kept in their continuous form in only 17 studies (23.9%). Regarding internal and external validation techniques, the most frequent internal validation method was split sample (21 studies, 29.6%), followed by bootstrapping (13 studies, 18.3%) and cross-validation (7 studies, 9.9%). External validation was reported in 23 studies (32.4%), while 17 studies (23.9%) did not perform any form of validation. The most utilized method of handling missing data during model development was complete case analysis (34 studies, 47.9%), followed by multiple (12 studies, 16.9%) and single (3 studies, 4.2%) imputation. Twenty studies (28.2%) did not report how missing data were handled. Most papers (53 studies, 74.6%) divided their sample into risk groups. In 21 studies (29.6%), the calculation of model outputs for each patient led to categorization into risk groups based on the distribution of the outputs (quartiles/tertiles). In four studies (5.6%), CART or OPT analyses led to the creation of the different groups, while three studies (4.2%) utilized specific software, such as X-tile, to obtain cut-offs. The optimal combination of sensitivity and specificity was used as the criterion for cut-offs in three studies (4.2%), while all possible scores derived from simplified model versions were used in three studies (4.2%). In 19 studies (26.8%), the details of how risk groups were created were unclear. A variety of model presentation methods were reported. Nomograms (28 studies, 39.4%) and risk scores with points assigned to each predictor (23 studies, 32.4%) were the most frequently used, followed by equations (8 studies, 11.3%), online calculators (5 studies, 7.0%), CART (3 studies, 4.2%) and OPT analysis (2 studies, 2.8%).
8. Model Performance
Performance of the prediction models developed in the included studies is summarized in Table 2. The discriminative ability was assessed with the AUC in 70 studies and with the Akaike’s Information Criterion (AIC) in 1 study. AUCs (after internal or external validation) mostly ranged between 0.60 and 0.70 (46.5%) and 0.70 and 0.80 (38%). Good discrimination (AUCs > 0.80) was reported in only eight studies (11.3%). Notably, calibration was not reported in 30 (42.3%) studies. The most frequently reported calibration measure was calibration curves, which allow for a visual inspection of the agreement between predicted and observed events. Calibration curves were presented in 31 (50.7%) studies, followed by the HL goodness of fit test (10 studies, 14.1%), calibration slope (4 studies, 5.6%) and calibration intercept (3 studies, 4.2%). DCA, which provides an overview of risk thresholds that are expected to be useful in clinical practice, was reported in 12 studies (16.9%).
Table 2.
Performance of clinical prediction models for prognosis of patients with colorectal liver metastases.
| First Author (Year) | Discrimination (AUC) | Calibration Measures | Calibration: Performance | DCA |
|---|---|---|---|---|
| Buisman (2022) [20] | 0.73 | Calibration curve | Good calibration (MSKCC model)/slight underprediction (Erasmus MC model) | NR |
| Bertsimas (2022) [21] | KRAS-variant: 0.76 (both training and testing)/external validation: 0.78/wild-type, training: 0.79/wild-type, testing: 0.57 | NR | NR | NR |
| Bao (2021) [22] | Mean time-dependent: 0.75 | NR | NR | NR |
| Lam (2023) [23] | 0.65 (both for OS and RFS) | NR | NR | NR |
| Reijonen (2023) [24] | 0.62 (OS) | NR | NR | NR |
| Margonis (2018) [25] | 0.625 | NR | NR | NR |
| Paredes (2020) [26] | Model without KRAS: 0.649–0.662 (validation cohort)/model with KRAS: 0.642–0.667 (validation cohort) | Calibration curve | No KRAS: good calibration/KRAS: fair | NR |
| Fruhling (2021) [27] | 1-, 3-, 5-year OS: 0.71, 0.67, 0.67/internal validation: 0.62 | Calibration curve | Excellent calibration in development cohort | NR |
| Taghavi (2021) [28] | Training: 0.64/validation: 0.71 | NR | NR | NR |
| Brudvik (2019) [29] | Development, 5 -y OS: 0.69/development: 5 y RFS: 0.66 | NR | NR | NR |
| Moaven (2023) [30] | GBT, OS: 0.77/GBT, recurrence: 0.63/LRB, OS: 0.64/LRB, recurrence: 0.57 | NR | NR | NR |
| Villard (2022) [31] | Development: 0.74/validation: 0.69/simplified model, development: 0.74, validation: 0.66 | Calibration curve, CITL, slope, HL test | CITL: 0.36, slope: 0.89 (validation), good overall fit | NR |
| Chen (2020) [32] | Development: 0.69 at 24 months and 0.65 at 33 months/internal validation: 0.63/cohort 2: 0.81 at 15 months | Calibration curve | Good calibration | NR |
| Chen (2022) [33] | 1-, 3-, 5-year OS: 0.828, 0.740, 0.700 in the solitary LM group; 0.747, 0.714, 0.753 in the 2–4 LM group; 0.728, 0.741, 0.792 in the ≥ 5 LM group | Calibration curve | Fair calibration only in the 2–4 LM group | NR |
| Dai (2021) [34] | Training: 0.866/validation: 0.792 | Calibration curve | Poor calibration in the validation cohort | Clinical utility with lift curves |
| Liu (2021) [35] | 0.707 | Calibration curve | Fair | NR |
| Liang (2021) [36] | Training: 0.742/validation: 0.773 | Calibration curve | Fair in both training and validation cohorts | NR |
| Wu (2021) [37] | 0.71 (both neoadjuvant and non-neoadjuvant groups) | NR | NR | NR |
| Sasaki (2022) [38] | Development: 0.61 (model as a continuous variable), 0.60 (model as a categorical variable)/Asian external validation cohort: 0.62 (model as a continuous variable), 0.60 (model as a categorical variable)/European external validation cohort: 0.57 (model as a continuous variable), 0.57 (model as a categorical variable) | NR | NR | NR |
| Huiskens (2019) [39] | Stage 1 model: 0.70/Stage 2 model: 0.72 | H-L test | Stage 1 model: chi-square: 3.5, p = 0.63/Stage 2 model: chi-square: 7.8, p = 0.18 | NR |
| Bai (2022) [40] | 5-year OS, development: 0.721/5-year OS, validation: 0.665/2-year RFS, development: 0.728/2-year RFS, validation: 0.640 | NR | NR | NR |
| Fang (2022) [41] | 0.715 | NR | NR | NR |
| Qin (2022) [42] | 1-, 2-, 3-year ihPFS: 0.695, 0.764, 0.782 | Calibration curve | Fair calibration | yes |
| Kawaguchi (2021) [43] | RAS mutant, development: 0.629/RAS mutant, validation: 0.644/wild type, development: 0.625/wild type, validation: 0.624 | Calibration curve | Fair calibration (development and validation cohort) | NR |
| Zhang (2023) [44] | Risk score: 1, 3, 5 years, training: 0.624, 0.630, 0.662/testing: 0.610, 0.646, 0.688/validation: 0.612, 0.622, 0.652/full model: 0.783, corrected: 0.772 | Calibration curve | Fair calibration | yes |
| Chen (2021) [45] | Complications: 0.658/PFS: 0.676/OS: 0.700 | Calibration curve, HL test | Complications: fair, HL test: chi-square 3.99, p = 0.91/PFS: fair/OS: good | yes (for complications) |
| Jin (2022) [46] | Training: 0.826/validation: 0.820/external validation: 0.763 | Calibration curve | Poor calibration (internal validation), fair (external validation) | yes |
| Zhai (2022) [47] | 0.659 | NR | NR | NR |
| Liu (2021) [48] | Development: 0.696/validation: 0.682 | Calibration curve | Development: fair/validation: poor | NR |
| Moro (2020) [49] | AIC: wtKRAS: 1356, mtKRAS: 1356 | Brier scores after bootstrapping | Brier: 0.1741 (wtKRAS), 0.1793 (mtKRAS) | NR |
| Chen (2021) [50] | Complications: 0.750/PFS: 0.663/OS: 0.684 | Calibration curves and HL test | Complications: fair/PFS: fair/OS: fair | yes |
| Yao (2021) [51] | Presence of LN metastases: 0.655/PFS: 0.656 | Calibration curves and HL test | Presence of LN metastases: fair/PFS: fair | NR |
| Kazi (2023) [52] | 0.692 | Calibration table | Good calibration (small group numbers) | NR |
| Meng (2021) [53] | 1 yr OS, training: 0.788/3 yr OS, validation: 0.702/3 yr OS, training: 0.752/3 yr OS, validation: 0.848 | Calibration curve | 1 yr OS: fair, 3 yr OS: good (small numbers) | NR |
| Imai (2016) [54] | 0.66 | Calibration curve | 3 and 5 yr OS: fair | NR |
| Chen (2022) [55] | Development: 0.754/validation: 0.882 | Calibration curve, HL test | HL: chi-square: 1.36, p = 0.998, calibration curve: good calibration in development and validation cohorts | yes |
| Cheng (2022) [56] | Training: 0.709/validation: 0.735 | Calibration curve | CSS: fair in training and validation/OS: fair in training and validation | NR |
| Kulik (2018) [57] | Preoperative: 0.716/preop- and perioperative: 0.761 | NR | NR | NR |
| Bai (2021) [58] | LDH-CRS: 0.674/mCRS: 0.681 | NR | NR | NR |
| Wang (2021) [59] | 1st score, 1, 3, 5 yr OS, training: 0.84, 0.73, 0.70/1, 3, 5 yr OS, int. validation: 0.75, 0.70, 0.70/1, 3, 5 yr OS, ext. validation: 0.77, 0.78, 0.72/2nd score, 3 yr OS, training: 0.76/5 yr OS, training: 0.75/3 yr OS, validation: 0.74/5 yr OS, validation: 0.66 | Calibration curve | Merged score: fair | NR |
| Xu (2021) [60] | Training: 0.746/validation: 0.764 | Calibration curve, slope, intercept | Validation: fair, calibration slope 1.09, intercept: −0.006 | NR |
| Sasaki (2018) [61] | 0.669 | NR | NR | NR |
| Wada (2022) [62] | Training: 0.83/validation: 0.81/mixed model: 0.85 | NR | NR | NR |
| Kim (2020) [63] | Training: 0.824/validation: 0.898 | H-L test | p = 0.831 | NR |
| Dupre (2019) [64] | Preoperative: 0.619/postoperative: 0.637 | NR | NR | NR |
| Qi (2023) [65] | SOF, 5 yr: 0.63/SOF, 8 yr: 0.74/combined, 5 yr: 0.69/combined, 8 yr: 0.79 | Calibration curve | Fair calibration | NR |
| Wu (2021) [66] | 0.705 | Calibration curve | Fair calibration | NR |
| Dasari (2023) [67] | Development, 1, 2, 3, 5 yr: 0.756, 0.745, 0.706, 0.698/validation, 1, 2, 3, 5 yr: 0.679, 0.659, 0.678, 0.732 | NR | NR | NR |
| Liu (2023) [68] | DEG risk score, development, 5 yr: 0.74/validation, 5 yr: 0.64/mixed model: 0.69 | Calibration curve | Good calibration | yes |
| Amygdalos (2023) [69] | 0.70 | NR | NR | NR |
| Chen (2023) [70] | 0.732 | Calibration curve | Fair | NR |
| Wu (2018) [71] | OS, 1 and 3 yr: 0.621,0.661/CSS, 1 and 3 yr: 0.621,0.660 | Calibration curve | Fair in training and validation, both for OS and CSS | NR |
| Deng (2023) [72] | Training: 0.720/validation: 0.740 | Calibration curve, HL test | Training: fair calibration, chi-square 4.97, p = 0.7612/validation: poor calibration, chi: 3.89, p = 0.8671 | yes (utility in a narrow range of thresholds) |
| Berardi (2023) [73] | Training: 0.68/validation: 0.60 | Calibration curve | Fair | NR |
| Liu (2019) [74] | Development: 0.675/validation: 0.77 | Calibration curve | Development: 1 yr poor, 3 yr good/validation: 1 yr poor, 3 yr poor, 5 yr poor | NR |
| Welsh (2008) [75] | 0.781 | Calibration plot, HL test | Validation: chi-square = 6.03, p = 0.196 | NR |
| Famularo (2023) [76] | RF model: 0.66 | NR | NR | NR |
| He (2023) [77] | Training: 0.801/validation: 0.739 | Calibration curve, slope, intercept | Development: good calibration/validation: fair calibration, slope: 1.0, intercept 0.0 | yes |
| Kattan (2008) [78] | Optimism-corrected: 0.612 | Calibration curve | Fair | NR |
| Wensink (2023) [79] | Optimism-corrected, 6 m: 0.643, 12 m: 0.641 | Calibration curve, slope | Fair at 6 and 12 months, optimism-corrected slope: 0.86 | yes |
| Fendler (2015) [80] | Training 0.81/validation: 0.83 | NR | NR | NR |
| Marfa (2016) [81] | Training: 0.903 | NR | NR | NR |
| Jiang (2023) [82] | CSS, training, 1 and 3 yr: 0.77, 0.70/validation, 1 and 3 yr: 0.72, 0.68/OS, training, 1 and 3 yr 0.78, 0.70/validation, 1 and 3 yr: 0.74, 0.70 | Calibration curve | Training: fair, validation poor | yes (superior to AJCC stage) |
| Endo (2023) [83] | OS-OPT, training: 0.68/testing: 0.69/RFS-OPT, training: 0.68/testing: 0.69 | NR | NR | NR |
| Rees (2008) [84] | Preoperative: 0.781/postoperative: 0.805 | H-L test | Preoperative: chi-square: 8.125; p = 0.087/postoperative: chi-square: 7.453, p = 0.114 | NR |
| Zakaria (2007) [85] | DSS: 0.61/recurrence: 0.58 | NR | NR | NR |
| Tan (2008) [86] | 0.59 | NR | NR | NR |
| Hill (2012) [87] | Apparent: 0.69/optimism-corrected: 0.67 | NR | NR | NR |
| Takeda (2021) [88] | Development: 0.65 | NR | NR | NR |
| Wang (2017) [89] | 0.642 | NR | NR | NR |
| Spelt (2013) [90] | ANN: 0.72/Cox model: 0.66 | NR | NR | NR |
AUC: area under the curve, DCA: decision curve analysis, MSKCC: Memorial Sloan Kettering Cancer Centre, KRAS: Kirsten rat sarcoma virus, NR: not reported, OS: overall survival, RFS: recurrence-free survival, GBT: gradient-boosted trees, LRB: logistic regression with bootstrapping, CITL: calibration-in-the-large, HL: Hosmer–Lemeshow, LM: liver metastases, ihPFS: intrahepatic progression-free survival, PFS: progression-free survival, AIC: Akaike information criterion, LN: lymph node, CSS: cancer-specific survival, LDH: lactate dehydrogenase, mCRS: modified clinical risk score, SOFs: spatial organization features, DEGs: differentially expressed genes, RF: random forest, AJCC: American Joint Committee on Cancer, OPT: optimal policy tree, DSS: disease-specific survival, ANN: artificial neural network.
9. Critical Appraisal of Published Models
Studies focusing on the prediction of patients with CRLM have increased in number in recent years. Traditional statistical techniques remain the pillar of model development and produce prognostic models that can be easily presented in detail and applied to new patients in the setting of an external validation study. ML models are apparently becoming popular; however, in the context of prognosis for CRLM, there is still a sparsity of studies when it comes to models using clinical predictors. In a recent review of AI-based models for CRLM, focusing both on diagnostic and on prognostic types of outcomes, the available models almost exclusively relied on radiomics and imaging-related predictors, while only two studies were based on clinical variables [4,26,90]. The present review focused on clinical, easily interpretable models that used predictors readily available for clinicians and documented in most contemporary databases. Even though imaging-based models, studies focusing on diagnostic outcomes and those that lacked reporting of performance measures were not presented in this review, the recent interest in predictive analytics and risk stratification of colorectal cancer patients led to the inclusion of a large number of papers solely targeting a variety of prognostic outcomes.
Several types of predictors were incorporated in the models, mostly related to the neoplastic disease or to the different treatment modalities chosen for each patient. The most influential predictors were associated with a high disease burden, like multiple and/or large metastatic lesions, originating from a node-positive primary located mostly in the ascending colon, combined with a high CEA level and a positive resection margin. These factors have already been identified in multiple studies as predictors of poor survival in CRLM patients [92,93,94]. The presence of a mutant RAS oncogene was also highlighted as a key predictor in this review, which confirms the findings of a meta-analysis showing the poor OS and RFS following resection of CRLM in mutant KRAS patients [95]. The recently emerged DEGs are promising new predictors, as components of multiple signalling pathways are shown to be correlated with a poor prognosis [96].
In the recently published Prediction model Risk Of Bias Assessment Tool (PROBAST), multiple details in the model development process are mentioned that can assist with the judgment of the risk of bias in each study [97]. Despite the fact that the majority of studies included in this review were published following the release of the TRIPOD and PROBAST guidance papers, several aspects assessed here would place most studies at a high risk of bias [5,97]. Such aspects include univariate screening of candidate predictors, poor handling of missing data, dichotomization of continuous predictors, inadequate reporting of model performance and poor assessment of optimism and overfitting. Regarding dichotomization, a large number of predictors included in the majority of studies, like number and size of CRLM, or laboratory predictors, like the CEA, were mostly dichotomized, and possible non-linear relationships between predictors and outcome in the development sample were not examined, leading to the loss of valuable information [97]. Assessment of optimism and overfitting was also problematic, due to the either frequent random splitting of the dataset, which is regarded as an inadequate method of quantifying optimism, or due to the complete absence of internal or external validation in many studies [97].
In this review, the focus was placed on assessment of model performance within a proposed framework [98]. The focus has now shifted from plain reporting of measures of discrimination to a thorough assessment with the aid of calibration metrics and DCA. The percentage of studies reporting calibration metrics was 57.7% in this review, which is relatively high compared to systematic reviews of prediction models, in which reporting is as low as 5.6%, especially when ML-based models are examined [99]. Regarding the discriminative performance of the models, the vast majority of studies reported moderate discrimination (AUC: 0.60–0.80) following internal or external validation. Similarly, a meta-analysis of prediction models for colorectal cancer patients presented pooled c-statistics between 0.57 to 0.74 for multiple survival outcomes [100]. Discrimination was significantly lower for external validation compared to development studies, indicating the need for better modelling and the proper assessment of overfitting [100].
Limitations of the present study include selection and analysis of a specific model type, namely those related to prognostic types of outcomes with clinical predictors. Exclusion of models based on radiological predictors and those aiming to promptly diagnose CRLMs was deliberate, since homogeneity in the included models was important for a proper overview and presentation. Studies incompletely reporting performance measures were also excluded, since the goal was to summarize available studies conforming to a proposed guidance. This may have led to the exclusion of studies utilizing proper model building and validation procedures if performance was not adequately assessed. Lastly, due to the fact that no formal systematic review was performed, no assessment of selection and publication bias was made, nor was there a formal assessment of within-study bias with the PROBAST tool.
10. Conclusions and Future Directions
Despite the rise of artificial intelligence and its popularity in all aspects of surgical oncology, research focusing on prognostication of CRLM patients is still dominated by models developed with conventional statistical techniques. The overview provided in this review can be utilized in future model development studies when selecting candidate predictors to be included in the model-building procedure. Predictors proven to be of relevance in multiple studies can be combined with other variables judged to be of clinical significance by physicians with experience in the management of CRLM patients. This method will assist in avoiding bias introduced with univariate predictor screening [97,101]. Another issue arising from this review is incomplete reporting, hampering the design of external validation studies and the proper quantification of pooled model performance in a future systematic review. Studies attempting to develop and/or externally validate prediction models for CRLM patients should adhere to the framework provided by TRIPOD [5]. Complying with such guidelines will improve the assessment of the generalizability and transportability of prediction models in a variety of different patient settings and populations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers16091645/s1, Supplementary methods: Details of the search strategy; Table S1: Characteristics of traditional clinical prediction models for prognosis of colorectal liver metastases; Table S2: Inclusion/exclusion criteria and median follow-up of studies developing clinical prediction models for prognosis of colorectal liver metastases; Table S3: Predictors included in clinical prediction models for prognosis of patients with colorectal liver metastases.
Author Contributions
Conceptualization, S.K., I.A.Z. and G.T.; literature search, S.K.; formal analysis, S.K.; data curation, S.K. and I.A.Z.; writing—original draft preparation, S.K.; writing—review and editing, S.K., I.A.Z., J.D.L.S., D.P.M. and G.T.; supervision, G.T. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Zeineddine F.A., Zeineddine M.A., Yousef A., Gu Y., Chowdhury S., Dasari A., Huey R.W., Johnson B., Kee B., Lee M.S., et al. Survival Improvement for Patients with Metastatic Colorectal Cancer over Twenty Years. NPJ Precis. Oncol. 2023;7:16. doi: 10.1038/s41698-023-00353-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cervantes A., Adam R., Roselló S., Arnold D., Normanno N., Taïeb J., Sel09igmann J., De Baere T., Osterlund P., Yoshino T., et al. Metastatic Colorectal Cancer: ESMO Clinical Practice Guideline for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2023;34:10–32. doi: 10.1016/j.annonc.2022.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Martin J., Petrillo A., Smyth E.C., Shaida N., Khwaja S., Cheow H., Duckworth A., Heister P., Praseedom R., Jah A., et al. Colorectal Liver Metastases: Current Management and Future Perspectives. World J. Clin. Oncol. 2020;11:761–808. doi: 10.5306/wjco.v11.i10.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rompianesi G., Pegoraro F., Ceresa C.D., Montalti R., Troisi R.I. Artificial Intelligence in the Diagnosis and Management of Colorectal Cancer Liver Metastases. World J. Gastroenterol. 2022;28:108–122. doi: 10.3748/wjg.v28.i1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moons K.G.M., Altman D.G., Reitsma J.B., Ioannidis J.P.A., Macaskill P., Steyerberg E.W., Vickers A.J., Ransohoff D.F., Collins G.S. Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): Explanation and Elaboration. Ann. Intern. Med. 2015;162:W1–W73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 6.Nordlinger B., Guiguet M., Vaillant J.-C., Balladur P., Boudjema K., Bachellier P., Jaeck D., de Chirurgie A.F. Surgical Resection of Colorectal Carcinoma Metastases to the Liver: A Prognostic Scoring System to Improve Case Selection, Based on 1568 Patients. Cancer. 1996;77:1254–1262. doi: 10.1002/(SICI)1097-0142(19960401)77:7<1254::AID-CNCR5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 7.Fong Y., Fortner J., Sun R.L., Brennan M.F., Blumgart L.H. Clinical Score for Predicting Recurrence after Hepatic Resection for Metastatic Colorectal Cancer: Analysis of 1001 Consecutive Cases. Ann. Surg. 1999;230:309–321. doi: 10.1097/00000658-199909000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka K., Shimada H., Fujii Y., Endo I., Sekido H., Togo S., Ike H. Pre-Hepatectomy Prognostic Staging to Determine Treatment Strategy for Colorectal Cancer Metastases to the Liver. Langenbeck’s Arch. Surg. 2004;389:371–379. doi: 10.1007/s00423-004-0490-y. [DOI] [PubMed] [Google Scholar]
- 9.Minagawa M. Simplified Staging System for Predicting the Prognosis of Patients with Resectable Liver Metastasis. Arch. Surg. 2007;142:269. doi: 10.1001/archsurg.142.3.269. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi T., Mori T., Takahashi K., Matsumoto H., Miyamoto H., Kato T. A New Classification System for Liver Metastases from Colorectal Cancer in Japanese Multicenter Analysis. Hepatogastroenterology. 2008;55:173–178. [PubMed] [Google Scholar]
- 11.Adam R., de Haas R.J., Wicherts D.A., Vibert E., Salloum C., Azoulay D., Castaing D. Concomitant Extrahepatic Disease in Patients with Colorectal Liver Metastases. Ann. Surg. 2011;253:349–359. doi: 10.1097/SLA.0b013e318207bf2c. [DOI] [PubMed] [Google Scholar]
- 12.Iwatsuki S., Dvorchik I., Madariaga J.R., Marsh J.W., Dodson F., Bonham A.C., Geller D.A., Gayowski T.J., Fung J.J., Starzl T.E. Hepatic Resection for Metastatic Colorectal Adenocarcinoma: A Proposal of a Prognostic Scoring System11No Competing Interests Declared. J. Am. Coll. Surg. 1999;189:291–299. doi: 10.1016/S1072-7515(99)00089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lise M., Bacchetti S., Da Pian P., Nitti D., Pilati P. Patterns of Recurrence after Resection of Colorectal Liver Metastases: Prediction by Models of Outcome Analysis. World J. Surg. 2001;25:638–644. doi: 10.1007/s002680020138. [DOI] [PubMed] [Google Scholar]
- 14.Nagashima I., Takada T., Matsuda K., Adachi M., Nagawa H., Muto T., Okinaga K. A New Scoring System to Classify Patients with Colorectal Liver Metastases: Proposal of Criteria to Select Candidates for Hepatic Resection. J. Hepatobiliary Pancreat. Surg. 2004;11:79–83. doi: 10.1007/s00534-002-0778-7. [DOI] [PubMed] [Google Scholar]
- 15.Schindl M. Prognostic Scoring in Colorectal Cancer Liver Metastases. Arch. Surg. 2005;140:183. doi: 10.1001/archsurg.140.2.183. [DOI] [PubMed] [Google Scholar]
- 16.Malik H.Z., Prasad K.R., Halazun K.J., Aldoori A., Al-Mukhtar A., Gomez D., Lodge J.P.A., Toogood G.J. Preoperative Prognostic Score for Predicting Survival After Hepatic Resection for Colorectal Liver Metastases. Ann. Surg. 2007;246:806–814. doi: 10.1097/SLA.0b013e318142d964. [DOI] [PubMed] [Google Scholar]
- 17.Ueno H., Mochizuki H., Hatsuse K., Hase K., Yamamoto T. Indicators for Treatment Strategies of Colorectal Liver Metastases. Ann. Surg. 2000;231:59. doi: 10.1097/00000658-200001000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee W.S., Kim M.J., Yun S.H., Chun H.K., Lee W.Y., Kim S.J., Choi S.H., Heo J.S., Joh J.W., Kim Y. Il Risk Factor Stratification after Simultaneous Liver and Colorectal Resection for Synchronous Colorectal Metastasis. Langenbeck’s Arch. Surg. 2008;393:13–19. doi: 10.1007/s00423-007-0231-0. [DOI] [PubMed] [Google Scholar]
- 19.Konopke R., Kersting S., Distler M., Dietrich J., Gastmeier J., Heller A., Kulisch E., Saeger H. Prognostic Factors and Evaluation of a Clinical Score for Predicting Survival after Resection of Colorectal Liver Metastases. Liver Int. 2009;29:89–102. doi: 10.1111/j.1478-3231.2008.01845.x. [DOI] [PubMed] [Google Scholar]
- 20.Buisman F.E., Grünhagen D.J., Verhoef C., Groot Koerkamp B. Response to Letter Entitled: Re: Predicting 10-Year Survival after Resection of Colorectal Liver Metastases; an International Study Including Biomarkers and Perioperative Treatment. Eur. J. Cancer. 2023;188:80. doi: 10.1016/j.ejca.2023.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Bertsimas D., Margonis G.A., Sujichantararat S., Boerner T., Ma Y., Wang J., Kamphues C., Sasaki K., Tang S., Gagniere J., et al. Using Artificial Intelligence to Find the Optimal Margin Width in Hepatectomy for Colorectal Cancer Liver Metastases. JAMA Surg. 2022;157:e221819. doi: 10.1001/jamasurg.2022.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao X., Wang K., Liu M., Li B., Wang H., Jin K., Yan X., Zhang H., Bao Q., Xu D., et al. Characterization of Genomic Alterations in Colorectal Liver Metastasis and Their Prognostic Value. Front. Cell Dev. Biol. 2022;9:760618. doi: 10.3389/fcell.2021.760618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam C.S.N., Bharwani A.A., Chan E.H.Y., Chan V.H.Y., Au H.L.H., Ho M.K., Rashed S., Kwong B.M.H., Fang W., Ma K.W., et al. A Machine Learning Model for Colorectal Liver Metastasis Post-Hepatectomy Prognostications. Hepatobiliary Surg. Nutr. 2023;12:495–506. doi: 10.21037/hbsn-21-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reijonen P., Nordin A., Savikko J., Poussa T., Arola J., Isoniemi H. Histopathological Helsinki Score of Colorectal Liver Metastases Predicts Survival after Liver Resection. Apmis. 2023;131:249–261. doi: 10.1111/apm.13305. [DOI] [PubMed] [Google Scholar]
- 25.Margonis G.A., Sasaki K., Gholami S., Kim Y., Andreatos N., Rezaee N., Deshwar A., Buettner S., Allen P.J., Kingham T.P., et al. Genetic and Morphological Evaluation (GAME) Score for Patients with Colorectal Liver Metastases. Br. J. Surg. 2018;105:1210–1220. doi: 10.1002/bjs.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paredes A.Z., Hyer J.M., Tsilimigras D.I., Moro A., Bagante F., Guglielmi A., Ruzzenente A., Alexandrescu S., Makris E.A., Poultsides G.A., et al. A Novel Machine-Learning Approach to Predict Recurrence after Resection of Colorectal Liver Metastases. Ann. Surg. Oncol. 2020;27:5139–5147. doi: 10.1245/s10434-020-08991-9. [DOI] [PubMed] [Google Scholar]
- 27.Frühling P., Urdzik J., Strömberg C., Isaksson B. Composite Score: Prognostic Tool to Predict Survival in Patients Undergoing Surgery for Colorectal Liver Metastases. BJS Open. 2021;5:zrab104. doi: 10.1093/bjsopen/zrab104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taghavi M., Trebeschi S., Simões R., Meek D.B., Beckers R.C.J., Lambregts D.M.J., Verhoef C., Houwers J.B., van der Heide U.A., Beets-Tan R.G.H., et al. Machine Learning-Based Analysis of CT Radiomics Model for Prediction of Colorectal Metachronous Liver Metastases. Abdom. Radiol. 2021;46:249–256. doi: 10.1007/s00261-020-02624-1. [DOI] [PubMed] [Google Scholar]
- 29.Brudvik K.W., Jones R.P., Giuliante F., Shindoh J., Passot G., Chung M.H., Song J., Li L., Dagenborg V.J., Fretland Å.A., et al. RAS Mutation Clinical Risk Score to Predict Survival after Resection of Colorectal Liver Metastases. Ann. Surg. 2019;269:120–126. doi: 10.1097/SLA.0000000000002319. [DOI] [PubMed] [Google Scholar]
- 30.Moaven O., Tavolara T.E., Valenzuela C.D., Cheung T.T., Corvera C.U., Cha C.H., Stauffer J.A., Niazi M.K.K., Gurcan M.N., Shen P. Machine Learning Models for Predicting the Outcomes of Surgical Treatment of Colorectal Liver Metastases. J. Am. Coll. Surg. 2023;236:884–893. doi: 10.1097/XCS.0000000000000573. [DOI] [PubMed] [Google Scholar]
- 31.Villard C., Abdelrafee A., Habib M., Ndegwa N., Jorns C., Sparrelid E., Allard M.A., Adam R. Prediction of Survival in Patients with Colorectal Liver Metastases- Development and Validation of a Prognostic Score Model. Eur. J. Surg. Oncol. 2022;48:2432–2439. doi: 10.1016/j.ejso.2022.06.021. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y., Chang W., Ren L., Chen J., Tang W., Liu T., Jian M., Liu Y., Wei Y., Xu J. Comprehensive Evaluation of Relapse Risk (CERR) Score for Colorectal Liver Metastases: Development and Validation. Oncologist. 2020;25:e1031–e1041. doi: 10.1634/theoncologist.2019-0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen F.L., Wang Y.Y., Liu W., Xing B.C. Prognostic Factors in Colorectal Liver Metastases Patients with Various Tumor Numbers Treated by Liver Resection: A Single-Center, Retrospective Study. World J. Surg. Oncol. 2022;20:237. doi: 10.1186/s12957-022-02700-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai S., Ye Y., Kong X., Li J., Ding K. A Predictive Model for Early Recurrence of Colorectal-Cancer Liver Metastases Based on Clinical Parameters. Gastroenterol. Rep. 2021;9:241–251. doi: 10.1093/gastro/goaa092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu W., Liu J.M., Wang K., Wang H.W., Xing B.C. Recurrent Colorectal Liver Metastasis Patients Could Benefit from Repeat Hepatic Resection. BMC Surg. 2021;21:327. doi: 10.1186/s12893-021-01323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang J.Y., Lin H.C., Liu J., Wang D.S., Yuan Y.F., Li B.K., Zheng Y., Wu X.J., Chen G., Wang F.H., et al. A Novel Prognostic Nomogram for Colorectal Cancer Liver Metastasis Patients with Recurrence after Hepatectomy. Cancer Med. 2021;10:1535–1544. doi: 10.1002/cam4.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y., Guo T., Xu Z., Liu F., Cai S., Wang L., Xu Y. Risk Scoring System for Recurrence after Simultaneous Resection of Colorectal Cancer Liver Metastasis. Ann. Transl. Med. 2021;9:966. doi: 10.21037/atm-21-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki K., Margonis G.A., Moro A., Wang J., Wagner D., Gagnière J., Shin J.K., D’Silva M., Sahara K., Miyata T., et al. Nontumor Related Risk Score: A New Tool to Improve Prediction of Prognosis after Hepatectomy for Colorectal Liver Metastases. Surgery. 2022;171:1580–1587. doi: 10.1016/j.surg.2022.01.030. [DOI] [PubMed] [Google Scholar]
- 39.Huiskens J., Schadde E., Lang H., Malago M., Petrowsky H., de Santibañes E., Oldhafer K., van Gulik T.M., Olthof P.B. Avoiding Postoperative Mortality after ALPPS–Development of a Tumor-Specific Risk Score for Colorectal Liver Metastases. HPB. 2019;21:898–905. doi: 10.1016/j.hpb.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 40.Bai L., Yan X.L., Lu Y.X., Meng Q., Rong Y.M., Ye L.F., Pan Z.Z., Xing B.C., Wang D.S. Circulating Lipid- and Inflammation-Based Risk (CLIR) Score: A Promising New Model for Predicting Outcomes in Complete Colorectal Liver Metastases Resection. Ann. Surg. Oncol. 2022;29:4308–4323. doi: 10.1245/s10434-021-11234-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang C., Huang Y., Chen C., Nie D., Lin J., Xiao Z., Li S., Liu S., Luo R., Lin H., et al. The Prognostic Value of Serum Apolipoprotein A-I Level and Neutrophil-to-Lymphocyte Ratio in Colorectal Cancer Liver Metastasis. J. Oncol. 2022;2022:9149788. doi: 10.1155/2022/9149788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin S., Hu H., Cui R., Lin J., Liu Y., Wang Y., Chen Y., Liu G. A Prognostic Nomogram for Intrahepatic Progression-Free Survival in Patients with Colorectal Liver Metastases after Ultrasound-Guided Percutaneous Microwave Ablation. Int. J. Hyperth. 2022;39:144–154. doi: 10.1080/02656736.2021.2023226. [DOI] [PubMed] [Google Scholar]
- 43.Kawaguchi Y., Kopetz S., Tran Cao H.S., Panettieri E., De Bellis M., Nishioka Y., Hwang H., Wang X., Tzeng C.W.D., Chun Y.S., et al. Contour Prognostic Model for Predicting Survival after Resection of Colorectal Liver Metastases: Development and Multicentre Validation Study Using Largest Diameter and Number of Metastases with RAS Mutation Status. Br. J. Surg. 2021;108:968–975. doi: 10.1093/bjs/znab086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X., Wu T., Zhou J., Chen X., Dong C., Guo Z., Yang R., Liang R., Feng Q., Hu R., et al. Establishment and Verification of Prognostic Model and CeRNA Network Analysis for Colorectal Cancer Liver Metastasis. BMC Med. Genom. 2023;16:99. doi: 10.1186/s12920-023-01523-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Q., Mao R., Zhao J., Bi X., Li Z., Huang Z., Zhang Y., Zhou J., Zhao H., Cai J. Nomograms Incorporating Preoperative RDW Level for the Prediction of Postoperative Complications and Survival in Colorectal Liver Metastases after Resection. Ann. Palliat. Med. 2021;10:4143–4158. doi: 10.21037/apm-20-2418. [DOI] [PubMed] [Google Scholar]
- 46.Jin X., Wu Y., Feng Y., Lin Z., Zhang N., Yu B., Mao A., Zhang T., Zhu W., Wang L. A Population-Based Predictive Model Identifying Optimal Candidates for Primary and Metastasis Resection in Patients with Colorectal Cancer with Liver Metastatic. Front. Oncol. 2022;12:899659. doi: 10.3389/fonc.2022.899659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhai Y., Bai W., Zhou J., Dong Q., Zhang J. Effect of Tumour Size Ratio on Liver Recurrence-Free Survival of Patients Undergoing Hepatic Resection for Colorectal Liver Metastases. BMC Cancer. 2022;22:103. doi: 10.1186/s12885-022-09199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu W., Zhang W., Xu Y., Li Y.H., Xing B.C. A Prognostic Scoring System to Predict Survival Outcome of Resectable Colorectal Liver Metastases in This Modern Era. Ann. Surg. Oncol. 2021;28:7709–7718. doi: 10.1245/s10434-021-10143-6. [DOI] [PubMed] [Google Scholar]
- 49.Moro A., Mehta R., Tsilimigras D.I., Sahara K., Paredes A.Z., Bagante F., Guglielmi A., Alexandrescu S., Poultsides G.A., Sasaki K., et al. Prognostic Factors Differ According to KRAS Mutational Status: A Classification and Regression Tree Model to Define Prognostic Groups after Hepatectomy for Colorectal Liver Metastasis. Surgery. 2020;168:497–503. doi: 10.1016/j.surg.2020.05.019. [DOI] [PubMed] [Google Scholar]
- 50.Chen Q., Mao R., Zhao J., Bi X., Li Z., Huang Z., Zhang Y., Zhou J., Zhao H., Cai J. Upgraded Nomograms for the Prediction of Complications and Survival in Patients with Colorectal Liver Metastases Treated with Neoadjuvant Chemotherapy Followed by Hepatic Resection. Ann. Transl. Med. 2021;9:265. doi: 10.21037/atm-20-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao J., Chen Q., Deng Y., Zhao J., Bi X., Li Z., Huang Z., Zhang Y., Zhou J., Zhao H., et al. Nomograms Predicting Primary Lymph Node Metastases and Prognosis for Synchronous Colorectal Liver Metastasis with Simultaneous Resection of Colorectal Cancer and Liver Metastases. Ann. Palliat. Med. 2021;10:4220–4231. doi: 10.21037/apm-20-2303. [DOI] [PubMed] [Google Scholar]
- 52.Kazi M., Patkar S., Patel P., Kunte A., Desouza A., Saklani A., Goel M. Simultaneous Resection of Synchronous Colorectal Liver Metastasis: Feasibility and Development of a Prediction Model. Ann. Hepato-Biliary-Pancreat. Surg. 2023;27:40–48. doi: 10.14701/ahbps.22-043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meng Q., Zheng N., Wen R., Sui J., Zhang W. Preoperative Nomogram to Predict Survival Following Colorectal Cancer Liver Metastasis Simultaneous Resection. J. Gastrointest. Oncol. 2021;12:556–567. doi: 10.21037/jgo-20-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imai K., Allard M.A., Castro Benitez C., Vibert E., Sa Cunha A., Cherqui D., Castaing D., Bismuth H., Baba H., Adam R. Nomogram for Prediction of Prognosis in Patients with Initially Unresectable Colorectal Liver Metastases. Br. J. Surg. 2016;103:590–599. doi: 10.1002/bjs.10073. [DOI] [PubMed] [Google Scholar]
- 55.Chen Q., Zhang Y., Li X., Huang Z., Zhao H., Cai J. Nomogram Incorporating Preoperative Testing Markers for the Prediction of Early Recurrence for Colorectal Liver Metastases with Neoadjuvant Chemotherapy Followed by Hepatectomy. J. Cancer. 2022;13:1758–1767. doi: 10.7150/jca.65677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng X., Li Y., Chen D., Xu X., Liu F., Zhao F. Nomogram Predicting the Survival of Young-Onset Patients with Colorectal Cancer Liver Metastases. Diagnostics. 2022;12:1395. doi: 10.3390/diagnostics12061395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kulik U., Plohmann-Meyer M., Gwiasda J., Kolb J., Meyer D., Kaltenborn A., Lehner F., Klempnauer J., Schrem H. Proposal of Two Prognostic Models for the Prediction of 10-Year Survival after Liver Resection for Colorectal Metastases. HPB Surg. 2018;2018:5618581. doi: 10.1155/2018/5618581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bai L., Lin Z.Y., Lu Y.X., Chen Q., Zhou H., Meng Q., Lin C.P., Huang W.L., Wan Y.L., Pan Z.Z., et al. The Prognostic Value of Preoperative Serum Lactate Dehydrogenase Levels in Patients Underwent Curative-Intent Hepatectomy for Colorectal Liver Metastases: A Two-Center Cohort Study. Cancer Med. 2021;10:8005–8019. doi: 10.1002/cam4.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., Chen X., Lin H., Sun X., Fong W.P., Wu X., Pan Z., Yuan Y., Liang J., Wang D., et al. A Prehepatectomy Circulating Exosomal Microrna Signature Predicts the Prognosis and Adjuvant Chemotherapeutic Benefits in Colorectal Liver Metastasis. Cancers. 2021;13:4258. doi: 10.3390/cancers13174258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu D., Wang Y.Y., Yan X.L., Li J., Wang K., Xing B.C. Development of a Model to Predict Pathologic Response to Chemotherapy in Patients with Colorectal Liver Metastases. J. Gastrointest. Oncol. 2021;12:1498–1508. doi: 10.21037/jgo-21-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sasaki K., Morioka D., Conci S., Margonis G.A., Sawada Y., Ruzzenente A., Kumamoto T., Iacono C., Andreatos N., Guglielmi A., et al. The Tumor Burden Score: A New “Metro-Ticket” Prognostic Tool for Colorectal Liver Metastases Based on Tumor Size and Number of Tumors. Ann. Surg. 2018;267:132–141. doi: 10.1097/SLA.0000000000002064. [DOI] [PubMed] [Google Scholar]
- 62.Wada Y., Shimada M., Morine Y., Ikemoto T., Saito Y., Baba H., Mori M., Goel A. A Transcriptomic Signature That Predicts Cancer Recurrence after Hepatectomy in Patients with Colorectal Liver Metastases. Eur. J. Cancer. 2022;163:66–76. doi: 10.1016/j.ejca.2021.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim W.J., Lim T.W., Kang S.H., Park P.J., Choi S.B., Lee S.I., Min B.W., Kim W.B. Development and Validation of Novel Scoring System for the Prediction of Disease Recurrence Following Resection of Colorectal Liver Metastasis. Asian J. Surg. 2020;43:438–446. doi: 10.1016/j.asjsur.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 64.Dupré A., Berhane S., Chan A.W.H., Rivoire M., Chong C.C.N., Lai P.B.S., Cucchetti A., Poston G.J., Malik H.Z., Johnson P.J. Multicentre Validation of a Clinical Prognostic Score Integrating the Systemic Inflammatory Response to the Host for Patients Treated with Curative-Intent for Colorectal Liver Metastases: The Liverpool Score. Eur. J. Surg. Oncol. 2019;45:999–1004. doi: 10.1016/j.ejso.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 65.Qi L., Liang J.Y., Li Z.W., Xi S.Y., Lai Y.N., Gao F., Zhang X.R., Wang D.S., Hu M.T., Cao Y., et al. Deep Learning-Derived Spatial Organization Features on Histology Images Predicts Prognosis in Colorectal Liver Metastasis Patients after Hepatectomy. iScience. 2023;26:107702. doi: 10.1016/j.isci.2023.107702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu H., Liu G.J., Zhang Z.Y., Wu W., Meng Y.F., Wang S., Yang W., Yan K. Nomogram Including Chemotherapy Response for Prediction of Intrahepatic Progression-Free Survival in Patients with Colorectal Liver Metastasis through Chemotherapy Followed by Radiofrequency Ablation. Int. J. Hyperth. 2021;38:633–639. doi: 10.1080/02656736.2021.1912415. [DOI] [PubMed] [Google Scholar]
- 67.Dasari B.V.M., Raptis D., Syn N., Serrablo A., Ramia J.M., Laurenzi A., Sturesson C., Pawlik T.M., Siriwardena A.K., Lesurtel M. Development and Validation of a Novel Risk Score to Predict Overall Survival Following Surgical Clearance of Bilobar Colorectal Liver Metastases. BJS Open. 2023;7:zrad085. doi: 10.1093/bjsopen/zrad085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu C., Lu Z., Yan J., Xue D., He X., Huang W., Sun Q., Zhao W., Li F. Construction of a Prognostic Signature Associated with Liver Metastases for Prognosis and Immune Response Prediction in Colorectal Cancer. Front. Oncol. 2023;13:1234045. doi: 10.3389/fonc.2023.1234045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amygdalos I., Müller-Franzes G., Bednarsch J., Czigany Z., Ulmer T.F., Bruners P., Kuhl C., Neumann U.P., Truhn D., Lang S.A. Novel Machine Learning Algorithm Can Identify Patients at Risk of Poor Overall Survival Following Curative Resection for Colorectal Liver Metastases. J. Hepatobiliary Pancreat. Sci. 2023;30:602–614. doi: 10.1002/jhbp.1249. [DOI] [PubMed] [Google Scholar]
- 70.Chen D., Li Q., Yu H. Prognosis of Resectable Colorectal Liver Metastases after Surgery Associated with Pathological Features of Primary Tumor. Front. Oncol. 2023;13:1181522. doi: 10.3389/fonc.2023.1181522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu Q., Wang W.J., Huang Y.Q., Fang S.Y., Guan Y.J. Nomograms for Estimating Survival in Patients with Liver-Only Colorectal Metastases: A Retrospective Study. Int. J. Surg. 2018;60:1–8. doi: 10.1016/j.ijsu.2018.10.032. [DOI] [PubMed] [Google Scholar]
- 72.Deng Y., Chen Q., Li C., Chen J., Cai J., Li Y., Zhao H. Nomogram Predicting Early Recurrence Defined by the Minimum P Value Approach for Colorectal Liver Metastasis Patients Receiving Colorectal Cancer Resection with Simultaneous Liver Metastasis Resection: Development and Validation. J. Gastrointest. Oncol. 2023;14:1279–1292. doi: 10.21037/jgo-22-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berardi G., Chou J., Gonen M., Balachandran V.P., Drebin J., Jarnagin W.R., Kingham T.P., Soares K.C., Wei A., D’Angelica M. A Model to Predict Treatment Failure in Patients Undergoing Upfront Surgery for Resectable Colorectal Liver Metastases. Ann. Surg. Oncol. 2023;30:2820–2827. doi: 10.1245/s10434-023-13113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu W., Wang K., Han Y., Liang J.Y., Li Y.H., Xing B.C. Nomogram Predicted Disease Free Survival for Colorectal Liver Metastasis Patients with Preoperative Chemotherapy Followed by Hepatic Resection. Eur. J. Surg. Oncol. 2019;45:2070–2077. doi: 10.1016/j.ejso.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 75.Welsh F.K.S., Tekkis P.P., O’Rourke T., John T.G., Rees M. Quantification of Risk of a Positive (R1) Resection Margin Following Hepatic Resection for Metastatic Colorectal Cancer: An Aid to Clinical Decision-Making. Surg. Oncol. 2008;17:3–13. doi: 10.1016/j.suronc.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 76.Famularo S., Milana F., Cimino M., Franchi E., Giuffrida M., Costa G., Procopio F., Donadon M., Torzilli G. Upfront Surgery versus Neoadjuvant Perioperative Chemotherapy for Resectable Colorectal Liver Metastases: A Machine-Learning Decision Tree to Identify the Best Potential Candidates under a Parenchyma-Sparing Policy. Cancers. 2023;15:613. doi: 10.3390/cancers15030613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He M., Jia Z., Hu L., Wu H. Development and Validation of a Nomogram to Predict Which Patients with Colorectal Cancer Liver Metastases Would Benefit from Primary Tumor Resection. Int. J. Color. Dis. 2023;38:144. doi: 10.1007/s00384-023-04426-5. [DOI] [PubMed] [Google Scholar]
- 78.Kattan M.W., Gönen M., Jarnagin W.R., DeMatteo R., D’Angelica M., Weiser M., Blumgart L.H., Fong Y. A Nomogram for Predicting Disease-Specific Survival after Hepatic Resection for Metastatic Colorectal Cancer. Ann. Surg. 2008;247:282–287. doi: 10.1097/SLA.0b013e31815ed67b. [DOI] [PubMed] [Google Scholar]
- 79.Wensink G.E., Bolhuis K., Elferink M.A.G., Fijneman R.J.A., Kranenburg O., Borel Rinkes I.H.M., Koopman M., Swijnenburg R.J., Vink G.R., Hagendoorn J., et al. Predicting Early Extrahepatic Recurrence after Local Treatment of Colorectal Liver Metastases. Br. J. Surg. 2023;110:362–371. doi: 10.1093/bjs/znac461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fendler W.P., Ilhan H., Paprottka P.M., Jakobs T.F., Heinemann V., Bartenstein P., Khalaf F., Ezziddin S., Hacker M., Haug A.R. Nomogram Including Pretherapeutic Parameters for Prediction of Survival after SIRT of Hepatic Metastases from Colorectal Cancer. Eur. Radiol. 2015;25:2693–2700. doi: 10.1007/s00330-015-3658-7. [DOI] [PubMed] [Google Scholar]
- 81.Marfà S., Marti J., Reyes A., Casals G., Fernández-Varo G., Carvajal S., García-Valdecasas J.C., Fuster J., Jiménez W. Metastatic Tissue Proteomic Profiling Predicts 5-Year Outcomes in Patients with Colorectal Liver Metastases. Transl. Oncol. 2016;9:445–452. doi: 10.1016/j.tranon.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang Y.J., Zhou S.C., Zhu Z.X., Chen J.H., Liang J.W. Survival Nomograms for Simultaneous Resection of Primary and Hepatic Lesions without Neoadjuvant Chemotherapy in Patients with Resectable Colorectal Liver Metastasis. Cancer Innov. 2023;2:240–252. doi: 10.1002/cai2.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Endo Y., Alaimo L., Moazzam Z., Woldesenbet S., Lima H.A., Yang J., Munir M.M., Shaikh C.F., Azap L., Katayama E., et al. Optimal Policy Tree to Assist in Adjuvant Therapy Decision-Making after Resection of Colorectal Liver Metastases. Surgery. 2024;175:645–653. doi: 10.1016/j.surg.2023.06.045. [DOI] [PubMed] [Google Scholar]
- 84.Rees M., Tekkis P.P., Welsh F.K.S., O’Rourke T., John T.G. Evaluation of Long-Term Survival after Hepatic Resection for Metastatic Colorectal Cancer: A Multifactorial Model of 929 Patients. Ann. Surg. 2008;247:125–135. doi: 10.1097/SLA.0b013e31815aa2c2. [DOI] [PubMed] [Google Scholar]
- 85.Zakaria S., Donohue J.H., Que F.G., Farnell M.B., Schleck C.D., Ilstrup D.M., Nagorney D.M. Hepatic Resection for Colorectal Metastases: Value for Risk Scoring Systems? Ann. Surg. 2007;246:183–191. doi: 10.1097/SLA.0b013e3180603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tan M.C.B., Castaldo E.T., Gao F., Chari R.S., Linehan D.C., Wright J.K., Hawkins W.G., Siegel B.A., Delbeke D., Pinson C.W., et al. A Prognostic System Applicable to Patients with Resectable Liver Metastasis from Colorectal Carcinoma Staged by Positron Emission Tomography with [18F]Fluoro-2-Deoxy-D-Glucose: Role of Primary Tumor Variables. J. Am. Coll. Surg. 2008;206:857–868. doi: 10.1016/j.jamcollsurg.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 87.Hill C.R.S., Chagpar R.B., Callender G.G., Brown R.E., Gilbert J.E., Martin R.C.G., McMasters K.M., Scoggins C.R. Recurrence Following Hepatectomy for Metastatic Colorectal Cancer: Development of a Model That Predicts Patterns of Recurrence and Survival. Ann. Surg. Oncol. 2012;19:139–144. doi: 10.1245/s10434-011-1921-y. [DOI] [PubMed] [Google Scholar]
- 88.Takeda Y., Mise Y., Matsumura M., Hasegawa K., Yoshimoto J., Imamura H., Noro T., Yamamoto J., Ishizuka N., Inoue Y., et al. Accuracy of Modern Clinical Risk Score Including RAS Status Changes Based on Whether Patients Received Perioperative Chemotherapy for Colorectal Liver Metastases. World J. Surg. 2021;45:2176–2184. doi: 10.1007/s00268-021-05976-x. [DOI] [PubMed] [Google Scholar]
- 89.Wang K., Liu W., Yan X.L., Li J., Xing B.C. Long-Term Postoperative Survival Prediction in Patients with Colorectal Liver Metastasis. Oncotarget. 2017;8:79927–79934. doi: 10.18632/oncotarget.20322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Spelt L., Nilsson J., Andersson R., Andersson B. Artificial Neural Networks-A Method for Prediction of Survival Following Liver Resection for Colorectal Cancer Metastases. Eur. J. Surg. Oncol. 2013;39:648–654. doi: 10.1016/j.ejso.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 91.Dindo D., Demartines N., Clavien P.-A. Classification of Surgical Complications. Ann. Surg. 2004;240:205–213. doi: 10.1097/01.sla.0000133083.54934.ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taylor A., Primrose J., Langeberg W., Kelsh M., Mowat F., Alexander D., Choti M., Poston G., Kanas G. Survival after Liver Resection in Metastatic Colorectal Cancer: Review and Meta-Analysis of Prognostic Factors. Clin. Epidemiol. 2012;4:283–301. doi: 10.2147/CLEP.S34285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ai X., Tao M., Wang H., Li J., Sun T., Xiu D. Analysis of Survival Factors after Hepatic Resection for Colorectal Cancer Liver Metastases: Does the R1 Margin Matter? Front. Surg. 2023;9:1020240. doi: 10.3389/fsurg.2022.1020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dueland S., Smedman T.M., Syversveen T., Grut H., Hagness M., Line P.-D. Long-Term Survival, Prognostic Factors, and Selection of Patients with Colorectal Cancer for Liver Transplant. JAMA Surg. 2023;158:e232932. doi: 10.1001/jamasurg.2023.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brudvik K.W., Kopetz S.E., Li L., Conrad C., Aloia T.A., Vauthey J.-N. Meta-Analysis of KRAS Mutations and Survival after Resection of Colorectal Liver Metastases. Br. J. Surg. 2015;102:1175–1183. doi: 10.1002/bjs.9870. [DOI] [PubMed] [Google Scholar]
- 96.Wu Q.-Q., Wang X.-Y., Wu W.-X., Chen Y.-X., Wang J., Zhang X., Qian Y., Du S.-S., Sun J., Zeng Z.-C. Molecular Mechanisms Investigation for Liver Metastasis of Colorectal Cancer by Combined Bioinformatic Gene Expression Profile Analysis. Cancer Treat. Res. Commun. 2023;35:100694. doi: 10.1016/j.ctarc.2023.100694. [DOI] [PubMed] [Google Scholar]
- 97.Moons K.G.M., Wolff R.F., Riley R.D., Whiting P.F., Westwood M., Collins G.S., Reitsma J.B., Kleijnen J., Mallett S. PROBAST: A Tool to Assess Risk of Bias and Applicability of Prediction Model Studies: Explanation and Elaboration. Ann. Intern. Med. 2019;170:W1. doi: 10.7326/M18-1377. [DOI] [PubMed] [Google Scholar]
- 98.Steyerberg E.W., Vickers A.J., Cook N.R., Gerds T., Gonen M., Obuchowski N., Pencina M.J., Kattan M.W. Assessing the Performance of Prediction Models: A Framework for Traditional and Novel Measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Andaur Navarro C.L., Damen J.A.A., van Smeden M., Takada T., Nijman S.W.J., Dhiman P., Ma J., Collins G.S., Bajpai R., Riley R.D., et al. Systematic Review Identifies the Design and Methodological Conduct of Studies on Machine Learning-Based Prediction Models. J. Clin. Epidemiol. 2023;154:8–22. doi: 10.1016/j.jclinepi.2022.11.015. [DOI] [PubMed] [Google Scholar]
- 100.He Y., Ong Y., Li X., Din F.V., Brown E., Timofeeva M., Wang Z., Farrington S.M., Campbell H., Dunlop M.G., et al. Performance of Prediction Models on Survival Outcomes of Colorectal Cancer with Surgical Resection: A Systematic Review and Meta-Analysis. Surg. Oncol. 2019;29:196–202. doi: 10.1016/j.suronc.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 101.Steyerberg E.W. Clinical Prediction Models. Springer International Publishing; Cham, Switzerland: 2019. Statistics for Biology and Health. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.